Abstract
Purpose
Adenitis for which no causative organism can be isolated is a common occurrence in patients with chronic granulomatous disease (CGD). Here we identify Acidomonas methanolica as a pathogen associated with adenitis in a patient with CGD.
Methods
The causative pathogen was obtained after prolonged incubation of an excised lymph node in thioglycolate broth. Identification was carried out by sequencing the 16s rRNA. Immunoblots were prepared utilizing protein extracts from the case patient’s A. methanolica isolate, an ATCC type strain of A. methanolica and G. bethesdensis.
Results
Fastidious gram-negative rods grew after prolonged incubation of an excised lymph node in thioglycolate broth. Sequencing of the 16s rRNA identified the organism as A. methanolica. Immunoblot confirmed the pathogen’s role in the patient’s adenitis by showing the patient’s specific immune response to the organism.
Conclusions
A. methanolica is the second member of the family, Acetobacteaceae to be associated with adenitis in patients with CGD.
Keywords: Chronic granulomatous disease, Acidomonas methanolica
Introduction
Chronic granulomatous disease (CGD) is an inherited primary immunodeficiency characterized by recurrent infections with a restricted range of bacteria and fungi. Adenitis is common in CGD, but no pathogen is isolated in up to 40 % of cases [1]. A number of emerging pathogens, which are difficult to isolate, have been implicated in a subset of these infections. Previously, we reported disseminated Trichosporon pullulans in a 19 year-old patient with CGD [2] and Granulibacter bethesdensis in CGD patients with necrotizing lymphadenitis [3, 4]. It is likely that other fastidious pathogens are responsible for cases of CGD-associated lymphadenitis in which no organism can be identified. Here we describe a previously unrecognized pathogen isolated from a patient with CGD-associated necrotizing lymphadenitis.
Case Report
A 10 year-old boy with CGD presented with a 10-month history of worsening cervical lymphadenopathy. He had been diagnosed with X-linked CGD based upon absent neutrophil reduction of nitroblue tetrazolium (NBT) and confirmed by CYBB gene sequencing to have the R226X mutation (GeneDx, Gaithersburg, MD) [5]. Although he experienced serious infections including perirectal abscess, pneumonia, and osteomyelitis, the family remained avid outdoor campers. At 9 years of age he developed cervical adenitis associated with Francisella tularensis seropositivity (titer 1:1280) (Quest Diagnostics, Madison, NJ). Despite therapy with gentamicin and doxycycline with subsequent resolution of F. tularensis seropositivity (titer <1:20), his cervical lymphadenopathy worsened. Fine needle aspiration and open biopsy failed to reveal a causative organism.
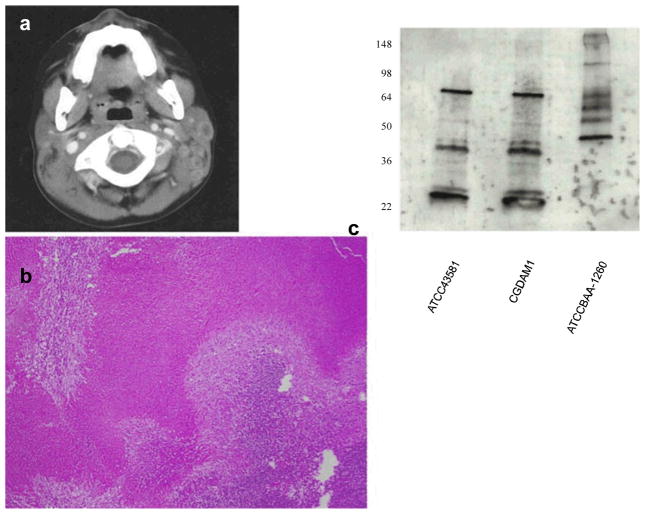
Separate courses of clindamycin, trimethoprim-sulfamethoxazole, vancomycin, cefepime and ampicillin-sulbactam failed to mitigate the adenitis. Computed tomography of the patient’s neck revealed necrotizing cervical adenitis (Fig. 1a). Excisional biopsy of several cervical lymph nodes showed necrotizing granulomatous lymphadenitis (Fig. 1b), but again failed to identify any pathogen.
Fig. 1.
a. Computed axial tomography of patient’s neck revealing necrotizing adenitis. b. Low-powered view of excisional lymph node biopsy revealing diffuse necrosis. c. Patient IgG response to A. methanolica extract. The first lane (ATCC 43581) contains protein extracts from A. methanolica type strain; the second lane (CGDAM1) contains protein extracts from the case patient’s A. methanolica isolate; the third lane (ATCC BAA-1260) contains protein extracts from another pathogenic methylotroph, G. bethesdensis. All are run on SDS-PAGE gel. Immunoblots were performed with patient sera at 1:1000 dilution. Identical banding patterns are seen with patient sera against the extracts of both A. methanolica strains (ATCC 43581 and CGDAM1). The banding pattern with G. bethesdensis extract (ATCC BAA-1260) is distinct. The numbers on the left display the molecular weights of particular protein bands in kDa
Further treatment with trimethoprim-sulfamethoxazole, ceftriaxone, and doxycycline therapy did not abate the adenitis. Therefore, removal of a three-centimeter axillary lymph node was performed.
Tailored combination therapy with trimethoprim-sulfamethoxazole, rifabutin, and gentamicin resulted in a partial clinical response. Matched unrelated donor peripheral blood hematopoietic stem cell transplantation (HSCT) resulted in 910 neutrophils/mm3 all of which reduced NBT by 6 weeks after transplantation. His adenopathy resolved and all antibiotics were discontinued.
Methods
Written and informed consents for investigative studies were obtained from the patient and his mother.
Isolation
Portions of the patient’s lymph node were cultured on sheep blood agar, MacConkey agar, chocolate agar, and thioglycolate broth. After 12-day incubation in thioglycolate broth at 35 °C, rare fastidious gram-negative rods grew. Culture containing this rare organism and frozen lymph node material were sent to the Microbiology Service, Department of Laboratory Medicine, Warren Grant Magnuson Clinical Center, National Institutes of Health (NIH) for identification.
The organism was planted onto sheep blood, chocolate agar, BCYE, and fastidious broth at 35 °C. After larger colonies grew from several of the above media, the organism was subcultured on BCYE, Middlebrook, Sabouraud and sheep blood agar at 30 °C. Lymph node tissue was planted on Sabouraud, BCYE, and sheep blood agar at 30 °C and Mid-dlebrook, sheep blood agar, chocolate agar, BCYE, and fastidious broth at 35 °C. Colonies that grew from sheep blood agar at 35 °C were sub-cultured onto BCYE at 30 °C. Identification was carried out by sequencing the 16 s rRNA gene, initially partial sequencing of about 500 base pairs (bp), followed by full gene length ofroughly1,500 bp (MicroSEQ®500 and Full Gene, Applied Biosystems, Foster City, CA, USA).
Protein Purification
Bacterial cells were streaked onto 5 % sheep blood plates (Remel) and grown at 37 °C with 5 % CO2. A single colony was inoculated into 50 mL of Mueller-Hinton Broth (Oxoid) and grown overnight at 37 °C with shaking. B-PER Protein Extraction Reagent (Thermo Scientific Pierce) was used on bacterial cell pellets per the manufacturer’s instructions. Total protein was measured by Coomassie Plus (Bradford) Protein Assay (Thermo Scientific Pierce).
Immunoblotting
Soluble protein samples (2 μg) were mixed with Laemmli Sample Buffer (Bio-Rad), heated (5 min, 95 °C), separated on Any kD Mini-PROTEAN TGX Precast Gels (Bio-Rad) and transferred to nitrocellulose membranes in 10x Tris/Glycine containing 20 % methanol (Bio-Rad). Membranes were blocked (TBS containing 0.05 % (v/v) Tween-20 (Fisher) with 5 % (w/v) Blotting-Grade Blocker (Bio-Rad) and incubated overnight at 4°C. Serum was diluted as indicated in TBS containing 0.05 % (v/v) Tween-20 (TBST) and membranes were incubated (1 h, RT), washed three times with TBST, then incubated with horseradish peroxidase-conjugated goat anti-human IgG (Amersham Biosciences) at 1:10,000 dilution. Signals were detected by enhanced chemiluminescence ECL Plus kit (Amersham Biosciences). Extracts prepared included the case patient’s A. methanolica isolate, an ATCC type strain of A. methanolica (ATCC 43581, American Type Culture Collection, Manassas, VA) and G. bethesdensis (ATCC BAA-1260).
Results
Sequencing of the 16s rRNA gene (~1413 bp) identified the isolate as A. methanolica with 99.6 % identity to the type strain (accession number X77468). The A. methanolica isolated from our patient was named CGDAM1. The 16S rRNA gene sequence of CGDAM1 has been deposited in GenBank under accession number JN256031.
To confirm specificity of our patient’s IgG to A. methanolica, immunoblots comparing extracts of our patient’s A. methanolica (CGDAM1), A. methanolica (ATCC 43581), and G. bethesdensis (ATCC BAA-1260) were probed with patient serum (Fig. 1c). Whereas identical banding patterns were noted on extracts from CGDAM1 and ATCC 43581, a different banding pattern was noted using the G. bethesdensis extract.
Discussion
A. methanolica belongs to the family Acetobacteraceae, members of which are known as “acetic acid bacteria.” Acetobacteraceae contains the genera, Acetobacter, Gluconobacter, Gluconacetobacter, Swaminathania, Neoasaia, Saccharibacter, Asaia, Kozakia, and Granulibacter [6]. Acetobacteraceae have been isolated from fruits and vegetables worldwide. They have industrial applications in the production of various pharmaceuticals and vinegar [7]. Several Acetobacteraceae have been implicated as opportunistic pathogens in humans. For instance, members of the genera, Asaia have infected patients with cystic fibrosis, Granulibacter has infected patients with CGD, and Gluconobacter infected a patient with a history of intravenous drug use, and another with cystic fibrosis [8]. A. methanolica is an acidotolerant, facultatively methylotrophic, aerobic, gram-negative, catalase-positive, urease-positive, and oxidase-positive, non-spore-forming, nonmotile, rod-shaped bacteria that is 0.8 to 1.0 by 1.5 to 3.0 micrometers, occurring singly, or rarely in pairs [9]. Initially categorized in 1986 as Acetobacter methanolicus, the organism, was reclassified to the genus Acidomonas in 1989 by Urakami et al. based upon its methyltrophic capabilities, which were absent in bacteria of the genus Acetobacter [10, 11].
The specificity of our patient’s immune reponse to A. methanolica is strongly supported by the identical banding patterns seen using our patient’s serum against protein extracts from two A. methanolica strains, CGDAM1 and ATCC 43581. The distinct banding pattern seen when his serum was used to probe protein extracts from G. bethesdensis indicates that this is not a non-specific reaction to another methylotroph isolated from CGD patients, G. bethesdensis. We did not exhaustively screen his plasma against all other methylotrophs, but the combination of isolating a rare organism in pure culture from a sterile site and specific immune response to its proteins is clear evidence that this organism was indeed involved in his lymphadenopathy and elicited an immune response.
This is the first reported case of human infection with A. methanolica, and the second methylotroph to be identified causing infection in CGD, after G. bethesdensis. This suggests that NADPH oxidase activity is central to host defense against this class of organisms, and that these fastidious organisms find a comfortable niche in CGD patients that is not available elsewhere. The fact that these organisms have not been recognized in other forms of neutropenia suggests that the critical defect may be more about NADPH activity than neutrophils per se. The identification of a rare and fastidious organism from frequently sampled lymph nodes in a CGD patient suggests that the reasons for the poor culture recovery from CGD infections is not due to recurrent bouts of sterile inflammation, but rather to organisms that are not easily detected in routine microbiology laboratories. We suggest that expanded culture techniques for fastidious organisms should be considered in all CGD patient samples.
Acknowledgments
Funding: Department of Laboratory Medicine, National Institutes of Health
Contributor Information
John M. Chase, Email: chasejm@gmail.com, Division of General Pediatrics, MS#75, Children’s Hospital Los Angeles and Keck School of Medicine, University of Southern California, 4650 Sunset Blvd., Los Angeles, CA 90027, USA
Steven M. Holland, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA
David E. Greenberg, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA
Kimberly Marshall-Batty, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.
Adrian M. Zelazny, Laboratory of Clinical Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA. Microbiology Service, Department of Laboratory Medicine, Clinical Center, National Institutes of Health, Bethesda, MD, USA
Joseph A. Church, Email: jchurch@chla.usc.edu, Division of Clinical Immunology and Allergy, MS#75, Children’s Hospital Los Angeles and Keck School of Medicine, University of Southern California, 4650 Sunset Blvd., Los Angeles, CA 90027, USA
References
- 1.Winkelstin JA, Marino MC, Johnston RB, Jr, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine(Baltimore) 2000;79(3):155–69. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Lestini BJ, Church JA. Trichosporon pullulans as a complication of chronic granulomatous disease in a patient undergoing immunosuppressive therapy for inflammatory bowel disease. Pediatr Infect Dis J. 2006;25(1):87–9. doi: 10.1097/01.inf.0000195641.69380.a0. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg DE, Ding L, Zelazny AM, et al. A novel bacterium associated with lymphadenitis in a patient with chronic granulomatous disease. PLoS Pathog. 2006;2(4):e28. doi: 10.1371/journal.ppat.0020028. Epub 2006 Apr. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg DE, Shoffner AR, Zelazny AM, et al. Recurrent Granulibacter bethesdensis infections and chronic granulomatous disease. Emerg Infect Dis. 2010;16(9):1341–8. doi: 10.3201/eid1609.091800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerard B, El Benna J, Alcain F, Gougerot-Pocidola MA, Grand-champ B, Chollet-Martin S. Characterization of 11 novel mutations in the X-linked chronic granulomatous disease (CYBB gene) Hum Mutat. 2001;18(2):163. doi: 10.1002/humu.1166. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg DE, Porcella SF, Zelazny AM, et al. Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. J Bacteriol. 2007;189(23):8727–36. doi: 10.1128/JB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sokollek SJ, Hertel C, Hammes WP. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int J Syst Bacteriol. 1998;48:935–40. doi: 10.1099/00207713-48-3-935. [DOI] [PubMed] [Google Scholar]
- 8.Alauzet C, Teyssier C, Jumas-Bilak E, et al. Gluconobacter as well as Asaia species, newly emerging opportunistic human pathogens among acetic acid bacteria. J Clin Microbiol. 2010;48(11):3935–42. doi: 10.1128/JCM.00767-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashita S, Uchimura T, Komagata K. Emendation of the genus Acidomonas Urakami, Tamaoka, Suzuki, and Komagata 1989. Int J Syst Evol Microbiol. 2004;54:865–70. doi: 10.1099/ijs.0.02946-0. [DOI] [PubMed] [Google Scholar]
- 10.Uhlig H, Karbaum K, Steudel A. Acetobacter methanolicus sp. nov., an acidophilic facultatively methylotrophic bacterium. Int J Syst Bacteriol. 1986;36:317–22. [Google Scholar]
- 11.Urakami T, Tamaoka J, Suzuki KI, Komagata K. Acidomonas gen. nov. Incorporating Acetobacter methanolicus as Acidomonas methanolica comb. nov. Int J Syst Bacteriol. 1989;39:50–5. [Google Scholar]