Abstract
Background
The xeroderma pigmentosum complementary group D (XPD) gene has been linked to the development of colorectal cancer (CRC) through disruption of DNA repair. Several studies have suggested that the XPD polymorphism Lys751Gln is associated with an increased risk of developing CRC. However, previous results remain inconclusive. Herein, we performed a meta-analysis to evaluate the potential for this relationship.
Methods
Relevant studies were retrieved from the PubMed database. Strict selection and exclusion criteria were determined, and the odds ratio with a 95% confidence interval was used to assess the strength of associations. The fixed or random effects model was selected on the basis of heterogeneity tests among studies. Publication bias was estimated using funnel plots and Egger’s regression test.
Results
The meta-analysis included 2,961 cases and 4,539 controls from eleven studies. The results indicated that the XPD Lys751Gln polymorphism had no association with CRC risk for all genetic models (Gln-Gln versus Lys-Lys, P=0.477; Lys-Gln versus Lys-Lys, P=0.283; Lys-Gln + Gln-Gln versus Lys-Lys, P=0.562), even when compared within subgroups based on ethnicity and source of controls.
Conclusion
Based on the results of our meta-analysis, there is no evidence of a link between the XPD Lys751Gln polymorphism and risk of CRC.
Keywords: XPD Lys751Gln polymorphism, colorectal cancer risk, meta-analysis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and is currently listed third under incidence and second under mortality worldwide.1 The National Center for Health Statistics in the USA estimates that, in 2014, 71,830 men and 65,000 women will be diagnosed with CRC and 26,270 men and 24,040 women will die of the disease.2 Worldwide, the prevention and treatment of CRC faces enormous challenges. Development of CRC is closely linked to many environmental and genetic factors, including lack of dietary fiber, overweight and obesity, physical inactivity, a short appendix vermiformis, a high-fat diet, smoking, and excessive alcohol consumption.3 Genetic factors play an important role, as susceptibility to CRC may result from inherited mutations in genes involved in carcinogen metabolism and DNA repair.4 It is now widely thought that the pathogenesis of CRC is related to environmental triggers and genetic susceptibility to multifactorial interactions.
Nucleotide excision repair is an important element of genome maintenance that is mainly responsible for repairing DNA adducts and other types of damage that cause helical distortions.5 A number of enzymes are involved in the nucleotide excision repair pathway, including xeroderma pigmentosum complementary groups A, C, D, and F (XPA, XPC, XPD, and XPF, respectively), replication protein A (RPA), and excision repair cross-complementing 1 (ERCC1).6 XPD consists of about 20 kb on chromosome 19q13.3 and contains 23 exons. An A→C polymorphism in XPD codon 751 of exon 23 leads to a Lys→Gln amino acid substitution (Lys751Gln) that is associated with a DNA damage repair phenotype.7 A difference in DNA repair capacity has been proposed to be a contributor to CRC susceptibility. Many studies have focused on the gene loci of XPD Lys751Gln.
To date, many studies focus on the genetic polymorphism of XPD and its contribution to CRC susceptibility; however, due to the interethnic heterogeneity of the disease, the limitations of statistical power in individual studies, small sample sizes, and other factors, the results are inconsistent. To reduce the research bias and improve the effectiveness of statistical correlation analysis, we conducted a comprehensive quantitative meta-analysis of previous results to evaluate the relationship between the XPD Lys751Gln polymorphism and the risk of CRC.
Materials and methods
Publication search
This meta-analysis adhered to the relevant criteria of the Preferred Reporting Items for Systemic Reviews and Meta-Analyses (PRISMA) statement.8 The PubMed database was searched through February 2014 for English material published between 1984 and 2014. Articles were sought with the following medical subject heading (MeSH) terms: XPD [All Fields] OR Lys751Gln [All Fields] AND (“colorectal neoplasms” [MeSH terms] OR (“colorectal” [All Fields] AND “neoplasms” [All Fields]) OR “colorectal neoplasms” [All Fields] OR (“colorectal” [All Fields] AND “cancer” [All Fields]) OR “colorectal cancer” [All Fields]). All references cited in the original studies or review articles concerning the relevant topic were retrieved to broaden the search for relevant publications.
Inclusion and exclusion criteria
In this meta-analysis, publications were included using the following criteria: case-control studies investigating the relationship between the XPD Lys751Gln polymorphism and CRC risk; patients with histologically confirmed CRC; sufficient genotype distribution information in cases and controls; and genotype distribution compliant with the Hardy-Weinberg equilibrium (HWE). The following exclusion criteria were used: abstracts and reviews; study designs other than case-control method; detailed genotype frequency not reported; and repeat or overlapping publications.
Data extraction
Data were independently extracted by two different investigators from all included studies: name of first author, publication year, country or area, characteristics of controls, sources of controls (population-based or hospital-based), genotyping method, fitness of HWE in controls, and genotype distribution. Any discrepancy was resolved through discussion or by a third person.
Statistical analysis
The crude odds ratio (OR) and 95% confidence interval (95% CI) were used to assess the strength of the association between the XPD Lys751Gln polymorphism and CRC risk. The pooled ORs were performed in homozygous (Gln-Gln versus Lys-Lys), heterozygous (Lys-Gln versus Lys-Lys), and dominant (Lys-Gln + Gln-Gln versus Lys-Lys) models. A chi square-based Q-test was used to test the assumption of heterogeneity. P>0.1 for the Q-test suggested a lack of heterogeneity among studies and required the fixed effects model (the Mantel-Haenszel method) to estimate the pooled OR of each study.9 Otherwise, the random effect model was used. Egger’s test and Begg’s funnel plot was plotted to examine the underlying publication bias among the included studies. An asymmetric plot suggested possible publication bias, while P>0.05 suggested no bias. Statistical analyses were performed using Stata statistical software (version 10.0, Stata Corporation, College Station, TX, USA). P<0.05 was considered to be statistically significant, and all P-values were two-sided.
Results
Characteristics of studies
In this meta-analysis, eleven case-control studies were included to evaluate the relationship between the XPD Lys751Gln polymorphism and CRC risk, which provided a total of 2,961 cases and 4,539 controls for the present meta-analysis.10–20 All cases were histologically confirmed as colon or rectal cancer. There were four studies from Asian populations10,13,18,20 and seven from Caucasian populations.11,12,14–17,19 Among the eleven studies, seven were hospital-based10,11,14,16,18–20 and four were population-based.12,13,15,17 Controls were mainly healthy populations and were matched for age and gender. Genotyping methods included polymerase chain reaction and restriction fragment length polymorphism and TaqMan in accordance with HWE (see Table 1).
Table 1.
Study characteristics
| Study | Ethnicity | Control source | Genotyping method | Case | Control | HWE |
|---|---|---|---|---|---|---|
| Yeh et al10 | Asian | HB | PCR-RFLP | 727 | 736 | Y |
| Skjelbred et al11 | Caucasian | HB | TaqMan-assay | 157 | 399 | Y |
| Hansen et al12 | Caucasian | PB | TaqMan-assay | 396 | 798 | Y |
| Stern et al13 | Asian | PB | TaqMan-assay | 303 | 1,163 | Y |
| Sliwinski et al14 | Caucasian | HB | PCR-RFLP | 100 | 100 | Y |
| Joshi et al15 | Caucasian | PB | TaqMan-assay | 380 | 381 | Y |
| Engin et al16 | Caucasian | HB | PCR-RFLP | 110 | 116 | Y |
| Jelonek et al17 | Caucasian | PB | PCR-RFLP | 123 | 153 | Y |
| Wang et al18 | Asian | HB | PCR-RFLP | 302 | 291 | Y |
| Procopciuc and Osian19 | Caucasian | HB | PCR-RFLP | 150 | 162 | Y |
| Ni et al20 | Asian | HB | TaqMan-assay | 213 | 240 | Y |
Abbreviations: HB, hospital-based; PB, population-based; PCR-RFLP, polymerase chain reaction and restriction fragment length polymorphism; HWE, Hardy-Weinberg equilibrium; Y, yes.
Quantitative synthesis
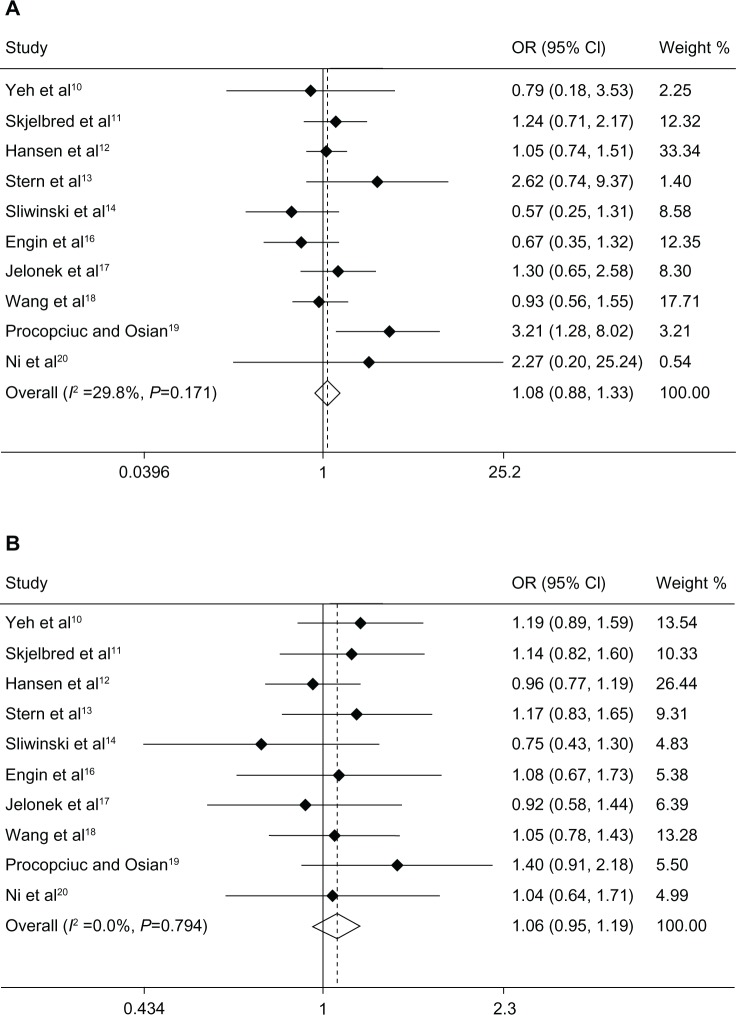
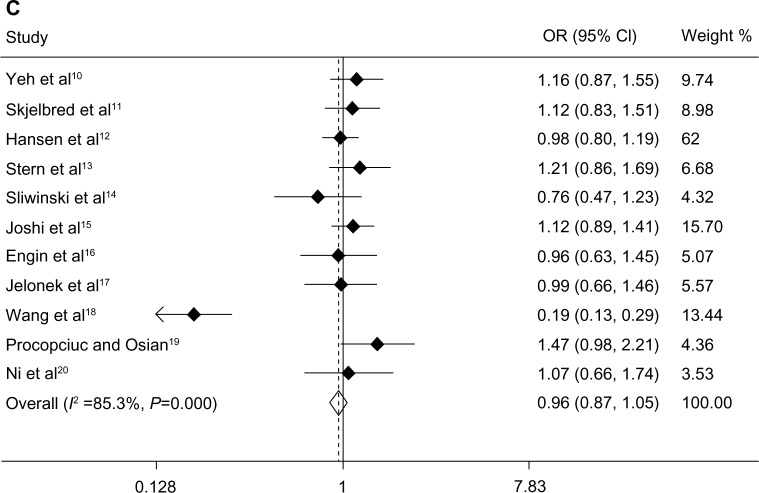
Overall, there was no significant association between the XPD Lys751Gln polymorphism and risk of CRC. The specific results were as follows: homozygous model (Gln-Gln versus Lys-Lys, OR 1.08, P=0.477, 95% CI 0.88–1.33), heterozygous model (Lys-Gln versus Lys-Lys, OR 1.06, P=0.283, 95% CI 0.95–1.19), and dominant model (Lys-Gln + Gln-Gln versus Lys-Lys, OR 0.96, P=0.562, 95% CI 0.87–1.05, Figure 1). We performed subgroup analysis, and the results suggested no significant association in any of the genetic models with ethnicity or source of controls (see Table 2).
Figure 1.
Association between the XPD Lys751Gln polymorphism and colorectal cancer risk. Forest plots of (A) homozygous model (Gln-Gln versus Lys-Lys), (B) heterozygous model (Lys-Gln versus Lys-Lys), and (C) dominant model (Lys-Gln + Gln-Gln versus Lys-Lys).(/p)(/p)Abbreviations: CI, confidence interval; OR, odds ratio; XPD, xeroderma pigmentosum complementary group D.
Table 2.
Subgroup analysis of correlation between polymorphism of XPD 751 and CRC risk
| Comparisons | Number of studies | Odds ratio
|
Test of heterogeneity
|
|||
|---|---|---|---|---|---|---|
| OR (95% CI) | P-value | Z-value | I2 (%) | P-value | ||
| Asians | ||||||
| Gln-Gln versus Lys-Lys | 4 | 1.05 (0.72–1.53) | 0.807 | 0.24 | 0.0 | 0.426 |
| Lys-Gln versus Lys-Lys | 4 | 1.12 (0.95–1.33) | 0.181 | 1.34 | 0.8 | 0.930 |
| Lys-Gln + Gln-Gln versus Lys-Lys | 4 | 0.74 (0.33–1.67) | 0.467 | 0.73 | 94.9 | <0.001 |
| Caucasians | ||||||
| Gln-Gln versus Lys-Lys | 6 | 1.07 (0.89–1.28) | 0.497 | 0.68 | 49.8 | 0.076 |
| Lys-Gln versus Lys-Lys | 6 | 1.02 (0.88–1.18) | 0.788 | 0.27 | 0.0 | 0.509 |
| Lys-Gln + Gln-Gln versus Lys-Lys | 7 | 1.05 (0.94–1.17) | 0.424 | 0.80 | 85.3 | <0.001 |
| PB | ||||||
| Gln-Gln versus Lys-Lys | 3 | 1.12 (0.87–1.43) | 0.369 | 0.90 | 3.5 | 0.355 |
| Lys-Gln versus Lys-Lys | 3 | 1.00 (0.84–1.18) | 0.969 | 0.04 | 0.0 | 0.794 |
| Lys-Gln + Gln-Gln versus Lys-Lys | 4 | 1.05 (0.93–1.20) | 0.423 | 0.80 | 0.0 | 0.657 |
| HB | ||||||
| Gln-Gln versus Lys-Lys | 7 | 1.02 (0.81–1.28) | 0.876 | 0.16 | 41.4 | 0.115 |
| Lys-Gln versus Lys-Lys | 7 | 1.11 (0.96–1.28) | 0.154 | 1.43 | 0.0 | 0.745 |
| Lys-Gln + Gln-Gln versus Lys-Lys | 7 | 0.84 (0.52–1.35) | 0.475 | 0.71 | 90.6 | <0.001 |
Abbreviations: CI, confidence interval; CRC, colorectal cancer; OR, odds ratio; HB, hospital-based; PB, population-based; XPD, xeroderma pigmentosum complementary group D.
Tests of heterogeneity and publication bias
Due to heterogeneity in the meta-analysis of association between the dominant model and CRC risk (Lys-Gln + Gln-Gln versus Lys-Lys, I2=85.3%, P=0.000), a random effects model was adopted for this analysis. Subgroup analysis indicated that ethnicity and source of controls were not significant sources of heterogeneity. The fixed effects model was used for the homozygous and heterozygous models because of the absence of heterogeneity (Gln-Gln versus Lys-Lys, I2=29.8%, P=0.171; Lys-Gln versus Lys-Lys, I2=0.0%, P=0.794).
Begg’s funnel plot and Egger’s test were used to assess the publication bias of the included articles. The shape of the funnel plot was not obviously asymmetric (see Figure 2). In addition, Egger’s test revealed no evidence of publication bias (P>0.05).
Figure 2.
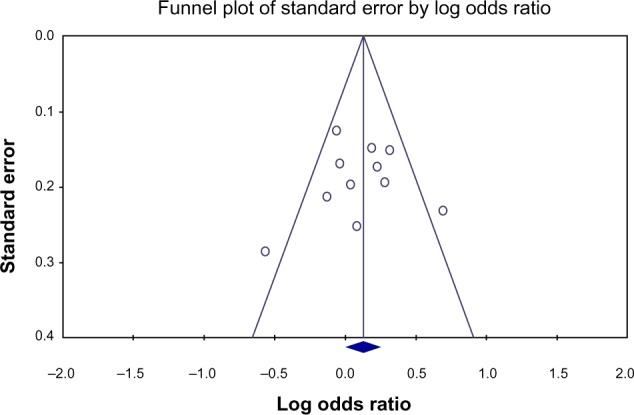
Funnel plot of standard error observed in each study by the log odds ratio.
Discussion
The pathogenesis of CRC has not been elucidated, but the following factors are thought to be contributory: diet and carcinogens; chronic colorectal inflammations such as ulcers, polyps, and schistosomiasis; tumor suppressor gene mutations or genetic instability; and colorectal adenomas and other precancerous lesions.21–24 Currently, it is generally thought that the occurrence of CRC is a gradual process involving multiple oncogene activations and tumor suppressor gene inactivation.25,26 DNA repair systems are the body’s main defense barrier for internal and external environmental factors that are sources of genome instability. Defects in these systems or low DNA repair capacity increase the risk of genetic mutations and cell carcinogenesis. Thus, individuals with compromised DNA repair capacity are susceptible to tumors.
XPD is an important DNA repair gene involved in base and nucleotide excision repair of DNA. Single nucleotide polymorphisms present in multiple sites of this gene have been linked to enhanced susceptibility to various cancers, including lung cancer, gastric cancer, and breast cancer.7,27,28 XPD participates in unwinding the DNA helix to allow excision of damaged DNA fragments.29 A study of the XPD gene found that a mutation at the codon 751 allele is most common.25 Many epidemiological studies have thereafter been indicating the role of Lys751Gln polymorphism on CRC susceptibility, but the result remains controversial. Thus, our meta-analysis, comprising 2,961 CRC patients and 4,539 controls from eleven studies, was performed to assess precisely the possible association of XPD Lys751Gln polymorphism with susceptibility to CRC. Our meta-analysis suggests that XPD Lys751Gln polymorphism was not associated with CRC risk. In the subgroup analysis, where studies were divided by ethnicity or source of controls, there was still no significant association detected in the homozygous, heterozygous, or dominant model.
Meta-analysis is reliant on available published data, and publication bias is common. Through the qualitative funnel plot and quantitative Egger’s linear regression, we have shown that there is no significant publication bias in the current study. However, there are some limitations to this meta-analysis. First, the studies included involve only Asian and Caucasian populations, without representation of other racial or ethnic groups, including the African population. Additionally, we could only carry out a subgroup analysis for a large geographical area despite the presence of different national characteristics within a region due to the lack of raw data for each study. Moreover, we could not carry out individual subgroup analysis for patients with colon and colorectal cancer due to the lack of availability of raw data. Finally, the potential effect of gene-gene or gene-environment interactions on the statistical results was not considered.
In summary, our meta-analysis pooled all available data related to potential links between XPD Lys751Gln and CRC, and found no evidence that the polymorphism is associated with CRC risk.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hoffmeister M, Jansen L, Stock C, et al. Smoking, lower gastrointestinal endoscopy, and risk for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2014;23(3):525–533. doi: 10.1158/1055-9965.EPI-13-0729-T. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 3.Cakmak YO, Ergelen R, Ekinci G, et al. The short appendix vermiformis as a risk factor for colorectal cancer. Clin Anat. 2014;27(3):498–502. doi: 10.1002/ca.22375. [DOI] [PubMed] [Google Scholar]
- 4.Qiu LX, Yao L, Mao C, et al. Lack of association of CYP1A2-164 A/C polymorphism with breast cancer susceptibility: a meta-analysis involving 17,600 subjects. Breast Cancer Res Treat. 2010;122(2):521–525. doi: 10.1007/s10549-009-0731-4. [DOI] [PubMed] [Google Scholar]
- 5.Mandal RK, Yadav SS, Panda AK. Meta-analysis on the association of nucleotide excision repair gene XPD A751C variant and cancer susceptibility among Indian population. Mol Biol Rep. 2014;41(2):713–719. doi: 10.1007/s11033-013-2910-y. [DOI] [PubMed] [Google Scholar]
- 6.Chiang CC, Tsai YY, Bau DT, et al. Pterygium and genetic polymorphisms of the DNA repair enzymes XRCC1, XPA, and XPD. Mol Vis. 2010;16:698–704. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Zhang C, Xu C, et al. Effects of selected genetic polymorphisms in xeroderma pigmentosum complementary group D on gastric cancer. Mol Biol Rep. 2011;38(3):1507–1513. doi: 10.1007/s11033-010-0258-0. [DOI] [PubMed] [Google Scholar]
- 8.Panic N, Leoncini E, de Belvis G, et al. Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses. PLoS One. 2013;8(12):e83138. doi: 10.1371/journal.pone.0083138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Zhang Y, Ying X, et al. The association between RAD23B Ala249Val polymorphism and cancer susceptibility: evidence from a meta-analysis. PLoS One. 2014;9(3):e91922. doi: 10.1371/journal.pone.0091922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh CC, Hsieh LL, Tang R, et al. MS-920: DNA repair gene polymorphisms, diet and colorectal cancer risk in Taiwan. Cancer Lett. 2005;224(2):279–288. doi: 10.1016/j.canlet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 11.Skjelbred CF, Saebø M, Wallin H, et al. Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study. BMC Cancer. 2006;6:67. doi: 10.1186/1471-2407-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen RD, Sørensen M, Tjønneland A, et al. XPA A23G, XPC Lys939Gln, XPD Lys751Gln and XPD Asp312Asn polymorphisms, interactions with smoking, alcohol and dietary factors, and risk of colorectal cancer. Mutat Res. 2007;619(1–2):68–80. doi: 10.1016/j.mrfmmm.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Stern MC, Conti DV, Siegmund KD, et al. DNA repair single-nucleotide polymorphisms in colorectal cancer and their role as modifiers of the effect of cigarette smoking and alcohol in the Singapore Chinese Health Study. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2363–2372. doi: 10.1158/1055-9965.EPI-07-0268. [DOI] [PubMed] [Google Scholar]
- 14.Sliwinski T, Krupa R, Wisniewska-Jarosinska M, et al. Common polymorphisms in the XPD and hOGG1 genes are not associated with the risk of colorectal cancer in a Polish population. Tohoku J Exp Med. 2009;218(3):185–191. doi: 10.1620/tjem.218.185. [DOI] [PubMed] [Google Scholar]
- 15.Joshi AD, Corral R, Siegmund KD, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30(3):472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engin AB, Karahalil B, Engin A, et al. Oxidative stress, helicobacter pylori, and OGG1 Ser326Cys, XPC Lys939Gln, and XPD Lys751Gln polymorphisms in a Turkish population with colorectal carcinoma. Genet Test Mol Biomarkers. 2010;14(4):559–564. doi: 10.1089/gtmb.2009.0195. [DOI] [PubMed] [Google Scholar]
- 17.Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single- nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51(3):343–352. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Zhao Y, Jiang J, et al. Polymorphisms in DNA repair genes XRCC1, XRCC3 and XPD, and colorectal cancer risk: a case-control study in an Indian population. J Cancer Res Clin Oncol. 2010;136(10):1517–1525. doi: 10.1007/s00432-010-0809-8. [DOI] [PubMed] [Google Scholar]
- 19.Procopciuc LM, Osian G. Lys751Gln XPD and Arg399Gln XRCC1 in Romanians. Association with sporadic colorectal cancer risk and different stages of carcinomas. Chirurgia (Bucur) 2013;108(5):711–718. [PubMed] [Google Scholar]
- 20.Ni M, Zhang WZ, Qiu JR, et al. Association of ERCC1 and ERCC2 polymorphisms with colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:4112. doi: 10.1038/srep04112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrör K, Rauch BH. Aspirin and prevention of colorectal carcinomas. Internist (Berl) 2013;54(7):884–891. doi: 10.1007/s00108-013-3311-y. [DOI] [PubMed] [Google Scholar]
- 22.Waldner MJ, Neurath MF. Master regulator of intestinal disease: IL-6 in chronic inflammation and cancer development. Semin Immunol. 2014;26(1):75–79. doi: 10.1016/j.smim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Win AK, Hopper JL, Buchanan DD, et al. Are the common genetic variants associated with colorectal cancer risk for DNA mismatch repair gene mutation carriers? Eur J Cancer. 2013;49(7):1578–1587. doi: 10.1016/j.ejca.2013.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conteduca V, Sansonno D, Russi S, et al. Precancerous colorectal lesions. Int J Oncol. 2013;43(4):973–984. doi: 10.3892/ijo.2013.2041. [DOI] [PubMed] [Google Scholar]
- 25.Juárez-Vázquez CI, Rosales-Reynoso MA. Colorectal cancer (CCR): genetic and molecular alterations. Gac Med Mex. 2014;150(2):154–164. Spanish. [PubMed] [Google Scholar]
- 26.Sameer AS. Colorectal cancer: molecular mutations and polymorphisms. Front Oncol. 2013;3:114. doi: 10.3389/fonc.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou M, Wan HY, Gao BL, et al. Genetic polymorphisms of XPD and CDA and lung cancer risk. Oncol Lett. 2012;4(2):247–251. doi: 10.3892/ol.2012.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathieu N, Kaczmarek N, Naegeli H. Strand and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc Natl Acad Sci U S A. 2010;107(41):17545–17550. doi: 10.1073/pnas.1004339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan Y, Liang H, Light M, et al. XPD Asp312Asn and Lys751Gln polymorphisms and breast cancer susceptibility: a meta-analysis. Tumour Biol. 2014;35(3):1907–1915. doi: 10.1007/s13277-013-1256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]