Abstract
The dynamic packaging of DNA into chromatin is a key determinant of eukaryotic gene regulation and epigenetic inheritance. Nucleosomes are the basic unit of chromatin, and therefore the accessible states of the nucleosome must be the starting point for mechanistic models regarding these essential processes. Although the existence of different unwound nucleosome states has been hypothesized, there have been few studies of these states. The consequences of multiple states are far reaching. These states will behave differently in all aspects, including their interactions with chromatin remodelers, histone variant exchange, and kinetic properties. Here, we demonstrate the existence of two distinct states of the unwound nucleosome, which are accessible at physiological forces and ionic strengths. Using optical tweezers, we measure the rates of unwinding and rewinding for these two states and show that the rewinding rates from each state are different. In addition, we show that the probability of unwinding into each state is dependent on the applied force and ionic strength. Our results demonstrate not only that multiple unwound states exist but that their accessibility can be differentially perturbed, suggesting possible roles for these states in gene regulation. For example, different histone variants or modifications may facilitate or suppress access to DNA by promoting unwinding into one state or the other. We anticipate that the two unwound states reported here will be the basis for future models of eukaryotic transcriptional control.
Introduction
The dynamic packaging of DNA into nucleosomes and higher-order chromatin structure is a key mechanism by which DNA accessibility is regulated. Transcription, recombination, replication, and repair all require DNA to become accessible to various proteins (1,2). Understanding the mechanism of nucleosome formation and disassembly is therefore essential if we are to understand how reversible DNA packaging can regulate these biological processes.
The structure of a nucleosome is known at atomic resolution and shows 147 bp of DNA wrapped around a histone octamer (3). Many researchers have posited that when the DNA unwinds, it leaves an intact octamer (4). However, alternate unwound states have also been proposed. Böhm et al. (5) used Förster resonance energy transfer (FRET) measurements in vitro to monitor nucleosome disassembly as a function of ionic strength. They observed evidence of dissociation between the tetramer/dimer interface 0.2–3% of the time at physiological ionic strength. Others have proposed a mechanism involving symmetric splitting of the octamer along the H3-H3 dimer interface. There is evidence that the cysteine residue at the H3-H3 dimer interface becomes accessible to thiol-modifying reagents in regions of active transcription and in hyperacetylated nucleosomes (6,7). It has also been proposed that certain histone variants, such as CENP-A (8,9) and H3.3 (10), have a greater propensity to induce nucleosome unwinding via octamer splitting than their canonical histone counterparts. In HeLa cells, for example, it has been reported that H3-H4 tetramer splitting is associated with the H3.3 variant (10).
The ability of the nucleosome to unwind into multiple states has profound implications for our understanding of nucleosome-mediated regulation of gene expression. In a system with multiple unwinding pathways, each pathway will have different unwinding/rewinding rates and be susceptible to different perturbations, which can be modulated as a means for gene regulation. In addition, each distinct structure of the unwound nucleosome will interact differently with cellular components. For example, evidence shows that the chromatin assembly factor anti-silencing function 1 (Asf1) binds only to the H3-H4 heterodimer, blocking the H3-H3 dimer interface (11), indicating that Asf1 can bind to the nucleosome only if the H3-H3 dimer interface has been disrupted.
Although a detailed mechanism of nucleosome unwinding has remained elusive, it is well established that a number of ATP-dependent molecular motors apply force on DNA and chromatin (12). RNA polymerase II, for example, can impart forces of 15–25 pN (13–15). In addition, many chromatin-remodeling complexes, such as Swi/Snf and RSC, can apply forces up to 12 pN (16). At a more global level, microtubules apply force to chromosomes through the nuclear membrane (17).
In an effort to understand the effects of these forces on nucleosome unwinding, researchers have conducted a variety of studies to investigate the behavior of nucleosomes under force (4,18–21). These studies led to a general model for nucleosome unwinding, shown as the top pathway in Fig. 1 (4). In this model of unwinding, the nucleosome begins in state 2 with DNA wrapped almost two full turns around the histone octamer. As force is applied, the nucleosome transitions to state 1, where the outer turn of DNA is unwound but the inner turn remains wound. As the applied force is increased still further, the nucleosome inner turn unwinds, leaving the nucleosome in state 0, in which the intact histone octamer is tangentially bound to the DNA. The nucleosome can then rewind when the force is lowered.
Figure 1.
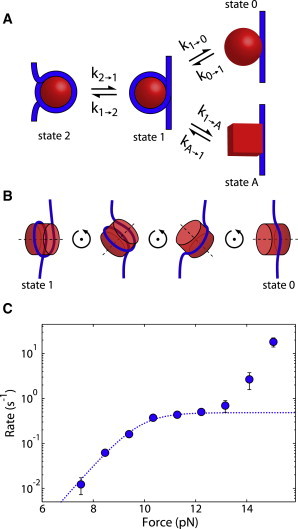
Model of nucleosome unwinding. (A) Kinetic diagram of nucleosome unwinding and rewinding. DNA is shown in blue and the octamer is in red. In state 2, the DNA is wrapped nearly two full times around the octamer. In state 1, the outer turn is unwrapped but the inner turn is still wound. From state 1, the nucleosome can unwind into one of two states, either state 0 (top) or state A (bottom). (B) Model of nucleosome unwinding into state 0. The nucleosome must first rotate before DNA can be removed. (C) Rate of nucleosome unwinding as a function of force. The theoretical unwinding rate versus force based on the mechanism in B is shown as a dotted line. The measured unwinding data are shown as blue circles. To see this figure in color, go online.
In this model of nucleosome unwinding, the octamer is assumed to be a solid cylinder that the DNA cannot pass through. Therefore, for the nucleosome to unwind, the nucleosome must first rotate as shown in Fig. 1 B. We and others have modeled this process and calculated the theoretical rate versus force (22,23), shown as the dotted line in Fig. 1 C.
We previously measured the rate versus force of nucleosome unwinding over a range of forces (24), and remeasure it here to higher accuracy, as shown in Fig. 1 C (blue circles). These data show a remarkable similarity to the theoretical predictions for forces below 13 pN (Fig. 1 C, dotted line), where the unwinding rate increases up until ∼10 pN, at which point the rate plateaus (22,23). This plateau corresponds to nucleosome rotation becoming the rate-determining step.
At forces above 13 pN, the unwinding rate increases again, contrary to the theory of a transition requiring nucleosome rotation. We hypothesize that this deviation from theory is the result of the nucleosome unwinding into an alternate unwound state that does not require nucleosome rotation. Fig. 1 shows our proposed alternative unwinding pathway, where the nucleosome can unwind into either state 0 or state A. It is important to note that at this point, we do not propose a specific structure for state A; however, an unwinding pathway that involves a rupture of the histone octamer would be consistent with our observations.
Here, we show the existence of an alternate state of the unwound nucleosome. The kinetic diagram in Fig. 1 A, which shows two different unwound states via two parallel pathways, makes two testable predictions. First, there will be two distinct rewinding rates depending on whether the nucleosome is first unwound to state 0 (k0→1) or state A (kA→1). Second, the unwinding pathways into state 0 and state A will pass through two different transition states, each of which may be differentially dependent on physical perturbations such as ionic strength. Here, we demonstrate that both of these predictions are realized experimentally and that the previously accepted nucleosome unwinding pathway must be changed to that shown in Fig. 1 A.
Materials and Methods
DNA preparation
DNA was a gift from Dr. Daniela Rhodes and was created by linearizing pUC18 plasmid containing an array of 12 601 nucleosome positioning sequences (NPS), each separated by a linker for a total of 200 bp per repeat, using AatII and AhdI (New England Biolabs, Ipswich, MA). The DNA was labeled with biotin (Roche, Nutley, NJ) on one end and an amine (Invitrogen, Carlsbad, CA) on the other end via a terminal transferase reaction (Roche). The amine-labeled end was covalently attached to a glass coverslip via a silane PEG N-hydroxysuccinimide linker (Nanocs, New York, NY), whereas the biotin end bound to a streptavidin-coated polystyrene bead (Spherotech, Lake Forest, IL). Detailed methods are described in Schlingman et al. (25).
Histone expression and purification
pET-11 plasmids containing H2A, H2B, H3, or H4 were a gift from Dr. Karolin Luger. Histones were expressed in BL21-Gold (DE3) E. coli, extracted from inclusion bodies using 7 M guanidine buffer (GuHCl) (20 mM Tris, pH 7.6, 10 mM DTT), and dialyzed into 8 M urea buffer (10 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol). Histones were passed over a Q-Sepharose column (Amersham Biosciences, Piscataway, NJ) and bound to a Hi-Trap SP-Sepharose column (Amersham Biosciences). Histones were eluted using 8 M urea buffer with 600 mM NaCl. Histones were dialyzed into double-distilled water and lyophilized. Detailed methods are described in Schlingman et al. (24).
Nucleosome reconstitution
Lyophilized nucleosomes were dissolved in 6 M GuHCl buffer (20 mM Tris-HCl, pH 7.5, 5 mM DTT), mixed in equimolar ratios, and dialyzed into 2 M NaCl buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5 mM β-mercaptoethanol) to remove the GuHCl and form octamer. Octamer was purified from unincorporated dimer and tetramer (Fig. S1 in the Supporting Material) using a Superdex S200 16/60 gel-filtration column (Amersham Biosciences). The octamer was mixed in equimolar amounts with sheared salmon sperm DNA (Invitrogen) as a carrier and was continuously dialyzed into no-salt buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5 mM β-mercaptoethanol) to form nucleosomes. Then 250 nM nucleosomes on carrier DNA were transferred in situ to the tethered DNA containing 12 repeats of the 601 NPS (12 × 601) at 680 mM NaCl (10 mM Hepes, pH 7.4, 1 mg/mL casein, 0.1% TWEEN) for 30 min. The buffer was then replaced with 100 mM NaCl (10 mM Hepes, pH 7.4, 1 mg/mL casein, 0.1% TWEEN). All force-clamp experiments were carried out on 12 × 601 DNA. Detailed methods are described in Mack et al. (24). Nucleosome formation was verified in a force-versus-extension experiment (Fig. S2) similar to those described previously (4,18–21).
Optical trapping experiments
We employed optical tweezers to apply a force clamp to the nucleosome arrays in an axial geometry as described previously (26). All experiments took place in a flow cell in 10 mM Hepes, pH 7.4, 1 mg/mL casein, 0.1% TWEEN. All buffers contained 100 mM NaCl unless stated otherwise. Nucleosome unwinding and rewinding was measured using the molecular yo-yo algorithm described in Mack et al. (27). In brief, the molecular yo-yo algorithm uses live jump detection to detect a nucleosome unwinding event. An event is detected when the mean of the last eight points exceeds the mean of the previous 40 points by a threshold of 15 nm. Once an unwinding event is detected, the force is immediately lowered to a force at which rewinding is possible. Using the molecular yo-yo, we only unwind and subsequently rewind a single nucleosome. An example trace is shown in Fig. S3. The time for each individual unwinding/rewinding event is calculated as the time from when the target force is reached to the time when the unwinding/rewinding event occurs, and then corrected for the number of nucleosomes bound to the DNA as described in the Supporting Material. The collected times for the population of unwinding/rewinding events are then assembled into the cumulative lifetime distribution for the force in question as described in the Supporting Material. The resultant cumulative distribution is composed of unwinding/rewinding times at a specific force from many different experiments, including data from multiple different tethers and slides. Using these cumulative distributions of lifetimes, we use a maximum-likelihood method to calculate the rates as described in the Supporting Material.
Results
Using optical tweezers and employing a powerful new method, the molecular yo-yo, we have measured over 2000 nucleosome transitions (27). The molecular yo-yo combines the use of a force clamp and a live jump detection algorithm that can detect the unwinding transition of a nucleosome and quickly (in less than 30 ms) lower the force, allowing the nucleosome to rewind. This method minimizes the time the nucleosome spends in the unwound state, decreasing the probability of the octamer unbinding from the DNA. Nucleosome unwinding at any force is completely reversible and nondestructive, allowing us to measure the same nucleosome repeatedly and thus verify our measurements. As described in detail in Mack et al. (27), the nucleosome is held at a constant unwinding force (Fig. S3). When the nucleosome unwinds, the extension increases by ∼25 nm. This unwinding event is detected and the force is then lowered to the desired rewinding force. The nucleosome then rewinds, causing a decrease in extension of ∼25 nm. Once the nucleosome rewinds, the force is increased again. To construct the unwinding lifetime distribution, the measured unwinding time of each nucleosome unwinding event is corrected by the number nucleosomes on the DNA, as described in the Supporting Material. This method allowed us to completely characterize the force-dependent unwinding and rewinding rates of the nucleosome, which in turn revealed the existence of multiple unwound states of the nucleosome.
Fitting the unwinding data to our model of unwinding into two states
Using the molecular yo-yo, we measure more than 1000 nucleosome unwinding events, determining a highly accurate rate of nucleosome unwinding as a function of force (Fig. 2 A). We use this unwinding rate as a function of force as the basis for our model of nucleosome unwinding into two states. Our data fit well to the theoretical unwinding rate into state 0 (22) for forces less than 13 pN (dotted line). We fit the data above 13 pN to an alternate unwinding pathway, forming state A, shown as a dashed line. By taking the ratio of the unwinding rate into state 0 to the sum of the two unwinding rates, state 0 and state A, at each force, we calculate the probability of unwinding into state 0, shown as the black line in Fig. 2 B.
Figure 2.
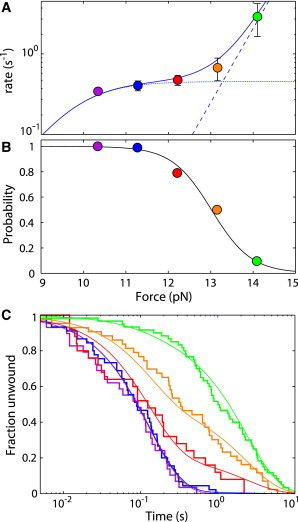
The rate of nucleosome rewinding depends on the force of unwinding. (A) Rate-versus-force plot for nucleosome unwinding. The dotted line corresponds to the predicted rate of the state 1 to state 0 transition and the dashed line corresponds to the predicted rate of the state 1 to state A transition as described in the text. The solid line is the sum of these two rates. (B) The probability of unwinding into state 0 with force calculated from the predicted rates in A is shown as a black line. The data correspond to the probability of rewinding from state 0 with force as calculated from the fits in C. (C) Cumulative fraction unwound for nucleosomes rewinding at 3.8 pN after being unwound at 10.3 (purple), 11.3 (blue), 12.2 (red), 13.2 (orange), and 14.1 (green) pN. Solid lines are fits of the sum of two exponential fits with the relative amplitudes determined using maximum-likelihood methods. To see this figure in color, go online.
Modeling the nucleosome unwinding into two states
We model the nucleosome unwinding into two states as depicted in Fig. 2 A. Nucleosome unwinding into state 0 has been theoretically predicted to increase approximately exponentially at low force and then reach an approximate plateau at higher force (22), as seen in our data below 13 pN. To capture this behavior with a simple analytic form to facilitate fitting, we represent the nucleosomes unwinding rate into state 0 with a force-dependent rate of
| (1) |
where CL, xL, and kplateau are fitting parameters; F is the applied force; and k1→0 is the unwinding rate from state 1 to state 0.
Above 13 pN, the nucleosome unwinds into an alternate state, state A. We model the rate of unwinding into state A as an exponentially increasing rate with force
| (2) |
where CH and xH are fitting parameters and k1→A is the transition rate from state 1 to the alternate state. The measured unwinding rate at any force is the sum of these two rates.
| (3) |
We fit the measured rates in Fig. 2 A using Eq. 3 with CL, kplateau, and CH varied with salt, whereas global values of xL and xH are used for all salt concentrations. The values for the fits shown in Fig. 4 B are given in Table S3.
Figure 4.
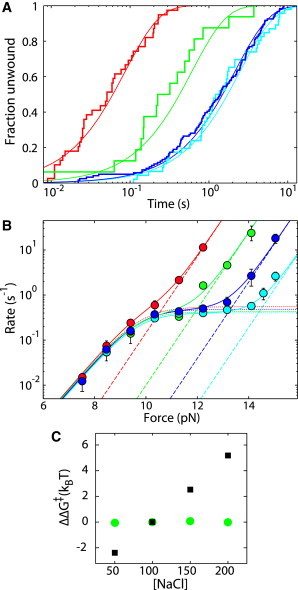
Nucleosome unwinding as a function of ionic strength. (A) Cumulative fraction unwound for nucleosome unwinding at 12.2 pN at 50 mM (cyan), 100 mM (blue), 150 mM (green), and 200 mM (red) NaCl. Solid lines are exponential fits with rates determined using maximum-likelihood methods. (B) Rates of nucleosome unwinding as a function of force at 50 (cyan), 100 (blue), 150 (green), and 200 (red) mM NaCl are shown as circles. The dotted lines correspond to our model of nucleosomes unwinding into state 0, and the dashed lines correspond to nucleosomes unwinding into state A. Solid lines are the sums of these two rates. (C) The change in transition state free energy for nucleosome unwinding as a function of salt concentration normalized to 100 mM NaCl, based on fits to the unwinding rate versus force data. The state 0 transition is shown as green circles, and state A is shown as black squares. To see this figure in color, go online.
By using a ratio of these rates at a particular force, as calculated above, we can determine the ΔΔG of the transition state, ΔΔG‡, using the following:
| (4) |
where k100 is the rate of unwinding in 100 mM NaCl and kx is the rate of unwinding at either 50, 150, or 200 mM NaCl.
Measuring nucleosome rewinding
Based on our model of the nucleosome unwinding into two states, we measure the rewinding rates from state 0 (k0→1) and state A (kA→1). Based on our fits to the unwinding data in Fig. 2 A and the resultant calculated probability of unwinding into state 0 in Fig. 2 B, unwinding the nucleosome at 10.3 pN should give nucleosomes unwinding into predominantly state 0. Additionally, unwinding at 14.1 pN should give unwinding into predominantly state A. Fig. 3 A shows the measured rewinding distributions for nucleosomes held at 3.8 (red), 4.2 (brown), and 4.7 pN (purple) after first being unwound at 10.3 pN, where we expect unwinding into state 0. These distributions are compared with Fig. 3 B, which shows the measured rewinding distributions for nucleosomes held at 2.8 (teal), 3.3 (blue), and 3.8 pN (red) after first being unwound at 14.1 pN, where we expect unwinding into state A (see also Fig. S7). The black lines in Fig. 3, A and B, correspond to single exponential fits with rates determined by maximum likelihood (24). Fig. 3 C summarizes these rates of rewinding versus force. Rewinding from state 0 is shown as orange triangles and rewinding from state A is shown as purple triangles (see also Table S2). In both cases, as force increases, the rewinding rate decreases. However, at all forces measured, rewinding from state A is about 10 times slower than rewinding from state 0.
Figure 3.
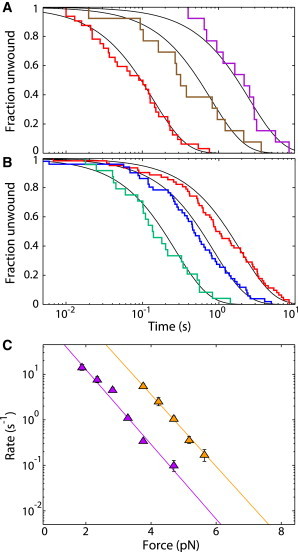
The rate of nucleosome rewinding depends on the force at which the nucleosome is unwound. (A) Cumulative fraction unwound for nucleosome rewinding at 3.8 (red), 4.2 (brown), and 4.7 (purple) pN after being unwound at 10.3 pN. The solid black lines correspond to single-exponential fits determined using maximum-likelihood methods. (B) Cumulative fraction unwound for nucleosome rewinding at 2.8 (teal), 3.3 (blue), and 3.8 (red) after unwinding at 14.1 pN. Black lines are the same as in A. (C) Rates of nucleosome rewinding as a function of force for nucleosome unwound at 10.3 and 14.1 pN are shown as orange and purple triangles, respectively. Orange and purple lines are single-exponential fits to the data. To see this figure in color, go online.
Having measured the rewinding rate for the nucleosome from state 0 and state A, we wanted to further verify our model of the nucleosome unwinding into two states as shown in Fig. 2 A. To do so, we measure nucleosome rewinding after unwinding at a range of forces. Fig. 2 C shows the rewinding distributions at 3.8 pN after unwinding at 10.3 (purple), 11.3 (blue), 12.2 (red), 13.2 (orange), and 14.1 pN (green) (see also Fig. S6 and Table S1). Again, based on the probability calculated from our model, we expect rewinding from state 0 after unwinding at 10.3 pN and rewinding from state A after unwinding at 14.1 pN. Indeed, these data fit well to a predominantly single exponential, indicating rewinding from a single state. However, after unwinding at 12.3 and 13.2 pN, the rewinding distributions are clearly not single exponentials. Instead, as shown by the red and orange curves in Fig. 2 C, the measured distributions at these forces are well described as a sum of two exponentials, indicating rewinding from two different states. Using χ2 analysis, we show that the fit to a sum of two exponentials is greater than 2 million times more likely than a single exponential fit (see Supporting Material and Table S1). We take the measured fraction rewinding from state 0, calculated from the fits in Fig. 2 C, and plot it over the calculated probability curve in Fig. 2 B. The data from these two experiments are in excellent agreement, verifying our model of nucleosome unwinding into two states.
Our model of unwinding into two states is the simplest model consistent with our data
Based on our data in Figs. 2 and 3, the simplest model in agreement with all of our data has the nucleosome unwinding into two different states, as described in Fig. 1. Here, we consider several other kinetic models and outline why they are inconsistent with our data.
In one possibility, the nucleosome unwinds via two parallel pathways into the same unwound state. If both pathways result in the same unwound state, then the rewinding rate would be independent of unwinding force, contrary to our results. Similarly, if the nucleosome unwinds into state 0 through a state A intermediate (1↔A↔0), we would again expect the rewinding rate to be independent of the unwinding force in our experiments, which is not the case. In addition, we would not expect the increase in unwinding rate at high force because the plateau region would continue to be the rate-limiting step.
Another possibility is that the nucleosome unwinds into state 0 and then transitions into state A with no apparent change in length (1↔0↔A) (Fig. S5). In our experiments, we can rewind in less than 30 ms; therefore, the rate of transitioning from state 0 to state A would have to be very fast. If this model were true, we would expect the fraction of state 0 and state A nucleosomes to be dependent on the amount of time spent in the unwound state, i.e., holding the nucleosome in state 0 for twice the time would result in twice the population in state A. We measure nucleosome unwinding with various delays after the unwinding event and observe that the rewinding rate is independent of the delay time (data not shown). In addition, there is no discernible length change between the putative state 0 and state A, and therefore it is unreasonable to expect the unwinding rate to have the strong force dependence we observe.
Nucleosome unwinding as a function of ionic strength
For a nucleosome unwinding via two pathways, there are two different transition states that will be differentially affected by outside perturbations, such as ionic strength. We investigate the effect of ionic strength on nucleosome unwinding by measuring the rate of unwinding versus force over a range of salt concentrations. All measurements reported so far were taken at 100 mM NaCl.
Fig. 4 A compares the distributions of nucleosome unwinding at 12.2 pN in 50 (cyan), 100 (blue), 150 (green) and 200 (red) mM NaCl (see also Fig. S8 and Table S3). At this force, the unwinding rate changes little from 50 to 100 mM NaCl, but above 100 mM NaCl, the rate increases dramatically as NaCl concentration increases. Fig. 4 B summarizes the rates of unwinding versus force at 50 (cyan circles), 100 (blue circles), 150 (green circles), and 200 mM (red circles) NaCl. The rate-versus-force data for each salt concentration are fit to our model of nucleosome unwinding into two states, with state 0 shown as dotted lines and state A shown as dashed lines (Table S4). It is clear that the behavior we model as unwinding into state 0 is insensitive to the concentration of NaCl within the range of concentrations investigated. In contrast, the behavior we model as unwinding into state A depends strongly on the concentration of NaCl.
To quantify the sensitivity of each state to ionic strength, we calculate the change in rate at a given force as a function of salt concentration normalized to 100 mM NaCl. We use this change in rate to calculate the change in transition-state free energy (ΔΔG‡), shown in Fig. 4 C. It is evident that the transition state for unwinding into state A (black squares) is highly dependent on ionic strength, whereas the transition state for unwinding into state 0 (green circles) is essentially unaffected. From this plot, we see that the state A transition-state free energy changes by ∼2 kBT (1.2 kcal/mol) per 50 mM change in NaCl concentration.
Discussion
In this article, we demonstrate the existence of two unwound states of the nucleosome, which we call state 0 and state A. Unwinding into state 0 occurs at lower forces, shows a fast rewinding rate, and has minimal dependence on ionic strength between 50 and 200 mM NaCl.
We hypothesize that this independence of ionic strength arises from the fact that the rate-limiting step of nucleosome unwinding into state 0 is primarily the bending of DNA, as shown by Kulić and Schiessel (22), which is independent of ionic strength within the concentration range used (28,29). In contrast, state A occurs at higher forces, shows a much slower rewinding rate, and is highly sensitive to ionic strength, indicating that the bending of DNA is no longer the transition state, but rather is most likely a rupture of the histone octamer.
Understanding the nucleosome and its mechanisms of unwinding, which allows access to DNA, is essential. Alternate states of the nucleosome, such as the one demonstrated here, could play an important role in DNA packaging and regulation of the processes that require DNA access. The vastly different rates of unwinding and rewinding between these two states could be a mechanism for gene regulation. In addition, different unwound structures would change the mechanism of DNA access for cellular components and could play a role in processes such as histone variant exchange (Fig. 5).
Figure 5.

Cartoon of possible nucleosome states and their implications in vivo. DNA is black, the nucleosome octamer is shown as an H3-H4 tetramer in magenta, and the H2A-H2B dimer is in yellow. (A) Nucleosome interactions with various proteins. The circular gray protein can only bind state A and the trapezoidal purple protein can only bind state 0. (B) Nucleosome structure and histone variant exchange. An open confirmation of state A provides a mechanism for histone variant exchange, as indicated by exchange with the green H3-H4 dimer. State 0 is inaccessible inhibiting variant exchange. To see this figure in color, go online.
Effects of ionic strength and a reevaluation of previous nucleosome measurements
The nucleosome contains positively charged amino acids that work to bind to the negatively charged backbone of DNA. The free energy of the nucleosome is therefore highly dependent on ionic strength (30). Our results reveal that in addition to affecting the free energy, the ionic strength can change the physical structure of the unwound nucleosome. It is imperative to take this effect into account when evaluating data under varying salt conditions. For example, in a previous study by Bintu et al. (31), nucleosomes were unwound at 40 mM KCl and 300 mM KCl. Based on our results, we think it is possible that the authors were measuring the kinetics of the two different states.
Structures of the unwound nucleosome
The structures of state 0 and state A are not known conclusively. However, based on our data and other theoretical predictions (22), we speculate as to the structures for both state 0 and state A. The structure of state 0 is most likely an intact octamer tangentially bound to the DNA. As shown in Fig. 1 B, the unwinding pathway into this structure requires a rotation of the nucleosome, which creates a plateau region in the unwinding rate (Fig. 1 C). This is further substantiated by the lack of salt dependence of the state 0 transition as discussed above.
We hypothesize that state A is most likely a symmetrically split octamer. The pathway into state A does not require the nucleosome to rotate, as signified by the rate increase above 13 pN, suggesting a rupture in the histone octamer. Specifically, the octamer is most likely splitting at the H3-H3 interface. We measure the unwinding of the nucleosome inner turn, which consists almost entirely of (H3-H4)2 tetramer contacts (3,32), and therefore it is unlikely that the H2A-H2B dimers play a large role in state A formation (5).
This hypothesis of a symmetrically split octamer is further supported by data from other groups (6–11,33–36). There is evidence of a symmetrically split octamer for nucleosomes in regions of active transcription (6,33). There is also evidence of a symmetrically split octamer in nucleosomes containing the H3 variant Cenp-A (8–10) and in nucleosomes with certain modifications (36).
Implications of alternate states
Nucleosomes interact with a variety of proteins in vivo, and both state 0 and state A will interact with these proteins differently (Fig. 5 A). For example, a split octamer explains the binding mechanism of the histone assembly proteins Asf1 and DAXX. Both of these proteins bind H3-H4 heterodimers (11,35), which are inaccessible from the state 0 structure. In addition, the existence of multiple states, such as the structure shown in Böhm et al. (5) and the split structure hypothesized here, provides a mechanism for histone variant exchange (Fig. 5 B) (37). With the increasing evidence for the existence of multiple unwound states, it becomes necessary to take these states into account when attempting to understand nucleosome regulation in vivo.
Possible mechanism for gene regulation
Our results show a large difference in unwinding/rewinding rates between state 0 and state A. We hypothesize that the formation of state A could act as a mechanism for gene regulation in vivo. Polymerase and chromatin remodelers such as SWI/SNF and RSC apply forces on DNA, and these forces are within the range measured in this study (13,14). Interestingly, these forces are near the force where the unwinding into state 0 changes to unwinding into state A, at ∼12 pN. Here, we show that at 12 pN, increasing the ionic strength from 100 mM NaCl to 150 mM NaCl shifts unwinding into state A, increasing the rate of nucleosome unwinding at 12 pN almost 10-fold. If histone modifications or variants in the cell could similarly change the force at which state 0 unwinding shifts to state A unwinding, these modifications could then act as a regulatory switch for DNA accessibility. For example, when a gene is not being transcribed, the forces on DNA are smaller and the nucleosome is stable. However, during transcription, polymerase is applying forces similar to those applied to unwind the nucleosome in these experiments. If the gene is active and contains nucleosomes with switch modifications that promote state A, the polymerase will quickly unwind nucleosomes, allowing transcription to proceed rapidly. However, if the gene is inactive and contains unmodified nucleosomes or nucleosomes modified to inhibit state A, nucleosome unwinding will be much slower and rewinding much faster, thereby decreasing the rate of transcription.
Evidence for a histone variant perturbing this equilibrium and acting as a switch can be seen in our previous study regarding the sin mutant H4-R45 (24). Studies have shown that nucleosomes at the HO promoter are removed using the SWI/SNF remodeling complex. If SWI/SNF is deleted, transcription is greatly diminished (38). However, the H4-R45H sin mutant was shown to allow transcription even when SWI/SNF was deleted (39,40). The rate-versus-force measurements of nucleosomes containing H4-R45H shown in Fig. 6 were fit with our model of unwinding into two states. These rates show remarkable similarities to wild-type nucleosome unwinding at increased ionic strengths. These measurements on H4-R45H and the existence of the alternate state shown here provide a possible mechanism for histone modifications and variants in gene regulation.
Figure 6.
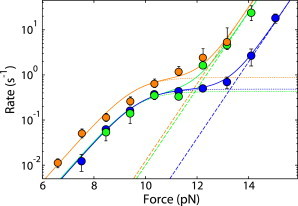
Nucleosome unwinding as a function of force for wild-type nucleosomes at 100 mM NaCl (blue) and 150 mM NaCl (green) compared with nucleosomes containing H4-R45H at 100 mM NaCl (orange). The dotted lines correspond to our model of nucleosomes unwinding into state 0 and the dashed lines correspond to nucleosomes unwinding into state A. Solid lines are the sums of these two rates. To see this figure in color, go online.
Acknowledgments
We thank members of S.G.J.M.’s and L.R.’s laboratories for valuable discussions.
This study was supported by the Raymond and Beverly Sackler Institute for Biological, Physical and Engineering Sciences; the Division of Physics, National Science Foundation (NSF, grant 1305509); and the Connecticut Department of Public Health (grant 2013-0194). D.J.S. is the recipient of an NSF Graduate Research Fellowship. M.K. was supported by an NSF Postdoctoral Research Fellowship in Biology.
Supporting Material
References
- 1.Saha A., Wittmeyer J., Cairns B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 2.Williams S.K., Tyler J.K. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 2007;17:88–93. doi: 10.1016/j.gde.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Luger K., Mäder A.W., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 4.Brower-Toland B.D., Smith C.L., Wang M.D. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. USA. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böhm V., Hieb A.R., Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior C.P., Cantor C.R., Allfrey V.G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983;34:1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- 7.Johnson E.M., Sterner R., Allfrey V.G. Altered nucleosomes of active nucleolar chromatin contain accessible histone H3 in its hyperacetylated forms. J. Biol. Chem. 1987;262:6943–6946. [PubMed] [Google Scholar]
- 8.Dalal Y., Wang H., Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conde e Silva N., Black B.E., Prunell A. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 10.Xu M., Long C., Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 11.English C.M., Maluf N.K., Tyler J.K. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3-H4 heterotetramer on DNA. Biochemistry. 2005;44:13673–13682. doi: 10.1021/bi051333h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavelle C., Victor J.-M., Zlatanova J. Chromatin fiber dynamics under tension and torsion. Int. J. Mol. Sci. 2010;11:1557–1579. doi: 10.3390/ijms11041557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 14.Yin H., Wang M.D., Gelles J. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 15.Cook P.R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Smith C.L., Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King M.C., Drivas T.G., Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennink M.L., Leuba S.H., Greve J. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 19.Simon M., North J.A., Poirier M.G. Histone fold modifications control nucleosome unwrapping and disassembly. Proc. Natl. Acad. Sci. USA. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gemmen G.J., Sim R., Smith D.E. Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J. Mol. Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 21.Mihardja S., Spakowitz A.J., Bustamante C. Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. USA. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulić I.M., Schiessel H. DNA spools under tension. Phys. Rev. Lett. 2004;92:228101. doi: 10.1103/PhysRevLett.92.228101. [DOI] [PubMed] [Google Scholar]
- 23.Mochrie S.G.J., Mack A.H., Regan L. Unwinding and rewinding the nucleosome inner turn: force dependence of the kinetic rate constants. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013;87:012710. doi: 10.1103/PhysRevE.87.012710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mack A.H., Schlingman D.J., Mochrie S.G.J. Kinetics and thermodynamics of phenotype: unwinding and rewinding the nucleosome. J. Mol. Biol. 2012;423:687–701. doi: 10.1016/j.jmb.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlingman D.J., Mack A.H., Regan L. A new method for the covalent attachment of DNA to a surface for single-molecule studies. Colloids Surf. B Biointerfaces. 2011;83:91–95. doi: 10.1016/j.colsurfb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mack A.H., Schlingman D.J., Mochrie S.G.J. Practical axial optical trapping. Rev. Sci. Instrum. 2012;83 doi: 10.1063/1.4757862. 103106–103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack A.H., Schlingman D.J., Mochrie S.G.J. The molecular yo-yo method: live jump detection improves throughput of single-molecule force spectroscopy for out-of-equilibrium transitions. Rev. Sci. Instrum. 2013;84:085119. doi: 10.1063/1.4819026. [DOI] [PubMed] [Google Scholar]
- 28.Wenner J.R., Williams M.C., Bloomfield V.A. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann C.G., Smith S.B., Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc. Natl. Acad. Sci. USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thåström A., Lowary P.T., Widom J. Measurement of histone-DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Bintu L., Ishibashi T., Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheinin M.Y., Li M., Wang M.D. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katan-Khaykovich Y., Struhl K. Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc. Natl. Acad. Sci. USA. 2011;108:1296–1301. doi: 10.1073/pnas.1018308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tagami H., Ray-Gallet D., Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 35.Elsässer S.J., Huang H., Patel D.J. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–565. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tropberger P., Pott S., Schneider R. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013;152:859–872. doi: 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Park Y.-J., Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg P.W., Stern M.J., Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 39.Kruger W., Peterson C.L., Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 40.Wechser M.A., Kladde M.P., Peterson C.L. Effects of Sin- versions of histone H4 on yeast chromatin structure and function. EMBO J. 1997;16:2086–2095. doi: 10.1093/emboj/16.8.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


