Abstract
Peters Plus syndrome (PPS) is a rare autosomal-recessive disorder characterized by Peters anomaly of the eye, short stature, brachydactyly, dysmorphic facial features, developmental delay, and variable other systemic abnormalities. In this report we describe screening of 64 patients affected with PPS, isolated Peters anomaly and PPS-like phenotypes. Mutations in the coding region of B3GALTL were identified in nine patients; six had a documented phenotype of classic PPS and the remaining three had a clinical diagnosis of PPS with incomplete clinical documentation. A total of nine different pathogenic alleles were identified. Five alleles are novel including one frameshift, c.168dupA, p.(Gly57Argfs*11), one nonsense, c.1234C>T, p.(Arg412*), two missense, c.1045G>A, p.(Asp349Asn) and c.1181G>A, p.(Gly394Glu), and one splicing, c.347+5G>T, mutations. Consistent with previous reports, the c.660+1G>A mutation was the most common mutation identified, seen in eight of the nine patients and accounting for 55% of pathogenic alleles in this study and 69% of all reported pathogenic alleles; while two patients were homozygous for this mutation, the majority had a second rare pathogenic allele. We also report the absence of B3GALTL mutations in 55 cases of PPS-like phenotypes or isolated Peters anomaly, further establishing the strong association of B3GALTL mutations with classic PPS only.
INTRODUCTION
Peters Plus syndrome (PPS) is a rare autosomal-recessive disorder characterized by anterior segment dysgenesis of the eye, typically Peters anomaly, and systemic defects. Patients with classic PPS display brachydactyly, short stature, developmental delay, and dysmorphic facial features, along with variable other systemic anomalies (1). The gene affected in PPS, B3GALTL, was identified in 2006 by Lesnik Oberstein and coauthors (2). B3GALTL acts as a glucosyltransferase which catalyzes the addition of glucose to O-linked fucose via a β-1,3 attachment on thrombospondin type-1 repeats (TSRs) (3). Nine additional reports have been published linking mutations in B3GALTL with classic PPS (4–12).
Peters anomaly can also occur as an isolated feature or be associated with a broad range of structural anomalies (present in up to 60%); structural anomalies often overlap those seen in PPS, but may be present as a single anomaly or in varying combinations of anomalies which do not meet the criteria for classic PPS (PPS-like) (13,14). Peters anomaly with or without systemic anomalies is genetically heterogeneous with mutations in PAX6, PITX2, FOXC1, FOXE3, and CYP1B1 identified in some cases (15); in many cases, the genetic etiology remains unknown. No pathogenic mutations have been identified in B3GALTL in cases of isolated Peters anomaly or PPS-like phenotypes, but a limited number of patients was examined so far (4,7,16,17).
In this manuscript we report B3GALTL screening results of 64 patients affected with PPS, isolated Peters anomaly and PPS-like phenotypes.
PATIENTS AND METHODS
Human subjects
This human study was approved by the Institutional Review Boards of the Children’s Hospital of Wisconsin and the University of Iowa with written informed consent obtained for every subject. Genomic DNA was extracted using standard procedures from blood or buccal samples. Patients were referred from numerous institutions; clinical descriptions were obtained from review of medical records. A total of 64 probands with classic PPS, PPS- like phenotypes, and isolated Peters anomaly were studied (Table 1 and Supplemental Table 1).
Table 1.
Genotype and phenotype summary of all reported B3GALTL-positive Peters plus syndrome cases.
| Reference # |
Race (Sex) |
Allelesa | #b | Eye | Short Stature/ IUGR |
Limb | DD | CL/P | CHD | UT | GT | Other systems |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peters | ASD | Other | BD | Other | |||||||||||
| This paper (Patient 1) |
U (M) |
c.660+1G>A; Deletion |
1 | + | + | − | + | NR | + | U | − | + | − | − | EA, FD |
| This paper (Patient 2) |
U (M) |
c.660+1G>A p.(Gly57Argfs*11) |
1 | + | + | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| This paper (Patient 3) |
U (M) |
c.660+1G>A; c.660+1G>A |
1 | + | + | + | + | NR | + | + | + | − | − | − | NR |
| This paper (Patient 4) |
C (M) |
c.459+1G>A; c.660+1G>A |
1 | + | + | − | + | + | − | + | − | + | − | + | CNS,EA FD,GI |
| This paper (Patient 5) |
C (F) |
c.660+1G>A; c.1065−1G>A |
1 | + | + | − | + | + | − | + | − | + | + | + | CNS,EA FD, GI, PD, SD |
| This paper (Patient 6) |
H/C (F) |
c.660+1G>A; p.(Arg412*) |
1 | + | + | + | + | + | + | + | − | + | + | − | CNS,EA FD, SD |
| This paper (Patient 7) |
A (F) |
c.347+5G>T; p.(Asp349Asn) |
1 | + | + | − | + | + | + | + | + | − | + | − | EA, FD |
| This paper (Patient 8) |
C (M) |
c.660+1G>A; c.660+1G>A |
1 | + | + | + | + | + | + | + | − | + | + | + | CNS,EA FD, GI |
| This paper (Patient 9) |
C (M) |
c.660+1G>A; p.(Gly394Glu) |
1 | + (R) |
+ (R) |
+ | + | + | − | + | − | − | + | − | EA, FD, GI |
| 4 | 15 C 1 A |
c.660+1G>A; c.660+1G>A |
16 (13) |
12/16 | 16/16 | NR | 16/16 | NR | NR | 11/15 | 5/16 | 3/15 | 2/15 | NR | NR |
| 4 | 2 C |
c.660+1G>A; c.347+5G>A |
2 (1) | 2/2 | 2/2 | NR | 2/2 | NR | NR | 2/2 | 1/2 | 0/2 | 0/2 | NR | NR |
| 4 | 2 C |
c.660+1G>A; Deletion |
2 (1) | 2/2 | 2/2 | NR | 2/2 | NR | NR | 2/2 | 2/2 | 2/2 | 0/2 | NR | NR |
| 6 | 1 C 1 H |
c.660+1G>A; c.660+1G>A |
2 | 2/2 | 2/2 | 1/2 | 2/2 | 2/2 | 2/2 | 2/2 | 2/2 | 1/2 | 0/2 | 1/2 | CNS,EA FD, GI |
| 6 | C |
c.459+1G>A; c.660+1G>A |
1 | + | + | − | + | + | + | + | + | + | − | − | EA, FD |
| 6 | C |
c.660+1G>A; c.230dupT |
1 | + | + | − | + | + | + | U | + | + | + | − | CNS, FD GI, SD |
| 7 | A |
c.660+1G>A; c.660+1G>A |
1 | + | + | − | + | + | + | + | − | + | − | − | EA, FD, SD |
| 8 | C |
c.660+1G>A; Deletion |
1 | + | + | NR | + | NR | + | U | − | + | + | + | CNS,EA FD, SD |
| 9 | U |
c.660+1G>A; p.(Gly393Glu) |
1 | + | + | NR | + | + | NR | + | − | + | − | − | FD |
| 9 | U |
c.459+1G>A; c.459+1G>A |
1 | + | + | NR | + | + | NR | + | − | + | − | − | FD |
| 10 | A |
p.(Tyr366*); p.(Tyr366*) |
1 | + | + | − | + | + | + | + | − | − | + | + | CNS,EA FD |
| 11 | C |
c.1065−1G>A; c.1065−1G>A |
1 | + | + | + | + | + | + | − | − | + | + | − | CNS,EA FD, SD |
| 12 | C |
c.597−2A>G; c.597−2A>G |
2 | 2/2 | 2/2 | 0/2 | 2/2 | 2/2 | 2/2 | 2/2 | 0/2 | 0/2 | 0/2 | 0/2 | CNS, FD |
| 13 | U |
c.660+1G>A; c.660+1G>A |
1 | + | + | − | + | − | − | − | − | − | − | + | CNS, SD |
| 14 | U |
c.660+1G>A; c.660+1G>A |
5 (2) | 1/4 | 4/4 | 2/4 | 5/5 | 3/3 | 0/3 | U | 3/5 | 0/5 | 2/5 | 1/5 | CNS, TAb |
| All patients | 47 | 39/46 85% |
46/46 100% |
8/21 38% |
46/46 100% |
20/21 95% |
15/22 68% |
31/37 84% |
17/46 37% |
18/45 40% |
13/45 31% |
8/26 31% |
|||
Abbreviations: A, Asian; ASD, Anterior segment dysgenesis; BD, brachydactyly; C, Caucasian; CHD, congenital heart defect; CL/P, cleft lip/palate; CNS, structural brain anomaly; DD, developmental delay; EA, Ear anomaly (hearing loss in some); FD, facial dysmorphism; GI, gastrointestinal; GT, genitourinary tract; H, Hispanic; IUGR, intrauterine growth restriction; NR, not reported; PD, premature death; R, right eye only; Tab, terminated pregnancy; U, unknown; UT (urinary tract)
novel alleles identified in this study are indicated in bold;
when siblings are included, the number of families is indicated in parentheses.
B3GALTL screening
The coding region of B3GALTL was amplified using the previously described specific primers (4) and HotMaster Taq (5 PRIME, Gaithersburg, MD) or Taq98®Hot Start 2× Master Mix (Lucigen, Middleton, WI). PCR products were sequenced as previously described (4); analysis was performed with Mutation Surveyor (SoftGenetics, State College, PA) using reference sequence NM_194318.3. Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/) and dbSNP (http://www.ncbi.nlm.nih.gov/snp) were used to determine variant frequency in the general population and SIFT (http://SIFT.jcvi.org/) and PolyPhen (http://genetics.bwh.harvard.edu/pph2/) were used to examine the effects of missense variants.
Variations in the copy number of B3GALTL were analyzed using TaqMan® Copy Number Assays (Applied Biosystems, Carlsbad, California) using the specific probes Hs02373786_cn and Hs03844228_cn, targeting B3GALTL intron 1 and exon 14, respectively, and previously described protocols (18).
RESULTS
In total, B3GALTL sequence was examined in 64 probands with classic PPS, PPS-like conditions, or isolated Peters anomaly. We identified recessive pathogenic B3GALTL mutations in all six patients with documented features consistent with classic PPS (anterior segment dysgenesis, short stature, brachydactyly, and variable other features) and three patients who were referred with a clinical diagnosis of PPS but incomplete clinical documentation (Table 1, Figure 2, Supplemental Figure 1); eight patients had two nucleotide mutations and one patient (Patient 1) had a nucleotide mutation paired with a deletion of the B3GALTL gene. Eight patients carried at least one copy of the common splicing mutation, c.660+1G>A, and two of these patients were homozygous for this allele (copy number analysis confirmed diploid copy number). The c.660+1G>A mutation is also present in the general population at a frequency of 0.08% (7/8598) for European American alleles and 0% (0/4404) for African American alleles in EVS.
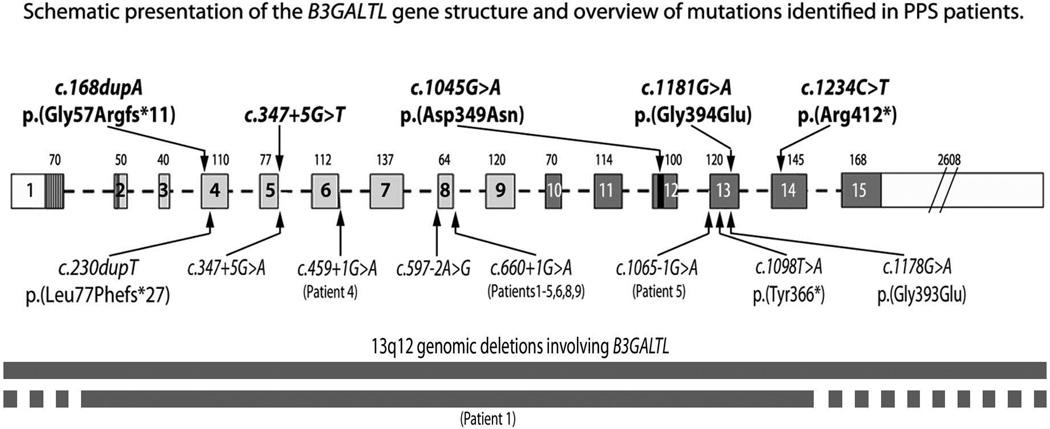
Figure 2.
Schematic presentation of the B3GALTL gene structure and overview of the mutations identified in patients with PPS. Novel mutations are indicated in bold above the gene diagram; previously reported mutations are indicated below the gene diagram. The B3GALTL transcript is 4254-bp in size (1497-bp for the coding region) and consists of 15 exons. Each exon is represented by a box with its size (bp) indicated on top. The N-terminal domain is shown as a vertically striped box, the stem region is shaded in light grey, the catalytic domain is shaded in dark grey and the catalytic core is indicated in black. Deletions of B3GALTL are indicated at the bottom of the drawing; previously reported deletions were identified using array-based comparative genomic hybridization approach and included B3GALTL and five additional genes, HSPH1, LGR8, LOC196545, FRY, and BRCA2, in one case (2) and B3GALTL and four additional gene, USPL1, ALOX5AP, C13orf23, HSPH1, in another study (6); the deletion in Patient 1 in this study was identified using B3GALTL-specific TaqMan copy number assays (Supplemental Figure 1), thus the full extent of the deletion is unknown.
Five of the mutations are novel variants that are predicted to be pathogenic (Figure 2; Supplemental Figure 1). Two of these pathogenic variants are truncating: the c.168dupA, p.(Gly57Argfs*11) change is the most N-terminal allele reported to date, while the c.1234C>T, p.(Arg412*) change is the most C-terminal mutation currently known; neither allele was observed in EVS. Two novel pathogenic missense mutations were identified, only the second and third missense mutations in B3GALTL reported to date. The c.1181G>A, p.(Gly394Glu) mutation was predicted to be deleterious/damaging by both SIFT and Polyphen and was found at a low frequency in EVS (1/4406 African American alleles; 0/8600 European American alleles). The c.1045G>A, p.(Asp349Asn) mutation was predicted to be deleterious/damaging by both SIFT and PolyPhen and was not present in EVS. Finally, a novel splicing mutation, c.347+5G>T, was identified at the same position as the previously reported c.347+5G>A mutation (2) and was not seen in EVS. Unfortunately, mRNA from the patient was unavailable for RT-PCR to confirm aberrant splicing; however, analysis of the sequences using MaxEntScan software (19), which estimates the probability for nucleotide sequences to be recognized as splice sites, resulted in a MaxEnt score of 6.49 for the reference 5' splice sequence of exon 5/intron 5 of B3GALTL (CAGgtacgt) and highly reduced negative scores of -2.43 for the previously reported c.347+5G>A (CAGgtacat) splice mutation, which was verified by RT-PCR as a splicing defect (2), and −2.13 for the novel c.347+5G>T (CAGgtactt) mutation.
Among these nine patients with pathogenic mutations, eight were affected with bilateral and one with unilateral Peters anomaly. Short stature was documented in all but one patient; birth length was available for three patients and was normal in all three (between 10th and 50th centile). Brachydactyly was documented in six of the nine cases. Additional ocular defects included microphthalmia in two patients and microcornea, chorioretinal degeneration, iris coloboma, and optic nerve coloboma in one patient each. Craniofacial, heart, kidney, and genital anomalies were common, consistent with previous reports of PPS. Developmental delay and facial dysmorphism were seen in seven patients (unreported in two); structural brain anomalies were seen in four patients including sphenoidal encephalocele in one. Feeding difficulties were noted in three patients: one patient required nasogastric feeds for the first year of life, another one had feeding intolerance with severe gastrointestinal dysmotility/pseudo-obstruction and was Total Parenteral Nutrition (TPN) dependent, and the third one had gastroesophageal reflux and oral dysphagia. One patient also had thrombocytopenia and chronic pancreatitis/pancreatic divisum; she passed away at 10 years of age from Systemic Inflammatory Response Syndrome.
In addition to the above described PPS cases, we screened 55 patients with Peters anomaly (45) or another ocular defect (10) with (37) or without (18) systemic anomalies overlapping with the PPS spectrum (Supplemental Table 1). No pathogenic variants in B3GALTL were identified in this group. Several heterozygous rare variants (<1% allele frequency in EVS or 1000 genomes) were identified in these cases, including three missense alleles, one synonymous variant, and four intronic variants located within 20 nucleotides of the exon (Supplemental Table 2); none of these changes was predicted to affect protein function and the majority are present in the general population.
DISCUSSION
In this report we describe nine patients with pathogenic mutations in the coding region of B3GALTL; six of these patients have a documented classic PPS phenotype that includes Peters anomaly, short stature, brachydactyly, dysmorphic facies, and variable other anomalies and three were referred with a clinical diagnosis of PPS and incomplete documentation. A total of nine different pathogenic alleles were identified in this study. Consistent with previous reports (2,4–12), the c.660+1G>A mutation was the most common mutation identified (Table 2). Overall, 86% of the reported pathogenic alleles are splicing mutations, 6% are truncating mutations, 4% are missense mutations, and 4% are whole gene deletions (Table 2).
Table 2.
Summary of B3GALTL pathogenic alleles and their frequencies in patient and general populations.
| PPS Allelea | Distribution in PPSb | Allele frequency in general populations | |||
|---|---|---|---|---|---|
| Occurrence | Frequency | EVS (EA)c | EVS (AA)d | dbVare | |
| B3GALTL deletion | 3 | 3.85% | - | - | 0.01% (4/32954) 4/16296 Asian 0/13627 Caucasian 0/3031 Other |
| c.168dupA,p.(Gly57Argfs*11) | 1 | 1.28% | 0 | 0 | - |
| c.230dupT, p.(Leu77Phefs*27) | 1 | 1.28% | 0 | 0 | - |
| c.347+5G>A | 1 | 1.28% | 0 | 0 | - |
| c.347+5G>T | 1 | 1.28% | 0 | 0 | - |
| c.459+1G>A | 4 | 5.13% | 0 | 0 | - |
| c.597−2A>G | 4 | 5.13% | 0 | 0 | - |
| c.660+1G>A | 54 | 69.23% | 0.08% (7/8598) | 0 | - |
| c.1045G>A,p.(Asp349Asn) | 1 | 1.28% | 0 | 0 | - |
| c.1065−1G>A | 3 | 3.85% | 0 | 0 | - |
| c.1098T>A, p.(Tyr366*) | 2 | 2.56% | 0 | 0 | - |
| c.1178G>A, p.(Gly393Glu) | 1 | 1.28% | 0 | 0 | - |
| c.1181G>A, p.(Gly394Glu) | 1 | 1.28% | 0 | 0.02% (1/4406) | - |
| c.1234C>T,p.(Arg412*) | 1 | 1.28% | 0 | 0 | - |
Novel alleles identified in this study are indicated in bold
European American Alleles; allele count ranged from 8592 to 8600;
African American Alleles; allele count ranged from 4402 to 4406;
Five novel pathogenic variants were identified, all predicted to result in complete loss-of-function alleles. The c.168dupA, p.(Gly57Argfs*11) and c.1234C>T, p.(Arg412*) truncating mutations are likely to result in nonsense mediated decay (NMD) (20). The two novel missense mutations, p.(Asp349Asn) and p.(Gly394Glu), affect highly conserved amino acids in or near the catalytic core of B3GALTL (Figure 2, Supplemental Figure 1). The final novel mutation, c.347+5G>T, affects the same position as the previously reported c.347+5G>A mutation (2) and is predicted to similarly disrupt splicing (19). The remaining three pathogenic alleles represent new occurrences of previously reported mutations. The whole gene deletion in Patient 1 represents the third family to be affected with B3GALTL deletion (2,6). Two other splicing mutations, c.1065−1G>A and c.459+1G>A, represent the second and third families to be reported with these mutations, respectively (4,7,9).
Several phenotypic features are of special interest within our B3GALTL-mutation positive group. Three of the nine patients reported here had feeding issues, in one case severe enough to require Total Parenteral Nutrition (TPN); while feeding problems were noted in a review of PPS (1) and at least one previously reported patient with B3GALTL mutations required a gastric tube (4), feeding problems had not been previously reported as a common feature associated with mutations in B3GALTL. This is the second report of encephalocele associated with mutations in B3GALTL; the previous case involved a meningoencephalocele seen in a fetus terminated at 18 weeks of gestation (12). Finally, one patient was affected with severe pancreatitis/pancreatic divisum, a condition not previously associated with PPS.
To date, all reported cases of PPS which have been explained by mutations in B3GALTL have been associated with ASD, short stature, dysmorphic facial features, and brachydactyly in all but one case where hands were examined; developmental delay is typically observed, but occasionally normal development is reported, and variable other features including cleft lip/palate, heart, genitourinary, ear, skeletal, and CNS anomalies are also seen (2,4–12). Our data bring the total number of PPS cases explained by mutations in B3GALTL to 47 cases from 39 independent families.
We also screened 55 additional cases affected with PPS-like conditions lacking the combination of anterior segment ocular defect, short stature, brachydactyly and facial dysmorphism, or isolated Peters anomaly; no pathogenic sequence variants were detected in B3GALTL in any of these cases. Five other reports have also described PPS-like phenotypes with no mutations in B3GALTL (4,7,9,16,17). Therefore, the data available in the literature as well as that presented here seem to indicate that loss of function of B3GALTL results in a classic PPS phenotype only. The PPS-like phenotypes are still awaiting molecular explanation. Identification of the genes involved in these PPS-like phenotypes is necessary for better classification and understanding of these complex conditions.
Supplementary Material
Supplemental Figure 1. (a) Novel B3GALTL mutations in PPS patients identified in this study. The position of each mutation is indicated with a black arrow. (b) Results of TaqMan Copy Number assays targeting 5’ (intron 1) and 3’ (exon 14) regions of B3GALTL that were performed on DNA samples from Patients 1, 3 and 8, who appeared homozygous for the common c.660+1G>A splicing mutation, alongside a control sample with diploid B3GALTL copy number; haploid copy number for both probes was detected for Patient 1 (arrow) while Patients 3 and 8 were diploid for both probes (c) Evolutionary conservation of amino acid residues affected by missense mutations identified in patients 7 and 9; note 100% conservation of both positions in vertebrates (box).
Figure 1.
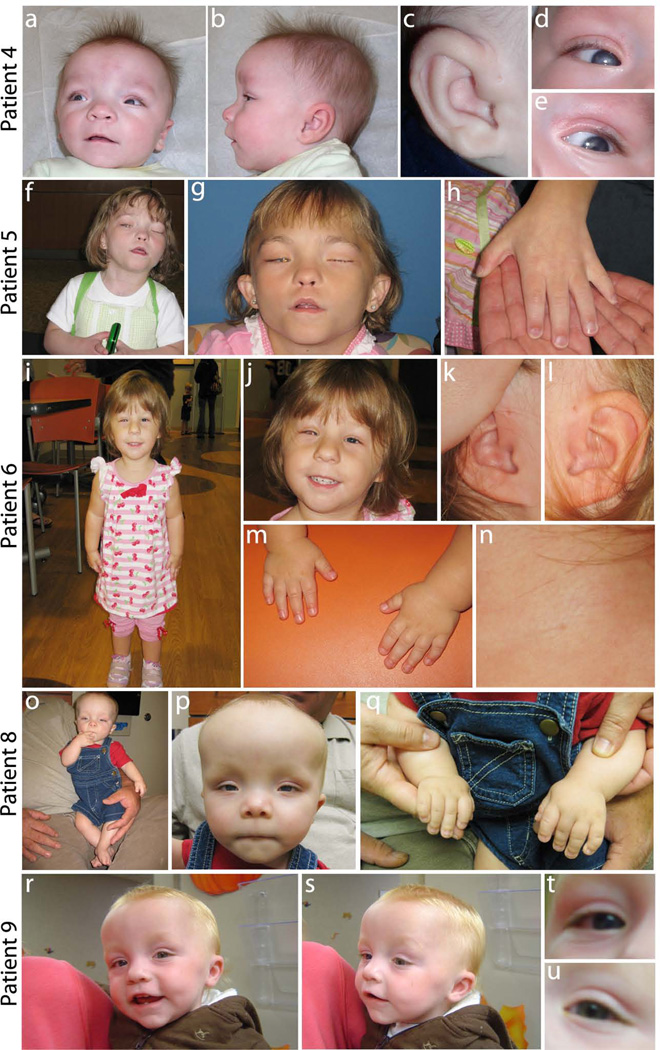
Photographs of patients documenting features of Peters Plus syndrome. (a–e) Patient 4 with relative macrocephaly, high forehead with sparse eyebrows, hypertelorism, broad nasal bridge, small nose, long, smooth philtrum, thin upper lip (a,b), bilateral preauricular pits (c), and bilateral Peters anomaly with partial clearing of the corneal opacity (d, e); (f–h) Patient 5 with right eye post corneal transplant, left eye enucleated, high forehead, hypertelorism, cupid bow upper lip, long smooth philtrum (f, g) and brachydactyly (h); (i–n) Patient 6 with short stature (i), left ocular prosthesis and right eye post sector iredectomy, microcephaly with micrognathia, a long philtrum (i, j), posteriorly rotated ears with preauricular pits (k, l), brachydactyly (m), and branchial fistula (n) ; (o–q) Patient 8 with short stature (o), broad forehead with sparse eyebrows, relative macrocephaly, a broad nasal bridge, hypertelorism, upslanting palpebral fissues, a simple philtrum with thin lips, low set ears (o,p), and brachydactyly (q); (r–u) Patient 9 with prominent forehead, prominent nose, protruding, thin upper lip, high arched palate, mildly receding chin, small, low set ears (r,s) and Peters anomaly of the right eye only (t,u).
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the patients and their families for their participation in research studies. This work was supported by the National Institutes of Health awards R01EY015518, R21DC010912 and funds provided by the Children’s Hospital of Wisconsin (EVS), and 1UL1RR031973 from the Clinical and Translational Science Award (CTSA) program.
Footnotes
THE AUTHORS DECLARE NO CONFLICT OF INTEREST.
REFERENCES
- 1.Maillette de Buy Wenniger-Prick LJ, Hennekam RC. The Peters' plus syndrome: a review. Ann Genet. 2002;45(2):97–103. doi: 10.1016/s0003-3995(02)01120-6. [DOI] [PubMed] [Google Scholar]
- 2.Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, den Dunnen JT, Breuning MH, Hennekam RC. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet. 2006;79(3):562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinonen TY, Maki M. Peters'-plus syndrome is a congenital disorder of glycosylation caused by a defect in the beta1,3-glucosyltransferase that modifies thrombospondin type 1 repeats. Ann Med. 2009;41(1):2–10. doi: 10.1080/07853890802301975. [DOI] [PubMed] [Google Scholar]
- 4.Reis LM, Tyler RC, Abdul-Rahman O, Trapane P, Wallerstein R, Broome D, Hoffman J, Khan A, Paradiso C, Ron N, Bergner A, Semina EV. Mutation analysis of B3GALTL in Peters Plus syndrome. Am J Med Genet A. 2008;146A(20):2603–2610. doi: 10.1002/ajmg.a.32498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapoor S, Mukherjee SB, Arora R, Shroff D. Peters plus syndrome. Indian J Pediatr. 2008;75(6):635–637. doi: 10.1007/s12098-008-0122-6. [DOI] [PubMed] [Google Scholar]
- 6.Haldeman-Englert CR, Naeem T, Geiger EA, Warnock A, Feret H, Ciano M, Davidson SL, Deardorff MA, Zackai EH, Shaikh TH. A 781-kb deletion of 13q12.3 in a patient with Peters plus syndrome. Am J Med Genet A. 2009;149A(8):1842–1845. doi: 10.1002/ajmg.a.32980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dassie-Ajdid J, Causse A, Poidvin A, Granier M, Kaplan J, Burglen L, Doummar D, Teisseire P, Vigouroux A, Malecaze F, Calvas P, Chassaing N. Novel B3GALTL mutation in Peters-plus Syndrome. Clin Genet. 2009;76(5):490–492. doi: 10.1111/j.1399-0004.2009.01253.x. [DOI] [PubMed] [Google Scholar]
- 8.Aliferis K, Marsal C, Pelletier V, Doray B, Weiss MM, Tops CM, Speeg-Schatz C, Lesnik SA, Dollfus H. A novel nonsense B3GALTL mutation confirms Peters plus syndrome in a patient with multiple malformations and Peters anomaly. Ophthalmic Genet. 2010;31(4):205–208. doi: 10.3109/13816810.2010.512355. [DOI] [PubMed] [Google Scholar]
- 9.Faletra F, Athanasakis E, Minen F, Fornasier F, Marchetti F, Gasparini P. Vertebral defects in patients with Peters plus syndrome and mutations in B3GALTL. Ophthalmic Genet. 2011;32(4):256–258. doi: 10.3109/13816810.2011.587082. [DOI] [PubMed] [Google Scholar]
- 10.Siala O, Belguith N, Kammoun H, Kammoun B, Hmida N, Chabchoub I, Hchicha M, Fakhfakh F. Two Tunisian patients with Peters plus syndrome harbouring a novel splice site mutation in the B3GALTL gene that modulates the mRNA secondary structure. Gene. 2012;507(1):68–73. doi: 10.1016/j.gene.2012.06.052. [DOI] [PubMed] [Google Scholar]
- 11.Corso-Diaz X, Borrie AE, Bonaguro R, Schuetz JM, Rosenberg T, Jensen H, Brooks BP, Macdonald IM, Pasutto F, Walter MA, Gronskov K, Brooks-Wilson A, Simpson EM. Absence of NR2E1 mutations in patients with aniridia. Mol Vis. 2012;18:2770–2782. [PMC free article] [PubMed] [Google Scholar]
- 12.Schoner K, Kohlhase J, Muller AM, Schramm T, Plassmann M, Schmitz R, Neesen J, Wieacker P, Rehder H. Hydrocephalus, agenesis of the corpus callosum, and cleft lip/palate represent frequent associations in fetuses with Peters' plus syndrome and B3GALTL mutations. Fetal PPS phenotypes, expanded by Dandy Walker cyst and encephalocele. Prenat Diagn. 2013;33(1):75–80. doi: 10.1002/pd.4012. [DOI] [PubMed] [Google Scholar]
- 13.Heon E, Barsoum-Homsy M, Cevrette L, Jacob JL, Milot J, Polemeno R, Musarella MA Peters' anomaly. The spectrum of associated ocular and systemic malformations. Ophthalmic Paediatr Genet. 1992;13(2):137–143. doi: 10.3109/13816819209087614. [DOI] [PubMed] [Google Scholar]
- 14.Traboulsi EI, Maumenee IH. Peters' anomaly and associated congenital malformations. Arch Ophthalmol. 1992;110(12):1739–1742. doi: 10.1001/archopht.1992.01080240079035. [DOI] [PubMed] [Google Scholar]
- 15.Reis LM, Semina EV. Genetics of anterior segment dysgenesis disorders. Curr Opin Ophthalmol. 2011;22(5):314–324. doi: 10.1097/ICU.0b013e328349412b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu R, Saito R, Hoshino K, Ogawa K, Negishi T, Nishimura J, Mitsui N, Osawa M, Ohashi H. Severe Peters Plus syndrome-like phenotype with anterior eye staphyloma and hypoplastic left heart syndrome: proposal of a new syndrome. Congenit Anom (Kyoto) 2010;50(3):197–199. doi: 10.1111/j.1741-4520.2010.00282.x. [DOI] [PubMed] [Google Scholar]
- 17.Al-Gazali L, Shather B, Kaplan W, Algawi K, Ali BR. Anterior segment anomalies of the eye, growth retardation associated with hypoplastic pituitary gland and endocrine abnormalities: Jung syndrome or a new syndrome? Am J Med Genet A. 2009;149A(2):251–256. doi: 10.1002/ajmg.a.32626. [DOI] [PubMed] [Google Scholar]
- 18.Volkmann BA, Zinkevich NS, Mustonen A, Schilter KF, Bosenko DV, Reis LM, Broeckel U, Link BA, Semina EV. Potential Novel Mechanism for Axenfeld-Rieger Syndrome: Deletion of a Distant Region Containing Regulatory Elements of PITX2. Invest Ophthalmol Vis Sci. 2011;52(3):1450–1459. doi: 10.1167/iovs.10-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11(2–3):377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 20.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14(10):1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. (a) Novel B3GALTL mutations in PPS patients identified in this study. The position of each mutation is indicated with a black arrow. (b) Results of TaqMan Copy Number assays targeting 5’ (intron 1) and 3’ (exon 14) regions of B3GALTL that were performed on DNA samples from Patients 1, 3 and 8, who appeared homozygous for the common c.660+1G>A splicing mutation, alongside a control sample with diploid B3GALTL copy number; haploid copy number for both probes was detected for Patient 1 (arrow) while Patients 3 and 8 were diploid for both probes (c) Evolutionary conservation of amino acid residues affected by missense mutations identified in patients 7 and 9; note 100% conservation of both positions in vertebrates (box).