Abstract
The wild tobacco species Nicotiana attenuata has been intensively used as a model plant to study its interaction with insect herbivores and pollinators in nature, however very little is known about its native pathogen community. We describe a fungal disease outbreak in a native N. attenuata population comprising 873 plants growing in an area of about 1500 m2. The population was divided into 14 subpopulations and disease symptom development in the subpopulations was monitored for 16 days, revealing a waxing and waning of visible disease symptoms with some diseased plants recovering fully. Native fungal N. attenuata pathogens were isolated from diseased plants, characterized genetically, chemotaxonomically and morphologically, revealing several isolates of the ascomycete genera Fusarium and Alternaria, that differed in the type and strength of the disease symptoms they caused in bioassays on either detached leaves or intact soil-grown plants. These isolates and the bioassays will empower the study of N. attenuata-pathogen interactions in a realistic ecological context.
Introduction
Pathogens from natural plant communities have largely been ignored for a long time, as Dinoor and Esched pointed out in their review in 1984, since phytopathologists have focused mainly on crop plants and plant ecologists have often failed to recognize the impact of phytopathogens on natural plant communities due to the general low incidence of disease in natural populations [1]. However, this changed as plant breeders soon started examining wild relatives of cultivated crop plants as potential sources of pathogen resistance traits [2]–[4] and wild plant pathosystems became valuable models for the study of plant-pathogen coevolution and evolutionary biologists and ecologists began to examine the influence of various biotic and abiotic factors, such as host and pathogen genetic diversity, habitat and community structures, and time scales on these important interactions [5]–[14].
Due to its synchronized germination from long-lived seed banks after sporadically occurring wildfires, Nicotiana attenuata typically forms monoculture-like populations exhibiting great genetic diversity and phenotypic plasticity [15]–[20]. These qualities make N. attenuata a very interesting model plant for studies of wild plant-pathogen interactions [21], [22]. However, in contrast to the tremendous repertoire of knowledge on N. attenuata ´s interactions with its native herbivores gained from the past two decades of field and laboratory research [23], reports on native phytopathogens of this native tobacco are scarce. One example is given by Rayapuram and Baldwin (2007) who observed disease symptoms caused by phytopathogenic Pseudomonas spp. on N. attenuata in nature [24]. Because native pathogens for bioassays on N. attenuata were lacking, common lab pathogen strains have been used, such as Pseudomonas syringae pv. tomato DC 3000 [25], the oomycete Phytophthora parasitica var. nicotianae and the ascomycete fungus Fusarium oxysporum f. sp. tuberosi [26]. The inferences from these studies with non-native pathogens may not be applicable to the real interactions that occur in nature [27]. Therefore the search for native N. attenuata pathogens has become more important in order to improve ecological relevance of the pathogen bioassay used to acquire ecologically relevant insights into the interaction of N. attenuata with its pathogens.
In the summer of 2011, populations of N. attenuata growing in the Great Basin Desert, Utah, USA suffered a dynamic disease outbreak likely caused by fungal pathogens. This study describes the dynamics of this fungal disease outbreak in a natural N. attenuata population, and how this outbreak was used to obtain native fungal isolates that were phylogenetically and biochemically characterized, and how candidate isolates with high pathogenicity potential were selected to establish laboratory bioassays with N. attenuata.
Results
The dynamics of disease symptoms a native Nicotiana attenuata population
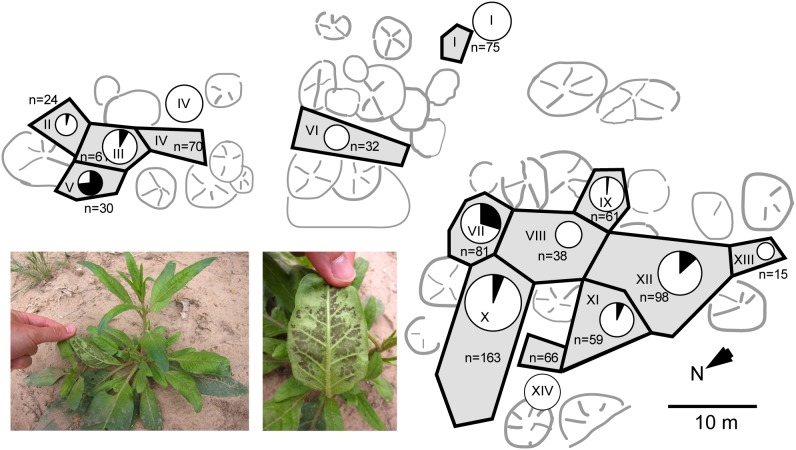
During 2011 summer, a native N. attenuata population comprising 873 plants growing in an area of approximately 1500 m2 suffered a disease outbreak. The occurrence and strength of disease symptoms (dark spots on abaxial leaf surfaces which resembled fungal mycelium or sporangium-like structures shown in Figure 1 and 2, chlorosis, necrosis and curly leaves) were monitored during a 16 day-time interval (Table S1). The population was divided into 14 subpopulations (“sections”) according to habitat characteristics (grassland, shrubs, trees and areas free of N. attenuata plants) to better assess the disease dynamics. To be able to track the disease development for each individual plant, its position was marked in a schematic map of the respective section and assigned a number (Figure 1). All plants were categorized into four different disease stages as described in detail in the Materials and Methods section.
Figure 1. Schematic map illustrating the spatial distribution of diseased N. attenuata plants in a natural population.
The plant number (n) is indicated for each section of the population (labeled I to XIV). The area of the circles represents the number of plants in each section. The black fractions represent the percentage of diseased plants in each section surveyed on the second monitoring (June 9th 2011).
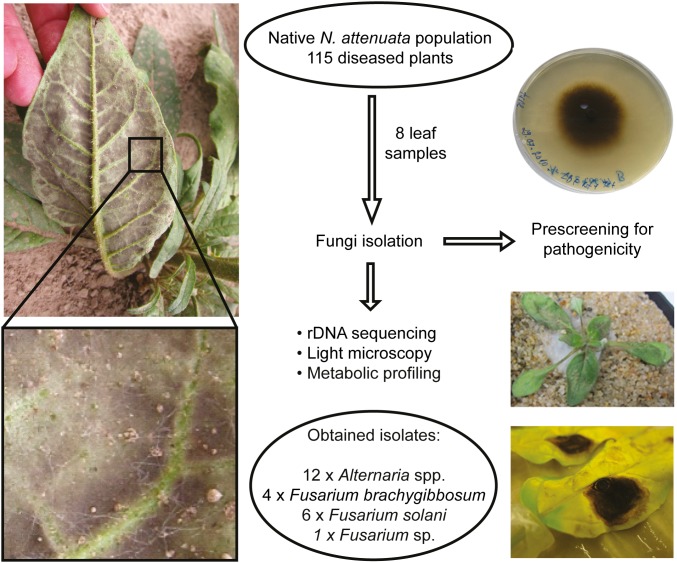
Figure 2. Work flow for the isolation of the native fungal pathogen isolates used in this study.
In a native N. attenuata population plants suffered from disease symptoms (chlorosis, curly leaves and dark mycelium-like structures on the abaxial side, illustrated on the left). Leaf samples were taken from diseased plants for fungi isolation. Pathogenicity of the isolates was evaluated using different infection methods. Fungi, which were able to cause severe disease symptoms on plants were re-isolated from infected plant material and further characterized by rDNA sequencing, microscopy and their polar metabolite profiles, resulting in 12 Alternaria spp., 4 Fusarium brachygibbosum, 6 F. solani and one Fusarium sp. isolates.
Even heavily diseased plants (disease stage category 3) with necrotic rosette leaves survived and attained the flowering stage. However, some of the diseased plants showed flower deformation with a shortened and bent corolla and protruding pistils. N. attenuata plants of different developmental stages (varying from young rosette stage to already flowering plants) occurred together in close association, but no stage-dependent disease prevalence was observed. Indeed, completely asymptomatic and heavily diseased plants of all plant developmental stages were found growing immediately next to each other. In some cases, the distance between diseased and asymptomatic plants was so small that their rosette leaves were touching each other.
Similar to the situation with neighboring plants, no disease symptom accumulation pattern could be observed on the scale of the different population sections. Sections with a relatively high percentage of diseased plants [section II: 41.7% (May 24th); section V: 66.7% (June 9th); section VII: 29.6% (June 9th)] or intermediate percentage [section III: 8.2% (June 9th); section X: 9.2% (May 24th); section XII: 13.3% (June 9th)] were not adjacent to each other and separated by nearly unaffected sections [section VIII: 2.6% (May 24th); section IX: 3.3% (May 24th); section XIII: 6.7% (May 24th)] or completely unaffected ones (sections I, IV, VI and XIV) (Figure 1 and Table S1). Only sections I and VI were spatially isolated from the other sections and entirely lacked diseased plants (Table S1).
With regard to the temporal pattern of disease symptom occurrence, 53.2% of the diseased plants recovered completely between the two surveys of the native N. attenuata population, while 6.5% of previously asymptomatic plants suddenly showed disease symptoms (Table S1). Remarkably, in section II, 90% of previously diseased plants recovered completely and no plant with progressing or newly occurring disease symptoms was found during the second survey (Table S1 and Figure S1). In contrast, in section VII, all plants were asymptomatic during the first survey and 29.6% suddenly showed disease symptoms during the second survey (Table S1 and Figure S1). In other sections (e.g. section X), the number of newly diseased (8.0%) and fully recovered (4.9%) plants was very similar and the overall number of diseased plants remained nearly unchanged (9.2% vs. 6.1%) (Table S1 and Figure S1). With regard to the entire population, the number of diseased N. attenuata plants increased slightly from 7.1% to 9.4%. Approximately 37.9% of all diseased plants showed a progression of disease symptoms, while 56.5% partly or fully recovered (Table S1). Overall, the disease outbreak turned out to be much more dynamic than could be inferred from only looking at the changes in the average values per section. In summary, diseased N. attenuata plants showed an astonishing capacity to fully recover.
Classification of the native fungal isolates by ribosomal DNA sequence analysis
In order to identify and characterize potential causal agents responsible for the disease outbreak in native N. attenuata populations, eight leaf samples showing characteristic disease symptoms were collected and used to obtain fungal isolates. The isolation and characterization strategy is summarized in Figure 2. The fungal isolates were morphologically different and tested for their capacity to cause disease symptoms using the following inoculation procedure: the hypocotyls of 24 day-old N. attenuata plants were pricked with a needle before the application of actively growing fungus cultures to the wound sites as described in Bonaventure et al. [23]. This infection method provided reliable infections even for pathogens such as Phytophthora parasitica var. nicotianae and Fusarium oxysporum f. sp. tuberosi which are barely able to infect N. attenuata via spore suspension, and was therefore used to identify fungal pathogen candidates amongst the obtained native fungal isolates.
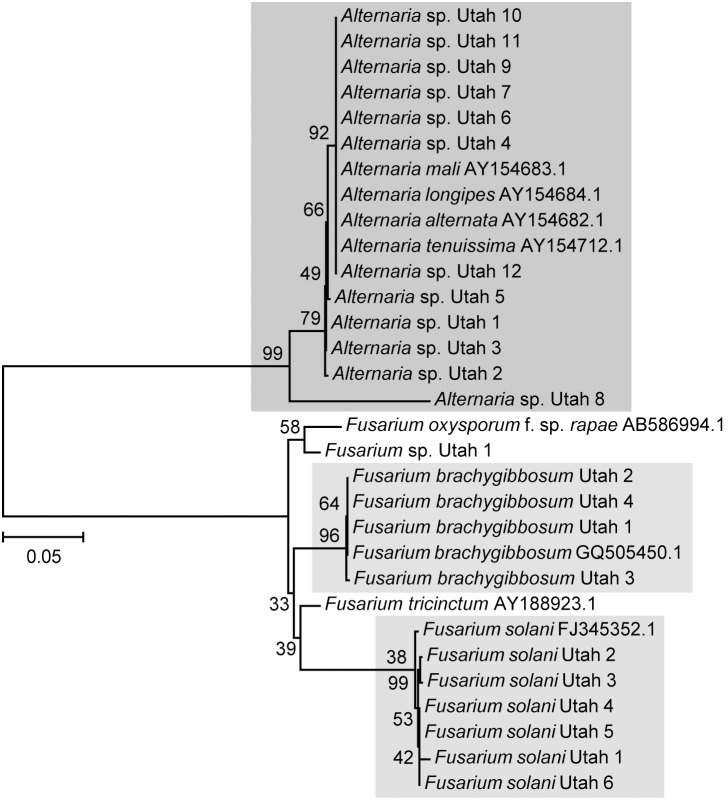
23 native fungal isolates that caused disease symptoms were used for phylogenetic analysis (genomic DNA extraction and sequencing of their nuclear ITS-LSU rDNA region). The length of the amplified DNA region varied between the different fungal isolates from 1090 to 1156 bp, except for a sequence of a particular Alternaria isolate (Alternaria sp. Utah 6) being only 585 bp big due to poor quality PCR amplification. The rDNA sequences were aligned with ClustalW algorithm together with sequences of known reference strains, and a phylogenetic tree was constructed using the MEGA5 software (neighbor-joining tree using Kimura-2-parameter with 1000 bootstrap replications). The assignment to certain fungal species/genera via sequence similarity was performed according to criteria used by Bosshard et al. (2003) [28]. If the percentage of similarity of the query sequence and the reference sequence was 99% or above, the unknown isolate was assigned to the reference species. When percentage similarity was between 95 and 99%, the unknown isolate was assigned to the corresponding genus. Four native fungal isolates revealed 99% sequence homology to Fusarium brachygibbosum strain NRRL 34033 (GenBank accession number GQ505450.1) and were therefore assigned to F. brachygibbosum Utah 1 to 4 (Figure 3). Six native fungal isolates had 99% sequence similarity to Fusarium solani strain ATCC 56480 (GenBank accession number FJ345352.1) and were thus renamed to F. solani Utah 1 to 6 (Figure 3). The sequence of one native fungal isolate showed 98% sequence similarity to Fusarium oxysporum f. sp. rapae (GenBank accession number AB586994.1) and was therefore classified only at genus level as Fusarium spp. Utah 1 (Figure 3). Twelve other native fungal isolates revealed 99% sequence similarity to Alternaria alternata (GenBank accession number AY154682.1), Alternaria tenuissima strain IA287 (GenBank accession number AY154712.1), Alternaria longipes (GeneBank accession number AY154684.1) and Alternaria mali (GeneBank accession number AY154683.1), indicating that these twelve native fungal isolates belong to the genus Alternaria. However, they were indistinguishable at species level by rDNA sequence comparison (Figure 3).
Figure 3. Phylogenetic relationship of native fungal pathogen isolates based on their rDNA sequence.
The tree was constructed using the neighbor-joining method based on the sequences of ITS regions plus LSU using MEGA5 software. Sequences were aligned using ClustalW algorithm implemented in the MegAlign software. The numbers on the nodes indicate bootstrap scores in NJ analyses (1000 steps). The number on the scale indicates distance level with relative units.
Chemotaxonomic characterization of the native fungal isolates
To support the fungal phylogeny data obtained by rDNA sequence analysis, we followed a polar metabolite profiling approach with the different native fungal isolates using liquid chromatography-time-of-flight-mass spectrometry (UPLC-ToF-MS). Fungal mycelium of the native pathogen isolates was scratched from the surface of 14 day-old fungal cultures and used for polar metabolite extraction. Negatively charged ions eluting from the column within a retention time of 3 to 481 s were analyzed by mass-spectrometry. Those having mass-to-charge values ranging from 107 to 1200 were selected for subsequent data analysis. Using the conditions mentioned above and after data processing, a total of 559 ions were identified. The cluster analysis of mass profiles with the MetaboAnalyst software was summarized as a dendrogram (Figure S2A) and showed a separation between the two fungal genera Fusarium and Alternaria. Within the genus Fusarium, a dichotomous clustering of F. solani and F. brachygibbosum isolates could be shown. The genus Alternaria clustered into three major subgroups, one containing Alternaria spp. Utah 4, 8 and 10, a second one with Alternaria spp. Utah 1, 11 and 12, and a third one with Alternaria spp. Utah 2, 3, 5, 6 and 9. To facilitate the graphical interpretation of the differences among the native fungal isolates, the data set corresponding to the differentially accumulating ions was subjected to a principal component analysis. The result is shown in Figure S2B for the separation of Alternaria spp. from F. solani and F. brachygibbosum isolates and in Figure S2C for the separation of the three different Alternaria isolate subgroups. Interestingly, the polar metabolite profiles of Alternaria spp. isolates revealed more within-taxon differences compared to both Fusarium species (F. solani and F. brachygibbosum) taken together (Figure S2B).
Since it is known that small-spored Alternaria species are hard to distinguish by rDNA sequence analysis [29], we also analyzed species-specific differences in the production of certain mycotoxins to further classify the native Alternaria isolates [30]–[34]. Common Alternaria mycotoxins are Alternariol (AOH), Alternariol monomethyl ether (AME), Altertoxin I (Alx I), Tentoxin (Ten) and Altenuene. AOH and Ten were found in A. alternata and A. tenuissima, but not in A. mali and A. longipes [31]–[33]. AME was found to be produced by A. alternata, A. tenuissima and A. longipes, but not A. mali [31]–[33]. Alx I is present in all four Alternaria species [31]–[33]. To identify these Alternaria-mycotoxins, mass-over-charge values and retention times of those compounds available at a public database were used to verify the UPLC-ToF-MS data for the presence of such compounds in the native Alternaria isolates [34]. AOH and Alx I were found to be produced by all native Alternaria isolates, as well as AME (except Alternaria sp. Utah 11) (Table S2). The presence of these metabolites indicates that all native Alternaria isolates belong either to A. alternata or A. tenuissima. However, Tentoxin typically produced by A. alternata and A. tenuissima could be detected for Alternaria spp. Utah 4 and 8 at only very low levels (Table S2). Even though this analysis was still not sufficient to exactly classify the native Alternaria isolates at the species level, it narrowed the choice to A. alternata and A. tenuissima.
Morphological characterization of the native fungal isolates
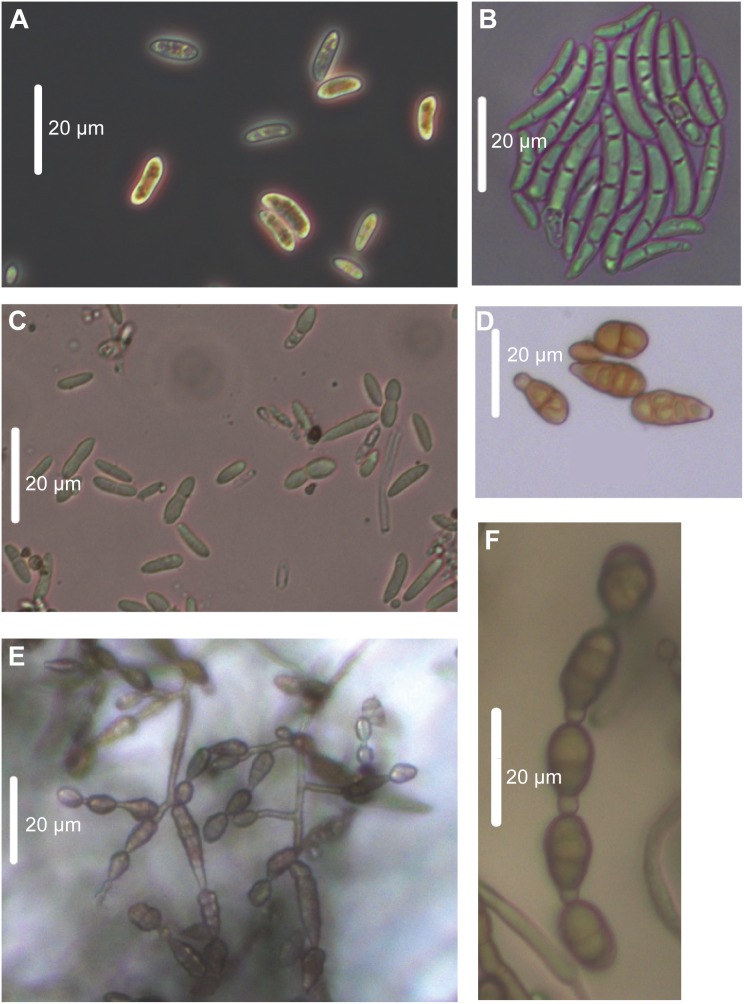
In addition, the native fungal isolates were also characterized morphologically by analyzing the sporulation structures and conidia shapes of 14 day-old fungal cultures under the light microscope (Figure 4). Typical conidia shape, color, length and septum number were compared with reference conidia pictures of Fusarium solani published online at http://tolweb.org/Sordariomycetes/29050/2008.01.14. Isolates of F. solani Utah 1 to 4 could be confirmed with typical banana-shaped 3- to 5-septate macroconidia (Figure 4B). Oval-shaped 1- to 2-celled Microconidia were abundant (Figure 4A). In contrast to Fusarium solani, very little information is published about F. brachygibbosum, especially regarding its morphological characteristics. Spore structures of F. brachygibbosum Utah 1 to 4 were observed under the microscope as colorless spores, 1- to 2-celled with ellipsoid shape and 4 to 13 µm in length and 2 to 4 µm in width (Figure 4C). All native Alternaria spp. isolates had the conidia shapes characteristic of this genus, i.e. mostly ovoid with short conical beak at the tip (or beakless) (Figure 4D–F) [35], [36]. They were multicellular with several vertical and transverse septa (Figure 4D). Conidia color was pale brown. Interestingly, Alternaria sp. Utah 10 was showing the typical sporulation pattern of A. alternata with a single suberect conidiophore and an apical cluster of conidia chains (Figure 4E). Such conidia structures are typical for A. alternata with branching chains of small conidia separated by short secondary conidiophores (Figure 4F). This similarity between Alternaria sp. Utah 10 and A. alternata references published by de Hoog et al. (2000) [35] supports the assumption that Alternaria sp. Utah 10 might be a native A. alternata isolate.
Figure 4. Morphology of native fungal pathogens observed by light microscopy.
Conidia of 14 day-old fungal cultures grown for 14 days on PDA in the dark at room temperature. A. Microconidia of F. solani Utah 2. B. Macroconidia of F. solani Utah 4. C. Fungal spores of F. brachygibbosum Utah 4. D. Alternaria-typical conidia. E. Alternaria sp. Utah 10 sporulation clump and conidia. F. Alternaria-typical conidia chain. The white bars represent 20 µm.
Development of bioassays using N. attenuata and its native fungal pathogens
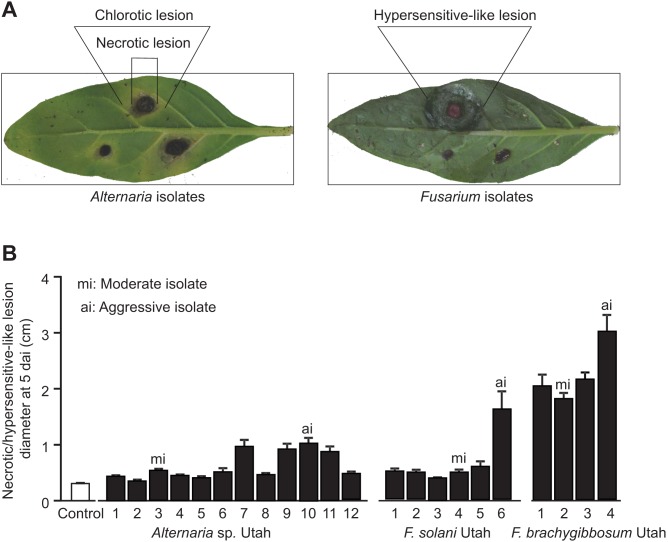
To assess the virulence of the individual fungal isolates on N. attenuata plants, a detached leaf assay was performed by placing agar plugs with fungal cultures on excised N. attenuata leaves (Figure 5). The differences in virulence of the native fungal isolates were evaluated based on average diameters of chlorotic, necrotic and hypersensitive-like lesions. Native F. brachygibbosum isolates were the first ones to cause visible symptoms in form of hypersensitive-like lesions after three days of incubation (Figure 5A). The lesions expanded rapidly from the inoculation points and started to overlap each other already two days later. Chlorotic lesions were not detected for F. brachygibbosum isolates. Only Alternaria spp. Utah 10 and 11 could induce chlorotic lesions already three days after inoculation, but after five days of incubation all detached N. attenuata leaves inoculated with native Alternaria isolates started to show chlorosis and necrosis (Figure 5A). Leaves inoculated with native F. solani developed chlorosis and necrosis at four to five days after inoculation. For leaves inoculated by native Alternaria and F. solani, necrotic lesions were measured at five days after inoculation and were used to evaluate the aggressiveness of the fungal isolates (Figure 5B). Among the twelve Alternaria isolates, Alternaria sp. Utah 10 which caused the largest necrotic lesions (1.03±0.09 cm diameter) appeared to be the most aggressive isolate, followed by Alternaria spp. Utah isolates 7, 9 and 11. Alternaria sp. Utah 3 was assigned as a moderate isolate since it was able to cause visible necrotic lesions which were significantly (P = 0.039) larger (0.54±0.02 cm) than the pure agar plug diameter from control leaves (0.31±0.01 cm), but significantly less aggressive than Alternaria spp. Utah isolates 7, 9, 10 and 11 (P<0.001) (Figure 5B). The necrotic lesions caused by Alternaria spp. Utah 1, 2, 4, 5, 6, 8 and 12 were not significantly different from the diameter of the pure agar plug (Figure 5B). Therefore these fungal strains were defined as being less aggressive isolates. Among the six native F. solani isolates, F. solani Utah 6 turned out to be an aggressive isolate since it caused the largest necrotic lesion (1.61±0.31 cm) at five days after inoculation compared to controls and the other native F. solani isolates (P<0.001) (Figure 5B). F. solani Utah 1, 2, 3, 4 and 5 were not significantly different from controls and therefore considered as less aggressive isolates (Figure 5B). Interestingly, F. brachygibbosum Utah caused hypersensitive-like lesions which were significantly bigger compared to the necrotic lesions caused by native Alternaria (P<0.001) and F. solani isolates (P = 0.002) (Figure 5B). At five days after inoculation on detached N. attenuata leaves, F. brachygibbosum Utah 4 was the most aggressive isolate with hypersensitive-like lesion diameters of 3.04±0.28 cm, followed by the rather moderate isolates F. brachygibbosum Utah 1 to 3 (Figure 5B). This experiment helped to assess the virulence of each native fungal isolate necessary for choosing the appropriate candidates for further bioassays.
Figure 5. Evaluation of fungal pathogenicity using detached leaf assay.
A. Disease symptoms caused by Alternaria and Fusarium on N. attenuata detached leaves. Agar plugs containing fungus culture were placed on detached leaves of N. attenuata and incubated at high humidity. Detached leaves incubated with pure PDA plugs were used as control. 12 biological replicates were used for each fungal isolate. Photos were taken at 3 and 5 days after inoculation for Fusarium and Alternaria isolates, respectively. B. Necrotic lesion diameters of Fusarium- and Alternaria-inoculated leaves at 5 days after inoculation (dai). The most aggressive isolates (ai) caused the largest necrotic/hypersensitive-like lesion on detached N. attenuata leaves while the moderate isolates (mi) caused visible necrotic/hypersensitive-like lesions only slightly larger than the diameter of pure agar plugs (control).
To be able to study the interaction between N. attenuata and its native pathogenic fungi in a more realistic way (i.e. mimicking natural infection conditions as much as possible), an infection assay using intact N. attenuata plants and fungal spore suspension needed to be developed for all three native fungal pathogen species. Based on the pathogenicity evaluation result from the detached-leaf assay, the most aggressive and a “moderate” isolate of native F. brachygibbosum, F. solani and Alternaria spp. were tested in intact-plant infection assays in order to identify the isolate with the most appropriate virulence for each native pathogen species. F. brachygibbosum Utah 2 was used as a moderate F. brachygibbosum isolate because it had the smallest necrotic lesion diameter (1.80±0.09 cm) compared to its conspecifics. For F. solani no moderate isolate was available, therefore the less virulent F. solani Utah 4 strain was randomly selected. Root-dip and spore-spraying methods were chosen to reflect the natural infection process of soil-borne and aerial pathogens as it would occur in the native habitat of N. attenuata. Development of disease symptoms on infected plants was observed and recorded for 20 days after inoculation by estimating the percentage of chlorosis and necrosis compared to the total leaf area. In contrast to diseased plants in natural N. attenuata populations, dark-spotted abaxial leaf sides, leaf curling and changes in flower morphology could not be observed within the 20 days after inoculation. Different plant ages (10 to 20 days after germination) were tested with regard to their suitability for infection. The work flow for the development of a suitable infection assay with native F. solani is described in detail in Figure 6 as an example of the workflow used. Inoculation of young N. attenuata seedlings (10 day-old) by spraying spore suspension of F. solani Utah 6 did not result in any diseased plant even at the highest spore concentration tested (107 spores mL−1) (Figure 6). This spore application method proved to be an ineffective inoculation procedure also for all of the other fungal isolates tested. In contrast, dipping seedling roots into spore suspension resulted in heavily diseased plants even with the lowest tested F. solani spore concentration (105 spores mL−1), indicating that the root-dip method was very effective for inoculation. However, inoculation of very young seedlings (10 days-old) resulted in disease symptoms that were too severe for experimentation and did not reflect the disease severity observed in the natural population. Therefore older plants (20 days-old) were also tested, but disease symptoms were still too severe in case of the aggressive F. solani Utah 6. F. solani Utah 4 caused much less severe and yet clearly visible disease symptoms, and provided the best bioassays reflecting a near-natural infection process. The infection assays for the two other native fungal species were tested and optimized in a similar manner. Inoculation with the most aggressive native F. brachygibbosum isolate (F. brachygibbosum Utah 4) to 20 day-old plants using the root-dip method did not result in any visible disease symptoms. Diseased plants could be obtained only when very young seedlings (10 day-old) were inoculated with the highest spore concentration (107 spores mL−1). F. brachygibbosum was barely infective when used on intact N. attenuata plants, which is in good agreement with the strong hypersensitive-like lesions on detached leaves pointing to a high degree of plant resistance to this pathogen [37]. A root-dip (105 spores mL−1) bioassay using 12 day-old N. attenuata seedlings and the most aggressive native Alternaria isolate (Alternaria sp. Utah 10) proved to be most appropriate experimental setup for this native fungal pathogen species.
Figure 6. Testing different infection setups using F. solani spore suspension and intact N. attenuata plants.
Two different infection methods (spore-spraying and root-dip), three different spore concentration (107, 106 and 105 spores mL−1), two different plant ages (10 and 20 days-old) and two different virulence levels (aggressive and moderate F. solani Utah isolate) were tested with intact soil-grown plants.
Discussion
Disease dynamics in a native N. attenuata population
Within only 16 days, disease symptom development in a native N. attenuata population appeared very dynamic, since more than half of all diseased plants recovered completely, while at the same time many previously asymptomatic plants started to show symptoms (Figures 1; Figure S1; Table S1). No clear spatial pattern was observed in the occurrence of disease symptoms within the native N. attenuata population at the two survey time points. The sections with high recovery rate (sections II, V and X) and sections with a high abundance of newly diseased plants (sections V, VII, X and XII) were spatially disjunct and distributed over almost the complete population, being separated by more or less unaffected population sections (Figure S1 and Table S1). The lack of a spatial pattern was also observed within population sections, since diseased plants were usually not clustered and surrounded by completely asymptomatic plants. Micro-spatial heterogeneity of spore density within the soil could be a potential explanation for the distribution of diseased plants [38]. However, our data seems to support another potential explanation for this phenomenon: native N. attenuata populations typically harbor a great deal of genetic diversity which could lead to high phenotypic plasticity even on very small spatial scale, likely due to their long-lived seed banks [16]–[18], [20], [39]. In contrast to domesticated crop plants, wild plant populations are genetically diverse and based on each plant´s individual genotype, the ability to cope with fungal pathogen attack may differ within a native N. attenuata population [16]–[18], [21], [22], [39]. This genetic variability within a population could augment the phenotypic plasticity that occurs within individuals and could explain the coexistence of individuals with different defense strategies, as has been shown for plant-herbivore interactions [16]–[18], [39].
Plant defense against pathogens implies signaling processes mediated by phytohormones such as salicylic and jasmonic acid, and depending on the individual plant´s genotype and this phytohormone signaling response to pathogen attack may vary even among neighboring plants [16], [18], [40]. Disease symptoms could be observed within a wide range of different plant developmental stages in the native N. attenuata population, suggesting a stage-independent disease prevalence. However, age-related resistance as reported for Nicotiana benthiamana to the oomycete pathogen, Phytophthora infestans, is not uncommon [41]. Indeed, the results from our bioassays suggest an age-related resistance of N. attenuata to native Alternaria spp., Fusarium solani and Fusarium brachygibbosum as well, since high infection efficiency was only observed when young plants (10 to 20 days after germination) were exposed to the pathogen. Therefore we speculate that plants from the native N. attenuata population were probably infected already at a very early developmental stage before the monitoring of disease symptoms started, and developed symptoms after a latency period which could vary among individuals. In addition to the proposed variability in this latency period, differences in the time of germination together with long-lasting conditions favorable for pathogen infection could also provide a plausible explanation for the wide range of plant developmental stages that were affected.
Such disease-promoting conditions could have been created by the weather situation a few weeks prior the first survey of diseased plants: for several days in a row, strong winds were blowing and sporadic rainfalls (sometimes in association with thunderstorms) created an unusually cool and moist environment in the native N. attenuata habitat in 2011. Even in the relatively dry environment of the Great Basin Desert, fungal fruiting bodies emerged out of the sandy soil surface between the native N. attenuata plants. The wind blew around a lot of sand and dust particles which could have caused small wounds on the plant surface and created potential entry sites for all sorts of pathogens. Such environmental conditions are thought to be conducive for infection for a variety of fungal pathogens [42]–[46].
Despite a population-wide slight increase in disease incidence and symptom severity (Table S1), none of the plants were observed to die from the disease. Together with the astonishing high capacity to recover from all symptoms, this suggests that the plants harbor effective and adaptive defense responses against the causal disease agent(s). This property makes N. attenuata an interesting plant species to study plant defense against pathogens. While the disease outbreak did not result in any mortality within the monitored native N. attenuata population, it clearly seriously reduced the Darwinian fitness of diseased plants by reducing growth (leaf damage) and reproduction (flower deformation).
A complex of Alternaria and Fusarium species as the causal agent for the disease outbreak observed in nature
From the wild N. attenuata population monitored during the disease outbreak, leaves showing typical symptoms were used to obtain native fungal isolates as potential causal disease agents (Figure 2). These isolates were identified as Alternaria spp., Fusarium solani and F. brachygibbosum (Figure 3), known from the published literature to be phytopathogens that occur in arid environments [47]–[51]. Ribosomal DNA sequence analysis commonly used for fungal phylogenetics was sufficient to distinguish the Fusarium, but not the Alternaria isolates at the species level [29], [52]. To support fungal species/genus identity of the native isolates and to further narrow the number of possible species for the Alternaria isolates, actively growing fungus cultures were characterized by light microscopy (Figure 4) and their polar metabolite profiles (Figure S2 and Table S2). Both methods were consistent with the results of the rDNA sequence analysis and pointed to an assignment of Alternaria spp. Utah 4, 8 and 10 to Alternaria alternata due to the sporulation pattern (Figure 4E) and the similarity of their polar metabolite profiles (Figure S2C). Alternaria spp. Utah 1–3, 5, 6, 9, 11 and 12 could be theoretically assigned either to A. alternata or A. tenuissima due to the presence of Alternariol, a metabolite indicative of the two Alternaria species in all of these native Alternaria isolates (Table S2) [30].
The native fungal isolates were originally picked from diseased N. attenuata samples (seedlings or leaf cuttings) according to their culture morphology. As a consequence, all obtained isolates differed already morphologically from each other, even if they later turned out to be closely related according to their rDNA sequence data (Figure 3). These morphological differences on culture medium were also reflected in virulence differences as shown with a bioassay using detached N. attenuata leaves (Figure 5). Natural variability of a certain fungal species in virulence is known to occur on a very small spatial scale (i.e. within a single plant) [14]. Co-infection events with different pathogen strains belonging to the same species might positively influence virulence evolution, transmissibility and the maintenance of pathogenicity polymorphism in local pathogen populations [14]. This would also explain why so many different Alternaria spp., F. solani and F. brachygibbosum isolates could be found in only a few leaf samples from diseased N. attenuata plants. Application of only a single native fungal isolate could not reproduce the full spectrum of symptoms observed on diseased N. attenuata plants in nature. Plant infection by multiple fungal isolates/species (either simultaneously or in a certain sequential order) might be necessary to mimic all symptoms observed in the natural N. attenuata population in 2011. We hypothesize that a combination of Alternaria and Fusarium species could form a disease complex leading to N. attenuata superinfection events in nature. Indeed, it is known for several plant diseases, e.g. pea root rot and hazelnut brown apical necrosis, that both of these fungal genera typically co-occur in such fungal disease complexes [53]–[55].
Establishment of bioassays for N. attenuata using its native fungal pathogens
As mentioned in the introduction, there was an increasing demand to find native pathogens for N. attenuata in order to improve pathogen bioassay quality and to study the interaction of N. attenuata with its pathogens in nature. This work yielded a large repertoire of native fungal pathogen isolates differing largely in their virulence, and enables N. attenuata researchers to choose the pathogen most appropriate for their experiments from this resource. In contrast to other common lab pathogens previously used for bioassays with N. attenuata [25], [26], the native fungal isolates are known to interact with N. attenuata in nature and therefore are particularly interesting for those experiments which aim to draw inferences to natural infections. To establish such a near-natural pathogen bioassay setup for N. attenuata, we used fungal isolates which can easily infect entire plants via spore suspension. Such bioassays were successfully developed for all three native pathogen groups (Alternaria spp., F. solani and F. brachygibbosum) by dipping the roots of young N. attenuata plants into fungal spore suspension (Figure 6). Small wounds at the roots generated by removing N. attenuata plants from their growth medium could have served as pathogen entry sites and facilitated plant infection by the native fungal pathogens. This root-dip method was sufficiently efficient to ensure infection and allowed the plants to gradually develop disease symptoms (chlorosis and necrosis) over a long time period. This gradual disease progression might be especially useful for pathogen bioassays with N. attenuata which require a long-term observation of the plant-pathogen interaction, e.g. in tripartite-interaction experiments involving a rather slow developing herbivore species. Further studies could also use the obtained fungal isolates for co-infection experiments to address the following questions: could the symptoms observed in natural N. attenuata populations in 2011 be explained by a disease complex comprising Alternaria and Fusarium species? How large is the genetic variability in native N. attenuata populations with regard to the susceptibility to these pathogens? How does N. attenuata respond to the attack by native Alternaria and Fusarium species, e.g. on the level of phytohormone signaling and defensive secondary metabolite production? Obviously this plant species possesses defense responses which are efficient enough to enable many diseased plants to fully recover from all symptoms in nature. Since it is known that Alternaria and Fusarium species even possess pathovar-characteristic supernumerary chromosomes enabling infection of a distinct host range, it appears likely that the obtained native fungal isolates are specialized on N. attenuata (and possibly related species) as well [56]–[58]. In addition, the obtained fungal isolates might well be locally adapted to the surveyed N. attenuata population due to the sympatric occurrence of host and pathogen species [8], [13]. For the native fungal isolates, the degree of host specialization and the extent of local adaption remain to be tested in future. This work will hopefully encourage other plant scientists still lacking a native pathosystem for their model plant to make similar attempts to search for ecologically relevant pathogens as well.
Materials and Methods
Ethics statement
N. attenuata Torrey ex S. Watson seeds were originally collected from a wild accession on private property (DI ranch) in south western Utah in 1988. Ian Baldwin had permission from the owner, Herbert Fletcher to do this collection, and Herbert Fletcher has since deceased. The monitored native population of N. attenuata plants was located on private property of the Lytle Ranch Preserve (N 37.1427 W 114.0229), 45 miles northwest of St. George, Utah, USA, which is run by the Brigham Young University of Utah. None of the work was done with endangered species
Nicotiana attenuata cultivation
N. attenuata seeds of a 31st inbred generation were cultivated as previously described [59]. Seeds were sterilized for 5 min in 5 mL aqueous solution containing 0.1 g dichloroisocyanuric acid (DCCA: Sigma, St. Louis, MO, USA), supplemented with 50 µL of 0.5% (v/v) Tween-20 (Merck, Darmstadt, Germany). Seeds were washed three times with sterile water before incubation for 1 h in 5 mL sterile liquid smoke (House of Herbs, Inc.; Passaic, New Jersey, USA) 50-fold diluted in deionized water and supplemented with 50 µL of 0.1 M gibberellic acid (Roth, Karlsruhe, Germany). After this treatment, seeds were washed three times with sterile water and 25 seeds were transferred individually to a petri dish containing germination medium [Gamborg’s B5 medium with minimal organics (Sigma) and 0.6% (w/v) phytagel (Sigma)]. Plates were maintained in a growth chamber (Snijders Scientific, Tilburg, the Netherlands) at 26°C for 16 h light (155 µmol s−1 m−2) and 24°C for 8 h dark.
Ten day-old N. attenuata seedlings, which were used for entire-plant bioassays with fungal isolates, were transferred to TEKU plastic pots (Pöppelmann GmbH & Co. KG, Lohne, Germany) with sterile sand and grown in climate chambers with a day/night cycle of 16 h (20°C) and 8 h (20°C) under supplemental light from Master Son-T PIA Agro 400 and a relative humidity of 60%. After 10 days, early rosette plants were transferred into 10 cm-diameter pots and grown in the same climate chambers. Each plant was fertilized with 50 mL of Ca(NO3)2×4 H2O and 0.4 g L−1 Flory B1.
Plants used for detached leaf assays were transferred after 10 days in the growth chamber into the TEKU plastic pots with Klasmann plug soil (Klasmann-Deilmann GmbH, Geesten, Germany). After 10 days in TEKU pots, seedlings were transferred to 1 L pots in soil [0.75 g Superphosphate, Multimix 14∶16–18 (Haifa Chemicals Ltd., Haifa Bay, Israel), 0.35 g MgSO4×7 H2O (Merck KGaA, Darmstadt, Germany), 0.05 g Micromax (Scotts Deutschland GmbH, Nordhorn, Germany) per 1 L Fruhstorfer Nullerde (Hawita GmbH, Vechta, Germany)]. Fertilization was done also by flood irrigation with addition of 3.0 g Borax (Nic Sosef BV, Honselersdijk, Netherlands) on the day of potting, 2.0 g Borax and 20 g Peters Allrounder 20∶20∶20 (Scotts International, Heerlen, Netherlands) from day seven on, 1.0 g Borax, 40 g Peters Allrounder from day 14 on and 25–30 g Peters Allrounder from day 21 on, each to a 400 L watering tank. From day one up to day five, plants were watered with 300 mL and later with 100 mL per day and grown in the glasshouse at 26–28°C under 16 h supplemental light from Master Sun-T PIA Agro 400 or Master Sun-T PIA Plus 600 W Na lights (Philips, Turnhout, Belgium).
Disease symptom monitoring of the native N. attenuata population
During the field season in 2011, plants in several native N. attenuata populations had been discovered showing symptoms apparently caused by a fungal disease. To assess the ecological impact of the disease outbreak on this plant species, a particular N. attenuata population on the Lytle Ranch Preserve comprising 873 plants distributed over an area of approximately 1500 m2 was monitored for a population-wide screening of disease incidence and symptom severity. The area was divided into 14 subpopulations ( = “sections”) of 15 to 163 plants according to habitat properties (e.g. grassland, shrubs, trees and larger areas without any N. attenuata plant). To mark the sections, colored flags were put into corners and connected via colored woolen threads. Then schematic maps (not absolutely to scale) were drawn by hand to record the spatial distribution. In each section the positions of the individual plants were assigned with a number to track disease development for each plant position within the population. Afterwards all plants were screened with regard to the occurrence and strength of disease symptoms. In more moderate cases diseased plants appeared almost asymptomatic at first glance, however, when inspected more carefully, showed dark spots at the abaxial leaf side having a rough surface (most likely fungal mycelium plus sporangia; Figures 1 and 2). Such plants were classified into disease stage category 1. In addition, some plants turned chlorotic and had curly leaves and were therefore classified into disease stage category 2. In severe cases, plants developed necrotic lesions and were classified into disease stage category 3. The percentage of chlorotic and necrotic leaf area was estimated for each plant relatively to its overall leaf area. This population-wide screening for disease symptoms from May 24th 2011 was repeated 16 days later on June 9th 2011 in order to see how disease symptoms develop over time at the individual N. attenuata plants. Completely asymptomatic plants were categorized into “disease stage 0”.
Fungi isolation and cultivation
In order to culture potentially pathogenic fungi from diseased N. attenuata plants, eight leaf samples clearly showing disease symptoms were brought from the above mentioned native N. attenuata population to the Max Planck Institute, Jena, Germany. The process of fungi isolation is illustrated in Figure 2. Sampling was performed from freshly diseased plants not randomly distributed across the population sections. Leaf samples came from sections III, VII and VIII (majority from sections VII and VIII) (Figure 1). Fungi were isolated using different strategies. Two leaf samples were thoroughly ground with a pestle and mortar and diluted in 5 mL sterile deionized water, and 20 µL of the leaf suspension was applied onto 10 day-old N. attenuata seedlings growing on Gamborg B5 medium. A few days after inoculation, most of the seedlings started to turn chlorotic and wilting. Diseased seedlings were either surface-sterilized [60 s in 0.1% (w/v) DCCA in deionized water with three subsequent washes in sterile deionized water] or just rinsed in sterile deionized water. In addition, six leaf samples were cut with a sterile scalpel into approximately 5 mm big pieces and the obtained leaf cuttings were either surface-sterilized (two minutes in 0.1% DCCA (w/v) in deionized water with three subsequent washes in sterile deionized water) or just rinsed in sterile deionized water. Surface-sterilized and non-sterilized leaf cuttings/seedlings were then transferred onto various media [Luria-Bertani agar containing 25 mg L−1 Rifampicin and 5 mg L−1 Tetracyclin, Kaefer medium [60] agar and potato dextrose agar (PDA; Fluka Analytical, Steinheim, Germany)] and incubated at 25°C in the dark until fungal hyphae emerged from the plant tissue. Based on morphological differences (color and shape of mycelium) fungal cultures were picked and cultured on PDA plates. The obtained morphological different fungal isolates were inoculated to 24 day-old N. attenuata plants as described in Bonaventure et al. (2011) for an initial pathogenicity test [26]. In brief, 14 day-old fungal cultures were transferred to the hypocotyl of sand-grown plants after pricking it with a sterile syringe needle to facilitate a potential infection. Those fungal isolates which were able to cause plant disease symptoms within seven days were chosen for rDNA sequencing, morphological and chemical characterization and further bioassays (Figure 2). The Alternaria and Fusarium fungal isolates used in this study were re-isolated from infected N. attenuata plants from time to time and confirmed by morphological comparison to the originally isolated and characterized fungal cultures (mycelium growth pattern, structure and color, and conidia-shape by light microscopy).
Phylogenetic analysis of fungal isolates based on their rDNA-ITS-LSU sequence
Fungal DNA was extracted from mycelium for PCR amplification using the following protocol: 14 day-old fungal culture grown on PDA plates was used to extract total fungal DNA according to a slightly modified DNA extraction protocol using the Cetyl trimethylammonium bromide (CTAB) method as described by Ben Bubner (2004) [61]. Approximately 300 mg fungal mycelium material was scraped directly from the surface of the plates and immediately frozen in liquid nitrogen. The samples were ground with a 2000 Geno/Grinder machine (SPEX Certi Prep, Metuchen, New Jersey, USA) and kept frozen until extraction with liquid nitrogen. Extraction was performed with 800 µL preheated (65°C) buffer [2% (w/v) CTAB, 100 mM Tris pH 8.0, 20 mM EDTA pH 8.0, 1.4 mM NaCl, 1% (w/v) Polyvinylpyrrolidone Mr 40000, 1% (v/v) 2-mercaptoethanol] and 500 µL of chloroform/isoamyl alcohol (24∶1; v/v). After being mixed and centrifuged, the supernatant was supplemented with 0.1 volume 10% (w/v) CTAB and further cleaned up using again phase-separation with one volume of chloroform/isoamyl alcohol (24∶1; v/v). The nucleic acids in the resulting supernatant were precipitated with an equal volume of ice cold isopropanol. After rehydrating the pellet in 500 µL high salt TE (10 mM Tris pH 8.0, 1 mM EDTA pH 8.0, 1 M NaCl, 100 ng µL−1 RNase A; Roth, Karlsruhe, Germany), the solution was incubated at 37°C for 15 min. Following a phase separation step with 500 µL chloroform/isoamyl alcohol (24∶1; v/v), the supernatant was taken for DNA precipitation by adding 1 mL ice cold isopropanol. The pellet resulting from centrifugation was washed with 80% (v/v) ethanol and rehydrated in 50 µL Milli-Q water (Milli-Q Advantage A10, Millipore, Massachusetts, USA). OD260 measurements on a Biophotometer (Eppendorf, Hamburg, Germany) indicated concentrations between 0.05 to 0.25 µg µL−1.
An approximately 1200 bp long genomic DNA sequence comprising the internal transcribed spacer (ITS) 1 and 2 regions, the 5.8S rDNA and part of the large ribosomal subunit (LSU) gene was amplified by PCR from fungal genomic DNA samples using the primer pair ITS1-F (5′-TTGGTCATTTAGAGGAAGTAA-3′) and TW13 (5′-GGTCCGTGTTTCAAGACG-3′) [52], [62]. PCR reagents were obtained from Fermentas GmbH, Germany. The PCR mix contained 0.5 µL of a 10 mM deoxynucleoside triphosphate mix, 0.5 U of Taq DNA polymerase, 0.2 µM of each primer, 1 µL 1∶10-diluted genomic DNA extract, 25 mM MgCl2 and 2.5 µL Taq buffer (10x) in a total volume of 25 µL supplemented with the appropriate amount of Milli-Q water. PCR amplification was conducted using a Mastercycler gradient thermocycler (Eppendorf, Hamburg, Germany) with the following program: initial denaturation for 3 min at 95°C, followed by 20 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 60°C and elongation for 3 min at 72°C, followed by a final extension step at 72°C for 10 min. Amplification products were visualized and quantified using horizontal gel electrophoresis and a 1% (w/v) agarose gel (Sigma-Aldrich Chemie GmbH, Hamburg, Germany) in 0.5x Tris-borate-EDTA-running buffer and ethidium bromide staining.
PCR products were sequenced from both strand ends using the same primers as described above. BigDye terminator cycle sequencing (Applied Biosystems, Foster City, USA) was employed as recommended by the manufacturer. All cycle sequencing reactions were performed on a Eppendorf Mastercycler gradient thermocycler using an initial denaturation step at 96°C for 5 min, followed by 35 cycles of 96°C for 10 s, 30°C for 20 s, and 60°C for 4 min. Products were purified using the Big Dye Purification Kit (DyeEx 2.0 Spin KIT, Qiagen, Hilden, Germany), dried in a vacuum centrifuge, and resuspended in template suppression reagent (Applied Biosystems, Foster City, USA). Products were then analyzed on a 16-capillary 3130xl Genetic Analyzer (Life Technologies GmbH, Darmstadt, Germany). The sequences were formatted and edited using Chromas Lite (Technelysium Pty. Ltd., Australia) and Multalin software (http://multalin.toulouse.inra.fr/multalin/). The appropriate sequences were then compared to sequences listed in the GenBank database using the BLASTn program (http://www.ncbi.nlm.nih.gov/BLAST/). DNA sequences of the native fungal pathogens and the according NCBI hits were then aligned using ClustalW algorithm in MegAlign (DNASTAR Inc., Madison, Wisconsin, USA). Phylogenetic and molecular evolutionary analyses were conducted using MEGA software version 5 using the neighbor-joining (NJ) method (1000 steps) [63]. Species identification was made based on a greater-than-99%-similarity between query and reference sequence according to [28]. The rDNA sequences used for this phylogenetic analysis can be found at the NCBI database under the following GenBank accession numbers: KJ541472, KJ541473, KJ541474, KJ541475, KJ541476, KJ541477, KJ541478, KJ541479, KJ541480, KJ541481, KJ541482, KJ541483, KJ541484, KJ541485, KJ541486, KJ541487, KJ541488, KJ541489, KJ541490, KJ541491, KJ541492, KJ541493 and KJ541494.
Chemotaxonomical analysis of fungal isolates by UPLC-ToF-MS
Fungal polar metabolite extraction was done using 14 day-old fungi cultures grown on PDA at 25°C in the darkness. The extraction protocol used was adapted from Andersen et al. (2005) [64]. Three agar plugs (6 mm diameter) were cut from the central, intermediate and marginal area of the fungal colony. The plugs were extracted with 1.0 mL of ethyl acetate containing 1% (v/v) formic acid by sonication for 60 min. The extract was transferred to a clean 2-mL HPLC vial and evaporated to dryness in a rotary vacuum concentrator (Eppendorf, Wesseling-Berzdorf, Germany). The residue was redissolved ultrasonically in 200 µL of methanol and centrifuged at 16,100 g for 15 min with Eppendorf centrifuge 5415 R (Eppendorf, Wesseling-Berzdorf, Germany). The supernatant was transferred into HPLC vials with insert and used for fungal metabolite analysis with a Ultraperformance LC-ToF-MS system (Agilent Technologies, Böblingen, Germany) using a modified version of a previously described LC-ToF-MS method for N. attenuata [65]. Four microliters of the fungal extract were injected onto a C18 column (Acclaim, 2.2 mm particle size, 150 mm 62.1 mm inner diameter (Dionex Corporation, Sunnyvale, USA) and separated using an RSLC system (Dionex). Solvent A was deionized water containing 0.1% (v/v) acetonitrile (Baker, HPLC grade) and 0.05% (v/v) formic acid. Solvent B was acetonitrile and 0.05% (v/v) formic acid. The gradient condition was applied as follows: 0 to 0.5 min 10% B, 0.5 to 6.5 min linear gradient 80% B, 6.5 to 10 min 80% B, and equilibration at 10% B for 3 min. The flow rate was 300 µL min−1. Eluted compounds were detected with a MicroToF mass spectrometer (Bruker Daltonic, Bremen, Germany) equipped with an electrospray ionization source in negative ion mode. Instrument settings were as follows: capillary voltage, 4500 V; capillary exit, 130 V; dry gas temperature, 200°C; drying gas flow, 8 liters min−1. Mass calibration was performed using sodium formeate clusters (10 mM solution of NaOH in isopropanol/water (50∶50; v/v) containing 0.2% (v/v) formic acid).
The raw data files were converted to netCDF format using the export function of Bruker software (Data Analysis v4.0) and processed using the XCMS package [66] and the R-package CAMERA (http://www.bioconductor.org/biocLite.R) as previously described [65]. Peak detection was performed using the centWave method [66] and the parameter settings ppm = 20, snthresh = 10, peak width = 5 to 20 s. Retention time correction was achieved using the parameter settings minfrac = 1, bw = 60 s, mzwid = 0.1 D, span = 1, and missing = extra = 0 [66]. The MetaboAnalyst 2.0 online software (http://www.metaboanalyst.ca/MetaboAnalyst/faces/Home.jsp) was used to perform multivariate analysis (principle component analysis). The data was normalized using Autoscaling. For Alternaria sp. Utah 7 metabolic profiling was not possible because of a mistake during extraction. Peak intensities of characteristic ions representing Alternaria mycotoxins were determined based on retention times and mass-over-charge values available on a public database [34].
Morphological characterization of fungal isolates by light microscopy
Mycelium and spore mats of 14 day-old fungal cultures were used for morphological observations under the light microscope (Axio Observer.D1, Carl-Zeiss Jena, Germany). Pictures were taken using AxioVision imaging software at 400-fold magnification. A description of Fusarium solani by de Hoog (2000) was used for confirmation of Fusarium solani Utah [35], together with reference pictures of its microconidia and macroconidia available online (http://tolweb.org/Sordariomycetes/29050/2008.01.14). Alternaria species were characterized using descriptions made by Simmons et al. (1999) together with a reference picture of Alternaria alternata conidia and conidiophores made by de Hoog (2000) [35], [36].
Evaluation of fungal isolate pathogenicity using a detached leaf bioassay
For a detached leaf assay plugs of fungal mycelium (3 mm diameter) were cut from the marginal regions of 14 day-old actively growing fungus culture and placed on fully expanded leaves collected from plants at rosette stage (30 day-old plants). The leaves were placed upside down on four layers of moist autoclaved tissue paper in square-shaped petri dishes. Three fungal agar plugs were placed on three distant spots on the abaxial side of a detached N. attenuata leaf. All plates were sealed twice with Parafilm to maintain high moisture conditions and kept at 25°C with a 14-hours photoperiod. After incubation for 5 days, the smallest and largest diameter of chlorotic, necrotic and hypersensitive-like lesions was measured using a caliper. Hypersensitive-like lesions as a typical consequence of an incompatible plant interaction with avirulent pathogens leading to disease resistance were distinguished from necrotic lesions caused by other disease-related cell death processes [37]. One-way analysis of variance (ANOVA) and mean value comparison by Fisheŕs LSD post-hoc test was calculated using the SPSS Statistics software version 17.0 (SPSS, Chicago, Illinois, USA). Twelve biological replicates were used for each fungal isolate. Detached leaves incubated with pure PDA were used as control and the agar plug diameter was referred to as “lesion” diameter of controls, although no necrotic and hypersensitive-like lesions occurred, to account for the fact that lesions could only be observed as soon as they exceeded the diameter of the inoculation plug.
Establishment of pathogen bioassays using the fungal isolates and intact soil-grown N. attenuata plants
For the bioassay with entire N. attenuata plants, fungal isolates with different virulence (aggressive and moderate isolates) were used with three different spore concentrations (105, 106 and 107 spores mL−1). Spores harvested from the surface of 14 day-old fungal cultures were filtered through miracloth (Calibiochem, UK) to remove mycelial fragments and centrifuged at 800 g for 20 min. The pellets were washed twice with 10 mM MgSO4 and the spore concentration was adjusted to the desired concentration with 10 mM MgSO4 using a Neubauer hemocytometer to count the number of spores under the light microscope. The spore suspensions were used to inoculate N. attenuata plants of different age (10 or 20 day-old) using two different infection methods. For the root-dip method, roots were dipped in spore suspension for 30 s and afterwards placed back into the substrate. Spraying with spore suspension was performed homogenously from about 20 cm distance to the plant with approximately 0.3 mL spore suspension per plant. Control plants were dipped into or sprayed with 10 mM MgSO4 solution.
Supporting Information
Schematic map illustrating changes in the percentage of diseased N. attenuata plants. The plant number (n) is indicated for each section (labeled I to XIV) of the native N. attenuata population. Bluish colors indicate an overall reduction, reddish colors an overall increase in the percentage of diseased plants over time (June 9th 2011 vs. May 24th 2011).
(TIF)
Chemotaxonomy of native fungal pathogen isolates. A. Dendrogram of native Fusarium and Alternaria species generated by hierarchical clustering in MetaboAnalyst using Ward’s linkage clustering method based on polar metabolite profiling data (UPLC-ToF-MS in negative ion mode). The numbers on the scale indicate the distance level with relative units. B. Two-dimensional distribution of Alternaria and Fusarium (F. brachygibbosum and F. solani) isolates according to the two major separating principal components (PCs). C. Principal componant analysis of polar metabolite profiles from Alternaria isolates. PC1 and PC2 explained together 44.2% of the variance of the samples (the explained variances per component are shown in brackets).
(TIF)
Development of disease symptoms in a native N. attenuata population within a 16 day-time interval.
(DOC)
Mycotoxins from native Alternaria spp. isolates assigned by mass-over-charge values.
(DOC)
Acknowledgments
We thank the Brigham Young University for the use of their field station in Utah, USA, and all the coworkers (Mario Kallenbach, Meredith Schuman, Son Truong Dinh, Melkamu Woldemariam, Danny Kessler, Lynn Ullmann-Zeunert, Youngjoo Oh, Felipe Yon, Mariana Stanton, Celia Diezel, Stefan Meldau and Matthias Erb) assisting in the surveys and sampling of diseased leaf material from the native N. attenuata population.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. GenBank accession numbers: [submitted under submission ID 1705568, see supplemental file].
Funding Statement
This work was funded by the Max Planck Society. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Dinoor A, Eshed N (1984) The role and importance of pathogens in natural plant communities. Annual Review of Phytopathology 22: 443–466. [Google Scholar]
- 2. Lenne JM, Wood D (1991) Plant diseases and the use of wild germplasm. Annual Review of Phytopathology 29: 35–63. [Google Scholar]
- 3. Leonards-Schippers C, Gieffers W, Salamini F, Gebhardt C (1992) The R1 gene conferring race-specific resistance to Phytophthora infestans in potato is located on potato chromosome V. Molecular and General Genetics. 233: 278–283. [DOI] [PubMed] [Google Scholar]
- 4. Van Der Vossen E, Sikkema A, Hekkert BtL, Gros J, Stevens P, et al. (2003) An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. The Plant Journal 36: 867–882. [DOI] [PubMed] [Google Scholar]
- 5. Burdon JJ, Jarosz AM (1991) Host-pathogen interactions in natural populations of Linum marginale and Melampsora lini: I. Patterns of resistance and racial variation in a large host population. Evolution 45: 205–217. [DOI] [PubMed] [Google Scholar]
- 6. Wills RT (1993) The ecological impact of Phytophthora cinnamomi in the Stirling Range National Park, Western Australia. Australian Journal of Ecology 18: 145–159. [Google Scholar]
- 7. Carlsson-Granér U, Thrall PH (2002) The spatial distribution of plant populations, disease dynamics and evolution of resistance. Oikos 97: 97–110. [Google Scholar]
- 8. Laine AL (2005) Spatial scale of local adaptation in a plant-pathogen metapopulation. Journal of Evolutionary Biology 18: 930–938. [DOI] [PubMed] [Google Scholar]
- 9. Lebeda A, Petrželová I, Maryška Z (2008) Structure and variation in the wild-plant pathosystem: Lactuca serriola-Bremia lactucae . European Journal of Plant Pathology 122: 127–146. [Google Scholar]
- 10. Barrett LG, Kniskern JM, Bodenhausen N, Zhang W, Bergelson J (2009) Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytologist 183: 513–529. [DOI] [PubMed] [Google Scholar]
- 11. Bernasconi G, Antonovics J, Biere A, Charlesworth D, Delph LF, et al. (2009) Silene as a model system in ecology and evolution. Heredity (Edinb) 103: 5–14. [DOI] [PubMed] [Google Scholar]
- 12. Soubeyrand S, Laine AL, Hanski I, Penttinen A (2009) Spatiotemporal structure of host-pathogen interactions in a metapopulation. The American Naturalist 174: 308–320. [DOI] [PubMed] [Google Scholar]
- 13. Nemri A, Barrett LG, Laine AL, Burdon JJ, Thrall PH (2012) Population processes at multiple spatial scales maintain diversity and adaptation in the Linum marginale-Melampsora lini association. PLoS One 7: e41366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tack AJ, Thrall PH, Barrett LG, Burdon JJ, Laine AL (2012) Variation in infectivity and aggressiveness in space and time in wild host-pathogen systems: causes and consequences. Journal of Evolutionary Biology 25: 1918–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baldwin IT (2001) An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiology 127: 1449–1458. [PMC free article] [PubMed] [Google Scholar]
- 16. Bahulikar RA, Stanculescu D, Preston CA, Baldwin IT (2004) ISSR and AFLP analysis of the temporal and spatial population structure of the post-fire annual, Nicotiana attenuata, in SW Utah. BMC Ecology 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu J, Hettenhausen C, Schuman MC, Baldwin IT (2008) A comparison of two Nicotiana attenuata accessions reveals large differences in signaling induced by oral secretions of the specialist herbivore Manduca sexta . Plant Physiology 146: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schuman MC, Heinzel N, Gaquerel E, Svatos A, Baldwin IT (2009) Polymorphism in jasmonate signaling partially accounts for the variety of volatiles produced by Nicotiana attenuata plants in a native population. New Phytologist 183: 1134–1148. [DOI] [PubMed] [Google Scholar]
- 19. Kessler D, Bhattacharya S, Diezel C, Rothe E, Gase K, et al. (2012) Unpredictability of nectar nicotine promotes outcrossing by hummingbirds in Nicotiana attenuata . The Plant Journal 71: 529–538. [DOI] [PubMed] [Google Scholar]
- 20. Bhattacharya S, Baldwin IT (2012) The post-pollination ethylene burst and the continuation of floral advertisement are harbingers of non-random mate selection in Nicotiana attenuata . The Plant Journal 71: 587–601. [DOI] [PubMed] [Google Scholar]
- 21. Burdon JJ, Chilvers GA (1982) Host density as a factor in plant disease ecology. Annual Review of Phytopathology 20: 143–166. [Google Scholar]
- 22. Mitchell CE, Tilman D, Groth JV (2002) Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology 83: 1713–1726. [Google Scholar]
- 23. Wang L, Wu J (2013) The essential role of jasmonic acid in plant-herbivore interactions using the wild tobacco Nicotiana attenuata as a model. Journal of Genetics and Genomics 40: 597–606. [DOI] [PubMed] [Google Scholar]
- 24. Rayapuram C, Baldwin IT (2007) Increased SA in NPR1-silenced plants antagonizes JA and JA-dependent direct and indirect defenses in herbivore-attacked Nicotiana attenuata in nature. The Plant Journal 52: 700–715. [DOI] [PubMed] [Google Scholar]
- 25. Hettenhausen C, Baldwin IT, Wu J (2012) Silencing MPK4 in Nicotiana attenuata enhances photosynthesis and seed production but compromises abscisic acid-induced stomatal closure and guard cell-mediated resistance to Pseudomonas syringae pv tomato DC3000. Plant Physiology 158: 759–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Allen C, Bent A, Charkowski A (2009) Underexplored niches in research on plant pathogenic bacteria. Plant Physiology 150: 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bosshard PP, Abels S, Zbinden R, Bottger EC, Altwegg M (2003) Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation). Journal of Clinical Microbiology 41: 4134–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andrew M, Peever TL, Pryor BM (2009) An expanded multilocus phylogeny does not resolve morphological species within the small-spored Altemrnaria species complex. Mycologia 101: 95–109. [DOI] [PubMed] [Google Scholar]
- 30. Weidenborner M (2001) Pine nuts: the mycobiota and potential mycotoxins. Canadian Journal of Microbiology 47: 460–463. [PubMed] [Google Scholar]
- 31. Bilgrami KS, Ansari AA, Sinha AK, Shrivastava AK, Sinha KK (1994) Mycotoxin production by some Indian Alternaria species. Mycotoxin Research 10: 56–59. [DOI] [PubMed] [Google Scholar]
- 32. Andersen B, Hansen ME, Smedsgaard J (2005) Automated and unbiased image analyses as tools in phenotypic classification of small-spored Alternaria spp. Phytopathology 95: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 33. Ostry V (2008) Alternaria mycotoxins: an overview of chemical characterization, producers, toxicity, analysis and occurrence in foodstuffs. World Mycotoxin Journal 1: 175–188. [Google Scholar]
- 34. Nielsen KF, Smedsgaard J (2003) Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography-UV-mass spectrometry methodology. Journal of Chromatography A 1002: 111–136. [DOI] [PubMed] [Google Scholar]
- 35.de Hoog GS, Guarro J, Gene J, Figueras MJ (2000) Atlas of clinical fungi, 2nd edition. Utrecht, The Netherlands: Centraal Bureau voor Schimmelcultures/Universitat Rovira i Virgili. 1141 p. [Google Scholar]
- 36.Simmons EG (1999) Alternaria themes and variations (236–243). Host-specific toxin producers. Mycotaxon. Ithaca, New York: Mycotaxon LTD. 46 p. [Google Scholar]
- 37. Morel J-B, Dangl JL (1997) The hypersensitive response and the induction of cell death in plants. Cell Death and Differentiation 4: 671–683. [DOI] [PubMed] [Google Scholar]
- 38. Giller PS (1996) The diversity of soil communities, the ‘poor man’s tropical rainforest’. Biodiversity and Conservation 5: 135–168. [Google Scholar]
- 39. Steppuhn A, Schuman MC, Baldwin IT (2008) Silencing jasmonate signalling and jasmonate-mediated defences reveals different survival strategies between two Nicotiana attenuata accessions. Molecular Ecology 17: 3717–3732. [DOI] [PubMed] [Google Scholar]
- 40. Glazebrook J (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43: 205–227. [DOI] [PubMed] [Google Scholar]
- 41. Shibata Y, Kawakita K, Takemoto D (2010) Age-related resistance of Nicotiana benthamiana against hemibiotrophic pathogen Phytophthora infestans requires both ethylene- and salicylic acid-mediated signaling pathways. Molecular Plant-Microbe Interactions 23: 1130–1142. [DOI] [PubMed] [Google Scholar]
- 42. Weber RW (2003) Meteorologic variables in aerobiology. Immunology and Allergy Clinics of North America 23: 411–422. [DOI] [PubMed] [Google Scholar]
- 43. Khan J, Qi A, Khan MF (2009) Fluctuations in number of Cercospora beticola conidia in relationship to environment and disease severity in sugar beet. Phytopathology 99: 796–801. [DOI] [PubMed] [Google Scholar]
- 44. Kriss AB, Paul PA, Madden LV (2010) Relationship between yearly fluctuations in Fusarium head blight intensity and environmental variables: a window-pane analysis. Phytopathology 100: 784–797. [DOI] [PubMed] [Google Scholar]
- 45. Fourie P, Schutte T, Serfontein S, Swart F (2013) Modeling the effect of temperature and wetness on Guignardia pseudothecium maturation and ascospore release in Citrus orchards. Phytopathology 103: 281–292. [DOI] [PubMed] [Google Scholar]
- 46. Pulimood TB, Corden JM, Bryden C, Sharples L, Nasser SM (2007) Epidemic asthma and the role of the fungal mold Alternaria alternata . The Journal of Allergy and Clinical Immunology 120: 610–617. [DOI] [PubMed] [Google Scholar]
- 47. Hadwiger LA (2008) Pea-Fusarium solani interactions contributions of a system toward understanding disease resistance. Phytopathology 98: 372–379. [DOI] [PubMed] [Google Scholar]
- 48. Rep M, Kistler HC (2010) The genomic organization of plant pathogenicity in Fusarium species. Current Opinion in Plant Biology 13: 420–426. [DOI] [PubMed] [Google Scholar]
- 49. Tsuge T, Harimoto Y, Akimitsu K, Ohtani K, Kodama M, et al. (2013) Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata . FEMS Microbiology Reviews 37: 44–66. [DOI] [PubMed] [Google Scholar]
- 50. Al-Mahmooli IH, Al-Bahri YS, Al-Sadi AM, Deadman ML (2013) First report of Euphorbia larica dieback caused by Fusarium brachygibbosum in Oman. Plant Disease 97: 687–687. [DOI] [PubMed] [Google Scholar]
- 51. Sun Y, Wang Q, Lu X, Okane I, Kakishima M (2012) Endophytic fungal community in stems and leaves of plants from desert areas in China. Mycological Progress 11: 781–790. [Google Scholar]
- 52.White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Shinsky J, White T, editors. PCR protocols: a guide to methods and applications: Academic Press. 315–322.
- 53. Belisario A, Maccaroni M, Corazza L, Balmas V, Valier A (2002) Occurrence and etiology of brown apical necrosis on Persian (English) walnut fruit. Plant Disease 86: 599–602. [DOI] [PubMed] [Google Scholar]
- 54. Xue AG (2003) Biological control of pathogens causing root rot complex in field pea using Clonostachys rosea strain ACM941. Phytopathology 93: 329–335. [DOI] [PubMed] [Google Scholar]
- 55. Belisario A, Maccaroni M, Coramusi A, Corazza L, Pryor BM, et al. (2004) First report of Alternaria species groups involved in disease complexes of hazelnut and walnut fruit. Plant Disease 88: 426–426. [DOI] [PubMed] [Google Scholar]
- 56. van der Does HC, Rep M (2007) Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Molecular Plant-Microbe Interactions 20: 1175–1182. [DOI] [PubMed] [Google Scholar]
- 57. Mehrabi R, Bahkali AH, Abd-Elsalam KA, Moslem M, M’Barek SM, et al. (2011) Horizontal gene and chromosome transfer in plant pathogenic fungi affecting host range. FEMS Microbiology Reviews 35: 542–554. [DOI] [PubMed] [Google Scholar]
- 58. Schmidt SM, Panstruga R (2011) Pathogenomics of fungal plant parasites: what have we learnt about pathogenesis? Current Opinion in Plant Biology 14: 392–399. [DOI] [PubMed] [Google Scholar]
- 59. Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183. [Google Scholar]
- 60.Pham G, Singh A, Malla R, Kumari R, Prasad R, et al. (2004) Interaction of Piriformospora indica with diverse microorganisms and plants. In: Varma A, Abbott L, Werner D, Hampp R, editors. Plant surface microbiology: Springer Berlin Heidelberg. 237–265.
- 61. Bubner B, Gase K, Baldwin IT (2004) Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnology 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gardes M, White TJ, Fortin JA, Bruns TD, Taylor JW (1991) Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Canadian Journal of Botany 69: 180–190. [Google Scholar]
- 63. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andersen B, Frisvad JC (2004) Natural occurrence of fungi and fungal metabolites in moldy tomatoes. Journal of Agriculture and Food Chemistry 52: 7507–7513. [DOI] [PubMed] [Google Scholar]
- 65. Gaquerel E, Heiling S, Schoettner M, Zurek G, Baldwin IT (2010) Development and validation of a liquid chromatography-electrospray ionization-time-of-flight mass spectrometry method for induced changes in Nicotiana attenuata leaves during simulated herbivory. Journal of Agriculture and Food Chemistry 58: 9418–9427. [DOI] [PubMed] [Google Scholar]
- 66. Tautenhahn R, Bottcher C, Neumann S (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9: 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic map illustrating changes in the percentage of diseased N. attenuata plants. The plant number (n) is indicated for each section (labeled I to XIV) of the native N. attenuata population. Bluish colors indicate an overall reduction, reddish colors an overall increase in the percentage of diseased plants over time (June 9th 2011 vs. May 24th 2011).
(TIF)
Chemotaxonomy of native fungal pathogen isolates. A. Dendrogram of native Fusarium and Alternaria species generated by hierarchical clustering in MetaboAnalyst using Ward’s linkage clustering method based on polar metabolite profiling data (UPLC-ToF-MS in negative ion mode). The numbers on the scale indicate the distance level with relative units. B. Two-dimensional distribution of Alternaria and Fusarium (F. brachygibbosum and F. solani) isolates according to the two major separating principal components (PCs). C. Principal componant analysis of polar metabolite profiles from Alternaria isolates. PC1 and PC2 explained together 44.2% of the variance of the samples (the explained variances per component are shown in brackets).
(TIF)
Development of disease symptoms in a native N. attenuata population within a 16 day-time interval.
(DOC)
Mycotoxins from native Alternaria spp. isolates assigned by mass-over-charge values.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. GenBank accession numbers: [submitted under submission ID 1705568, see supplemental file].