Abstract
Previous study indicated that the multi-resistance gene cfr was mainly found in gram-positive bacteria, such as Staphylococcus and Enterococcus, and was sporadically detected in Escherichia coli. Little is known about the prevalence and transmission mechanism of cfr in E. coli. In this study, the presence of cfr in E. coli isolates collected during 2010–2012 from food-producing animals in Guangdong Province of China was investigated, and the cfr-positive E. coli isolates were characterized by PFGE, plasmid profiling, and genetic environment analysis. Of the 839 E. coli isolates, 10 isolates from pig were cfr positive. All the cfr-positive isolates presented a multi-resistance phenotype and were genetically divergent as determined by PFGE. In 8 out of the 10 strains, the cfr gene was located on plasmids of ∼30 kb. Restriction digestion of the plasmids with EcoRI and sequence hybridization with a cfr-specific probe revealed that the cfr-harboring fragments ranged from 6 to 23 kb and a ∼18 kb cfr-carrying fragment was common for the plasmids that were ∼30 kb. Four different genetic environments of cfr were detected, in which cfr is flanked by two identical copies of IS26, which may loop out the intervening sequence through homologous recombination. Among the 8 plasmids of ∼30 kb, 7 plasmids shared the same genetic environment. These results demonstrate plasmid-carried cfr in E. coli and suggest that transposition and homologous recombination mediated by IS26 might have played a rule in the transfer of the cfr gene in E. coli.
Introduction
Encoding a methyltransferase that modifies 23S rRNA at A2503, the cfr gene confers resistance to five chemically distinct classes of antimicrobials, including Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins and Streptogramin A [1], and also reduces the susceptibility to selected 16-member-ring macrolides such as josamycin and spiramycin [2]. Since its first identification on plasmid pSCFS1 from Staphylococcus sciuri [3], the cfr gene has been identified on plasmid or chromosome in other staphylococcal species [4], [5], and subsequently in other genera of gram-positive bacteria such as Bacillus [6]–[8], Enterococcus [9]–[12], Streptococcus [13], Macrococcus and Jeotgalicoccus [14]. In those bacteria, plasmids and various insertion sequences have played an important role in the dissemination of the cfr gene between species and genera [15]. In gram-negative bacteria, the cfr gene has been sporadically detected in Escherichia coli and Proteus vulgaris [16]–[18], and little is known about the prevalence and transmission mechanism of the cfr gene in these bacteria. Here, we present the first study on the prevalence of the cfr gene in E. coli isolated from food animals in China. In addition, the main transmission mechanism of the cfr gene in E. coli was characterized by plasmid and genetic environment analysis.
Materials and Methods
Ethics statement
This study protocol was reviewed and approved by the South China Agriculture University Animal ethics committee. The owners of the farm animals from which faecal swabs were taken gave permission for their animals to be used in this study.
Bacterial strains and Antimicrobial susceptibility testing
A total of 839 E. coli isolates were isolated from faecal swabs of diseased food-producing animals submitted to the Veterinary Research Institute, Guangdong Academy of Agricultural Sciences, in Guangdong Province of China during 2010 and 2012 (Table 1). Between three and five herds were sampled from each farm, and all the samples were from 225 farms all over Guangdong province. Bacterial DNA was extracted by a DNA extraction kit (Omega, USA) following the manufacturer's instructions. The presence of the cfr gene in E. coli was determined by PCR amplification and sequence analysis with the primers described in a previous study [4]. The susceptibilities of cfr-positive isolates to ceftiofur, ampicillin, cefotaxime, streptomycin, kanamycin, gentamicin, amikacin, florfenicol, chloramphenicol, tetracycline, ciprofloxacin, enrofloxacin, and trimethoprim-sulfamethoxazole were tested by agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [19]. Escherichia coli ATCC 25922 was used as the control strain.
Table 1. Information on the E. coli isolates used in this study.
| Year of isolation | Pig | Chicken | Duck | ||||||
| No. of faecal swabs | No. of farms | No. of isolates | No. of faecal swabs | No. of farms | No. of isolates | No. of faecal swabs | No. of farms | No. of isolates | |
| 2010 | 264 | 61 | 255 | 105 | 30 | 103 | 96 | 23 | 94 |
| 2011 | 89 | 25 | 87 | 57 | 17 | 56 | 42 | 10 | 38 |
| 2012 | 124 | 34 | 116 | 41 | 10 | 41 | 51 | 15 | 49 |
PFGE
Pulsed field gel electrophoresis analysis of XbaI-digested genomic DNA of all cfr-positive strains was performed using the CHEF-MAPPER System (Bio-Rad Laboratories) as described previously [20]. The PFGE patterns were analyzed with BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice similarity coefficient with a cut-off at 80% of the similarity values to indicate identical PFGE types.
Assay of cfr transfer
Mating experiments were performed as previously described [21], using azide-resistant E. coli J53 or streptomycin-resistant E. coli C600 as recipient strain. Transconjugants were selected on tryptic soy agar plates containing florfenicol (10 mg/L) and azide (100 mg/L) or streptomycin (512 mg/L). Plasmid DNA of cfr-positive strains was extracted by QIAGEN Plasmids Midi Kit, and was then transformed by electroporation into the recipient strain E. coli DH10B. Putative transformants were selected on brain heart infusion agar plates containing florfenicol (10 mg/L).
Plasmid characterization
The size of cfr-carrying plasmid in every parental strain and their transformants was determined using S1-treated genomic DNA followed by PFGE and southern hybridization with probe specific for cfr gene. All plasmids from transformants were further analysed by restriction fragment length polymorphism (RFLP) using EcoRI (TaKaRa Biotechnology, Dalian, China) restriction enzyme followed by hybridization performed on RFLP gels. Because of the failure in electrotransformation, the cfr-carrying plasmid in strain FS13Z3C was analyzed with plasmid extracted from host cell. To investigate the genetic environment of the cfr gene, 3 pairs of primers used in previous studies [4], [17], [22] were used for PCR mapping, inverse PCR and sequencing based on the known structure in earlier studies [16], [17].
Results
Bacterial strains and Antimicrobial susceptibility testing
Among the 839 E. coli isolates, 10 isolates from pig were positive for the cfr gene as determined by PCR, which was further confirmed by sequencing the PCR product. Sampling information showed that all the cfr-carrying isolates were from different farms except strains 8ZB6D and 8ZG6D which were isolated from different animals of the same farm. Susceptibility testing showed that all the cfr-positive E. coli strains presented a multiresistance phenotype, including resistance to chloramphenicols, quinolones, ampicillin, kanamycin, gentamicin, tetracycline and trimethoprim-sulfamethoxazole (Table 2).
Table 2. Characteristics of the cfr-positive E. coli strains and the corresponding cfr-carrying plasmids.
| Strain | Year of isolation | Resistance profilea | Phylogroup | cfr-carrying plasmid | |||
| transformation | approximate size (kb) | EcoRI fragment(kb) | genetic environment of cfr b | ||||
| FS-P54 | 2010 | CEF,AMP,CTX,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | B | + | 30 | 18 | A |
| 1ZF13D | 2011 | AMP,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | D | + | 30 | 18 | A |
| 2ZX7S | 2011 | AMP,STR,KAN,GEN,AMK,FFC,CHL,TET,CIP,ENR,SXT | A | + | 45 | 23 | B |
| 3ZX12D | 2011 | AMP,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | F | + | 30 | 18 | A |
| 8ZG1D | 2011 | AMP,STR,KAN,GEN,AMK,FFC,CHL,TET,CIP,ENR,SXT | G | + | 9,75 | 9,23 | C |
| 8ZB6D | 2011 | AMP,CTX,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | A | + | 30 | 18 | A |
| 8ZG6D | 2011 | AMP,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | F | + | 30 | 18 | A |
| 8ZG8D | 2011 | CEF,AMP,CTX,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | E | + | 30 | 18 | A |
| 8ZG12D | 2011 | CEF,AMP,CTX,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | C | + | 30 | 21 | D |
| FS13Z3C | 2012 | CEF,AMP,CTX,STR,KAN,GEN,FFC,CHL,TET,CIP,ENR,SXT | H | - | 30 | 18 | A |
CEF, ceftiofur; AMP, ampicillin; CTX, cefotaxime; STR, streptomycin; KAN, kanamycin; GEN, gentamicin; AMK, amikacin; FFC, florfenicol; CHL, chloramphenicol; TET, tetracycline; CIP, ciprofloxacin; ENR, enrofloxacin; SXT, Trimethoprim-sulfamethoxazole.
PFGE
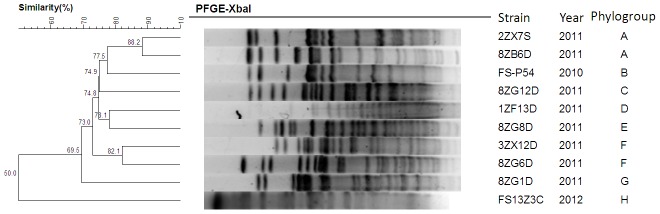
The dendrogram of the cfr-positive strains generated from cluster analysis of the PFGE profiles is shown in Figure 1. The 10 strains displayed different PFGE patterns and belonged to 8 phylogenetic groups (designated A to H). Most of the phylogenetic groups comprised a single strain, while Group A and F were represented by 2 strains, respectively.
Figure 1. UPGMA dendrogram, PFGE patterns, phylogenetic group and isolation date of cfr-positive E. coli.
Transfer of resistance
The conjugation experiment of cfr-positive strains using E. coli C600 or E. coli J53 as recipient strain failed, but electrotransformation was achieved in all the strains except FS13Z3C. Susceptibility testing of the 9 transformants revealed drastically increased florfenicol MICs (range, 16 to >256 mg/L) compared with DH10B (1 mg/L). Co-transfer of resistance to at least one other antimicrobial was observed in 8 transformants except 8ZG1D-21, which was only resistant to florfenicol. Transformant 8ZG1D-21 was moderately resistant to florfenicol with MIC of 16 mg/L and sensitive to chloramphenicol with MIC of 8 mg/L.
Plasmid characterization
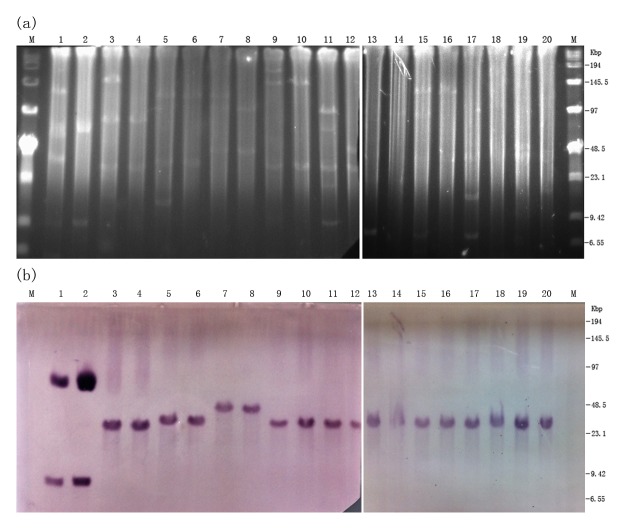
The result of S1-PFGE revealed that the ten cfr-positive strains possessed multiple plasmids of varying sizes, and at least two plasmids were electroporated into the recipient strain. Southern hybridization with cfr-specific probe identified the cfr gene located on an approximately 30 kb plasmid in all cfr-positive strains except 8ZG1D and 2ZX7S (Figure 2 and Table 2). Strain 2ZX7S had a cfr-carrying plasmid of ∼45 kb and strain 8ZG1D harbored two cfr-positive plasmids that were ∼9 kb and ∼75 kb in size, respectively. The RFLP profiles of cfr-carrying plasmids from 9 transformants and the cfr-positive strain FS13Z3C were obtained with EcoRI digestion and are presented in Figure 3. The plasmids of 5 transformants and the cfr-positive strain FS13Z3C showed similar pattern (lanes 3, 5, 6, 8, 9 and 10 in Fig. 3). Although hybridization bands of undigested plasmids due to incomplete enzyme digestion were observed in 3 lanes (Figure 3 lanes 1, 2 and 4), southern hybridization performed on the RFLP gel revealed that the plasmids yielded cfr-harboring fragments ranging from 9 to 23 kb, in which a fragment of ∼18 kb was observed in 7 out of the 8 plasmids of ∼30 kb. Additionally, even though the 2 cfr-carrying plasmids in 8ZG1D-21 cannot be characterized respectively by RFLP, hybridization showed that the 2 cfr-positive plasmids of ∼9 and ∼75 kb yielded cfr-carrying fragments of ∼9 and ∼23 kb separately.
Figure 2. Location of the cfr gene in the 10 E. coli strains and their corresponding transformants.
(a) S1-PFGE of the cfr-positive strains and their transformants, (b) subsequent southern hybridization with cfr-specific probe. Lanes: M, Low Range PFG Marker; 1, 8ZG1D; 2, 8ZG1D-21; 3, 8ZG12D; 4, 8ZG12D-50; 5, 1ZF13D; 6, 1ZF13D-22; 7, 2ZX7S; 8, 2ZX7S-41; 9, 8ZG6D; 10, 8ZG6D-59; 11, 3ZX12D; 12, 3ZX12D-6; 13, 8ZG8D; 14, 8ZG8D-81; 15 FS-P54; 16,FS-P54-2; 17,8ZB6D; 18, 8ZB6D-30; and 19,FS13Z3C.
Figure 3. RFLP and hybridization profiles of cfr-carrying plasmids in the 9 transformants and strain FS13Z3C.
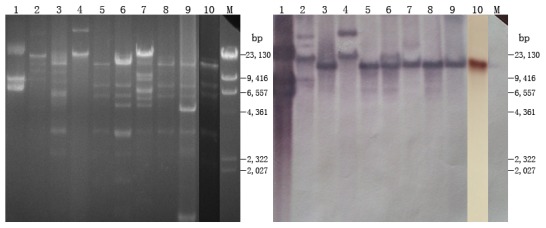
Lanes: M, λ-HindIII marker; 1, 8ZG1D-21; 2, 8ZG12D-50; 3 1ZF13D-22; 4, 2ZX7S-41; 5, 8ZG6D-59; 6, 3ZX12D-6; 7, 8ZB6D-30; 8, FS-P54-2; 9, FS13Z3C; and 10 8ZG8D-81.
Genetic environment of cfr
The result of PCR mapping and sequencing revealed the presence of 4 different genetic environments (Figure 4a-d; GenBank accession number KJ453116, KJ453117, KJ453114 and KJ453115) with the cfr gene flanked by two copies of IS26 located in the same orientation. Among the 8 plasmids of ∼30 kb, 7 plasmids shared the similar genetic environment, in which the cfr gene was oriented in the opposite direction of IS26 (Figure 4a). In contrast, the other 3 strains showed different environments, in which cfr was in the same orientation with IS26 (Figure 4b–d). Structural comparison of the genetic environments showed localized high homology (>98%) with plasmid pEC-01 from E. coli LYP-C-BCTb11 and chromosomal fragment from P. vulgaris PV-01 [16], [17]. To determine the stability of these structures, inverse PCR was performed, and amplicons of approximately 1570 bp (Figure 4a), 1562 bp (Figure 4b), 2520 bp (Figure 4c) and 4412 bp (Figure 4d) were obtained. Sequence analysis of these amplicons further confirmed the structure of these genetic environments obtained by PCR mapping.
Figure 4. Genetic environment of the cfr gene in this study, and structural comparison with plasmids pEC-01 (accession number JN982327) from E. coli LYP-C-BCTb11 and Chromosomal fragment from P. vulgaris PV-01 (accession number JF969273).
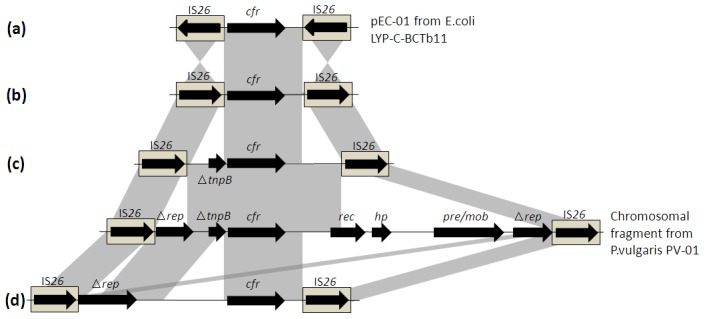
The arrows represent the positions and transcriptional direction of the ORFs. The IS26 elements are shown as light grey boxes. Regions with homology of over 98% are indicated by grey shading. Bacteria corresponding to each genetic environment are as follows: structure a (FS-P54, 1ZF13D, 3ZX12D, 8ZB6D, 8ZG6D, 8ZG8D and FS13Z3C), structure b (2ZX7S), structure c (8ZG1D), structure d (8ZG12D) (see Tables 2).
Discussion
In our study, 10 cfr-positive isolates were detected from 839 E. coli isolated between 2010 and 2012, and all the positive isolates were from swine farms where florfenicol is extensively used to prevent and cure diseases caused by a variety of bacterial pathogens in China [23]. PFGE analysis of the 10 cfr-positive E. coli revealed that these strains were genetically divergent. Most of the strains form a distinct phylogenetic group in the dendrogram based on genetic similarity, which suggested that the spread of the cfr gene in E. coli was not due to clonal dissemination but horizontal transfer.
S1 nuclease PFGE and hybridization showed that the cfr gene was located on an approximately 30 kb plasmid in all but two cfr-positive strains. Strains 2ZX7S and 8ZG1D harbored cfr-carrying plasmids of different sizes. The failure of the conjugation assays using cfr-positive strains suggests that the cfr gene was carried by nonconjugative plasmids in these strains. Recently cfr was identified on a conjugative plasmid in E. coli [18], which may further accelerate the dissemination of the cfr gene among different Gram-negative bacteria. Although we were not successful in obtaining transformants with a single plasmid after several attempts, RFLP and hybridization profiles of the plasmids showed that 7 cfr-carrying plasmids yielded the same-sized cfr-harboring fragment of ∼18 kb after digestion with EcoRI, and all the 7 plasmids are ∼30 kb in size with the same genetic environments confirmed by PCR mapping and inverse PCR, implying that the 7 plasmids are likely identical and originated from the same source. Interestingly, compared with the 9 strains in which a single cfr-carrying plasmid was harbored, strain 8ZG1D and the transformant 8ZG1D-21 have two cfr-positive plasmids. Considering the presence of IS26 flanking the cfr gene, IS-mediated recombination may account for the transfer of cfr between plasmids in the strain. Interestingly, the result of antimicrobial susceptibility testing showed that 8ZG1D-21 was moderately resistant to florfenicol and sensitive to chloramphenicol, suggesting that the cfr gene may confer low-level resistance to chloramphenicols in E. coli.
Four different genetic environments were detected surrounding the cfr gene, all of which have two copies of IS26 of the same orientation flanking the cfr gene. Previous studies have suggested that insertion element IS26 can mediate the transfer of cfr gene [16], [17]. In our study, structural comparison of the genetic environments showed that part of the segment between IS26 shares high homology (>98%) with plasmid pEC-01 from E. coli LYP-C-BCTb11 and chromosomal fragment from P. vulgaris PV-01 [16], [17], further suggesting that IS26 may have played an important role in the transfer of the cfr gene. Furthermore, inverse PCR performed on all of the cfr-positive strains can obtain an amplicon, and subsequent sequencing analysis showed that the pair of intact IS26 flanking the cfr gene can loop out the intervening sequence through homologous recombination, which can further accelerate the transfer of cfr gene.
To conclude, we present the first study on the prevalence of the cfr gene in E. coli from food producing animals. The identified cfr-positive E. coli strains were limited to pigs, coinciding with the extensive use of florfenicol for swine production. PFGE analysis showed that the cfr-positive E. coli strains were genetically diverse; however, plasmid and genetic environment analysis suggested that most of these strains harbored the same cfr-carrying plasmid of ∼30 kb. Additionally, the results suggest that transposition and homologous recombination mediated by IS26 as well as transformation of cfr-carrying plasmids may be the main mechanism for horizontal spread of the cfr gene in E. coli. Further surveillance and investigation are necessary to facilitate the control of cfr spread in gram-negative bacteria.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The sequences identified in the study (see Fig. 4) have been deposited in GenBank under accession numbers KJ453116 (Fig. 4a), KJ453117 (Fig. 4b), KJ453114 (Fig. 4c) and KJ453115 (Fig. 4d) respectively.
Funding Statement
This work was supported by the National Science Fund for Distinguished Young Scholars (Grant No. 31125026), the National Natural Science Foundation (Grant No. 31272609), Program for Changjiang Scholars and Innovative Research Team in University of Ministry of Education of China (Grant No. IRT13063) and Science and Technology Program of Guangzhou, China (Grant No. 2011J2200054). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B (2006) The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob Agents Chemother 50: 2500–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smith LK, Mankin AS (2008) Transcriptional and Translational Control of the mlr Operon, Which Confers Resistance to Seven Classes of Protein Synthesis Inhibitors. Antimicrob Agents and Chemotherapy 52: 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwarz S, Werckenthin C, Kehrenberg C (2000) Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri . Antimicrob Agents Chemother 44: 2530–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kehrenberg C, Schwarz S (2006) Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50: 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toh SM, Xiong L, Arias CA, Villegas MV, Lolans K, et al. (2007) Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol Microbiol 64: 1506–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dai L, Wu CM, Wang MG, Wang Y, Huang SY, et al. (2010) First report of the multidrug resistance gene cfr and the phenicol resistance gene fexA in a Bacillus strain from swine feces. Antimicrob Agents Chemother 54: 3953–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y, Schwarz S, Shen Z, Zhang W, Qi J, et al. (2012) Co-location of the multiresistance gene cfr and the novel streptomycin resistance gene aadY on a small plasmid in a porcine Bacillus strain. J Antimicrob Chemother 67: 1547–1549. [DOI] [PubMed] [Google Scholar]
- 8. Zhang WJ, Wu CM, Wang Y, Shen ZQ, Dai L, et al. (2011) The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J Antimicrob Chemother 66: 1174–1175. [DOI] [PubMed] [Google Scholar]
- 9. Liu Y, Wang Y, Wu C, Shen Z, Schwarz S, et al. (2012) First report of the multidrug resistance gene cfr in Enterococcus faecalis of animal origin. Antimicrob Agents Chemother 56: 1650–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y, Wang Y, Schwarz S, Li Y, Shen Z, et al. (2013) Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother 57: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Wang Y, Schwarz S, Wang S, Chen L, et al. (2014) Investigation of a multiresistance gene cfr that fails to mediate resistance to phenicols and oxazolidinones in Enterococcus faecalis . J Antimicrob Chemother 69: 892–898. [DOI] [PubMed] [Google Scholar]
- 12. Diaz L, Kiratisin P, Mendes RE, Panesso D, Singh KV, et al. (2012) Transferable plasmid-mediated resistance to linezolid due to cfr in a human clinical isolate of Enterococcus faecalis . Antimicrob Agents Chemother 56: 3917–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Li D, Song L, Liu Y, He T, et al. (2013) First report of the multiresistance gene cfr in Streptococcus suis . Antimicrob Agents Chemother 57: 4061–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Y, Schwarz S, Shen Z, Zhou N, Lin J, et al. (2012) Detection of the staphylococcal multiresistance gene cfr in Macrococcus caseolyticus and Jeotgalicoccus pinnipedialis . J Antimicrob Chemother 67: 1824–1827. [DOI] [PubMed] [Google Scholar]
- 15. Shen J, Wang Y, Schwarz S (2013) Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68: 1697–1706. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, He T, Schwarz S, Zhou D, Shen Z, et al. (2012) Detection of the staphylococcal multiresistance gene cfr in Escherichia coli of domestic-animal origin. J Antimicrob Chemother 67: 1094–1098. [DOI] [PubMed] [Google Scholar]
- 17. Wang Y, Wu CM, Schwarz S, Shen Z, Zhang W, et al. (2011) Detection of the staphylococcal multiresistance gene cfr in Proteus vulgaris of food animal origin. J Antimicrob Chemother 66: 2521–2526. [DOI] [PubMed] [Google Scholar]
- 18. Zhang WJ, Xu XR, Schwarz S, Wang XM, Dai L, et al. (2014) Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr . J Antimicrob Chemother 69: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI (2012) Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. CLSI document M100-S22 Wayne, PA: Clinical and Laboratory Standards Institute.
- 20. Gautom RK (1997) Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol 35: 2977–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen L, Chen ZL, Liu JH, Zeng ZL, Ma JY, et al. (2007) Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J Antimicrob Chemother 59: 880–885. [DOI] [PubMed] [Google Scholar]
- 22. Dhanji H, Patel R, Wall R, Doumith M, Patel B, et al. (2011) Variation in the genetic environments of bla (CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66: 1005–1012. [DOI] [PubMed] [Google Scholar]
- 23. Wang Y, Zhang W, Wang J, Wu C, Shen Z, et al. (2012) Distribution of the multidrug resistance gene cfr in Staphylococcus species isolates from swine farms in China. Antimicrob Agents Chemother 56: 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The sequences identified in the study (see Fig. 4) have been deposited in GenBank under accession numbers KJ453116 (Fig. 4a), KJ453117 (Fig. 4b), KJ453114 (Fig. 4c) and KJ453115 (Fig. 4d) respectively.