Abstract
We provide an overview of lipid-dependent polytopic membrane protein topogenesis, with particular emphasis on Escherichia coli strains genetically altered in their lipid composition and strategies for experimentally determining the transmembrane organization of proteins. A variety of reagents and experimental strategies are described including the use of lipid mutants and thiol-specific chemical reagents to study lipid-dependent and host-specific membrane protein topogenesis by substituted cysteine site-directed chemical labeling. Employing strains in which lipid composition can be controlled temporally during membrane protein synthesis and assembly provides a means to observe dynamic changes in protein topology as a function of membrane lipid composition.
Keywords: Membrane protein, Cysteine scanning, Maleimides, Phospholipid, Topogenesis, Phosphatidylethanolamine, Protein topology
1. Introduction
Polytopic membrane proteins account for 25–30% of all open reading frames in sequenced genomes, and they are responsible for a wide range of cellular functions such as solute transport, biosynthesis, energy production, intracellular signaling, and cell–cell communication. Moreover, 50% of all drug targets are membrane proteins. High-resolution crystal structures of only about 100 membrane proteins have been determined. Although, the methodology for obtaining high-resolution structures is improving, the need to determine low-resolution organizational information on membrane proteins in a native membrane will continue. Purification, crystallization, and structure determination still remain formable tasks. Crystal structures are static and may be distorted due to purification and crystallization constraints and loss of information on interactions with other proteins and the lipid environment. Dynamic aspects of protein structure as a function of physiological state of the cell are best probed in whole cells or membranes. Determination of membrane protein organization has mainly relied on in silico approaches of predicting extramembrane domains and transmembrane (TM) segments based on the hydrophobicity of the component amino acids. However, these hydropathy plots are only 60–70% accurate in predicting topological organization and therefore, only provide a starting point for the design of experimental approaches to arrive at a final topological map of a protein in a membrane.
Due to the importance of determining membrane protein topological organization, an increasing number of reports over the last few years have utilized diverse reagents and methodologies to address protein topology. There are no comprehensive reviews covering the latest approaches. We will first review factors that may determine membrane protein topological organization, and then focus on the methodologies used to establish topological organization with emphasis on the use of the substituted cysteine accessibility method (SCAM) as applied to determining TM segment orientation (SCAM™). In SCAM, cysteine is introduced as a single amino acid replacement in a protein engineered to remove all native cysteines. When applied to polytopic membrane proteins containing TM segments spanning the membrane bilayer (SCAM™), cysteine replacements are positioned in the putative extramembrane domains predicted by hydropathy plots and other information about the target protein. The orientation with respect to the plane of the bilayer of each TM segment in a native membrane or reconstituted vesicle system is determined by using a combination of reagents directed at cysteine under conditions of membrane impermeable or membrane permeable to the reagent. Cysteines placed within TM segments are generally inaccessible to thiol-specific reagents and combined with the labeling pattern of extramembrane domains defines the length and orientation of TM segments.
1.1. Membrane protein topogenesis
The biogenesis of polytopic membrane proteins with high helical content involves the proper positioning of TM helices, coordinated folding of extramembrane domains, and helical packing within the lipid bilayer. The vast majority of prokaryotic and eukaryotic integral transmembrane proteins are co-translationally inserted into the membrane in a signal recognition particle-dependent manner [1]. Insertion occurs continuously or in a step-wise manner either by lateral translocation of the TM segments from the translocon complex into the lipid bilayer or by retrograde recruitment back to the translocon pore for orientation or proofreading [2]. With few exceptions [3], this results in a unique topology determined during membrane insertion by interaction between topogenic signals within the nascent protein and extra-protein factors that are only partially understood.
A fundamental aspect of the structure of polytopic membrane proteins is the membrane topology, i.e., the number and orientation of TM segments. For transport proteins, channels, and pores the organization of the TM segments determines function. However, for most proteins with catalytic capacity or involvement in signaling and recognition processes, more relevant to function is the disposition of the extramembrane domains with respect to the plane of the membrane bilayer. It is generally accepted that the topology of most polytopic membrane proteins is established co-translationally during membrane insertion and once established is maintained during subsequent steps of biogenesis, cellular trafficking, and function. However, several recent studies provide an exception to this rule and demonstrate that initial membrane topology in the bacterial cytoplasmic membrane [4,5] or the eukaryotic cell endoplasmic reticulum membrane [6] is dynamic and may be modified by subsequent lipid-dependent or translocon-dependent folding events, respectively.
These findings have significant implications for the molecular mechanisms by which polytopic proteins acquire their topology in the membrane. The dynamic aspect of membrane protein topological organization and the possibility that topology may be a function of membrane location in the cell or post-synthetic temporal factors necessitates effective methods for determining membrane protein topological organization in native membranes.
1.2. Lipid mutants as “biological reagents”
The ability to manipulate membrane lipid composition in living cells has made possible the study of the role of the membrane lipid environment in a broad spectrum of cellular processes including determination of membrane protein topology. Genetic manipulation of Escherichia coli and other bacteria, yeast, and mammalian cells with resulting controlled changes in membrane lipid composition has been reviewed in detail elsewhere [7,8].
The major phospholipids of E. coli are phosphatidyl-ethanolamine (PE, 70–75%), phosphatidylglycerol (PG, 20–25%), and cardiolipin (CL, 5–10%). Viable strains are available with the following changes in these major lipid classes: 10-fold reduced level of CL [9]; complete lack of PG and CL with PE comprising 90% of the total phospholipid and the remainder being primarily phosphatidic acid and CDP-diacylglycerol [10]; complete lack of PE with the remainder being primarily PG and CL [11]. In addition, strains have been engineered in which the steady state level of PG plus CL [12] or PE [4] can be regulated in a dose dependent manner as a function of extracellular regulation of the biosynthetic enzymes responsible for their synthesis. Foreign lipids have been introduced into E. coli either in addition to or in the place of native lipids. The foreign lipids that have been introduced into E. coli are phosphatidylcholine [13], phosphatidylinositol [14], and monoglucosyl diacylglycerol [15]. Use of strains with altered lipid composition has established a defined role for phospholipids in: SecA-dependent, TAT-dependent, and FtsY-dependent translocation of proteins across the cytoplasmic membrane [16–24]; DnaA protein-dependent initiation of DNA replication [25–27]; sugar transport by the phosphotransfer system [28,29]; cell viability [30–33]; protein translocation across the inner membrane [34]; efficient electron transport [35]; cell division [36,37]; formation of distinct lipid domains in the cell membrane [38]; and signal transduction via the Cpx system [39]. These strains also have been used to: uncover the role of lipids as lipo-chaperones [40–42]; establish the role of lipids as factors controlling final membrane protein topology [4,5,23,43] and uncover lipid-dependent topological switches within membrane proteins [4,5].
Yeast, being a eukaryote, has a more complex lipid composition and possesses organelles. All phospholipid biosynthetic genes have been identified and cloned and yeast strains are available lacking phosphatidylserine, lacking phosphatidylcholine, or containing very low levels of PE [44]. In addition, the level of the mitochondrial-specific phospholipids PG and CL has also been genetically manipulated to eliminate CL [45] alone or both PG and CL [46]. Strains lacking CL have functional but compromised mitochondrial-dependent energy transducing systems [45] and appear to have reduced or less stable [45,47] interactions between complexes of the electron transport chain that are normally organized into supermolecular complexes. Strains lacking both PG and CL have severely dysfunctional mitochondria and defects in synthesis of electron transport chain components [48].
At the moment bacteria and yeast are the most tractable organisms for genetic manipulation of lipid metabolism to study the role of lipids in supporting cell function. The genetics of lipid metabolism in mammalian cells is well advanced [49] and the prospects for developing viable somatic cell lines with altered lipid content are good.
1.3. Lipid-dependent topogenesis
Simultaneous development of reagent strains of E. coli with altered phospholipid composition and more sophisticated methods for determining protein topology, in particular SCAM™, revealed that membrane lipid composition is a critical determinant of topology. Lactose permease (LacY) [4] and phenylalanine permease (PheP) [5] expressed in E. coli mutants lacking PE are defective in active transport but still carry out facilitated transport. These secondary transporters actively accumulate substrate against a concentration gradient by coupling uphill transport of substrate with downhill movement of a H+. They contain 12 TM segments with the N- and C-termini facing the cytoplasmic side of the membrane but belong to different families of secondary transporters. By using SCAM™ these proteins were shown to display significantly different topological organization in PE-containing cells than in PE-lacking cells. For LacY the six N-terminal TM segments and for PheP the two N-terminal TM segments assume an inverted topology with respect to the plane of the membrane bilayer in PE-lacking membranes. Moreover, for LacY the final topological organization appears to be determined solely by the phospholipid composition independent of cellular protein assembly based on reconstitution into proteoliposomes followed by SCAM™ [43]. Even more interesting was the observation that induction of PE synthesis after membrane insertion and folding of LacY and PheP resulted in a return of native topological organization and transport function. These results clearly demonstrated that the lipid composition is a determinant of TM segment orientation and challenged the dogma that once TM orientation is established during assembly it is static and not subject to change.
According to this new topology paradigm, it is possible that specific regions of a membrane protein can undergo reversible conformational or TM segment reorganizations in vivo, dictated not only by phospholipids [4,5,43], but also by components of the insertion machinery, substrates, or other effectors [6,50–52]. The N-terminal signal sequence of a polytopic membrane protein can undergo reorientation after entering into the translocon [50]. If downstream topogenic sequences override the initial topology of a TM segment, the TM segment may be able to re-enter the translocon to reorient itself [2]. Post-translational topological reorientation of viral proteins can be facilitated by the host endoplasmic reticulum translocon [53] or molecular chaperones [54]. The E. coli translocon component SecG shows an unusual property of inverting its orientation in the membrane, which is tightly coupled to the SecG function and linked with the ATP-driven insertion–deinsertion cycle of SecA [55]. The dynamic topological changes of the Tat protein insertion apparatus might be coupled to the translocation of folded proteins across the cytoplasmic membrane [56].
The direct interaction of positively charged protein residues with negatively charged lipids can be dominant in retaining these protein domains on the cytoplasmic side of the membrane providing a structural basis for the “positive inside” rule which is based on the observation that positively charged residues are four-times more abundant in the cytoplasmic domains than in translocated loops of membrane proteins [23]. At low anionic phospholipid content a higher positive charge is required to prevent translocation of cytoplasmic domains while increasing anionic phospholipid content results in increased retention for domains with a lower positive charge. Placing a negatively charge amino acid within six residues from the end of a TM segment can increase its potential for translocation across the membrane [57]. The head groups of zwitterionic phospholipids may contribute to the retention of negatively charged residues on the cytoplasmic side of the membrane by charge pairing between the phospholipid amine and the amino acid car-boxylate. PE also dilutes the high negative charge density of the anionic phospholipids PG and CL that would increase the protonated form of acidic amino acids thus favoring their translocation across the membrane. The cytoplasmic domains mis-oriented when LacY [4] or PheP [5] are expressed in cells lacking the zwitterionic phospholipid PE contain acidic amino acid residues, which may have a higher potential for translocation in the absence of PE. Therefore, phospholipids inXuence membrane protein topology either independently or in cooperation with components of the translocon.
1.4. Topological isoforms, mixed topologies, and topological disorders
In recent years, it has become evident that certain naturally occurring polytopic proteins exhibit variations in TM topology. There are an increasing number of examples of proteins that are expressed in different topological forms with different functions. For example, ductin was found in two different orientations in membranes, one of which serves as the subunit of the vacuolar H+-ATPase and the other serves as a component of the microsomal connexin channel of gap junctions [58]. A microsomal epoxide hydroxylase is found with a different topology in the endoplasmic reticulum than in the sinusoidal plasma membrane, where it mediates bile acid transport [59]. However, current dogma assumes that the initial topology of a protein in the endoplasmic reticulum membrane accurately reXects the topology of the protein elsewhere in the cell. Do these topological differences originate co-translationally during membrane insertion or are they induced by changes in lipid composition as proteins move through different organelles to their final destination?
Studies examining the synthesis and translocation of the prion protein (PrP) at the endoplasmic reticulum have revealed that it is capable of being made in three topological forms from the same pool of nascent chains. The majority is completely translocated into the lumen, but another fraction is integrated into the endoplasmic reticulum membrane as single-spanning proteins with either the N- or the C-terminus in the lumen (NtmPrP or CtmPrP, respectively) [60]. Remarkably, mutations that increase the hydrophobicity of residues adjacent to or within TM segments result in complete reversal of the (Ntm) PrP topology and cause neurodegenerative disease in either transgenic mice or in some naturally occurring inheritable prion diseases [60,61].
The three N-terminal TM helices of glutamate/aspar-tate transporter (GLAST) are encoded by exons 2, 3, and 4, respectively. The loss of exon 3 converts the three TM domains into two and reverses the whole membrane topology of this protein [62]. Moreover, this splice variant of the transporter (GLAST-1a) encodes a functional transporter with inverted orientation within the plasma membrane. Remarkably, in some neurological disorders the release of glutamate due to anoxia was found to be largely due to an inverse operation of the glutamate transporter.
It was recently elegantly demonstrated by SCAM™ that the preexisting, membrane-bound anti-apoptotic protein Bcl-2 changes membrane topology upon induction of apoptosis. Moreover, this topological change converts Bcl-2 into a suicide inhibitor of the Bcl-2 family of proteins, notably Bax and Bak, to mediate anti-apoptotic activity through formation of a dead end non-active supermolecular complex. This complex formation prevents both loss of mitochondrial membrane potential and release of pro-apoptotic proteins such as cyto-chrome c into the cytosol [63].
It is also important to note that some aspects of protein topology may be expression system dependent. The P-glycoprotein is localized to mammalian cytoplasmic membranes, and in its native host the protein exhibits 12 TM segments with both the N- and C-termini exposed to the cytoplasm. When expressed in E. coli, the N-terminal half of the protein assumes the same topology as in the native host. However, TM segment VII no longer spans the membrane and TM segments VIII–XII assume an inverted orientation [64]. Similarly, a citrate carrier of Klebsiella pneumoniae displays 11 TM segments when inserted into dog pancreas endoplasmic reticulum membranes but only nine TM segments when expressed in E. coli [65]. One intriguing possibility is that certain polytopic proteins may require individual phospholipids as specialized membrane components to achieve their proper topology in addition to the basic translocon components needed for protein insertion and translocation. The topogenic information present throughout a polytopic membrane protein might be interpreted differently in prokaryotic and eukaryotic cells, suggesting that problems encountered when trying to express eukaryotic membrane proteins in prokaryotic hosts may in some cases be related to incorrect topological organization.
2. Experimental strategies for TM topology assessment
2.1. Overview
The difficulties encountered in the crystallization of integral membrane proteins have led to the development of several alternative approaches for investigation of their structural organization in the membrane. Given the enormous number of sequences that are produced in genome-sequencing projects, it is not realistic to assume that the structures of all the encoded proteins will be generated by crystallographic approaches, especially for membrane proteins. The physico-chemical constraints imposed by the lipid environment and the known hydrophobicity of individual amino acids provide a method using hydropathy plots to predict the topology of a membrane protein [66–69]. Two databases of TM topologies, MPtopo [70] and TMPDB [71], are available and can be used to evaluate the reliability of predicted topologies. The TM topologies in these databases were determined experimentally by means of X-ray crystallography, NMR, gene fusions, SCAM™, insertion of glycosylation sites, and other biochemical methods [147].
However, very often predictive methods generate a misleading topology and, due to the simplicity of generating predicted topologies, are often cited without further veriWcation. Long range-interactions between TM helices, unanticipated inter- (translocon and subunits) and intra-protein (salt bridges between charged residues within the hydrophobic core of the bilayer) interactions, and specific lipid protein interaction are some of the variables not addressed by predictive methods [72]. Therefore, hydropathy analysis of the sequence of polytopic membrane proteins may only reveal potential TM segments and their relative orientation as a starting point for designing biochemical experiments to establish topological organization.
To verify predicted membrane protein topology models, the existence of all the putative TM domains must be veriWed and the hydrophilic loops must be localized to one side or the other of the membrane. Strategies employed are quite varied but utilize the impermeability of the membrane bilayer to hydrophilic molecules, the difference in properties between the compartments separated by the membrane, and incorporation into proteins of a large variety of reporter groups whose orientation is presumed to reflect the topology of the protein [73,74]. Reporter groups can be as simple as a single amino acid substitution as in SCAM™, insertion of a proteolysis site [75], insertion of foreign antigenic reporter epitopes [76], insertion of a glycosylation motif or as complex as fusions of truncated target proteins to reporter proteins.
2.2. Fusions with reporter proteins
An early and still used approach for proteins expressed in E. coli is to construct a chimeric protein between successively C-terminal truncated target proteins and the N-terminus of a mature reporter protein. The reporters are typically molecules whose properties (for example, enzymatic activity, resistance to protease, antibiotic resistance, and antigenicity) depend on their subcellular location. Reporter domains should ideally lack intrinsic topogenic information, be readily identified, and passively and efficiently follow topogenic information presented by the nascent target protein fragment. A major drawback of the approach is that it is difficult to fulWll these requirements without compromising the topological information of the target protein especially when long range and cooperative interactions are involved.
The most extensively used single reporter systems in E. coli have been fusion proteins with alkaline phosphatase (PhoA) and β-galactosidase (LacZ) [77]. Protein translational fusions are obtained by progressively deleting the target protein gene from its 3′ end and ligating the truncated gene to phoA that lacks the promoter and coding information for its membrane targeting leader sequence. A complementary approach is to fuse the same target protein coding regions to lacZ. Reporter function is based on the activation of PhoA activity only in the periplasm where it forms a dimer, acquires Zn+, forms an intrachain disulWde, and in cell lysates is highly resistant to proteolysis; PhoA is inactive in the cytoplasm and sensitive to proteolysis. LacZ fusions provide complement information since fusions to cytoplasmic domains are active and fusions to periplasmic domains show decreased activity. The characterization of a series of membrane protein-PhoA and -LacZ chimeric proteins yields a reciprocal activity pattern for the reporter proteins that is in agreement with the predicted TM topology of many membrane proteins. Initial screening for topological location can be done in whole cells based on conversion of substrate analogues of each reporter expressed in cells to a colored product on agar plates.
The third group of commonly used reporter molecules is proteins that confer antibiotic resistance [78]. The mature form of β-lactamase (bla gene product), when in the periplasm, confers resistance to antibiotics such as ampicillin. Since the antibiotic targets are cell wall biosynthetic enzymes, cells expressing cytoplasmic β-lactamase are sensitive to ampicillin. Chloramphenicol acetyltransferase (cat gene product) only confers resistance to chloramphenicol in the cytosol where there is a supply of acetyl CoA for inactivation of the antibiotic [79]. Thus, only cells expressing fusion proteins in which the mature form of β-lactamase is fused to a periplasmic domain of a membrane protein or chloramphenicol acetyltransferase is fused to a cytoplasmic domain will be resistant to the respective antibiotics.
The major assumption in the fusion approach for determining membrane protein topology is that truncation of a membrane protein does not affect its native topology. However, because the complete structure of the protein of interest is not used and functional tests are not then available, the effects of long-range interactions between domains of the protein are not detected by this method and can lead to incorrect conclusions. The use of “sandwich” PhoA fusions, in which the reporter is inserted into the membrane protein rather than replacing the C terminus, have been shown to remedy this problem and may give a more accurate picture of the topology [80]. The entire membrane protein is present and the approach does not suffer from the drawbacks associated with the C-terminal deletion fusion approach. However, inserting fusions into a whole membrane protein may alter the way the protein folds or inserts into membrane. Therefore, reliance on topological information is safest if the chimeric protein retains the original activity.
In general active PhoA is a more reliable marker of periplasmic location than active LacZ is of cytoplasmic location. The former must reach the periplasm to be activated while the latter when fused to a periplasmic domain may form the active tetramer in the cytoplasm and fail to translocate due to its size. The latter problem has been addressed by joining mature PhoA and the α-fragment of LacZ into a single dual reporter [81]. The α-fragment represents 6% of full length LacZ and is active in the cytoplasm of cells encoding the remaining inactive co-fragment of LacZ. The dual reporter, when fused to periplasmic domains, produces fusions with high PhoA activity and, when fused to cytoplasmic domains, produces fusions with high LacZ activity in E. coli strains capable of α-complementation. Dual indicator plates containing a blue PhoA-activity dependent chromogenic substrate and a red LacZ-activity dependent chromogenic substrate in conjunction with these reporters allows for initial discrimination between non-informative fusions (white), cytoplasmic fusions (red), periplasmic fusions (blue) or fusions within TM domains (purple).
Interpretation of reporter activities in terms of topological information is often complicated by variable expression of the fusion proteins. For example, the low expression of periplasmic PhoA may lead to low alkaline phosphatase activity characteristic of cytoplasmic fusions and hence to misinterpretation of experimental data unless reporter activities are normalized to the rate of protein synthesis. Therefore, the single reporter fusion approach involves time-consuming experiments employing pulse labeling, immunoprecipitation, and quantification of incorporated radioactivity. Using dual reporters allows for a simple alternative to normalization of PhoA and LacZ activities. It is assumed that the specific activities of both the LacZ and PhoA portions of the dual reporter are characteristic of a given fusion point and independent of the level of expression of the fusion protein; that is, the level of expression of the fusion protein will affect the absolute activity but not the ratio of the two reported activities. By normalizing the PhoA and LacZ activities of each fusion to the maximal activity observed for each reporter enzyme in the set of fusions, a correction can be made for the intrinsically higher PhoA activity and the resulting ratio of normalized PhoA to LacZ activities provides readily interpretable information about subcellular localization of the fusion point. Although the utilization of dual reporters alleviates most of the above-mentioned problems, this most advanced genetic approach suffers from lack of detection of long-range effects on topology.
2.3. Fusions with a glycosylation epitope
An affective approach to mapping the topology of proteins present in the endoplasmic reticulum is the lumen specific glycosylation machinery. This approach has become the eukaryotic counterpart of the bacterial PhoA fusion and sandwich techniques. In eukaryotic cells, glycosylation activity is found in the lumen of the endoplasmic reticulum and is carried out by an oligosaccharyl transferase, which catalyzes addition of oligosaccharides to the amino group of asparagine residues within the consensus sequence Asn-X-Thr/Ser. N-glycosylation is a common feature of eukaryotic membrane proteins, and the consensus sequence is usually found in the largest luminal exposed loops of the protein. Since modification of the glycosylation site occurs in a com-partment-specific manner, the presence of glycosylation provides information for topological assignment [74,82]. Addition of the oligosaccharide chain to a single site results in an increase in the apparent molecular mass on SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) by about 2.5 kDa. Glycosylation can be manipulated after synthesis by in situ treatment with glycosidases (Endo H and N-glycosidase F) or by in vitro expression of the protein in the presence of glycosylation inhibitors (tunicamycin) or competitive acceptor peptides (Ac-Asn-Tyr-Thr and N-benzoyl-Asn-Leu-Thr-N-methylamide) [83].
In the glycosylation scanning mutagenesis approach, consensus glycosylation sites or domains bearing a glycosylation site are introduced into membrane proteins devoid of glycosylation sites. Plasmids encoding these engineered proteins are transfected into mammalian cells and localization of the insertion site on the luminal or cytoplasmic side of the membrane is inferred from the presence or absence of glycosylation of the engineered protein, respectively. In the glycosylation fusion approach, a domain bearing one or more glycosylation sites is fused behind different C-terminal deletion mutants of a membrane protein [84]. The Asn in acceptor sites is glycosylated only in loops larger than 25 residues, and a sharp cutoff is observed in glycosylation of sites positioned less than 12 residues upstream or 14 residues downstream of a TM segment [85]. The distance constraints imposed on glycosylation sites by the oligosaccharyl transferase were used to map the ends of the TM segments on the luminal extramembrane domains of several polytopic membrane proteins [86].
The glycosylation scanning technique assumes that the initial topology of a protein in the endoplasmic reticulum membrane accurately reflects the topology of the protein elsewhere in the cell, which, as was mentioned above, is not always true. Because each strategy has its own inherent strengths and limitations, a topological model that is based on the results of several approaches is likely to be more informative.
3. Principles of SCAM™
3.1. Overview
Modification of thiol groups in proteins has become a powerful technique used to analyze protein structure. In the original substituted cysteine accessibility method (designated as SCAM by Karlin and co-workers [87–92]) single cysteine substitutions within a target protein coupled with covalent cysteine Modification by hydrophilic thiol-specific reagents was used to study structure–function relationships and dynamics of membrane protein function (mapping of channel gating residues, identification of residues lining a membrane channel, identification of residues involved in substrate or ligand binding, etc.). SCAM provides an approach to systematically map the residues on the water-accessible surface of membrane proteins either at steady state or related to protein function. SCAM can probe conformational changes that result in changes in steric constraints and electrostatic potential within the vicinity of the substituted cysteines by comparing the rates of reaction with reagents of varying size and charge.
By introducing and modifying cysteines in the polytopic membrane protein LacY of E. coli, Kaback and co-workers [92] identified and defined the structural relationship between the 12 TM-spanning helices of this protein. The movement of TM helices relative to each other, changes in accessibility of certain residues upon binding of substrate, and the residues critical to function were also demonstrated. The conclusions drawn from these extensive biochemical studies resulted in a model for the three-dimensional structure of LacY that is largely supported by the recently determined X-ray crystal structure [93].
We have adapted SCAM to map and assign TM topology of polytopic membrane proteins (designed SCAM™) as a function of membrane lipid composition. In this approach, cysteine residues replace individual amino acids that reside in the putative extracellular or intracellular loops connected to TM segments of a membrane protein. The use of SCAM™ was introduced and elegantly applied for first time by [88,94] to establish membrane protein topology. The orientation with respect to the membrane is determined using membrane-impermeable thiol reagents with intact membranes or with membranes that have been permeabilized or disintegrated. Alternatively, a combination of thiol reagents can be used that are membrane-impermeable or membrane-permeable due to differences in their physical properties but with the same chemical reactivity under different conditions. This method can be used in whole cells [4,94–100], uniformly oriented membrane vesicles isolated from cells [4,51,65,101–105], intact organelles (vacuoles and mitochondria) [106,107], or in reconstituted proteoliposomes containing uniformly oriented target proteins [43,108,109]. A major advantage of SCAM™ over methods reviewed above is that minimal perturbation of protein structure results from introducing single cysteine residues into a protein. In addition the whole protein is analyzed, and retention of native function in vivo can be used to verify structural integrity of the engineered protein.
Exact protocols and detailed descriptions of methodology will not be presented since SCAM™ must be tailored to specific proteins, cell hosts, and host membranes. Therefore, a more global approach will be presented describing the properties of thiol groups and reagents, membrane permeability properties with respect to different reagents, methods of accessing thiols in different compartments, methods of detection and analysis, and advantages and limitations of SCAM™.
3.2. Properties of the cysteinyl thiol
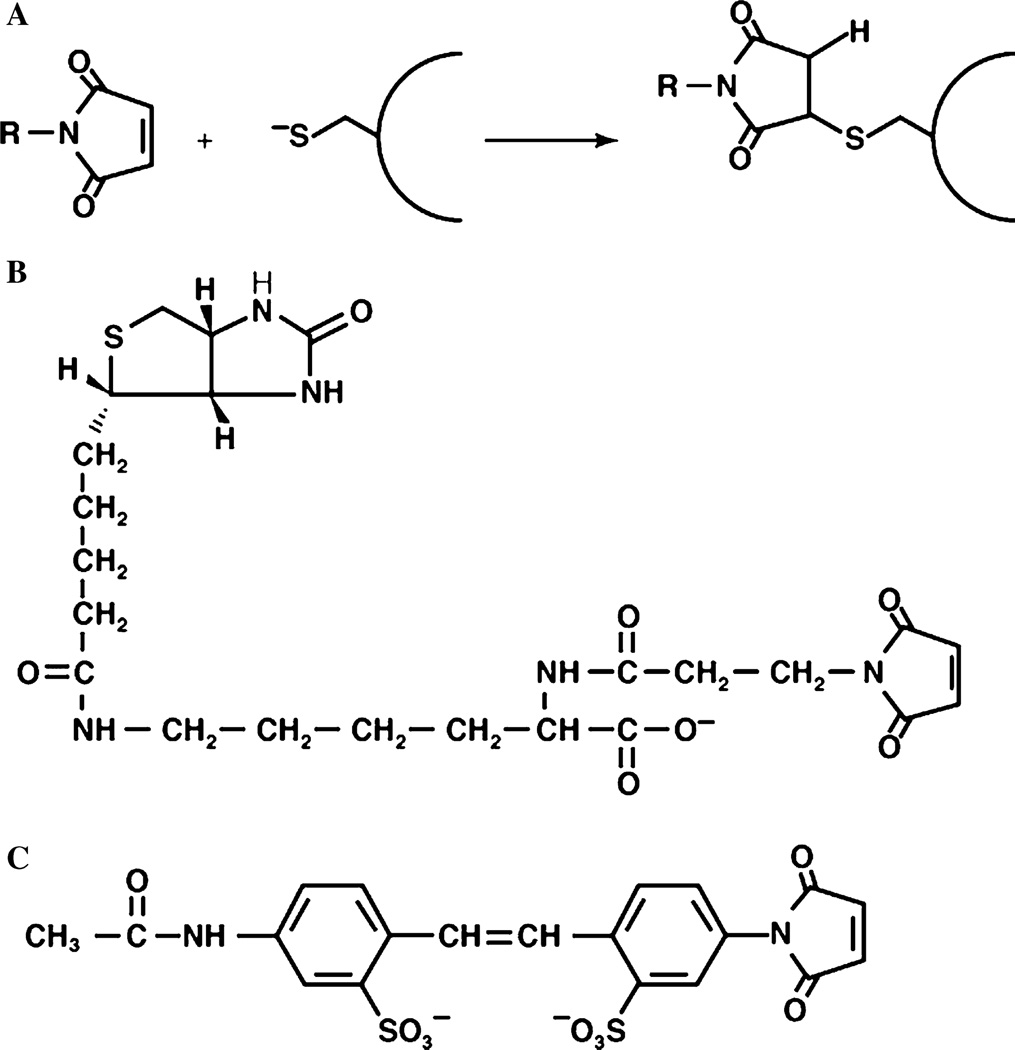
The chemical nature of the reactive portion of a labeling reagent should be highly reactive with and selective for thiol groups and form a stable non-exchangeable or non-hydrolysable derivative. Maleimides, which are available in a wide variety of forms, are particularly suited for SCAM™. Maleimide reacts with the ionized form of a thiol group (Fig. 1A), and this reaction requires a water molecule as a proton acceptor [88,110]. Maleimides are virtually unreactive until they encounter an available thiol group. The pKa of the thiol of cysteine in a water milieu is around 9 and in the hydrocarbon core of the bilayer is around 14 characteristic of cysteine in a nonpolar environment [111]. Therefore, the labeling characteristics of intramembrane (unreactive) and extramembrane (reactive) cysteines would be consistent with their localization either in a polar or nonpolar environment, respectively [88,94,96,99,112,113]. This is an important chemical feature of cysteine residues that is the main basis of SCAM™. Examples (discussed later) exist of highly hydrophobic maleimide derivatives that react with thiols in a hydrocarbon environment, which can be selectively applied to map thiols in TM segments. A possible explanation for the reactivity of the protonated thiol group in a lipid environment may be the high concentration of a hydrophobic maleimide in the hydrocarbon core of the bilayer due to its favorable partitioning coefficient. However, conclusions based on reactivity should be made with caution, since cysteines may be positioned facing other helices and therefore might be inaccessible but not in a hydrocarbon environment. Local secondary structure or properties of neighboring amino acids may restrict access by thiol reagents. Cysteine residues facing a hydrophilic pore or near a substrate-binding site maybe within a TM segment but chemically reactive due to water channels or pockets [114].
Fig. 1.
Thiol-modifying reagents widely used in SCAM™. (A) The reaction of the thiolate anion of cysteine with maleimide by nucleophilic addition to the double bond of the maleimide ring. (B) The structure of the maleimide- and biotin-containing labeling reagent MPB. (B) The structure of the blocking reagent AMS.
Since the formation of cysteinyl thiolate anions is favored by increasing the solution pH (optimum pH 8.0–8.5), increasing the pH during labeling should favor the reaction [115]. However, maleimides are known to react with primary amines at pH values above 7.5 [116]. Therefore, attempts to increase efficiency of labeling by raising the pH of the assay should be thoroughly controlled in order to ensure that the Modification is confined to cysteine. An effective control to rule out non-thiol modifications is to use a cysteineless target protein. Thus far the most extreme thiol labeling reaction conditions (millimolar concentration of reagent, room temperature, and pH 8.0–8.5) have been utilized without significant modification of additional reactive groups on target proteins [94]. Generally, pH 7.0–7.5 is sufficient for efficient labeling [4,65,88,94,95,102–104,107,113] and reduces background labeling especially in experiments with oriented inside-out membrane vesicles (ISOV) containing target proteins with cytoplasmic loops very often containing lysine residues.
3.3. Thiol reactivity in proteins
Cysteine is a relatively hydrophobic, small amino acid, and its introduction at most positions in a membrane protein is likely to be tolerated. Furthermore, cysteine has little preference for a particular secondary structure [117,118]. For most water-exposed cysteine residues in proteins, the thiol pKa lies in the range of 8–9 and formation of cysteinyl thiolate anions is optimum in aqueous rather in a non-polar environment. The process of choosing suitable residues for replacement by cysteine is often empirically determined, and the rationale for deciding which residues to alter is aided by the following considerations. Secondary structure predicted by computer-aided hydropathy analysis [66–69] (thus far 60–70% reliable) [119] is an initial starting point for the likelihood that a particular residue is in an extramembrane domain. Replacement of charged residues is generally not advised because these have a high probability of being topogenic signals or may be involved in long-range interactions. Consideration should be given to whether the replacement will be well tolerated based on structural and functional information about the protein. If the protein contains stretches of residues of intermediate hydrophobicity that cannot unambiguously be identified as membrane spanning, substitutions should be made approximately every 10 residues.
Ideally the protein under study should be devoid of all native cysteine residues because these residues may also react with thiol-modifying reagents or they may form disulfide bonds with the engineered cysteines and prevent their interaction with modifying thiol-specific reagents. The cysteineless protein serves as the starting template for introducing single cysteine residues at desired positions as well as a negative labeling control to assure that residues such as lysine are not labeled by the reagents. Alternatively, templates containing natural cysteines can be utilized in this assay if they do not react with the thiol-specific reagents. Very often the native cysteine residues present within TM segments are inaccessible to thiol reagents making it unnecessary to remove these cysteines. However, the possibility always exists that engineered cysteines may form disulWdes or changes in protein structure may expose the native cysteines. A limitation of SCAM™ is with proteins where cysteine pairs form disulWdes critical to the folding of the protein. Removal of these cysteines or introduction of additional cysteines might cause misfolding. Therefore, a prerequisite for each cysteine replacement is retention of function that provides assurance of retention of near native structure.
The native cysteine residues are usually changed into alanine or serine residues which are small, commonly found in membrane proteins and appear to be tolerated at most positions thus rendering an active protein. For example the eight native cysteine residues of LacY were simultaneously replaced to yield a cysteineless template that retained at least 50% of its wild-type activity. Of the 417 single cysteine replacements in LacY, only four disrupted transport function [92] supporting a near native structure for the other replacements. However, loss of function may not result in significant structural changes if these substitutions only affect substrate binding or catalytic processes. Thus, cysteine-scanning mutagenesis permits topology assessment under conditions in which the proteins are active, strongly suggesting the protein structure is not seriously altered.
To obtain a minimal topological map a single cysteine replacement in each of the putative extramembrane loops should be expressed from a plasmid and analyzed in appropriate host cells. In practice, several cysteine replacements or complete cysteine scanning across extramembrane loops and into TM segments is required for a more precise mapping of topology. Secondary structure, as discussed in Section 9, may sterically prevent access to cysteines in extramembrane loops, which requires analysis of several cysteine replacements along a loop. Cysteine residues closer to the membrane interface generally react slower than those near the center of extramembrane loops [94] and these differences can be used to assign residues at the membrane-aqueous interface. The host strain for plasmid expression should be deleted of the target protein gene if it contains native cysteines and is expressed at levels high enough to be detected in the assay. Since LacY expression is high in cells induced for lac operon expression, it was deleted in SCAM™ analysis [4]. However, the level of chromosomal expression of PheP was not high enough to be detected in the assay so deletion was not necessary [5].
The physical and chemical properties of thiol-specific reagents and basic properties of membrane proteins provide a rational basis for conclusions about exposure and positioning of residues that become modified. The hydrocarbon core of a model phosphatidylcholine bilayer is about 27Å wide with the phospholipid head groups occupying about 5Å on either side of the hydrophobic domain [120]. The majority of TM spanning regions are α-helical so that a minimum of 18 hydrophobic amino acids (1.5Å per turn of the helix) are sufficient to span the hydrocarbon core of the bilayer and another 2–3 amino acids are needed to bridge the phospholipid head group region on either side of the membrane. Recent crystal structure data indicates that TM segments of 18–20 amino acids in length are common but segments longer than 20 amino acids exist and are obliquely oriented to the plane of the bilayer. The cysteinyl thiol group lies some 8–10Å from the peptide backbone [111] so that maleimides derivatized with bulky residues would have access to cysteine residues only about 5–6 amino acids into a TM segment [110]. Therefore, for a 20 amino acid TM segment about 8–10 residues (12–15Å) at the center of the TM segment would not be accessible to these reagents. For TM segment 7 of UhpT, an E. coli sugar phosphate transporter, accessibility of cysteines by bulky maleimides Wts the above dimensions while smaller probes reacted with residues more that 10Å into the hydrocarbon core [91,114].
4. Application of SCAM™
4.1. Overview
Thiol-specific reagents are available in a large variety of sizes, polarity, and monitoring features, as will be discussed in Section 4.3. Membrane permeable or membrane impermeable thiol-reagents can be employed to selectively label the residues from either both sides of the membrane (hydrophilic maleimides) and the residues facing the lipid core of the bilayer (hydrophobic maleimides) or only residues exposed to the outer surface of the membrane, respectively. The outer membrane of E. coli allows small molecules under about 600 Da passage to the periplasm and access to the inner membrane so these reagents can be used in whole cells without the need to disrupt membrane structure [121]. The double bond of the maleimide group reacts with the thiol-group of cysteine to form a thioether bond (Fig. 1A) that is stable to reducing agents such as β-mercaptoethanol (βME) or dithiothreitol. The reaction rate of different thiols is controlled primarily by their surface exposure and proximal environment. For example cysteines buried within the structure of native proteins are essentially unmodified by N-ethyl maleimide (NEM, although it can cross the membrane) or fluorescein-5-maleimide (FM, membrane impermeable) over a time scale of 10min. However, when the cysteines are exposed by SDS denaturation of proteins, they became accessible to the aqueous phase and are rapidly modified by both reagents within minutes [88].
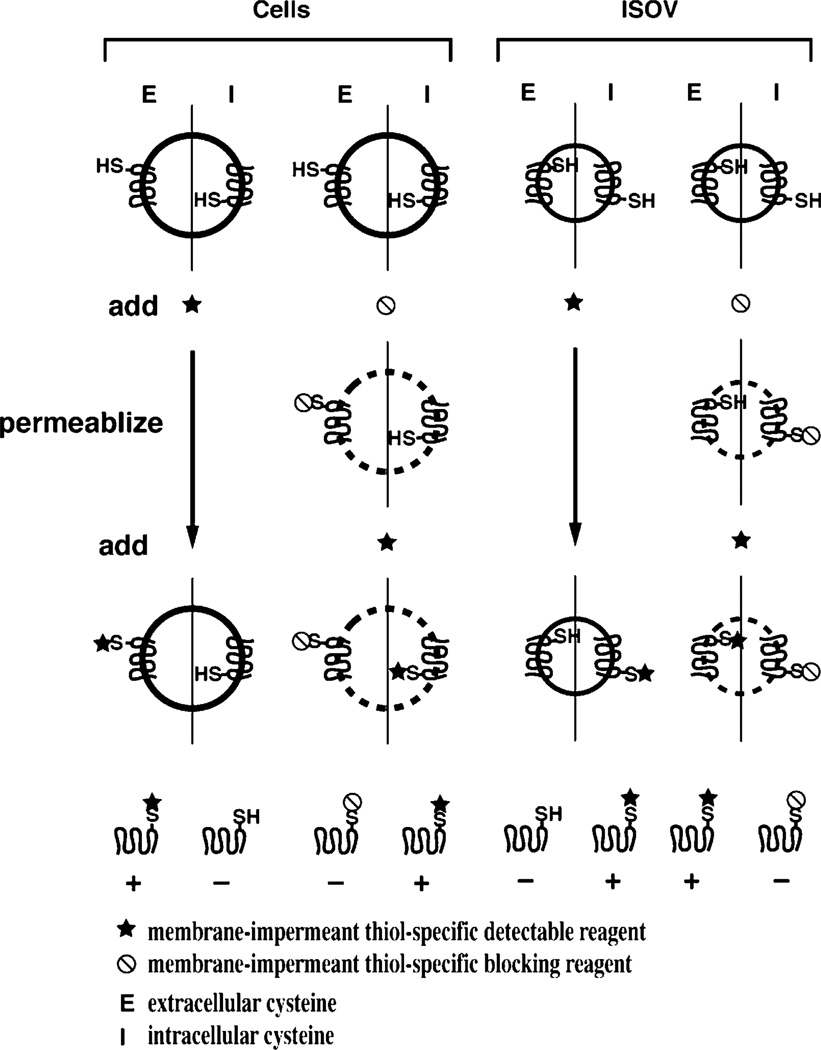
The general design of labeling experiments to distinguish between cysteines located in an extracellular or intracellular domain is outlined in Fig. 2. Extracellular (periplasmic for E. coli) residues are those that are labeled in intact cells but not in ISOV unless they are permeabilized. Intracellular (cytoplasmic) residues are those that are labeled in ISOV but not in intact cells unless they are permeabilized. Pre-blocking intact cells or ISOV with a thiol reagent that is transparent in the detection phase of the procedure allows selective labeling of luminal cysteines after permeabilization. The results of this approach are valid only if the modifying reagent is thiol-specific and membrane impermeable, orientation of ISOV is uniform and opposite to that of cells, and permeabilization does not expose sterically hindered or water inaccessible cysteine residues. The latter can generally be assumed if cells and ISOV are permeabilized before reaction with thiol reagents and then labeled with a detectable thiol reagent before and after reaction with a non-detectable blocking reagent. The blocking reagent should completely prevent labeling and the labeling without blocking should be to the same extent for external cysteines as seen in the protocol outlined in Fig. 2. Cysteine residues that are not labeled under any conditions are either inaccessible due to being located in a TM segment embedded in the hydrocarbon core of the bilayer (intramembrane) or restricted by secondary structure. More details concerning each step, different reagents, considerations for controls, dealing with inaccessible cysteines, and interpretation of results will be expanded in the following sections.
Fig. 2.
General strategy for SCAM™ using impermeable thiol reagents. A target membrane protein containing a single cysteine replacement exposed either to the extracellular (periplasmic, left half of circle) or intracellular (cytoplasmic, right half of circle) side of the membrane is expressed in cells. Preparation of uniformly oriented ISOV results in an opposite orientation for the same cysteine residues. Half the cells or ISOV is reacted with a detectable thiol reagent to specifically label the externally exposed cysteine and the other half is reacted with a non-detectable thiol reagent to protect external cysteines in subsequent labeling steps. The latter half of cells or ISOV is permeabilized to expose the interior cysteine and reacted with a detectable thiol reagent to specifically label internal cysteine residues. Cells and ISOV are then analyzed for labeling of the target protein by the detectable thiol reagent. Note that the cysteine labeling (+) and blocking (−) patterns in ISOV are the mirror image of the patterns in whole cells.
4.2. General protocol for SCAM™
Single cysteine replacements are expressed in the appropriate host, and the cells are harvested and suspended in modification buffer or ISOV are prepared [4] and suspended in modification buffer. Cells or ISOV are treated under a variety of conditions and with different thiol reagents in order to establish where the cysteine residues are located as described in Fig. 2. A maleimide-based thiol reagent is added and the modification reaction terminated by 5–10-fold dilution with buffer alone [107] or by adding a 50–100-fold-excess of either PME, dithiothreitol [94,103] or cysteine [105,122] to destroy the unreacted maleimide. The concentration of thiol reagent and time of reaction is empirically determined by using the most vigorous conditions that do not result in labeling of a cytosolic protein or luminal thiol scavenger as described in Sections 5 or 12, respectively. Termination of the reaction is followed immediately by several cycles of centrifugation and washing with thiol quencher [94,123] to remove excess labeling reagent. Dilution rather than centrifugation was employed [65,107] to eliminate additional variability in yield associated with pelleting and resuspending of cells. Excess reagent can also be removed by centrifuging the terminated reaction mixture through small columns of gel filtration resin [4]. The last two procedures should be used when sequential treatments of the sample is required such as pre-blocking with one thiol reagent followed by reaction with a second thiol reagent [4] or to avoid lysis of fragile preparations like intact spheroplasts, vacuoles [65,107] and proteoliposomes [43].
After labeling, cells or ISOV are solubilized with the appropriate detergent or detergent mixture such as SDS alone [4,104], Triton X-100 alone [65,105] SDS and Triton-X-100 [113,124], Chaps [124,125], octylglucoside, deoxycholate, cholate, and Tween 20 [102], octylglucoside [111], P-D-dodecylmaltoside [95], or nonidet P-40 and sodium deoxycholate [112,126]; use of detergents other than SDS may require lysis of cells by sonication prior to solubilization. Conditions must be empirically determined that yield a non-aggregated soluble target protein throughout the remainder of the procedure. For example many membrane proteins aggregate if boiled in SDS, and LacY forms irreversible polydisperse aggregates if solubilized by Triton X-100 alone.
The thiol reagents react with cysteine residues present in all other proteins in the membrane. Immunoprecipitation of the membrane protein of interest or a rapid purification step is necessary to eliminate other labeled proteins. A biotin-maleimide labeled protein can be recovered from cell lysates directly with streptavidin–agarose beads [100], and can then be detected by Western blotting using a target-specific antibody. For immunoprecipitation of labeled protein from solubilized samples, polyclonal and monoclonal antibodies [4,94,102,107,110,113] have been widely utilized. Antigen–antibody complexes can be isolated using precipitation with Pansorbin (Staphylococcus aureus cells) [4,113], protein A–agarose [102], or protein A or G–Sepharose beads [98,107,125,126]. If antibodies specific to the protein under study are not available, then epitope tags such as myc [124,126] or affinity tags such as His6 [124] can be incorporated at the C-terminus of the target protein for either immunoprecipitation [124] or isolation by Ni2+ chelated affinity resin packed into micro-columns or attached to agarose beads [65,103–105,111,116,127,128]. Use of affinity methods with His-tagged proteins and small-scale batch purification procedures is becoming the method of choice since the labeled protein can be directly extracted from the resin with SDS-containing buffers followed by SDS–PAGE [104]. Of course protein function or topology should not be compromised by the presence of the tag.
Following Modification and isolation, the target protein is resolved by SDS–PAGE, transferred to a solid support, and detected by Western blotting or one of the following techniques. Thiol reagents are available that contain a biotin group [4,94,98], a fluorescent group [51,88,103,116,122], or a radiolabel [113,129], allowing detection of labeled proteins by avidin linked to horse radish peroxidase (avidin-HRP) and indirect chemiluminescence detection, fluorescence, or autoradiography, respectively. Signals can be quantified using available Imaging systems and software.
A major problem is the variability between samples due to mechanical loss during the work up or due to differences in expression level of individual replacements. For example, of the 41 cysteine replacements within the ABC multidrug transporter LmrA, 40 were expressed and present in the membrane, but at different levels [103]. Variability can be corrected for by using a His-tagged target protein. The thiol derivative can be detected based on its properties, and the amount of target protein present can be detected using an antibody directed against the His tag. Since these can be done on the same blot, thiol signal can be normalized for the amount of target protein. Alternatively, Western blots of duplicate samples or of the same blot can be probed by antibody specific for the target protein to normalize the signal [4,100,111,112]. After recording a fluorescence or radioactive profile of a derivatized target protein resolved by SDS–PAGE, the signal can be normalize to the target protein recovered as determined by staining the same gel with Coomassie brilliant blue [111,129] or silver stain [123]. If no labeling occurs with the thiol reagent, it is important to verify that the target protein was expressed and is present on the blot.
4.3. Properties of thiol-specific reagents
The choice of thiol-specific reagents is influenced by physical–chemical properties, size, chemical reactivity, and monitoring features suitable for cysteine modification followed by detection. Following is a description of the various thiol-specific reagents that have been used.
4.3.1. Labeling with detectable thiol-specific reagents
Impermeable thiol reagents that can be easily detected after Modification of target proteins are essential for successful application of SCAM™. Biotin-linked maleimides such as 3-(N-maleimidylpropionyl) biocytin (MPB, Fig. 1B) and (+)-biotinyl 3-maleimidopropionomidyl-3,6-dioxaoctanediamine (MPEOB) are particularly useful due to their low membrane permeability properties and formation of stable non-hydrolysable bonds with thiols. After SDS–PAGE of target proteins isolated by immunoprecipitation or affinity tags, the biotinylated target proteins are easily detected using avidin-HRP and chemiluminescence [94,110]. Owing to the relatively long hydrophilic spacer between the biotinyl group and the reactive maleimide, MPEOB is fully soluble in aqueous solution and less likely to penetrate the E. coli cytoplasmic membrane [5]. This reagent was selected to study reversible TM orientation of PheP in response to a change in phospholipid composition of E. coli membranes [5] and establish TM topology of LacY reconstituted in proteoliposomes [43]. MPB is more hydrophobic than MPEOB and must be added dissolved in dimethyl sulfoxide so conditions must be established for minimal membrane permeability by MPB. MPB has been extensively utilized in SCAM™ under conditions of low membrane permeability for detection of external surface exposed cysteines [94,97,99,102,107,109,127] and under conditions of membrane permeability (higher concentration and longer incubation times) where MPB modifies water accessible thiols on both sides of the membrane [65,95,98,130].
FM, which at neutral pH is a dianion and membrane-impermeable, has also been used in SCAM™ [103–105] and has the advantage of being fluorescent and directly detectable in gels after SDS–PAGE. Cysteines in putative TM segments were found to be inaccessible to FM, whereas cysteines in polar loop regions were readily labeled [103,104]. High pH in the assay buffer can help to reduce the membrane permeability of FM by promoting the ionization of the carboxyl groups in the molecule [104] and was utilized along with a cysteineless control protein to demonstrate that the labeling was specific to cysteine. Oregon Green 488 maleimide carboxylic acid (OGM), another highly negatively charged fluorescent maleimide, has also been successfully used in SCAM™ [96,111,116]. N-Biotinylaminoethyl methanethiosulfonate (MTSEA-biotin) was used to label water exposed cysteine residues of the serotonin transporter expressed in Intestine 407 cells [131] or HeLa cells [100] and to map the extracellular boundary for TM segments of human reduced folate carrier in Chinese hamster ovary cells [126]. MTSEA-biotin was also successfully used to map the membrane topology of the renal Na+/K+-ATPase α-subunit expressed in baculovirus transformed insect cells [125]. This reagent has never been tested in bacterial systems.
4.3.2. Membrane impermeable thiol specific blocking reagents
Also required for SCAM™ is a set of impermeable blocking reagents that effectively react with thiols exposed to solvent but are transparent in the detection phase of the procedure. These reagents are useful in several ways. They can be used with membrane impermeable labeling reagents as outlined in Fig. 2 to restrict detectable labeling to luminal cysteine residues. Alternatively, they can be used with membrane permeable reagents to identify internal cysteines. NEM readily crosses the membrane bilayer but still labels primarily water accessible cysteines. MPB and FM are generally considered membrane impermeable, but at high concentrations and with long incubation times, luminal water exposed cysteines can be labeled [94,127]. Cells or ISOV can first be modified with a hydrophilic blocking reagent and then treated with radiolabeled NEM or high concentrations of MPB or FM to access internal water exposed cysteines. Even under the most stringent conditions some of the more hydrophobic labeling reagents (MPB) have limited membrane permeability. Pre-blocking external cysteines before labeling intact cells or ISOV provides a means of estimating the degree of labeling due to this low permeability.
A widely used blocking reagent is 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS, Fig. 1C) since it possesses two charged sulfonate groups and is highly soluble in water. The size and the charged nature of this reagent results in its demonstrated inability to cross the cytoplasmic membrane of bacteria or the plasma membrane of mammalian cells [65,94,95,107,113,124,127]. AMS treatment of E. coli cells and spheroplasts modified a cysteine residue exposed to the periplasmic side of the membrane but did not modify a cytoplasmic protein, elongation factor Tu [132]. Confirmation of labeling of external water-exposed cysteines by MPB was achieved by first blocking putative external cysteines with AMS in intact E. coli cells, oriented membrane vesicles, proteoliposomes [4,43,65,95,107,127], intact vacuoles [107], and mammalian cells [94,99,126].
The membrane impermeable, non-fluorescent, quaternary amine [2-(trimethylammonium)ethyl]methanethiosulfonate bromide (MTSET) showed a side-specific membrane modification pattern [103,111,116,123]. External cysteines were identified by initial preincubation with membrane impermeable and strongly acidic p-chloromercuribenzosulfonate (pCMBS) and subsequent labeling with fully membrane permeable radiolabeled NEM [101]. Both pCMBS and MTSET react with periplasmically exposed cysteines of the E. coli Na+/H+ antiporter 10-fold faster in right-side out membrane vesicles (RSOV) than in ISOV. Anionic lucifer yellow iodoacetamide (LYIA) and positively charged bromotrimethylam-moniumbimane bromide (BTMB) have also been used as blocking reagents in MPB labeling experiments [98,112]. The difference in the labeling between intact cells and ISOV with FM was further confirmed in protection experiments with 2-[(4′-maleimidyl)anilino]naphthalene-6-sulfonic acid (MIANS) [105]. The two positively charged and highly water-soluble thiol-specific reagents MTSET and 2-(aminoethyl)methane-thiosulfonate hydrobromide (MTSEA) have been used as blocking agents in combination with the membrane impermeable maleimides OGM and FM to verify the membrane topology of cysteines strategically placed in different proteins [96,103,111,116,123]. However, since MTSEA is a weak base with a pK of around 8.5, it is possible that it equilibrates across the membrane in its undissociated form. Therefore, this reagent should be avoided in topology assays [97,101]. The labeling of external residues with FM and NEM [103,104] or 4-acetamido-4′-[(iodoacetyl)amino]stilbene-2,2’-disulfonic acid (IASD) [123] was also verified by prior blocking with MTSET. Finally, FM and MIANS are fluorescent so they can be used as primary detectable labeling reagents with AMS as a blocking agent or as blocking reagents if a biotinylated-labeling reagent is used.
4.3.3. Labeling thiols within the hydrophobic core of the bilayer
Although the protonated thiol located in the hydrophobic core of the bilayer should display low reactivity with thiol reagents, there are several highly hydrophobic thiol reagents that react at appreciable rates with cysteine residues clearly in TM segments in a hydrophobic environment. These can be used to distinguish between inaccessibility due to secondary structure and inaccessibility due to location in a TM segment. Since these hydrophobic reagents will also modify water-exposed residues on both sides of the membrane, they must first be blocked after membranes are permeabilized and prior to labeling. Identification of intramembrane cysteine replacements in a protein was done by using a combination of hydrophilic FM as a blocking reagent and hydrophobic benzophe-none-4-maleimide (BM) as a labeling agent [106,133]. BM is a lipid soluble thiol regent that is large enough to cause an increase in the molecular weight of relatively small membrane proteins or polypeptide fragments, which is detectable by a shift in mobility during SDS–PAGE. Evidence for intramembrane location of engineered cysteine residues in the dog kidney Na+/K+-ATPase was obtained by exploiting the hydrophobic properties of CPM (7-diethylamino-3-(4-maleimidylphenyl)-4-methylcoumarin) and the hydrophilic and membrane impermeable properties of AMS [51]. Since CPM is highly fluorescent and AMS is not fluorescent, specific labeling by the former can be followed by visualizing the fluorescent target protein. BM and CPM also react with cysteines exposed to the aqueous phase; therefore, to identify thiols within the hydrophobic core of the membrane bilayer, water exposed thiols should first be blocked on both sides of the membrane with a hydrophilic thiol reagent.
5. Membrane permeability
SCAM™ is based on the controlled membrane permeability by sulfhydryl reagents. Membranes either in their native state or due to experimental manipulation can be slightly permeable to labeling reagents. Membrane permeability of some reagents such as MPB, FM, and OGM are time and concentration dependent. Cell lysis during a labeling experiment will also result in labeling of intracellular cysteines. Therefore, optimal labeling conditions must be established for each reagent, cell, membrane vesicle, or proteoliposome used.
The membrane permeability of a reagent can be tested by quantification of the degree of labeling of an abundant cytoplasmic protein that is rich in surface exposed cysteine residues. In E. coli LacZ is ideal for this purpose due to its content of cysteine, mobility in a region on SDS–PAGE devoid of other major proteins, availability of mutants lacking the enzyme as a control, and availability of antibody against LacZ. In this case a labeling experiment with both intact cells and permeabilized cells is carried out except that soluble proteins rather than the membrane fraction are analyzed after immunoprecipitation.
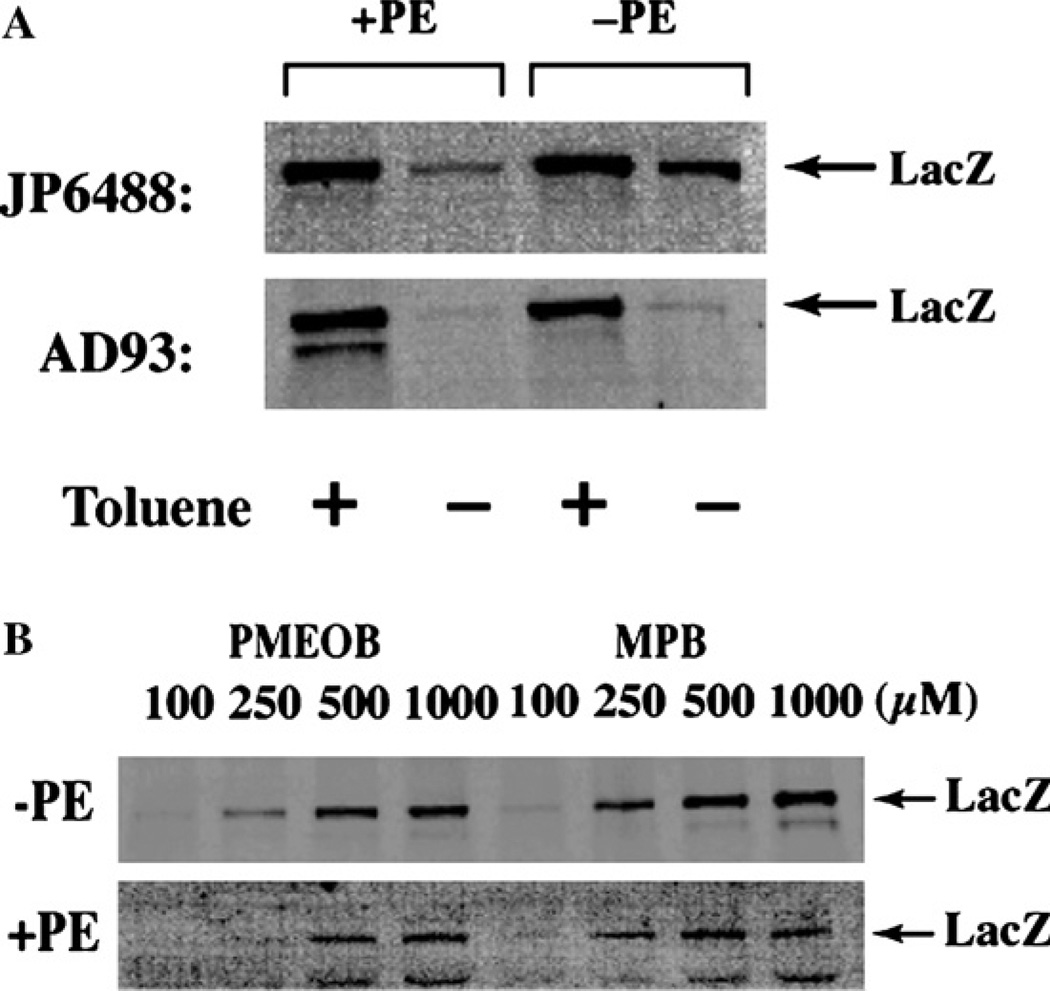
Based on protocols described above, both PE-con-taining and PE-lacking cells derived from different strains were labeled with and without pretreatment with toluene (Fig. 3). As discussed in Section 7, controlled treatment of cells with toluene permeabilizes the inner membrane without lysing cells. All cell types exhibited strong labeling of LacZ after permeabilization with toluene (Fig. 3A, lanes 1 and 3). Parental JP6488 cells (PE-containing) showed weak labeling (about 10%) of LacZ without pretreatment with toluene (lane 2), while PE-lacking cells derived from JP6488 showed almost 50% labeling of LacZ in the absence of toluene (lane 4). In contrast, AD93 cells showed essential no labeling of LacZ in the absence of toluene irrespective of the membrane lipid composition. This experiment clearly shows a significant difference in the permeability of different host strains and emphasizes the need to screen host strains for reagent permeability prior to initiating experiments.
Fig. 3.
Control experiments demonstrating membrane impermeability of labeling reagents. PE-containing and PE-deficient cells derived from two different host backgrounds (JP6488 and AD93) were labeled with 100 µM MPB (A) or in the presence of varying concentrations of MPB or PMEOB (B) at room temperature for 5 min. In (A), cells were treated as indicated (+) with a final concentration of 0.5% toluene prior to MPB treatment. The cells were lysed with detergent and the cytoplasmic fraction retained, after removal of the membrane fraction by centrifugation, for immunoprecipitation with anti-LacZ polyclonal antibody. The immunoprecipitates were subjected to SDS-PAGE and biotinylated protein was detected using avidin-HRP and chemiluminescence.
A similar experimental approach can be used to optimize the reaction conditions to maximize labeling of external cysteines and minimize labeling of internal cysteines. Cells or membranes should be treated with various concentrations of reagent from 10µM to 1 mM, at temperatures from 0 to 25 °C, and for various lengths of time from 5 min to 1 h. The following experiment compares treatment of strain AD93 with two commercially available biotinylated maleimides. MPB is usually made up as a stock solution in dimethyl sulfoxide or dimethyl-formamide due to its low solubility in water [4,94,110] while MPOEB can be used directly from a water stock solution [5]. Both reagents showed increased permeable of the inner membrane with increasing reagent concentration, as indicated by the increase in the amount of LacZ labeling in PE-containing or PE-lacking cells (Fig. 3B). Membranes were more permeable to MPB than MPOEB and PE-lacking cells were more permeable than PE-containing cells to both reagents. However, no significant labeling of LacZ occurred in all cell types when either reagent was used at 100 µM for 5 min.
Other cytosolic bacterial markers such a glutathione [88] or elongation factor Tu [132] have been used to access membrane permeability. The permeability of the eukaryotic cytoplasmic membrane can be tested by monitoring Modification of the Ca2+-ATPase, which is localized in the endoplasmic reticulum with its nucleotide-binding domain (containing 13 cysteines) facing the cytoplasm [94].
6. Membrane orientation
Most of thiol-specific reagents utilized in TM topology assays (FM, OGM, and MPB) in intact E. coli cells have intrinsic permeability with respect to the outer membrane and therefore preparations of spheroplasts or RSOV is usually not required [95,96,105]. In one report the periplasmic exposure of cysteine residues to MPB was enhanced by labeling of E. coli cells in the presence of 50 µM polymyxin B, which permeabilizes the outer membrane [102]. However, there is utility in obtaining an independent assessment of TM topology in ISOV. The thiol labeling and protection patterns in whole cells and ISOV should be mirror images of each other. If not, replacement positions inaccessible to reagent in whole cells but accessible in ISOV may be sterically hindered by cellular components and not truly buried in the membrane. Many initial reports using SCAM™ concluded that cysteines were in intracellular extramembrane domains based on the lack of labeling by membrane impermeable reagents or were in intramembrane domains by lack of labeling by permeable reagents in whole cells rather than on a positive result. Comparing intact cells or RSOV with ISOV gives a positive signal with the same reagent for each cysteine replacement in two oppositely oriented membrane preparations, thus avoiding the problem of diVerences in reactivity of different reagents, scavenging of the label by cytoplasmic thiol groups, or interference by interaction of the target protein with other cellular components. Therefore, obtaining topological results with ISOV consistent with conclusions drawn from whole cell experiments strengthens conclusions on topological organization particularly for unexpected results.
Conclusions drawn from analysis of ISOV are valid only if it can be shown that the ISOV are sealed, impermeable to the reagents, and have a uniform orientation opposite to that in whole cells. Fortunately, passing E. coli cells through a French press at a relatively low pressure of 8000 psi results in a yield of over 90% sealed ISOV [4]. Conditions for preparation of membrane vesicles varies among strains and may be significantly different for cells with changes in membrane lipid composition or mutations in membrane function. SCAM™ is sensitive to contamination of the population of ISOV by RSOV, unsealed membrane vesicles or unbroken cells. Therefore, control experiments should be carried out as outlined below to establish generation of sealed ISOV.
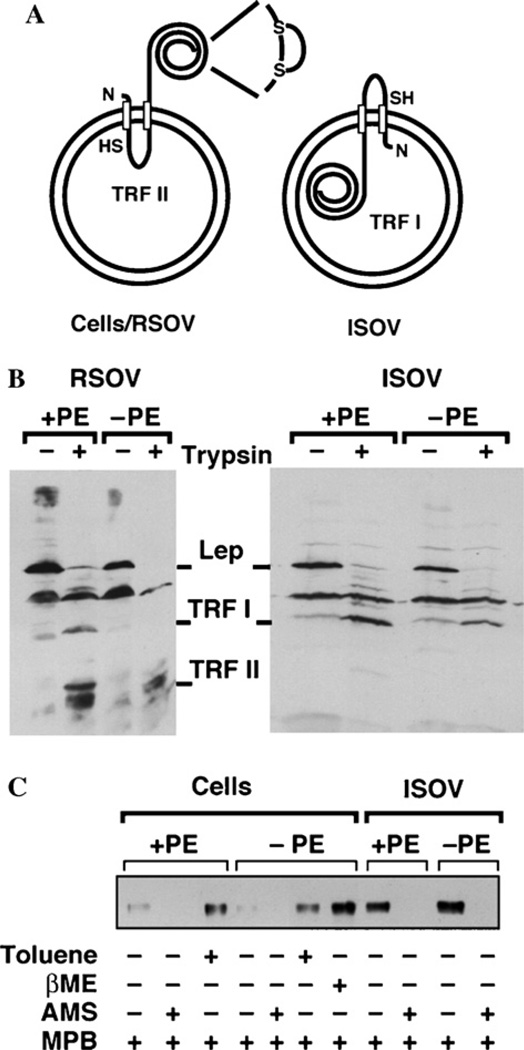
Leader peptidase (Lep) of E. coli is a typical bitopic membrane protein whose membrane topology has been extensively studied using various chemical approaches. The orientation of Lep has been established in both PE-containing [134] and PE-deWcient cells [34] with its larger catalytic domain oriented to the periplasm for processing of translocated proteins. Since misorientation of the catalytic domain would be lethal, orientation of Lep is a direct measure of orientation of membranes analyzed from any viable cell. Lep has only one free cysteine residue accessible from the cytoplasm, and the disulWde-linked cysteines in the periplasm are cryptic unless reduced (see Fig. 4A). Moreover trypsin cleavage sites in both extramembrane domains (large globular P2 in the periplasm and small P1 in the cytoplasm) of Lep provide sites for limited trypsinolysis that yield unique sized degradation products. Trypsin treatment of RSOV made by lysing spheroplasts produces a small cytoplasmic trypsin-resistant fragment (TRF II) while proteolysis of ISOV produces the larger TRF I. Leaky vesicles would allow access of trypsin to both sides of the membrane and result in formation of a 5-kDa fragment. A polyclonal antibody recognizes all the above forms of Lep on Western blots. As shown in Fig. 4B, trypsin treatment of RSOV made from PE-containing and PE-lacking membranes yields TRF II. The small amount of TRF I-like material results from autohydrolysis of Lep even without trypsin addition. Trypsin treatment of ISOV results in only TRF I. In both cases no 5-kDa fragment was observed so RSOV and ISOV made from PE-containing and PE-lacking cells are uniformly oriented and sealed.
Fig. 4.
Lep as a vehicle for determining intactness and sidedness of oriented vesicles. (A) The orientation of leader peptidase (Lep) in wild type E. coli for RSOV, whole cells (Cells), and ISOV. The larger C-ter-minal domain of Lep is exposed to the periplasm in whole cells or the exterior in RSOV and contains two cysteine residues in disulfide linkage. The smaller cytoplasmic loop that connects the two TM segments of Lep contains a single cysteine. Trypsin treatment of RSOV digests the periplasmic domain of Lep leaving the smaller TRF II. Trypsin treatment of ISOV digests the cytoplasmic loop leaving the larger TRF I. (B) The fragmentation pattern generated by trypsin treatment of Lep in either RSOV or ISOV from either +PE or −PE cells. (C) The results of various treatments of either whole cells or ISOV from either +PE or −PE strains with various thiol-specific reagents or toluene prior to immunoprecipitation with Lep-specific antibody followed by SDS–PAGE and detection of biotinylated protein using avidin-HRP and chemiluminescence. This figure was reproduced with permission from [4]. (Copyright 2002 EMBO) where experimental details can be found.
Analysis of Lep in these vesicles using SCAM™ validated the method using a known membrane protein and veriWed membrane orientation and lack of membrane permeability by the labeling reagent (Fig. 4C). Lep was not biotinylated in whole cells using MPB unless perme-abilized by toluene. Pretreatment with βME increased biotinylation due to reduction of the periplasmic disulfide bond. The small amount of labeling observed was variable and was blocked by AMS, which is less membrane permeable than MPB, suggesting this labeling was due to some cell lysis. With ISOV, biotinylation occurred without toluene treatment and was completely blocked by AMS. The same results were obtained for PE-containing and PE-lacking membranes.
Thus Lep, an essential cytoplasmic membrane protein of E. coli, appears to tolerate drastic changes in phospholipid composition [4,23,34] and has natural cysteine positions that are clearly extracellular or intracellular. For all these reasons, analysis of Lep can serve as a simple diagnostic tool and powerful control for determining the sealed state and orientation of whole cells and membrane vesicles as well as for establishing favorable biotinylation conditions for extramembrane residues.
7. Labeling internal cysteine residues by disrupting the membrane barrier
Inability of a membrane impermeable reagent to label a cysteine residue in intact cells or vesicles may be due to steric constraints by the local structure, residency within a TM segment, or residency in a luminal extramembrane domain. Therefore, in order to conclude that a position is on the luminal side of the membrane, it is necessary to acquire positive labeling of the residue by performing a complementary experiment using ISOV as described in Section 6 or by showing that the protein can be labeled in a non-compartmentalized system, i.e., in permeabilized cells or after membrane disintegration. Although results using ISOV as a complementary approach to using whole cells are very convincing, the approach suffers from being time consuming and the requirement to establish uniform inverted orientation of the vesicles. Direct permeabilization of cells is simpler and can be used in conjunction with ISOV as shown in Section 6. However, the approach must be optimized for each cell type and assumes that only cysteines in luminal domains are exposed by the treatment. Permeabilization of cells, organelles or oriented vesicles can be combined with prior blocking of external surface exposed cysteine residues with membrane impermeable reagents so that subsequent labeling will only reflect luminal residues (Fig. 2).
The membrane orientations of the cysteines introduced into the predicted extracellular or cytoplasmic loops of a fully functional cysteineless human reduced folate carrier expressed in Chinese hamster ovary cells were determined by treatments with the MPB, NEM, and AMS combined with the Streptococcus pyogenes cholesterol-specific pore-forming toxin streptolysin O (SLO) [126] which permeabilizes host plasma membranes and facilitates the labeling of internal cysteines without affecting intracellular membranes [97]. In these experiments cells expressing constructs with the cysteine substitutions within loops predicted to face the cytosol were treated with SLO (0.5 µg/ml) prior to treatment with MPB. A sample of cells was also pretreated with AMS before incubation with SLO to further verify cysteine accessibilities in the absence or in the presence of permeabilization. Biotinylation of cysteines placed in the predicted intracellular loops was only detected after cell permeabilization with SLO. Since these cysteines were not accessible to AMS during the pretreatment, no competition for labeling was observed with this reagent. However, the labeling of these residues was abolished upon pretreatment with the membrane-permeable reagent NEM. The labeling with MTSEA-biotin was used to define the accessibility of internal and external residues within the loop flanking TM segment 2 of rat serotonin transporter in intact Intestine 407 or HeLa cells and cells treated with digitonin (0.0025%) for 4 min to permeabilize the plasma membrane [100,131].
The extracellular residues should be accessible to the labeling agent to the same extent both in intact and permeabilized cells since in the presence of permeabilizing agent all the extramembrane cysteine residues regardless of their location should be available for modification. However, this is not always the case as was observed with using toluene to permeabilize bacterial cells. Toluene has been employed extensively to supply enzymes within cells with substrate by making holes in the membrane without lysing cells [135]. Theoretically, treating with high concentrations of toluene would make cells more permeable and expose cytoplasmic cysteines to thiol probes. However, higher levels of toluene might seriously alter membrane protein architecture and the solvent accessibility of cysteine residues. Therefore, it is important to determine a critical concentration of toluene, which allows exposure of cytoplasmically oriented cysteines without interfering with their labeling.
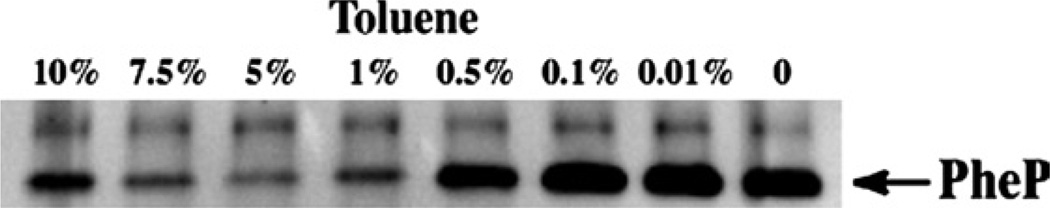
Cells expressing PheP with a single cysteine located in the periplasmic loop P5 were pretreated with an increasing amounts of toluene and the labeling of the protein with MPEOB was compared to a parallel sample without treatment with toluene (Fig. 5). Since P5 is a peri-plasmic domain, labeling of the single cysteine should not be influenced by toluene treatment. Unexpectedly, the higher levels of toluene did affect labeling by MPEOB (lanes 1, 2, 3, and 4) as compared with no toluene treatment (lane 8). The optimal concentration was found to be around 0.5% (lane 5). Accordingly, all toluene-related experiments were then conducted under this optimal concentration, which is enough to permeabilize the cell but not too high to affect labeling efficiency of extramembrane domains of the protein of interest. The optimal toluene concentration is strain dependent and was found to be ineffective as a permeabilizing agent (Jun Xie, personnel communication) in E. coli cells lacking PE in which the foreign lipid monoglucosyl diacyl-glycerol was introduced at 30–50 mol% of total lipid [15]. Therefore, toluene may not be universally effective in bacteria with lipid compositions significantly different from E. coli.
Fig. 5.
Determination of critical concentration of toluene to permeabi-lize cells. E. coli cells expressing PheP with a single cysteine in periplas-mic domain P5 [5] were incubated for 5 min at room temperature in the presence of varying concentrations of toluene prior to reaction with 100 µM of MPEOB. The cells were lysed and solubilized membrane proteins were subjected to immunoprecipitation with anti-PheP polyclonal antibody. The immunoprecipitates were subjected to SDS-PAGE and biotinylated proteins were detected using avidin-HRP and chemiluminescence.
Cell lysis approaches have also been used to gain access to internal cysteines. Several reports have used solubilization of membranes with detergents [104,106,107,114]. Some cysteine mutants were not labeled with the thiol-specific probe applied from either side of the membrane, indicating that the residues were either located within the membrane or buried within a tightly folded pocket of the protein. However, these cysteines were readily modified after protein denaturation or membrane solubilization with detergent making it is difficult to distinguish between these two possibilities [103,128,129]. Use of detergents to compromise the membrane barrier will not be further reviewed because such treatment has the high potential of exposing sites buried within the protein structure or the membrane bilayer.
Two effective methods of cell disruption are osmotic shock of spheroplasts [96] and disintegration of cells by sonication. The rate of labeling of cysteine replacements in the E. coli MotA protein (a component of the flagella) by FM as a function of time was measured in intact spheroplasts and spheroplasts lysed by dilution into hypotonic medium. Reaction with the reagent in intact spheroplasts was very slow relative to the rapid reaction in disrupted spheroplasts confirming a cytoplasm location for the cysteines [122]. To simultaneously label cysteines exposed to both surfaces of the membrane, membrane ghosts were prepared directly from E. coli cells expressing the formate transporter from Oxalob-acter formigenes by dilution of spheroplasts into hypotonic media containing OGM [111]. Periplasmically or cytoplasmically oriented cysteines in the E. coli osmo-protectant symporter ProP were also identified by a similar approach. Intact cells were treated with or without pre-blocking by membrane impermeable MTSET followed by reaction with membrane impermeable OGM before or after osmotic shock of spheroplasts [116].
Sonication is a rapid technique for cell disruption that is not labor intensive and can be used to label water-exposed cysteines on both sides of the membrane without regard for their normal disposition. LacY with a single cysteine replacement in either the C6 domain (F205C), which in PE-containing cells faces the cytoplasm, or TM segment VII (I230C), which residues within the hydrophobic core of the bilayer, was expressed in PE-containing and PE-lacking E. coli cells (Fig. 6). Samples of each cell type were split. One set was sonicated in the presence of MPEOB to simultaneously label cysteines exposed to both sides of the membrane. Samples were incubated at room temperature for another 4 min and then quenched by addition of 20 mM βME. The other set was treated with same amount of MPEOB for 5 min at room temperature, quenched with βME, and washed free of reagents by two cycles of cen-trifugation and resuspension before being subjected to sonication as above. As shown in Fig. 6, LacY was biotinylated only during sonication of PE-containing cells (consistent with cytoplasmic location of C6) while equal biotinylation occurred at C6 with or without sonication of PE-lacking cells (consistent with periplasmic location). It is important to note that the extent of biotinylation was the same before and after sonication of the PE-lacking sample indicating no alteration of the exposure of the cysteine occurred with sonication. This method does not appear to expose cysteines within TM segments to modification as indicated by the lack of labeling of a cysteine in TM segment VII, which should not be accessible to MPEOB. The labeling of membranes during sonication minimizes many problems for the detection of cysteine in luminal extramembrane domains. Intracellular cysteines can be selectively labeled by first blocking external cysteines followed by labeling during sonication (substituting of sonication for permeabilization in Fig. 2). This procedure does not require the preparation of oriented membrane vesicles or spheroplasts or the use of chemicals to compromise the membrane barrier.
Fig. 6.
Disintegration of the cell membrane by sonication. Cells of AD93 either containing (+) or lacking (−) PE were used to express LacY carrying a single cysteine replacement in extramembrane domain C6 (cytoplasmic in PE-containing cells) or intramembrane TM segment VII. Cells were labeled with 100 µM MPEOB either prior to or during sonication using a Branson sonicator (tapered probe, set at 15% of maximum output). The labeling reaction was quenched with excess β ME, and the whole cells disrupted by sonication as above. The membrane fraction was isolated, solubilized with detergent, imm-umnoprecipitated with LacY specific antibody, subjected to SDS-PAGE, and biotinylated protein was detected using avidin-HRP and chemiluminescence.
8. Use of SCAM™ to analyze lipid-dependent topological organization
SCAM™ was used to demonstrate the change in topological organization of LacY when expressed in a mutant of E. coli completely lacking PE [4]. A single cysteine replacement (F208C) in domain C6, which is cytoplasmic in PE-containing cells, was expressed in either a PE-containing (wild type) or PE-lacking (lipid mutant) strain of E. coli. Whole cells and ISOV from both cell types were treated with MPB either after or without toluene permeabilization and analyzed for biotinylation of LacY. As shown in Fig. 7, the cysteine is only labeled if whole PE-containing cells are permeabilized, while in ISOV made from PE-containing cells the cysteine is modified to the same extent with or without toluene treatment. These results are consistent with a cytoplasmic location for the cysteine in domain C6. The cysteine in C6 probed in PE-lacking cells and ISOV displayed a mirror image labeling pattern when compared to the results from PE-containing cells consistent with a periplasmic location. Although single cysteine replacements potentially could affect protein structure, the conclusions were based on comparison of the same protein in two different lipid environments strongly supporting a role for lipids as topological determinants. After the work up of the samples, each was analyzed by Western blotting using anti-LacY antibody to normalize the signal intensity.
Fig. 7.
TM orientation of the C6 loop of LacY depends of the presence of PE. A single cysteine replacement (208C) in the C6 loop of LacY was expressed in PE-containing or PE-lacking E. coli. Intact cells and ISOV were labeled with MPB either after or without permeabilization with toluene. After MPB treatment, LacY was immunoprecipitated and resolved by SDS-PAGE, and biotinylated protein was detected using avidin-HRP and chemiluminescence (upper panels). An identical blot (lower panel) was analyzed with anti-LacY antibody 4B11 to determine the amount of LacY immunoprecipitated [4].
9. Effects of secondary structure and determining TM boundaries
Since amino acid side chains are usually packed in defined structures, replacement with cysteines as specific targets may yield further insights regarding elements of secondary structure and conformational changes. Lack of labeling can be due to diVerences in local accessibility rather than by diVerences in topology making SCAM analysis useful to explore the secondary structure in putative extramembrane or transmembrane segments. To conWrm local accessibility more than one residue should be tested, and the protein should be assayed for reactivity of unreactive replacements after denaturation [128,129]. Thiol reagents that diVer in charge, size, and hydrophilicity can be also utilized for this purpose.
NEM has been shown to react periodically with cysteine residues engineered into a TM segment and partially exposed to an aqueous environment. Cysteines facing the hydrophobic core of the bilayer were inaccessible over a continuous sequence region. This approach has been used to identify the face of a TM α-helix that lines a channel or pore through the membrane based on a periodicity of every third to fourth position (one turn of the helix) being reactive [136]. Several single cysteines in the predicted TM segment 6 of the Lactococcus lactis ABC multidrug transporter LmrA could be labeled by the relatively bulky FM molecule, and the 3-fold periodicity of FM accessibility strongly suggests that the region spans the membrane as an α-helix with one face of the helix accessible to an aqueous cavity [103]. MPB-reactive and non-reactive cysteine replacements showed a periodicity of every second residue from Val828 to Leu835, which is consistent with a β-sheet conformation for one of the TM segments of the human plasma membrane anion exchanger AE1 [98]. In the same study Phe806 to Lys814 displayed a 3-fold periodicity that is suggestive of an α-helix. Local secondary structure for membrane proteins has also been inferred from the periodicity of cysteine modification by PCMBS [91].
The membrane-aqueous boundaries are generally inferred indirectly from the transition between accessible and inaccessible cysteine residues using a large hydrophilic thiol reagent and SCAM over the putative transition from aqueous to hydrocarbon environment. Bulky, hydrophilic, membrane impermeable thiol reagents such as FM [103,105,122], OGM [96,111,116], or MTSEA-biotin [100,125,126] have been used to map the extracellular boundary for TM segments. As indicated in Section 3.3, the thiol of cysteine extends 8–10Å from the peptide backbone so that these reagents have access, with decreasing efficiency, to five to six residues into the hydrophobic core of the bilayer. MPB at low concentrations and under mild reaction conditions was also used to identify indirectly the ends of TM segments [4,94,97,99,100,102,108–110,125].
10. Use of SCAM™ to monitor topological dynamics
Once inserted TM segments should not oscillate readily back and forth across the membrane due to the large free energy barrier to transfer hydrophobic sequences out of, and polar sequences back through the bilayer. Most membrane protein topology studies are consistent with a static and permanent location of extramembrane domains facing either one side or the other of the membrane. However, the actual structure of a membrane is very dynamic. Therefore, is the lipid bilayer really a non-flipping zone for integral membrane proteins?
The ability to change lipid composition in a regulated manner (Section 1.2) coupled with monitoring LacY topology as a function of changes in membrane phospholipid composition has shown that TM organization of membrane proteins is potentially more dynamic than previously assumed and responsive to changes in lipid environment after insertion and assembly. Placing the pssA gene under control of the regulatable araB promoter results in repressed gene expression in the presence of glucose with PE levels of less than 2%. Full gene expression in the presence of arabinose results in wild type levels of PE. LacY with a single cysteine replacement in either domain C6 (cytoplasmic in PE-containing cells) or in domain P7 (periplasmic irrespective of phospholipid composition) was expressed with isopropyl-1-thio-d-galactopyranoside (+IPTG) in the presence of glucose resulting in less than 2% PE in the membranes. Under these conditions (Fig. 8, −PE, Glu) both C6 and P7 in whole cells were biotinylated by MPB either with (+) or without (−) toluene treatment consistent with exposure of these domains to the periplasm [4]. New LacY synthesis was stopped (−IPTG) and glucose was replaced by arabinose to induce synthesis of PE for 60min in the absence of new LacY synthesis. MPB still labeled P7 in whole cells (+PE, Ara) with and without toluene treatment to the same extent as observed in the presence of glucose consistent with its periplasmic location in PE-containing cells. Remarkably, MPB only labeled C6 after cells were permeabilized by toluene consistent with the C6 domain returning to the cytoplasm with the induction of PE synthesis. A similar restoration of topological organization for the mis-assembled N-terminal helical hairpin (TM segments I and II) of PheP was observed when PE synthesis was turned on after initial insertion of PheP into membranes lacking PE [5]. For both transporters topology and active transport function were restored by exposure to PE after membrane insertion and assembly in the absence of PE.
Fig. 8.
Post-insertional topological reorganization detected by SCAM™. LacY with a cysteine replacement in either the cytoplasmic domain C6 or periplasmic domain P7 was expressed in an E. coli strain in which PE synthesis can be repressed by glucose (Glu) and induced by arabinose (Ara) [4]. LacY expression was under IPTG control. Accessibility of the single cysteine replacements was analyzed by treatment of whole cells with MPB either after or without toluene treatment. Cells were first grown under conditions of induction of LacY expression but repression of PE synthesis (+IPTG and −PE, Glu). After removing IPTG (+/−) to stop new synthesis of LacY, PE synthesis (+PE, Ara) was induced for 60 min. After reaction with MBP, bio-tinylated LacY was detected as in Fig. 7. This figure was reproduced with permission from [4]. (Copyright 2002 EMBO) where experimental details can be found.
These results challenge the dogma that once topological organization of a membrane protein is established, TM segments are stably maintained and do not flip. This result is consistent with the high degree of flexibility of LacY and other membrane proteins and suggests that the orientation of individual TM segments may not be fixed during assembly, after native structure has been attained, or during catalytic cycles. The lipid induced conformational and topological changes observed for LacY and PheP do not occur in wild type cells, but may be a consequence of their functional properties or reXect intermediates present during folding of these proteins.
SCAM™ has also been used to monitor topological changes induced by substrate binding and release during enzyme turnover [51,125]. In order to identify cysteine residues located in the extracellular domains of the α-subunit of the Na+/K+-ATPase and to determine whether these residues change orientation during the pump cycle, the sodium pump in RSOV was stabilized in two different conformations (the phosphorylated form or cation-occluded form). Topological changes in cysteines exposed to an aqueous or hydrophobic environment were assessed using AMS or CPM, respectively. Removal of cation from membrane preparations was accompanied by release (reaction with AMS) from the membrane of the hairpin formed by TM segment 5 and TM segment 6. Under these conditions some target residues within the C-terminal TM segment 7 to TM segment 10 fragment became exposed to lipophilic CPM labeling while others relocated outside of the membrane and became accessible to membrane impermeable AMS. Such reorganization occurs only in the C-terminal half of protein, since other changes in AMS or CPM accessibility were not observed.
Plasticity in the membrane location of the TM segment 8 of the Ca2+-ATPase may be the cause of the uncertainty in identifying the organization of this TM segment. At least two possible arrangements for TM segment 8 were suggested [137]. Competitive binding between thiol specific reagents and substrate revealed a mobile element within the TM structure of glutamate transporter GLT-1 [97]. The reactivity of a single cysteine placed in the loops flanking TM segment III of the E. coli tetracycline/H+ antiporter was drastically changed upon binding tetracycline, indicating that cytoplasmic and periplasmic loop regions undergo substrate-induced topological changes in opposite directions [52].
11. Topology of β-barrel proteins
SCAM™ can also be used to map the topology of membrane proteins that adopt a β-barrel architecture composed of anti-parallel TM β-strands. Site-directed fluorescence labeling by OGM was used to assign the topology of E. coli outer membrane protein PapC that appears to consist of 32 TM β-strands [121]. OGM readily crossed the outer membrane and labeled cysteine residues exposed to both the cell exterior and the periplasmic space. To differentiate cell exterior from periplasmic cysteines, Alexa Fluor 594 C5 was used as a blocking reagent. This reagent has a molecular weight of 909 Da that exceeds the 600 Da limit of outer membrane OmpF porin thus restricting the reagent access to the periplasmic space. By varying reaction conditions the modification of cysteines exposed to the cell exterior relative to periplasmic cysteines was reduced 5-fold by the blocking reagent. The single cysteine replacements within PapC that were not labeled with OGM in intact cells were subsequently labeled by OGM after protein solubilization with dodecyl-maltoside or denaturation with SDS suggesting they faced the hydrophobic core of the bilayer or were sterically hindered.
12. Use of SCAM™ to assess topology of proteins reconstituted into liposomes
Topological analysis of membrane proteins reconstituted into proteoliposomes is subject to the same constraints as in whole cells and membrane vesicles. What is the degree of lipid bilayer permeability to the reagents and is the protein uniformly oriented in the liposome? The topological organization of LacY, UhpT, and equ-inatoxin II has been successfully determined using MPB after reconstitution of the proteins with a uniform topology in proteoliposomes [43,109,138]. The topological orientation of TM segments of LacY was found to be dependent on the lipid composition of the proteoliposomes. Cytoplasmic domain C6 and periplasmic domain P7 were on opposite sides of liposomes containing PE and on the same side in liposomes lacking PE, which is consistent with their relative orientation in whole cells [4,43].
In general liposomes are more permeable to the commonly employed thiol reagents and permeability may vary with lipid composition. To assess whether these reagents are indeed membrane impermeable, proteoliposomes can be encapsulated with a thiol reporter compound and then probed with the thiol-specific reagent from the outside. Thionitrobenzoate (TNB) is an effective reporter compound since its absorbance at 412 nm is significantly reduced when the thiol group is blocked [139]. Moreover, appropriate thiol scavengers can eliminate undesired trans Modification. Modification of luminal cysteines within a target protein due to reagent permeability can be significantly reduced by inclusion of charged thiol scavengers inside liposomes. Cysteine is an effective thiol scavenger, which is strictly membrane impermeable due to its zwitterionic nature.
Uniform orientation of proteins in the liposome bilayer can usually be concluded from the labeling results. By using cysteine replacements in extramembrane domains that exhibit opposite orientations in vivo, both accessibility and inaccessibility should be observed by membrane impermeable reagents in a pattern consistent with in vivo orientation. The inaccessible cysteines should be modified after disruption of the proteoliposomes [43].
13. SCAM™ limitations
As is true of most approaches to determine membrane protein topological organization, there are limitations to using SCAM™. Caution must be used in assigning an intramembrane location to a cysteine residue because it is unreactive to hydrophilic thiol reagents in both intact and permeabilized membranes. Lack of or low levels of labeling may result from any of the following reasons: (1) steric hindrance due to local secondary structure; (2) internalization into the compact fold of the protein; (3) lack of ionization of the thiol group due to a hydrophobic environment; (4) local environment with the same charge as the thiol reagent; (5) increased pKa of the thiol due to the high negative charge density of neighboring residues or anionic lipids. For this reason several membrane-impermeable reagents should be tested in order to find the most appropriate one, including ones that are smaller and with different charge properties.
The secondary structure of extramembrane loops and local steric hindrance will influence whether or not the cysteine residue is accessible. Periplasmic extramembrane domains tend to be shorter (sometimes only three amino acids in length) than cytoplasmic domains. Therefore, there may be little or no protrusion of these loops into the extracellular space, thus preventing reaction of the cysteine residues in these locations with bulky maleimides. Biotinylated reagents appear to react better with cysteines towards the middle of extended hydrophilic loops than near the TM segment interfacial domain [4,94,112]. The lower reactivity of some of the residues in hydrophilic loops may reflect steric hindrance if clearly its neighbors react quantitatively. Due to the possibility of misidentifying residues as intramembrane because they are buried in an extracellular location, it is essential to analyze more than one position in each region of interest. All of these problems can usually be addressed by cysteine scanning replacement over a putative extramembrane domain that will show periodicity of accessibility, a short stretch usually more accessible by smaller thiol reagents, or a gradient of accessibility as the TM interface is approached. An extra membrane domain and an aqueous pore are not easily distinguished by SCAM™. Substitution at positions crucial to overall protein structure and stability cannot be used, but such substitutions often result in low levels of the protein and are informative in themselves.
Local electrostatic and ionic conditions may affect the pKa of an extramembrane position substituted by cysteine [140]. A local concentration of negatively charged amino acids or proximity to an anionic phospholipid domain would increase the pKa of the thiol due to the high negative charge density [130,141]. Negatively charged side chains proximal to a cysteine influence cysteinyl thiol pKa’s primarily via solvent-mediated coulombic interactions [142]. If negatively charged thiol reagents are employed, charge repulsion would further reduce the rate of Modification of the thiol. Reduced yield due to the reagent properties can be addressed by using reagents with different chemical properties.
TM topology studies assume that all copies of the target protein have the same orientation. The labeling patterns are the result of end-point titrations and assume a relatively fixed conformation for extramembrane loops, ignoring the presence of regions with heightened mobility and flexibility or the possibility of topological inversions on the time scale of the labeling. Topological models derived from accessibility patterns depict a static TM topology whereas the actual structure in a membrane is more likely to be dynamic. Through the application of SCAM™ some examples exist of dynamic changes in topological organization induced post-assembly of membrane proteins as well as some proteins that appear to exist with multiple topological organizations. Therefore, the static nature of methods that measure topological organization may have missed more dynamic properties of membrane proteins. In particular low yield of Modification due to slow rate of reaction may be due to dynamic movement of a domain into and out of an accessible region. As discussed below there are examples of cryptic intramembrane regions that become exposed to the aqueous phase and extramembrane domains that translocate to the opposite side of the membrane. Therefore, labeling studies in isolated membranes should complement topology studies in intact cells where the protein is turning over or metabolic conditions influence organization.
Even with optimal assays and reagents, membrane proteins that assume multiple conformations either within the same or between different membranes may yield confusing and conflicting results. Prenesilin 1 exists in distinctly different conformational states, one with eight TM topology retained within cells and the other transported to the cell surface in a seven TM organization. This would explain the contradictory observations concerning topological organization made by several groups [143]. Several examples of mixed topology or different topologies for the same protein in different membranes were noted above. It is not clear whether these topological diVerences occurred during or post assembly of these proteins. However, using SCAM™ topological reorganization was demonstrated for both LacY [4] and PheP [5] of E. coli post assembly of the proteins. Such results would suggest that diVerences in lipid environment could be a contributing factor to diVerences in organization of the same protein in different membranes of eukaryotic cells.
14. SCAM™ advantages
This approach is based on introduction of cysteine residues one at a time into putative extracellular or intracellular loops of a cysteineless membrane protein of interest followed by chemical Modification with membrane impermeable thiol-specific probes either before or after compromising membrane integrity to determine cysteine sidedness. Together, these tools document residue accessibility, and therefore topology of a membrane protein. Cysteine-scanning mutagenesis is the most useful technique thus far developed for topology studies and has the following advantages: (1) analysis can be done on the complete protein; (2) cysteine replacements are well tolerated with retention of topology and function; (3) analysis is done by chemical modification by a broad range of commercially available reagents with different physical-chemical properties; (4) detection of engineered cysteine modifications is simple; (5) only small quantities of the protein are required; (6) no complicated protein purification procedures are required; (7) chemical modification can be done using intact cells, thereby avoiding problems related to the conversion of cells into membrane vesicles with a uniform orientation.
SCAM™ is not only an alternative approach to determination of membrane protein structure, but also constitutes an attractive independent approach to structural and dynamic studies of membrane proteins. A quantitative analysis of the surface accessibility of individual cysteines at various stages of assembly of S. aureus α-hemolysin was carried by cysteine scanning mutagenesis and targeted chemical modification in order to map the structural changes which this polypeptide undergoes during pore formation [144]. The placement of cysteine residues in putative extracellular or intracellular domains of polytopic membrane proteins has proven very useful in defining the folding pattern and TM conformational changes of the polypeptide in the membrane. Labeling of single cysteine replacements of a major component (SecG) of the translocon with AMS either at rest or during protein translocation clearly demonstrated that a cytoplasmic region of SecG undergoes a change in membrane sidedness upon preprotein translocation, indicating that SecG undergoes topology inversion [132].
Although the labeling patterns derived from SCAM™ assay usually reflect end-point topology of a membrane protein, semi-quantitative analysis of the surface accessibility of individual cysteines introduced into extramembrane loops can be carried out at various stages of protein assembly. Cysteine accessibility during bacteriorhodopsin translation was monitored by pulsechase radiolabeling and modification by AMS to determine the order and timing of insertion of TM segments into the membrane of Halobacterium salinarum [145]. In this in vivo translocation assay, insertion of TM segments into the H. salinarum cytoplasmic membrane was monitored by rapid modification of unique cysteines in extracellular domains of the protein with AMS resulting in a shift in mobility of the protein in SDS-PAGE. SCAM™ also provided topological information at the time orientation was established during the biosynthesis of the protein and its insertion into the membrane.
Although X-ray crystallography produces highly detailed structural information of membrane proteins and even lipids resolved at high resolution within the structure, the structures only provide a static picture of lipid–protein interactions. The recent determination of a high resolution structure for LacY revealed a complex protein with TM segments of varied length and tilt angle [93]. However, the exact boundaries between TM segments and loop domains remain largely unknown. Where lipids have been resolved in crystal structures, considerable hydrogen bonding between polar residues at the ends of the TM helices and lipid polar head groups has been observed [146], indicating that these helices extend beyond the hydrophobic core of the bilayer. For most of the highly resolved membrane proteins, hydrophobic thicknesses of TM segments do not seem to match the lipid bilayer thickness expected or experimentally determined from the chain length of the surrounding lipids. significant mismatch between the hydrophobic thicknesses of a membrane protein and the lipid bilayer implies either that the structure of the protein when crystallized from detergent is different from that in the native membrane or that the lipid bilayer is distorted around the protein in the membrane. SCAM™ can be used to define the boundaries between membrane-embedded regions and the loop regions exposed to the aqueous phase of membrane proteins in their native environment thus supplementing high-resolution structural information. The precise boundaries of TM segments of the tetracycline/H+ antiporter TetA [113] and the human erythrocyte anion exchanger AE1 [112] were established by the reactivity of cysteine replacements with NEM and MPB, respectively.
15. Concluding remarks
Due to the lack of structural information on very hydrophobic membrane proteins determined by X-ray diffraction or NMR, other lower resolution methods have been employed to understand topological arrangement of proteins in the membrane. Several methods for determining TM organization are available and have been extensively reviewed. However, SCAM™, which has not been reviewed, has emerged over the past few years as the method of choice due to its relative simplicity for determining the TM structure of membrane proteins. SCAM™ has the advantage that structural perturbation of introduced cysteine mutations is milder than in other methods commonly used to determine topology. Although most examples of application have been from bacterial systems, several examples were outlined for TM mapping of eukaryotic proteins in their native environment, indicating that the method is generally applicable to different membrane systems.
This method can be used to assess secondary structure, membrane topology, and TM conformational changes as well as provide topological information during membrane insertion, folding, and assembly of proteins. This approach is capable of distinguishing accessibility of residues separated by only three or four residues and is useful in precise mapping of the ends of TM segments to define membrane aqueous boundaries of membrane proteins. By applying SCAM along extramembrane or TM segments, information of secondary structure and patterns of hydrophobic and aqueous exposure of domains can be obtained. Topological and other structural changes can be monitored during the catalytic cycle of enzymes.
By combining SCAM™ with mutants of E. coli in which membrane phospholipid composition can be systematically controlled, the role of phospholipids as determinants of membrane protein topological organization was established. In addition, the potential for polytopic membrane proteins to change their topological organization after insertion and assembly in the membrane was demonstrated. Combining of these techniques provides a system to study the role of lipid–protein interactions in the structure, assembly, and function of membrane proteins. The ability to regulate lipid composition temporally provides a powerful means to investigate molecular details of the dynamic topogenesis process.
A new algorithm (TMDET) has been developed to predict membrane embedded protein sequences by using their atomic coordinates. The method based on the frequency with which given amino acids residues are found in highly resolved protein transmembrane segments. This algorithm can be accessed at http://tmdet.enzim.hu/.
Acknowledgments
The work from the authors’ laboratory was supported by National Institutes of General Medical Science Grant GM20478 and the John S. Dunn, Sr. Research Foundation awarded to W.D. Phil Heacock assisted in drawing the figures.
References
- 1.White SH, von Heijne G. Curr. Opin. Struct. Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Higy M, Junne T, Spiess M. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 3.Levy D. Essays Biochem. 1996;31:49–60. [PubMed] [Google Scholar]
- 4.Bogdanov M, Heacock PN, Dowhan W. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. J. Biol. Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 6.Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Mol. Biol. Cell. 2000;11:2973–2985. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowhan W. Biochim. Biophys. Acta. 1998;1376:455–466. doi: 10.1016/s0304-4157(98)00013-6. [DOI] [PubMed] [Google Scholar]
- 8.Dowhan W, Mileykovskaya E, Bogdanov M. Biochim. Biophys. Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tropp BE, Ragolia L, Xia W, Dowhan W, Milkman R, Rudd KE, Ivanisevic R, Savic DJ. J. Bacteriol. 1995;177:5155–5157. doi: 10.1128/jb.177.17.5155-5157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi S, Shibuya I, Matsumoto K. J. Bacteriol. 2000;182:371–376. doi: 10.1128/jb.182.2.371-376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeChavigny A, Heacock PN, Dowhan W. J. Biol. Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 12.Heacock PN, Dowhan W. J. Biol. Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- 13.Sohlenkamp C, de Rudder KE, Rohrs V, Lopez-Lara IM, Geiger O. J. Biol. Chem. 2000;275:18919–18925. doi: 10.1074/jbc.M000844200. [DOI] [PubMed] [Google Scholar]
- 14.Xia W, Dowhan W. J. Bacteriol. 1995;177:2926–2928. doi: 10.1128/jb.177.10.2926-2928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Hea-cock P, Wieslander Å, Dowhan W. J. Biol. Chem. 2004;279:10284–10292. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 16.Mikhaleva NI, Santini CL, Giordano G, Nesmeyanova MA, Wu LF. FEBS Lett. 1999;463:331–335. doi: 10.1016/s0014-5793(99)01661-0. [DOI] [PubMed] [Google Scholar]
- 17.de Vrije T, de Swart RL, Dowhan W, Tommassen J, de Kruijff B. Nature. 1988;334:173–175. doi: 10.1038/334173a0. [DOI] [PubMed] [Google Scholar]
- 18.Kusters R, Dowhan W, de Kruijff B. J. Biol. Chem. 1991;266:8659–8662. [PubMed] [Google Scholar]
- 19.Kusters R, Breukink E, Gallusser A, Kuhn A, de Kruijff B. J. Biol. Chem. 1994;269:1560–1563. [PubMed] [Google Scholar]
- 20.Cabelli RJ, Dolan KM, Qian LP, Oliver DB. J. Biol. Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 21.Phoenix DA, Kusters R, Hikita C, Mizushima S, de Kruijff B. J. Biol. Chem. 1993;268:17069–17073. [PubMed] [Google Scholar]
- 22.Ulbrandt ND, London E, Oliver DB. FASEB J. 1992;6:481. ABS. [PubMed] [Google Scholar]
- 23.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Klompenburg W, Ridder AN, van Raalte AL, Killian AJ, von Heijne G, de Kruijff B. FEBS Lett. 1997;413:109–114. doi: 10.1016/s0014-5793(97)00888-0. [DOI] [PubMed] [Google Scholar]
- 25.Crooke E. Biochimie. 2001;83:19–23. doi: 10.1016/s0300-9084(00)01224-4. [DOI] [PubMed] [Google Scholar]
- 26.Zheng W, Li Z, Skarstad K, Crooke E. EMBO J. 2001;20:1164–1172. doi: 10.1093/emboj/20.5.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia W, Dowhan W. Proc. Natl. Acad. Sci. USA. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aboulwafa M, Hvorup R, Saier MH., Jr. Arch. Microbiol. 2004;181:26–34. doi: 10.1007/s00203-003-0623-7. [DOI] [PubMed] [Google Scholar]
- 29.Aboulwafa M, Saier MH., Jr. Res. Microbiol. 2002;153:667–677. doi: 10.1016/s0923-2508(02)01376-1. [DOI] [PubMed] [Google Scholar]
- 30.Rietveld AG, Verkleij AJ, de Kruijff B. Biochim. Biophys. Acta. 1997;1324:263–272. doi: 10.1016/s0005-2736(96)00232-5. [DOI] [PubMed] [Google Scholar]
- 31.Cullis PR, de Kruijff B. Biochim. Biophys. Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 32.Rietveld AG, Killian JA, Dowhan W, de Kruijff B. J. Biol. Chem. 1993;268:12427–12433. [PubMed] [Google Scholar]
- 33.Rietveld AG, Chupin VV, Koorengevel MC, Wienk HL, Dowhan W, de Kruijff B. J. Biol. Chem. 1994;269:28670–28675. [PubMed] [Google Scholar]
- 34.Rietveld AG, Koorengevel MC, de Kruijff B. EMBO J. 1995;14:5506–5513. doi: 10.1002/j.1460-2075.1995.tb00237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mileykovskaya EI, Dowhan W. J. Biol. Chem. 1993;268:24824–24831. [PubMed] [Google Scholar]
- 36.Mileykovskaya E, Sun Q, Margolin W, Dowhan W. J. Bacteriol. 1998;180:4252–4257. doi: 10.1128/jb.180.16.4252-4257.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mileykovskaya E, Fishov I, Fu X, Corbin BD, Margolin W, Dowhan W. J. Biol. Chem. 2003;278:22193–22198. doi: 10.1074/jbc.M302603200. [DOI] [PubMed] [Google Scholar]
- 38.Mileykovskaya E, Dowhan W. J. Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mileykovskaya E, Dowhan W. J. Bacteriol. 1997;179:1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogdanov M, Dowhan W. J. Biol. Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 41.Bogdanov M, Sun J, Kaback HR, Dowhan W. J. Biol. Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanov M, Umeda M, Dowhan W. J. Biol. Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Bogdanov M, Dowhan W. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowhan W, Bogdanov M. In: Biochemistry of Lipids. Vance DE, Vance JE, editors. Elsevier, Amsterdam: Lipoproteins and Membranes; 2002. pp. 1–35. [Google Scholar]
- 45.Zhang M, Mileykovskaya E, Dowhan W. J. Biol. Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 46.Ostrander DB, Zhang M, Mileykovskaya E, Rho M, Dow-han W. J. Biol. Chem. 2001;276:25262–25272. doi: 10.1074/jbc.M103689200. [DOI] [PubMed] [Google Scholar]
- 47.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. J. Biol. Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 48.Chang SC, Heacock PN, Clancey CJ, Dowhan W. J. Biol. Chem. 1998;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 49.Vance DE. In: Biochemistry of Lipids. Vance DE, Vance JE, editors. Elsevier, Amsterdam: Lipoproteins and Membranes; 2002. pp. 205–231. [Google Scholar]
- 50.Goder V, Spiess M. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutsenko S, Daoud S, Kaplan JH. J. Biol. Chem. 1997;272:5249–5255. doi: 10.1074/jbc.272.8.5249. [DOI] [PubMed] [Google Scholar]
- 52.Kimura T, Shiina Y, Sawai T, Yamaguchi A. J. Biol. Chem. 1998;273:5243–5247. doi: 10.1074/jbc.273.9.5243. [DOI] [PubMed] [Google Scholar]
- 53.Lambert C, Prange R. J. Biol. Chem. 2001;276:22265–22272. doi: 10.1074/jbc.M100956200. [DOI] [PubMed] [Google Scholar]
- 54.Lambert C, Prange R. Proc. Natl. Acad. Sci. USA. 2003;100:5199–5204. doi: 10.1073/pnas.0930813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nishiyama K, Suzuki T, Tokuda H. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 56.Gouff K, Gerard F, Santini CL, Wu LF. J. Biol. Chem. 2004;279:11608–11615. doi: 10.1074/jbc.M313187200. [DOI] [PubMed] [Google Scholar]
- 57.Rutz C, Rosenthal W, Schulien R. J. Biol. Chem. 1999;274:33757–33763. doi: 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 58.Dunlop J, Jones PC, Finbow ME. EMBO J. 1995;14:3609–3616. doi: 10.1002/j.1460-2075.1995.tb00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Q-S, von Dippe P, Xing W, Levy D. J. Biol. Chem. 1999;274:27898–27904. doi: 10.1074/jbc.274.39.27898. [DOI] [PubMed] [Google Scholar]
- 60.Kim SJ, Hegde RS. Mol. Biol. Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prus-iner SB, Lingappa VR. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- 62.Huggett J, Vaughan-Thomas A, Mason D. FEBS Lett. 2000;485:13–18. doi: 10.1016/s0014-5793(00)02175-x. [DOI] [PubMed] [Google Scholar]
- 63.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. Mol. Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 64.Linton KJ, Higgins CF. Mol. Membr. Biol. 2002;19:51–58. doi: 10.1080/09687680110103622. [DOI] [PubMed] [Google Scholar]
- 65.van Geest M, Lolkema JS. J. Biol. Chem. 1999;274:29705–29711. doi: 10.1074/jbc.274.42.29705. [DOI] [PubMed] [Google Scholar]
- 66.Nilsson J, Persson B, von Heijne G. FEBS Lett. 2000;486:267–269. doi: 10.1016/s0014-5793(00)02321-8. [DOI] [PubMed] [Google Scholar]
- 67.Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Persson B, Argos P. J. Protein Chem. 1997;16:453–457. doi: 10.1023/a:1026353225758. [DOI] [PubMed] [Google Scholar]
- 69.Jayasinghe S, Hristova K, White SH. J. Mol. Biol. 2001;312:927–934. doi: 10.1006/jmbi.2001.5008. [DOI] [PubMed] [Google Scholar]
- 70.Jayasinghe S, Hristova K, White SH. Protein Sci. 2001;10:455–458. doi: 10.1110/ps.43501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ikeda M, Arai M, Okuno T, Shimizu T. Nucleic Acids Res. 2003;31:406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ott CM, Lingappa VR. J. Cell Sci. 2002;115:2003–2009. doi: 10.1242/jcs.115.10.2003. [DOI] [PubMed] [Google Scholar]
- 73.Traxler B, Boyd D, Beckwith J. J. Membr. Biol. 1993;132:1–11. doi: 10.1007/BF00233047. [DOI] [PubMed] [Google Scholar]
- 74.van Geest M, Lolkema JS. Microbiol. Mol. Biol. Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimon MB, Goldshleger R, Karlish SJ. J. Biol. Chem. 1998;273:34190–34195. doi: 10.1074/jbc.273.51.34190. [DOI] [PubMed] [Google Scholar]
- 76.Toyoda T, Masunaga K, Ohtsu Y, Hara K, Hamada N, Kashiwagi T, Iwahashi J. Curr. Protein Pept. Sci. 2000;1:303–308. doi: 10.2174/1389203003381360. [DOI] [PubMed] [Google Scholar]
- 77.Manoil C, Boyd D, Beckwith J. Trends Genet. 1988;4:223–226. doi: 10.1016/0168-9525(88)90154-0. [DOI] [PubMed] [Google Scholar]
- 78.Broome-Smith JK, Tadayyon M, Zhang Y. Mol. Microbiol. 1990;4:1637–1644. doi: 10.1111/j.1365-2958.1990.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 79.Zelazny A, Bibi E. Biochemistry. 1996;35:10872–10878. doi: 10.1021/bi960815d. [DOI] [PubMed] [Google Scholar]
- 80.Ehrmann M, Boyd D, Beckwith J. Proc. Natl. Acad. Sci. USA. 1990;87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alexeyev MF, Winkler HH. J. Mol. Biol. 1999;285:1503–1513. doi: 10.1006/jmbi.1998.2412. [DOI] [PubMed] [Google Scholar]
- 82.Liu XY, Matherly LH. Biochim. Biophys. Acta. 2002;31:333–342. doi: 10.1016/s0005-2736(02)00467-4. [DOI] [PubMed] [Google Scholar]
- 83.Schwalbe RA, Rudin A, Xia SL, Wingo CS. J. Biol. Chem. 2002;277:24382–24389. doi: 10.1074/jbc.M201668200. [DOI] [PubMed] [Google Scholar]
- 84.Olivares L, Aragon C, Gimenez C, Zafra F. J. Biol. Chem. 1997;272:1211–1217. doi: 10.1074/jbc.272.2.1211. [DOI] [PubMed] [Google Scholar]
- 85.Nilsson IM, von Heijne G. J. Biol. Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- 86.Popov M, Tam LY, Li J, Reithmeier RA. J. Biol. Chem. 1997;272:18325–18332. doi: 10.1074/jbc.272.29.18325. [DOI] [PubMed] [Google Scholar]
- 87.Akabas MH, Stauffer DA, Xu M, Karlin A. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- 88.Falke JJ, Dernburg AF, Sternberg DA, Zalkin N, Milli-gan DL, Koshland DE., Jr. J. Biol. Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- 89.Karlin A, Akabas MH. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- 90.Karlin A. J. Gen. Physiol. 2001;117:235–238. doi: 10.1085/jgp.117.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan RT, Maloney PC. Cell. 1993;75:37–44. [PubMed] [Google Scholar]
- 92.Frillingos S, Sahin-Toth M, Wu J, Kaback HR. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 93.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 94.Loo TW, Clarke DM. J. Biol. Chem. 1995;270:843–848. doi: 10.1074/jbc.270.2.843. [DOI] [PubMed] [Google Scholar]
- 95.Jung H, Rubenhagen R, Tebbe S, Leifker K, Tholema N, Quick M, Schmid R. J. Biol. Chem. 1998;273:26400–26407. doi: 10.1074/jbc.273.41.26400. [DOI] [PubMed] [Google Scholar]
- 96.Ye L, Jia Z, Jung T, Maloney PC. J. Bacteriol. 2001;183:2490–2496. doi: 10.1128/JB.183.8.2490-2496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grunewald M, Menaker D, Kanner BI. J. Biol. Chem. 2002;277:26074–26080. doi: 10.1074/jbc.M202248200. [DOI] [PubMed] [Google Scholar]
- 98.Zhu Q, Lee DW, Casey JR. J. Biol. Chem. 2003;278:3112–3120. doi: 10.1074/jbc.M207797200. [DOI] [PubMed] [Google Scholar]
- 99.Cao W, Matherly LH. Biochem. J. 2004;378:201–206. doi: 10.1042/BJ20031288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sato Y, Zhang YW, Androutsellis-Theotokis A, Rudnick G. J. Biol. Chem. 2004;279:22926–22933. doi: 10.1074/jbc.M312194200. [DOI] [PubMed] [Google Scholar]
- 101.Olami Y, Rimon A, Gerchman Y, Rothman A, Padan E. J. Biol. Chem. 1997;272:1761–1768. doi: 10.1074/jbc.272.3.1761. [DOI] [PubMed] [Google Scholar]
- 102.Wada T, Long JC, Zhang D, Vik SB. J. Biol. Chem. 1999;274:17353–17357. doi: 10.1074/jbc.274.24.17353. [DOI] [PubMed] [Google Scholar]
- 103.Poelarends GJ, Konings WN. J. Biol. Chem. 2002;277:42891–42898. doi: 10.1074/jbc.M206508200. [DOI] [PubMed] [Google Scholar]
- 104.Valiyaveetil FI, Fillingame RH. J. Biol. Chem. 1998;273:16241–16247. doi: 10.1074/jbc.273.26.16241. [DOI] [PubMed] [Google Scholar]
- 105.Meuller J, Rydstrom J. J. Biol. Chem. 1999;274:19072–19080. doi: 10.1074/jbc.274.27.19072. [DOI] [PubMed] [Google Scholar]
- 106.Joseph-Liauzun E, Delmas P, Shire D, Ferrara P. J. Biol. Chem. 1998;273:2146–2152. doi: 10.1074/jbc.273.4.2146. [DOI] [PubMed] [Google Scholar]
- 107.Leng XH, Nishi T, Forgac M. J. Biol. Chem. 1999;274:14655–14661. doi: 10.1074/jbc.274.21.14655. [DOI] [PubMed] [Google Scholar]
- 108.Knol J, Veenhoff L, Liang WJ, Henderson PJ, Leblanc G, Poolman B. J. Biol. Chem. 1996;271:15358–15366. doi: 10.1074/jbc.271.26.15358. [DOI] [PubMed] [Google Scholar]
- 109.Fann MC, Maloney PC. J. Biol. Chem. 1998;273:33735–33740. doi: 10.1074/jbc.273.50.33735. [DOI] [PubMed] [Google Scholar]
- 110.Matos M, Fann MC, Yan RT, Maloney PC. J. Biol. Chem. 1996;271:18571–18575. doi: 10.1074/jbc.271.31.18571. [DOI] [PubMed] [Google Scholar]
- 111.Fu D, Maloney PC. J. Biol. Chem. 1998;273:17962–17967. doi: 10.1074/jbc.273.28.17962. [DOI] [PubMed] [Google Scholar]
- 112.Fujinaga J, Tang XB, Casey JR. J. Biol. Chem. 1999;274:6626–6633. doi: 10.1074/jbc.274.10.6626. [DOI] [PubMed] [Google Scholar]
- 113.Kimura T, Ohnuma M, Sawai T, Yamaguchi A. J. Biol. Chem. 1997;272:580–585. doi: 10.1074/jbc.272.1.580. [DOI] [PubMed] [Google Scholar]
- 114.Yan RT, Maloney PC. Proc. Natl. Acad. Sci. USA. 1995;92:5973–5976. doi: 10.1073/pnas.92.13.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gregory JD. J. Am. Chem. Soc. 1955:3922–3923. [Google Scholar]
- 116.Culham DE, Hillar A, Henderson J, Ly A, Vernikovska YI, Racher KI, Boggs JM, Wood JM. Biochemistry. 2003;42:11815–11823. doi: 10.1021/bi034939j. [DOI] [PubMed] [Google Scholar]
- 117.Creighton TE. Proteins: Structures and Molecular Properties. New York: W.H. Freeman and Co; 1983. [Google Scholar]
- 118.Kumarevel TS, Gromiha MM, Ponnuswamy MN. Prep. Bio-chem. Biotechnol. 2001;31:163–183. doi: 10.1081/PB-100103382. [DOI] [PubMed] [Google Scholar]
- 119.Taylor PD, Attwood TK, Flower DR. Nucleic Acids Res. 2003;31:3698–3700. doi: 10.1093/nar/gkg554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.White SH, Ladokhin AS, Jayasinghe S, Hristova K. J. Biol. Chem. 2001;276:32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 121.Henderson NS, Shu Kin Sou S, Martin C, Kulkarni R, Thanassi DG. J. Biol. Chem. 2004;279:53747–53754. doi: 10.1074/jbc.M409192200. [DOI] [PubMed] [Google Scholar]
- 122.Zhou J, Fazzio RT, Blair DF. J. Mol. Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]
- 123.Jockel P, Di Berardino M, Dimroth P. Biochemistry. 1999;38:13461–13472. doi: 10.1021/bi990303+. [DOI] [PubMed] [Google Scholar]
- 124.Fujihira E, Tamura N, Yamaguchi A. J. Biochem. (Tokyo) 2002;131:145–151. doi: 10.1093/oxfordjournals.jbchem.a003069. [DOI] [PubMed] [Google Scholar]
- 125.Hu YK, Kaplan JH. J. Biol. Chem. 2000;275:19185–19191. doi: 10.1074/jbc.M000641200. [DOI] [PubMed] [Google Scholar]
- 126.Cao W, Matherly LH. Biochem. J. 2003;374:27–36. doi: 10.1042/BJ20030301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Long JC, Wang S, Vik SB. J. Biol. Chem. 1998;273:16235–16240. doi: 10.1074/jbc.273.26.16235. [DOI] [PubMed] [Google Scholar]
- 128.Long JC, DeLeon-Rangel J, Vik SB. J. Biol. Chem. 2002;277:27288–27293. doi: 10.1074/jbc.M202118200. [DOI] [PubMed] [Google Scholar]
- 129.Mordoch SS, Granot D, Lebendiker M, Schuldiner S. J. Biol. Chem. 1999;274:19480–19486. doi: 10.1074/jbc.274.27.19480. [DOI] [PubMed] [Google Scholar]
- 130.Jewell JE, Orwick J, Liu J, Miller KW. J. Bacteriol. 1999;181:1689–1693. doi: 10.1128/jb.181.5.1689-1693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen JG, Liu-Chen S, Rudnick G. J. Biol. Chem. 1998;273:12675–12681. doi: 10.1074/jbc.273.20.12675. [DOI] [PubMed] [Google Scholar]
- 132.Nagamori S, Nishiyama K, Tokuda H. J. Biochem. (Tokyo) 2002;132:629–634. doi: 10.1093/oxfordjournals.jbchem.a003266. [DOI] [PubMed] [Google Scholar]
- 133.Jones PC, Sivaprasadarao A, Wray D, Findlay JB. Mol. Membr. Biol. 1996;13:53–60. doi: 10.3109/09687689609160575. [DOI] [PubMed] [Google Scholar]
- 134.Dalbey RE, Wickner W. J. Biol. Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- 135.Jackson RW, DeMoss JA. J. Bacteriol. 1965;90:1420–1425. doi: 10.1128/jb.90.5.1420-1425.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kimura-Someya T, Iwaki S, Konishi S, Tamura N, Kubo Y, Yamaguchi A. J. Biol. Chem. 2000;275:18692–18697. doi: 10.1074/jbc.M000354200. [DOI] [PubMed] [Google Scholar]
- 137.Moller JV, Ning G, Maunsbach AB, Fujimoto K, Asai K, Juul B, Lee YJ, Gomez de Gracia A, Falson P, le Maire M. J. Biol. Chem. 1997;272:29015–29032. doi: 10.1074/jbc.272.46.29015. [DOI] [PubMed] [Google Scholar]
- 138.Anderluh G, Barlic A, Podlesek Z, Macek P, Pungercar J, Gubensek F, Zecchini ML, Serra MD, Menestrina G. Eur. J. Biochem. 1999;263:128–136. doi: 10.1046/j.1432-1327.1999.00477.x. [DOI] [PubMed] [Google Scholar]
- 139.Holmgren M, Liu Y, Xu Y, Yellen G. Neuropharmacology. 1996;35:797–804. doi: 10.1016/0028-3908(96)00129-3. [DOI] [PubMed] [Google Scholar]
- 140.Krishnasastry M, Walker B, Braha O, Bayley H. FEBS Lett. 1994;356:66–71. doi: 10.1016/0014-5793(94)01240-7. [DOI] [PubMed] [Google Scholar]
- 141.Bulaj G, Kortemme T, Goldenberg DP. Biochemistry. 1998;37:8965–8972. doi: 10.1021/bi973101r. [DOI] [PubMed] [Google Scholar]
- 142.Snyder GH, Cennerazzo MJ, Karalis AJ, Field D. Biochemistry. 1981;20:6509–6519. doi: 10.1021/bi00526a001. [DOI] [PubMed] [Google Scholar]
- 143.Kim J, Schekman R. Proc. Natl. Acad. Sci. USA. 2004;101:905–906. doi: 10.1073/pnas.0307297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Walker B, Bayley H. J. Biol. Chem. 1995;270:23065–23071. doi: 10.1074/jbc.270.39.23065. [DOI] [PubMed] [Google Scholar]
- 145.Dale H, Angevine CM, Krebs MP. Proc. Natl. Acad. Sci. USA. 2000;97:7847–7852. doi: 10.1073/pnas.140216497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jones MR, Fyfe PK, Roszak AW, Isaacs NW, Cogdell RJ. Biochim. Biophys. Acta. 2002;1565:206–214. doi: 10.1016/s0005-2736(02)00570-9. [DOI] [PubMed] [Google Scholar]
- 147.Tusnady GE, Dosztanyi Z, Simon I. Bioinformatics. 2005;21:1276–1277. doi: 10.1093/bioinformatics/bti121. [DOI] [PubMed] [Google Scholar]