Abstract
Proteins function in cellular environments that are crowded with biomolecules, and in this reduced available space, their biophysical properties may differ from those observed in dilute solutions in vitro. Here, we investigated the effects of a synthetic macromolecular crowding agent, dextran 20, on the folded states of hyperthermophilic (S16Thermo) and mesophilic (S16Meso) homologs of the ribosomal protein S16. As expected for an excluded-volume effect, the resistance of the mesophilic protein to heat-induced unfolding increased in the presence of dextran 20, and chemical denaturation experiments at different fixed temperatures showed the macromolecular crowding effect to be temperature-independent. Förster resonance energy transfer experiments show that intramolecular distances between an intrinsic Trp residue and BODIPY-labeled S16Meso depend on the level of the crowding agent. The BODIPY group was attached at three specific positions in S16Meso, allowing measurements of three intraprotein distances. All S16Meso variants exhibited a decrease in the average Trp-BODIPY distance at up to 100 mg/mL dextran 20, whereas the changes in distance became anisotropic (one distance increased, two distances decreased) at higher dextran concentrations. In contrast, the two S16Thermo mutants did not show any changes in Trp-BODIPY distances upon increase of dextran 20 concentrations. It should be noted that the fluorescence quantum yields and lifetimes of BODIPY attached to the two S16 homologs decreased gradually in the presence of dextran 20. To investigate the origin of this decrease, we studied the BODIPY quantum yield in three protein variants in the presence of a tyrosine-labeled dextran. The experiments revealed distinct tyrosine quenching behaviors of BODIPY in the three variants, suggesting a dynamic local interaction between dextran and one particular S16 variant.
Introduction
It has been known for decades that macromolecular crowding may have thermodynamic and kinetic effects on protein properties (1). However, the impact of macromolecular crowding on protein structure, stability, folding, and activity is not clear, since most biophysical experiments are performed in dilute in vitro conditions. The high concentrations of different macromolecules within cells (50–400 mg/mL (2,3)) introduce excluded-volume effects and increase both specific and nonspecific interactions, as well as the macroscopic viscosity. Excluded volume due to the steric effect that two molecules cannot occupy the same space simultaneously leads to a decrease in entropy of the system as the available volume decreases. This phenomenon favors processes that result in a reduction of the total volume, such as formation of macromolecular complexes and compaction of conformations (3–6). Since protein folded states are smaller than the extended unfolded conformers, the excluded volume is thought to stabilize folded proteins indirectly due to unfavorable compaction on the unfolded ensemble. To experimentally mimic the crowded environment in vivo, large amounts of inert synthetic or natural macromolecules, so-called crowding agents, can be used for in vitro experiments. The most common synthetic crowding agents are dextrans and Ficoll 70. These polymers are useful as crowding agents, because they are uncharged and are believed not to interact with proteins, and because they are soluble at high concentrations. Using these agents, macromolecular crowding effects on protein stability and folding dynamics have been reported (5,7,8). Recently, in addition to excluded-volume effects, the presence of weak nonspecific interactions between crowding agent and protein has been proposed (9–11).
Experimental in vitro and computational in silico studies suggest that macromolecular crowding induces more compact unfolded states of proteins (12–15). We recently reported to our knowledge the first direct demonstration of unfolded-state compaction in vitro of a protein (S16Thermo) in the presence of the crowding agent dextran 20 by using Förster resonance energy transfer (FRET) (15). Although they are neglected in excluded-volume theory, structural changes of folded states in the presence of macromolecular crowding have been reported for a few other proteins (6,16–18). Using far-ultraviolet (UV) circular dichroism (CD) and coarse-grained simulations, Stagg et al. (16) demonstrated that upon addition of crowding agents, the secondary-structure content of folded apoflavodoxin increased to the level found in the crystal structure. Homouz et al. (6) used the same method to monitor changes in the folded structure of the aspherical protein Borrelia burgdorferi VlsE. It was found that upon gradual addition of the crowding agent, the protein obtained more helical structure followed by collapse of its spherical shape, in parallel with the formation of β-like structure. Dhar et al. (17) performed computer simulations of phosphoglycerate kinase (PGK) in combination with in vitro FRET experiments. Their data suggested that PGK populated a more compact folded state in the presence of the crowding agent Ficoll 70. In the presence of macromolecular crowding, Akabayov et al. (18) used small-angle x-ray scattering to reveal a compaction of the two domains in the human translation initiation factor eIF4A.
To follow up on our recent FRET study of unfolded S16Thermo (15), we here investigate the effects of macromolecular crowding on the folded states of S16Thermo and its mesophilic homolog, S16Meso, which share 33% sequence identity. The crystal structure of S16Thermo (19) is shown in Fig. 1. We report a combination of far-UV CD experiments and FRET measurements of N-((4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-yl)methyl)iodoacetamide (BODIPY)-labeled variants, as well as complementary studies with tyrosine-labeled dextran. Taken together, our data demonstrate explicitly that steric effects due to macromolecular crowding agents can modulate folded protein structures. Moreover, short-range dynamic interactions between crowding agent and protein are evident from BODIPY quenching data for particular variants. Our work emphasizes that folded structures of marginally stable proteins may be tuned by the crowded cellular environment. It also demonstrates that, in addition to global excluded volume effects, local dynamic interactions between the protein and the crowding agent may occur.
Figure 1.
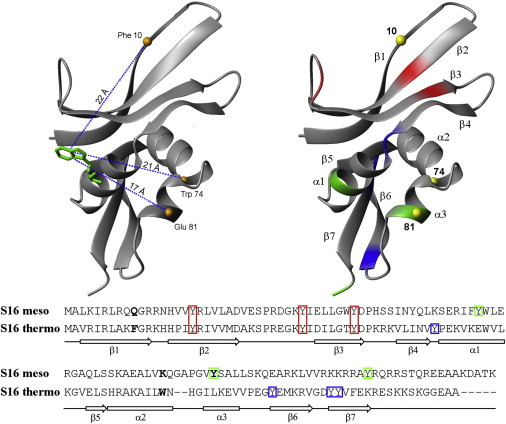
Crystal structure of wild-type S16Thermo (19) and the corresponding primary sequences for S16Thermo and S16Meso. (Left) Measured intramolecular distances in the prepared variants. The FRET donor Trp58 is green and the three possible acceptor sites are orange. The dashed lines show the intramolecular distances as determined from the x-ray structure. (Right) Tyrosine residues present within the thermophile (blue), mesophile (green), and both variants (red). The three possible acceptor sites are represented by yellow spheres. Residues that were substituted with cysteine and linked with BODIPY are marked in bold.
Materials and Methods
Chemicals
BODIPY was purchased from Invitrogen (Carlsbad, CA). Technical grade dextran 20 was purchased from Pharmacosmos (Holbaek, Denmark), NaCl pro analysis from Merck (Whitehouse Station, NJ), NaAc anhydrous pro analysis from Riedel-de Haën (Seelze, Germany), and DL-Tyrosine puriss from Fluka (Buchs, Switzerland). Tyrosine-labeled dextran was ordered and purchased from TdB Consultancy AB (Uppsala, Sweden). Water was run through a Milli-Q water purification system (Merck Millipore, Billerica, MA) with a resistivity of ≥18 MΩ cm−1.
The primary sequence of S16Thermo consists of 112 amino acid residues (13.0 kDa), whereas S16Meso contains 117 residues (13.9 kDa). Dextran 20 has an average molecular mass of 20 kDa and the tyrosine-labeled dextran molecules have an average molecular mass of 15.6 kDa. The tyrosine content was 53 μg/g, which yields ∼4.5 tyrosine residues per dextran molecule.
Protein expression and purification
The S16Meso mutants were created using the QuickChange method (Stratagene, La Jolla, CA), subcloned and expressed in a pET-3a vector, and verified by DNA sequencing (MWG Operon, Ebersberg, Germany). Plasmids were transformed into Rosetta 2 (DE3) competent cells and grown at 37°C in lysogeny broth medium supplemented with 100 μg/mL carbencillin and 34 μg/mL chloramphenicol until OD600 ∼ 0.6. Protein expression was then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside, and the cells were further grown overnight. The cells were centrifuged for 30 min at 5000 rpm, and resuspended in 50 mM Tris, 10 mM dithiothreitol (DTT), pH 7.5, followed by sonication on ice and centrifugation at 15,000 rpm for 30 min. The protein pellet was refolded in a buffer containing 30 mM NaPi, 50 mM NaCl, 6 M GuaHCl, 10 mM DTT at pH 6, then centrifuged at 15,000 rpm for 30 min. The supernatant was dialyzed overnight against 50 mM Tris and 5 mM DTT at pH 7.5, then centrifuged at 15,000 rpm for 30 min. The supernatant fraction was loaded on a cation-exchange column (SP Sepharose, GE Healthcare, Fairfield, CT) and the protein was eluted with a linear NaCl gradient. The binding buffer was 50 mM Tris and 5 mM DTT, pH 7.5, and the elution buffer was 50 mM Tris, 5 mM DTT, and 2 M NaCl, pH 7.5. The protein was further purified on a gel filtration column (HiLoad 16/60 Superdex 75, GE Healthcare) equilibrated with 30 mM NaAc, 50 mM NaCl, and 2 mM DTT, pH 5.5. The protein purity was verified with SDS-PAGE, and protein concentration was verified using the absorption at 280 nm. The S16Thermo mutants were purified directly from the soluble fraction after sonication and centrifugation. The supernatant was heated to 70°C for 30 min and centrifuged at 15,000 rpm for 30 min, and the soluble fraction was loaded on a cation-exchange column and finally purified by gel filtration, using the buffers and columns described above.
Sample preparation
The buffer was composed of 30 mM NaAc and 50 mM NaCl at pH 5.5, to which varying amounts of dextran 20 were added. All samples were left to equilibrate for 30 min before measurements were performed at 20°C, unless stated otherwise.
Absorption and fluorescence
All absorption spectra were measured on a Cary 5000 UV-VIS-NIR spectrophotometer (Varian, Sydney, Australia). The steady-state fluorescence measurements were recorded on a Fluorolog-3 spectrofluorometer (Jobin Yvon, Edison, NJ). Spectra were corrected and recorded using a spectral bandwidth of 2 nm. The absorbance of BODIPY was always kept at <0.08, and the protein concentration was ∼4 μM for all types of fluorescence measurements.
Time-resolved fluorescence
Time-resolved fluorescence relaxation was studied by means of the time-correlated single-photon counting technique, using a PRA 3000 system (PRA, Victoria, BC, Canada). Excitation source for tryptophan was a NanoLED-15 (IBH, Glasgow, United Kingdom) with excitation-peak maximum centered at 280 nm and operating at 800 kHz. BODIPY was excited with a PicoQuant pulsed diode laser with peak maximum at 470 nm, operating at 2.5 MHz. Neutral density filters in combination with an interference filter (Melles Griot, Didam, The Netherlands) centered around 470 nm (full width at half-maximum (FWHM) = 9.3 nm) were inserted into the excitation path to decrease the high intensity of the PicoQuant pulsed diode laser. The emission wavelengths were selected by using interference filters centered at 334 nm (FWHM = 8.9 nm) for tryptophan and at 520 nm (FWHM = 28 nm) for BODIPY. The count rate was kept at <1.5% of the repetition rate to avoid pile-up distortion. All lifetime measurements were collected under magic-angle conditions with a resolution of 26 ps/channel and collected over 2048 channels with 20,000 photons in the peak maximum.
Far-UV CD
Far-UV CD data were collected on a J-810 Spectropolarimeter (Jasco, Tokyo, Japan) equipped with a Peltier element for temperature control. In experiments, the protein concentration was 20 μM in a buffer consisting of 30 mM NaAc, 50 mM NaCl, pH 5.5, supplemented with different concentrations of dextran 20 (1 mm cell). Thermal unfolding of S16 was performed using a scan rate of 1°C/min, and data were obtained between 20°C and 90°C, using a fixed wavelength of 220 nm.
For equilibrium unfolding experiments in urea, ΔGu values for the S16Meso wild-type were obtained in the buffer described above using far-UV CD. The samples were incubated in appropriate concentrations of dextran 20 and urea overnight before taking measurements. The chosen temperatures (10°C, 25°C, and 40°C) were verified by inserting a thermometer in the cuvette for calibration. Equilibrium data were recorded between 0 and 7 M urea, using a fixed CD signal at 220 nm and 25 μM of sample. Data were fitted to a two-state model using Eq. 1 (20).
Theoretical prerequisites and data analysIs
Chemical unfolding
Equilibrium parameters from CD data can be fitted to Eq. 1, yielding the free energy of unfolding and the dependence of denaturation:
| (1) |
Here, CDobs denotes the observed CD data. The fitted parameters are , extrapolated to native conditions, and mN and mU which corresponds to the denaturant dependence of the folded and unfolded states, respectively. Here, meq is the denaturant dependence of in the direction of unfolding, and CDN and CDU are the CD amplitudes of the native and unfolded states, respectively. The denaturant concentration is denoted [D].
Thermal unfolding
The midpoint of thermal unfolding (Tm) can be obtained by fitting the data to the equations (21)
| (2a) |
where
| (2b) |
Here CDnorm(T) is the normalized CD signal; Sf, Su, a, and b are the CD signals for folded and unfolded conditions and the slopes for the folded and unfolded baselines, respectively. ΔHm corresponds to the enthalpy value at Tm.
FRET
The rate of energy transfer (ω) between the excited donor (D) and acceptor (A) in its electronic ground state is given by:
| (3) |
Here, , , , and R denote the averaged square of the angular part of the dipole-dipole interaction, the radiative lifetime of the donor, the Förster radius, and the distance between the center of masses of the donor and acceptor, respectively. The isotropic average of 2/3 is used for (22,23) and the radiative lifetime of tryptophan is 18.9 ns (24). The assumption of is supported by the low steady-state anisotropy of BODIPY in protein mutants . This is also expected, since two electronic transitions in Trp are overlapping at the excitation wavelength (25), which causes a wide angular spread in the orientation distribution of Trp dipoles (22,23). Furthermore, these dipoles interact in the energy transfer process via the S2 state of BODIPY and end up in the S1 state (26). The latter step also implies depolarization. Moreover, local internal reorientations of Trp (fast) and BODIPY (slower) contribute to randomizing . Thus, between the events of excitation of Trp and the emission of BODIPY, several depolarizing steps contribute to the randomization of the interacting transition dipoles. For each sample at each condition, the Förster radius was calculated according to
| (4a) |
where the overlap integral is defined by
| (4b) |
In Eqs. 4a and 4b, n, NA, FD(λ), and εA(λ) stand for the refractive index of the medium, Avogadro’s constant, the normalized fluorescence spectrum of the donor, and the extinction coefficient spectrum of the acceptor, respectively. For each sample, the refractive index was measured with a refractometer at 589 nm and the spectral overlap (J) was determined. The analysis of the time-resolved fluorescence of the acceptor has previously been described (27). The donor-acceptor distances were modeled both as a single distance and as a Gaussian distribution. The Gaussian distance distribution analysis yields an average D-A distance and a variance according to the function
| (5a) |
The variance (σ) is related to the FWHM of the distribution by
| (5b) |
The experimental data were analyzed by using a common deconvolution procedure of nonlinear least-squares analyses, which is based on the Levenberg-Marquardt algorithm (28). The statistical best fit to the data were judged by the χ2 and Durbin-Watson (D.W.) parameters.
Fluorescence quantum yield
The fluorescence quantum yield is the ratio of the number of photons emitted to the number absorbed. To determine the quantum yield of a fluorophore, a standard of known quantum yield is generally used. The quantum yield of the unknown compound is calculated using
| (6) |
Here F, A, and n stand for the integrated fluorescence intensity, the absorbance at the excitation wavelength, and the refractive index of the medium. The subscript ref refers to the reference fluorophore of known quantum yield. The reference compound used was fluorescein in 0.1 M NaOH with (29).
Results and Discussion
By means of time-resolved FRET, changes in intramolecular distances due to macromolecular crowding in variants of two homologous S16 proteins were investigated. Selected amino acids were replaced by cysteines and labeled with BODIPY. In combination with the natural Trp58, different Trp-BODIPY distances were determined. The remaining Trp residue (W39 in S16Meso and W74 in S16Thermo) was replaced by phenylalanine. For S16Thermo, two mutants, W74C and W74F/F10C, were prepared for probing the distances Trp58-Cys74 and Trp58-Cys10. For S16Meso, three mutants, W39F/Q10C, W39F/K74C and W39F/Y81C, were prepared for probing the distances Trp58-Cys10, Trp58-Cys74, and Trp58-Cys81 (the donor and the three acceptor sites are indicated in Fig. 1). Taken together, five S16 variants were created and used in combination with the wild-type protein in our studies.
Intramolecular distances from FRET
The intramolecular distances between Trp58 and five different positions labeled with BODIPY (three in S16Meso and two in S16Thermo) were determined using time-resolved BODIPY fluorescence upon excitation of the donor (Trp). Distances were probed for the proteins in the presence of increasing amounts of dextran 20. Two models were used for data analysis: one that assumes a fixed distance (R) between the donor and acceptor, and one that assumes a distribution with an average distance and a variance (σ) (Table 1). With the exception of W39F/Y81C S16Meso, a single distance is compatible with the experimental data for all mutants. For W39F/Y81C S16Meso, only a distance distribution provides acceptable statistics.
Table 1.
Distances between Trp and BODIPY as determined by FRET
| S16Thermo | [Dextran] (mg/mL) | R (Å) | (Å) | σ (Å) | χ2 | D.W. |
|---|---|---|---|---|---|---|
| W74F/F10C | 0 | 21.3 | 20.9 | 1.6 | 1.04 | 1.97 |
| 50 | 21.6 | 21.1 | 1.9 | 1.11 | 1.93 | |
| 100 | 21.8 | 21.3 | 2.1 | 1.09 | 2.00 | |
| 200 | 21.8 | 21.0 | 2.3 | 1.10 | 2.03 | |
| 300 | 22.1 | 21.0 | 2.8 | 1.07 | 1.94 | |
| W74C |
0 | 22.4 | 22.0 | 1.8 | 1.12 | 1.82 |
| 50 | 22.1 | 21.8 | 1.4 | 1.11 | 1.87 | |
| 100 | 22.1 | 21.9 | 1.3 | 1.09 | 1.93 | |
| 200 | 22.1 | 22.0 | 1.2 | 1.12 | 1.98 | |
| 300 | 22.3 | 22.1 | 1.6 | 1.13 | 1.95 | |
| S16Meso | ||||||
| W39F/Q10C | 0 | 20.6 | 20.6 | 2.3 | 1.15 | 1.80 |
| 50 | 20.7 | 19.5 | 3.3 | 1.20 | 1.84 | |
| 100 | 20.7 | 18.9 | 3.4 | 1.23 | 1.86 | |
| 200 | 20.1 | 20.5 | 3.0 | 1.12 | 1.81 | |
| 300 | 21.8 | 21.5 | 3.4 | 1.17 | 1.88 | |
| W39F/K74C | 0 | 17.3 | 17.4 | 1.3 | 1.16 | 1.86 |
| 50 | 17.3 | 17.0 | 0.8 | 1.20 | 1.89 | |
| 100 | 16.3 | 16.1 | 0.9 | 1.08 | 1.91 | |
| 200 | 15.6 | 15.9 | 0.7 | 1.15 | 1.90 | |
| 300 | 13.6 | 13.9 | 0.5 | 1.12 | 1.94 | |
| W39F/Y81C | 0 | — | 22.1 | 8.9 | 1.17 | 1.87 |
| 50 | — | 21.5 | 8.9 | 1.14 | 1.90 | |
| 100 | — | 21.2 | 9.2 | 1.14 | 1.91 | |
| 200 | — | 19.9 | 5.9 | 1.19 | 1.87 | |
| 300 | — | 20.0 | 6.7 | 1.03 | 1.96 | |
Values represent average distances between the centers of mass of Trp and BODIPY. The distances were analyzed assuming one fixed distance (R) or as a Gaussian distribution of distances with an average distance and a variance (σ). The qualities of the fittings were judged by the statistical χ2 and Durbin-Watson (D.W.) parameters. The errors in distances were all within ±0.6 Å.
An inspection of the data for the two S16Thermo mutants (see Table 1) reveals that the obtained distances are invariant to the concentration of dextran 20 (Fig. 2). In contrast, with increasing crowding agent concentration up to 100 mg/mL, the average D-A (donor-acceptor) distances of the mesophilic variants decrease. Above 100 mg/mL of dextran 20, the change in distances for the three D-A pairs becomes anisotropic. In W39F/Q10C S16Meso, the D-A distance at 200 mg/mL dextran has increased to the initial value, and at even higher dextran concentrations, the distance continues to increase. For W39F/K74C and W39F/Y81C S16Meso, the average D-A distance continuously decreases with higher dextran concentrations. Interestingly, the probed distance in W39F/Y81C S16Meso exhibits a large variance, which indicates that the BODIPY group is initially located in a flexible region of the protein. The variance decreases with increasing dextran concentration, which suggests a reduced flexibility of this region under crowded conditions. On the other hand, the distance variance of W39F/K74C S16Meso is narrow for all data points, which is consistent with a well-defined location of BODIPY within the structure, regardless of crowding. The difference in behavior between S16Thermo and S16Meso suggests that the structural influence of macromolecular crowding on the folded state depends on intrinsic protein stability. S16Meso (a mesophilic protein) is less stable than S16Thermo (a hyperthermophilic protein) at room temperature (19). The anisotropic distance changes for the three BODIPY-labeled S16Meso variants as a function of crowding-agent levels imply that at high concentrations, the crowding-induced effects on the folded protein become unique depending on the positions probed. We speculate that the unsymmetrical distance changes found among the measured FRET pairs at high crowding levels may be a way for the protein to accommodate a more spherical overall shape. The shape of S16 in the crystal structure is somewhat elongated, and upon twisting toward a more spherical structure (which would have the least excluded volume), some residues may come closer to each other, whereas others move further apart.
Figure 2.
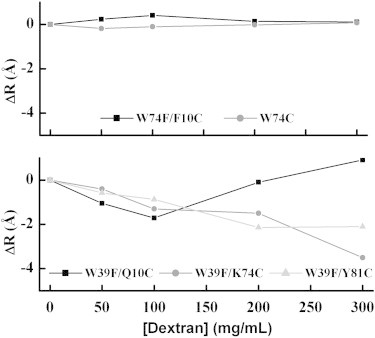
Change in Trp-BODIPY distances (ΔR = Rdextran − RBuffer) for the different mutants of S16Thermo (upper) and S16Meso (lower) at varying concentrations of dextran.
Thermal and chemical stability by CD
Far-UV CD spectra were recorded for the three S16Meso variants at different dextran concentrations (Fig. S1 in the Supporting Material). For these proteins, no significant changes are observed in the secondary structure content upon addition of crowding agent. Thus, changes in FRET distances (of 1–4 Å) in S16Meso due to macromolecular crowding cause no perturbations of the protein secondary structure detectable by far-UV CD. The midpoint of thermal unfolding was determined using far-UV CD changes as a function of temperature for the three S16Meso mutants and wild-type S16Meso in the presence of different fixed dextran 20 concentrations. Due to the high resistance toward thermally induced unfolding of the S16Thermo variants (19), the Tm for this protein in buffer without or with dextran could not be determined. For all S16Meso variants, thermally induced unfolding is reversible up to 200 mg/mL dextran, as judged from the recovery of the initial CD signal at 220 nm upon cooling. At 300 mg/mL dextran, only the wild-type and W39F/Q10C S16Meso showed a reversible thermal unfolding reaction; transitions for the others were irreversible and therefore not analyzed. The midpoints of the reversible thermal unfolding reactions of the S16Meso variants are summarized in Table 2. As expected for an excluded-volume effect, the Tm values increase with increasing dextran 20 concentration. Complete thermal melting curves are shown in Fig S2.
Table 2.
Thermally induced unfolding midpoints for the wild-type and three S16Meso mutants
| [Dextran] |
Thermal midpoint (°C) |
|||
|---|---|---|---|---|
| (mg/mL) | Wild-type | W39F/Q10C | W39F/K74C | W39F/Y81C |
| 0 | 66.7 ± 0.4 | 68.6 ± 0.4 | 57.6 ± 0.7 | 54.7 ± 0.5 |
| 100 | 66.8 ± 0.4 | 69.8 ± 0.4 | 59.9 ± 0.7 | 55.8 ± 0.5 |
| 200 | 68.2 ± 0.5 | 72.1 ± 0.4 | 62.1 ± 1.2 | 58.1 ± 0.5 |
| 300 | 72.1 ± 0.8 | 72.9 ± 0.7 | — | — |
Unfolding was probed by CD changes at 220 nm, and thermal midpoints were evaluated from the best fit of experimental data to Eq. 2. Reported errors are standard curve-fitting errors.
Chemical stability of wild-type S16Meso was probed by far-UV CD at three different fixed temperatures with and without 200 mg/mL dextran 20, using urea as the chemical denaturant. Chemical denaturation was reversible at all conditions. Analysis of the urea-induced unfolding curves reveals that S16Meso is stabilized by dextran at all temperatures by ∼2 kJ/mol (Table S1). In our earlier study, we found that S16Thermo was also stabilized toward chemical denaturation at 20°C by the addition of 200 mg/mL dextran 20 (15). A lack of temperature dependence in the crowding effect on S16Meso equilibrium unfolding constants is thought to be a hallmark of an entropic effect (11). If there were an enthalpic component in the protein-crowder interactions, it would reveal itself as a temperature dependence in since (9,10,30). As will be shown below, the fact that the dominant force in the presence of crowding agent is an excluded-volume effect does not prohibit the presence of dynamic, local interactions between crowder and protein.
Quantum yield measurements
BODIPY-dextran 20
Fluorescence quantum yields and average lifetimes of BODIPY were determined for all BODIPY-labeled mutants at varying concentrations of dextran 20 (Table S2 and Fig. S3). We find that the BODIPY group in all mutants shows a minor decrease in average lifetime as the dextran concentration increases. This is compatible with the presence of some dynamic quenching of BODIPY caused by the increased concentration of dextran. The BODIPY quantum yields change differently depending on the variant, but a small decrease in quantum yield with increasing dextran is apparent in most cases (Table S2). The quantum yields of W39F/Q10C S16Meso and W39F/K74C S16Meso are high and independent of dextran concentration. However, W74F/F10C S16Thermo and W39F/Y81C S16Meso exhibit substantial quenching, independent of the crowding level, which is most likely due to local intramolecular quenching by nearby amino acids. Tryptophan and tyrosine are known quenchers of BODIPY (26,31). The BODIPY group in W74F/F10C S16Thermo is located at the cysteine in position 10, which is part of β-strand 1 (see Fig. 1). In close proximity to β1 is β2, which contains a tyrosine that may be able to quench the BODIPY at position 10. The amino acid sequence in S16Meso is not identical to that in S16Thermo, which might explain why W39F/Q10C does not exhibit the same quenching effect as W74F/F10C. The BODIPY group in W39F/Y81C S16Meso is located in α-helix 3, which is close to α-helix 1. Helix 1 contains a tryptophan and a tyrosine, which could explain the observed strong quenching. Under dilute conditions, W74C S16Thermo is not quenched, but the quantum yield decreases with increasing dextran concentration. The BODIPY group is attached to position 74, at the end of α-helix 2. A potential quencher is the Tyr51 residue, located nearby at the interface between β4 and α1. One may speculate that in the presence of dextran 20, the conformational dynamics of the folded protein decrease, whereby higher chances of quenching of this labeling site are possible.
BODIPY-tyrosine-labeled dextran
Although we find evidence for excluded-volume forces in our mixtures, the quantum yield data indicate additional effects due to the presence of dextran 20 that depend on the attachment position of BODIPY. To gain more information about local interactions between dextran and the protein, we performed fluorescence experiments using dextran labeled with tyrosine. We speculated that if the observed quenching found for some protein-dextran mixtures is due to interactions between protein segments and the crowder, the quenching should be enhanced by Tyr-labeled dextran. The concentration of 50 mg/mL of Tyr-dextran corresponds to a Tyr-dextran/protein ratio of 1000, which under isotropic conditions yields significant quenching of the BODIPY fluorescence due to collisional quenching. Thus, some quenching in these experiments is expected without implications of specific interactions.
The quenching of three BODIPY-labeled S16 mutants was determined by steady-state and time-resolved fluorescence experiments in the presence of increasing concentrations of Tyr-dextran. The two S16Meso variants W39F/Q10C and W39F/K74C, and S16Thermo W74C were selected, first because W74F/F10C and W39F/Y81C exhibit rather low quantum yields independent of crowding, and second because the quenching of W74C depends on the crowding concentration. Interestingly, the three variants respond differently to the added Tyr-dextran (Fig. 3 and Table S3).
Figure 3.
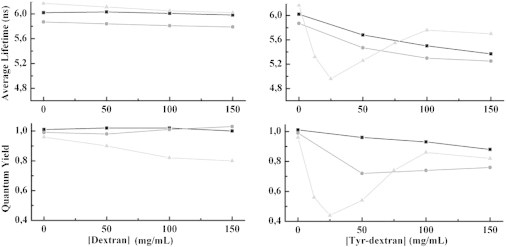
Average fluorescence lifetime (upper) and quantum yield (lower) of BODIPY at varying concentrations of dextran (left) and Tyr-dextran (right) for the three S16 variants W39F/K74C (squares), W39F/Q10C (circles) and W74C (triangles).
Depending on how the Tyr-dextran and protein interacts, different outcomes are expected.
-
1.
In the absence of specific protein-dextran interactions, the observed BODIPY quenching occurs in accordance with dynamic collisions, i.e., and decrease to the same extent with Tyr-dextran concentration.
-
2.
In the presence of attractive interactions between the Tyr-dextran and a protein region that includes the BODIPY site, the quenching pattern of and differs from pure collisional quenching. Since there are on average 4.5 Tyr residues per Tyr-dextran, the probability of finding a tyrosine in proximity to the BODIPY is random (Fig. 4). If a tyrosine group from the dextran is located close to BODIPY, the quenching will be faster compared to that for pure collisional quenching. For local and rapid quenching, as compared to the rate of unquenched BODIPY, the observed fluorescence may appear as static quenching. On the other hand, if no tyrosine is present at the interaction site of the dextran, the BODIPY group is shielded from other quenchers, and hence exhibits the fluorescence lifetime of unquenched BODIPY (see Fig. 4).
Figure 4.
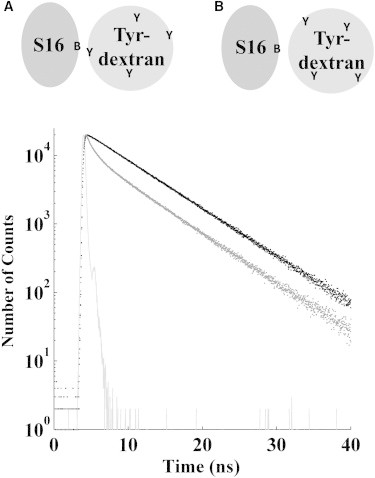
(Upper) Schematic illustration of the possible BODIPY (B)-to-Tyr (Y) distances upon interactions between S16 and Tyr-dextran. (A) BODIPY and Tyr are positioned close enough for quenching to occur. (B) No Tyr is close enough for BODIPY to be quenched. Here dextran shields the BODIPY-group from other quenchers. Note that the illustrations of the two macromolecules do not reflect their true size and shape. (Lower) Time-resolved fluorescence decay of BODIPY in S16Meso W74C in the presence of 0 (black circles) and 25 (gray circles) mg/mL Tyr-dextran is shown. Also shown is the instrumental response function (gray line). The biphasic decay is clearly seen at 25 mg/mL, with one fast component and one longer component. At longer times (>15 ns), the two decays are parallel, strongly indicating a fraction of unquenched fluorophores. The mutant W39F/Q10C displays similar biphasic decays.
For W39F/K74C S16Meso, the quantum yields and fluorescence lifetimes decrease as a function of the concentration of Tyr-dextran. This indicates collisional quenching (case 1 above), which is also supported by a Stern-Volmer plot (Fig. S4).
The BODIPY lifetime of W39F/Q10C S16Meso decreases with increasing concentration of Tyr-dextran in a manner similar to that observed for W39F/K74C S16 Meso. However, the quantum yield decreases to 0.7 at low Tyr-dextran concentration and stays approximately constant at higher Tyr-dextran concentrations. This is consistent with simultaneous dynamic and static quenching of BODIPY by the Tyr-dextran. Furthermore, the BODIPY fluorescence decay becomes biphasic (Fig. 4), with one fast (0.4 ns) and one slow component (5.8 ns). This behavior is atypical for collisional quenching in solution, and it is compatible with the presence of two BODIPY populations (32,33). One population is subjected to fast dynamical quenching (case 2 above), whereas the other population is unaffected by the quencher. The observation can be explained by a local dynamic interaction of a protein segment near the BODIPY and a Tyr from the Tyr-dextran. The Stern-Volmer plot (Fig. S4) is in agreement with this interpretation (25).
The third variant, W74C S16Thermo, shows an unusual behavior: the lifetime and quantum yield drop significantly at low Tyr-dextran concentrations, whereas both values increase at higher Tyr-dextran concentrations (Fig. 3). Also, this mutant exhibits a biphasic BODIPY fluorescence decay with a fast and a slow component (Fig. 4). We find that the fraction of the fast lifetime component decreases with increasing Tyr-dextran concentration. The high degree of quenching at low Tyr-dextran concentrations indicates that dextran is attracted to the region of the protein close to position 74 (and BODIPY) and dynamic complexes can form. The increase in quantum yield and lifetime at higher concentrations of Tyr-dextran for W74C S16Thermo is puzzling but indicates that protein-crowder interactions decrease at higher dextran concentration. One possibility is that the molecular shape of dextran changes as the concentration is increased, whereby the spatial distribution of tyrosines changes. Another possibility is that at higher dextran levels, local interactions with the crowders on short timescales are not favored, since the protein becomes more stable (due to the excluded-volume effect).
The slow BODIPY lifetime component (10–30 ns), τlong, for W74C S16Thermo, is found to be independent of Tyr-dextran concentration (Table S3). This supports the idea that one population of BODIPY-labeled protein is inaccessible to quenching by Tyr-dextran.
For W74C S16Thermo, the influence of dynamics was tested by varying the temperature at two different concentrations of Tyr-dextran, as well as in pure buffer (Table 3). The quantum yield and lifetime decrease with increasing temperature at 100 mg/mL Tyr-dextran, as might be expected. On the other hand, at 12.5 mg/mL Tyr-dextran, the quantum yield and lifetime increase with temperature. This is compatible with a decreasing fraction of fast component in the fluorescence decay upon increasing temperature. This suggests that the local interaction (resulting in fast quenching) between W74C S16Thermo and Tyr-dextran decreases with increasing temperature. It is important to note that BODIPY itself showed no temperature dependence of its fluorescence parameters in the absence of Tyr-dextran (Table 3).
Table 3.
Quantum yield and fluorescence lifetime of BODIPY at varying temperature
| Temp (°C) | 0 mg/mL |
12.5 mg/mL |
100 mg/mL |
|||
|---|---|---|---|---|---|---|
| <τ> (ns) | <τ> (ns) | <τ> (ns) | ||||
| 10 | 1.01 | 6.2 | 0.56 | 5.4 | 0.89 | 5.7 |
| 20 | 0.98 | 6.1 | 0.55 | 5.3 | 0.84 | 5.7 |
| 30 | 0.99 | 6.1 | 0.66 | 5.6 | 0.86 | 5.6 |
| 40 | 1.02 | 6.1 | 0.59 | 5.7 | 0.76 | 5.5 |
| 50 | 1.00 | 6.0 | 0.65 | 5.7 | 0.78 | 5.4 |
The quantum yield and average fluorescence lifetime (<τ>) for the S16Thermo variant W74C. Measurements were performed in buffer and in the presence of tyrosine-labeled dextran (Tyr-dextran). The errors of the quantum yield measurements are within 10%. The errors of <τ> are within 0.05 ns.
In W74C S16Thermo and W39F/K74C S16Meso, BODIPY is located at the same position, residue 74, which is in a helical region. Despite this, the effects of Tyr-dextran on BODIPY at this position are completely different between the proteins. Inspection of the amino acid sequence of the proteins around residue 74 reveals that the mesophilic S16 has two extra residues and a proline in this region (Fig. 1). Thus, the determined structure of the thermophilic S16 cannot provide information on the structure of the mesophile in this particular region. To investigate structural differences around residue 74, interresidue 1H-15N HSQC-NOESY contacts were compared between the mesophile and thermophile by means of previously recorded data (19). The data suggest that α2 is slightly different in the mesophile around this site, as indicated by stronger HN-HN contacts for residues 74–76 in the mesophilic protein compared to the corresponding residues in the thermophilic protein (Fig. S5). A difference in local structure is compatible with the finding that dextran interacts in the case of the thermophilic protein, but not in mesophilic S16.
The quantum yields and lifetimes were also measured for W74C-BODIPY in the presence of 1.5 mM free tyrosine (i.e., a limit of solubility in 30 mM NaAc and 50 mM NaCl buffer at pH 5.5), and in the presence and absence of 50 mg/mL unlabeled dextran (Table S3). This concentration of tyrosine (1.5 mM) is comparable to 25 mg/mL Tyr-dextran, assuming that one tyrosine at a time per Tyr-dextran can act as quencher. The quantum yields for BODIPY here are 0.98 and 0.93 in the absence and presence, respectively, of 50 mg/mL dextran. Taken together, this control experiment strongly implies that the differential quenching effects observed for some of the protein variants in the presence of Tyr-dextran cannot simply be ascribed to collisional quenching due to the high tyrosine concentration.
Although the major effects of dextran 20 on the S16 variants appear to be excluded volume effects, our investigation using the Tyr-dextran reveals that the molecular details of the protein-crowder interactions are more subtle. For some S16 variants, we find that local dynamic interactions between the protein and Tyr-dextran take place. This does not imply that there are enthalpic interactions between the protein and the crowder. In fact, our temperature study of the crowding-induced stabilization effect on wild-type S16Meso indicates a lack of enthalpic contribution. Moreover, the observed interactions with Tyr-labeled dextran 20 are dependent on the position in the protein where BODIPY is attached and thus on local protein structure/chemistry.
Some recent reports suggest that macromolecular crowding agents may exhibit significant attractive/enthalpic interactions with proteins in addition to excluded-volume effects (9–11). If such attractive interactions are nonspecific, as is proposed to be the general case, these favor protein destabilization, since the unfolded state would offer more opportunities for interactions. Another possibility to consider is nonspecific enthalpic repulsion interactions (such as charge-charge repulsion), which would then, like the excluded-volume effect, stabilize the folded state of the protein. However, in our results, there is no support of either attractive nonspecific interactions (since then the proteins should be destabilized) or repulsive nonspecific enthalpic interactions (since then there should be a temperature dependence for the stabilizing effects) between dextran and protein.
Nonetheless, there are two NMR-based studies with findings similar to ours: Pielak et al. (34) reported that the crowding agent polyvinyl pyrrolidone stabilized the protein CI2 by weakly interacting with its folded state. However, in contrast to those of our study, the interactions in that study were proposed to be nonspecific, although they targeted the folded state. Moreover, Ficoll 70 was reported to stabilize CI2 by purely enthalpic effects, which was explained by strong binding or preferential hydration in the folded state (35).This work clearly emphasizes the need for careful interpretation of macromolecular crowding studies and consideration of both structural and local interaction effects in addition to excluded-volume forces. Our experiments can only suggest the presence of dominant effects; they cannot rule out weak attractive and/or repulsive nonspecific interactions.
Conclusions
In this study, we analyzed folded-state structural effects in two homologs of the ribosomal protein S16 using direct intramolecular distance measurements in two (S16Thermo) or three (S16Meso) directions. Distances were calculated from time-resolved FRET between an intrinsic Trp and a covalently attached BODIPY-group. Although the distances are not affected by the crowding agent (dextran 20, up to 300 mg/mL) in the thermophilic S16 variants, in the mesophilic variants (up to 100 mg/mL dextran), there is an initial decrease in FRET distances, followed by further decreases in the distances in one direction, but increases in distance in a perpendicular direction (based on the crystal structure) at higher dextran concentrations (>100 mg/mL). This finding indicates that to adapt to the surrounding excluded-volume forces that occur in vivo, anisotropic (in terms of the analyzed residue-pair distances) structural tuning may occur if the protein has marginal stability. Despite the lack of a common trend in the distance changes among the studied residue pairs, we speculate that the alteration induced by crowding in terms of overall protein shape is toward a more compact spherical structure. If the protein instead comes from a thermophile, such as S16Thermo, it will have a more rigid fold at room temperature and structural tuning due to crowding may not be possible. Our findings are in agreement with reports on the marginally stable proteins VlsE and apoflavodoxin, both of which were found to change folded-state secondary structure and/or overall shape in response to macromolecular crowding (6,16), and with reports on the more stable proteins apoazurin and cytochrome c, neither of which was structurally affected by the presence of crowding agents (36,37).
Acknowledgments
Financial support for this work was provided by the Swedish Natural Research Council (P.W.S. and L.J.), the Knut and Alice Wallenberg Foundation (P.W.S.), the Göran Gustafsson Foundation (P.W.S.), and Umeå University (P.W.S.).
Contributor Information
Pernilla Wittung-Stafshede, Email: pernilla.wittung@chem.umu.se.
Lennart B.-Å. Johansson, Email: lennart.johansson@chem.umu.se.
Supporting Material
References
- 1.Laurent T.C. Interaction between polysaccharides and other macromolecules. 5. The solubility of proteins in the presence of dextran. Biochem. J. 1963;89:253–257. doi: 10.1042/bj0890253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimmerman S.B., Trach S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 3.Minton A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 4.Minton A.P. Implications of macromolecular crowding for protein assembly. Curr. Opin. Struct. Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 5.Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 6.Homouz D., Perham M., Wittung-Stafshede P. Crowded, cell-like environment induces shape changes in aspherical protein. Proc. Natl. Acad. Sci. USA. 2008;105:11754–11759. doi: 10.1073/pnas.0803672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H.X., Rivas G.N., Minton A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christiansen A., Wang Q., Wittung-Stafshede P. Effects of macromolecular crowding agents on protein folding in vitro and in silico. Biophys. Rev. 2013;5:137–145. doi: 10.1007/s12551-013-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao M., Li H.T., Liang Y. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys. J. 2010;99:914–923. doi: 10.1016/j.bpj.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Sarkar M., Pielak G.J. Macromolecular crowding and protein stability. J. Am. Chem. Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- 11.Fodeke A.A., Minton A.P. Quantitative characterization of temperature-independent and temperature-dependent protein-protein interactions in highly nonideal solutions. J. Phys. Chem. B. 2011;115:11261–11268. doi: 10.1021/jp2049266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Coeur C., Teixeira J., Longeville S. Compression of random coils due to macromolecular crowding: scaling effects. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;81:061914. doi: 10.1103/PhysRevE.81.061914. [DOI] [PubMed] [Google Scholar]
- 13.Johansen D., Jeffries C.M., Goldenberg D.P. Effects of macromolecular crowding on an intrinsically disordered protein characterized by small-angle neutron scattering with contrast matching. Biophys. J. 2011;100:1120–1128. doi: 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong J., Gierasch L.M. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J. Am. Chem. Soc. 2010;132:10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikaelsson T., Adén J., Wittung-Stafshede P. Direct observation of protein unfolded state compaction in the presence of macromolecular crowding. Biophys. J. 2013;104:694–704. doi: 10.1016/j.bpj.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stagg L., Zhang S.Q., Wittung-Stafshede P. Molecular crowding enhances native structure and stability of α/β protein flavodoxin. Proc. Natl. Acad. Sci. USA. 2007;104:18976–18981. doi: 10.1073/pnas.0705127104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar A., Samiotakis A., Cheung M.S. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akabayov S.R., Akabayov B., Wagner G. Molecular crowding enhanced ATPase activity of the RNA helicase eIF4A correlates with compaction of its quaternary structure and association with eIF4G. J. Am. Chem. Soc. 2013;135:10040–10047. doi: 10.1021/ja404404h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallgren M., Adén J., Wolf-Watz M. Extreme temperature tolerance of a hyperthermophilic protein coupled to residual structure in the unfolded state. J. Mol. Biol. 2008;379:845–858. doi: 10.1016/j.jmb.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Santoro M.M., Bolen D.W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 21.Consalvi V., Chiaraluce R., Ladenstein R. Thermal unfolding and conformational stability of the recombinant domain II of glutamate dehydrogenase from the hyperthermophile Thermotoga maritima. Protein Eng. 2000;13:501–507. doi: 10.1093/protein/13.7.501. [DOI] [PubMed] [Google Scholar]
- 22.Valeur B., Weber G. Resolution of the fluorescence excitation spectrum of indole into the 1La and 1Lb excitation bands. Photochem. Photobiol. 1977;25:441–444. doi: 10.1111/j.1751-1097.1977.tb09168.x. [DOI] [PubMed] [Google Scholar]
- 23.Haas E., Katchalski-Katzir E., Steinberg I.Z. Effect of the orientation of donor and acceptor on the probability of energy transfer involving electronic transitions of mixed polarization. Biochemistry. 1978;17:5064–5070. doi: 10.1021/bi00616a032. [DOI] [PubMed] [Google Scholar]
- 24.Privat J.P., Wahl P., Auchet J.C. Rates of deactivation processes of indole derivatives in water-organic solvent mixtures—application to tryptophyl fluorescence of proteins. Biophys. Chem. 1979;9:223–233. doi: 10.1016/0301-4622(79)85005-x. [DOI] [PubMed] [Google Scholar]
- 25.Lakowicz J.R. 3rd ed. Springer; New York: 2006. Principles of Fluorescence Spectroscopy. [Google Scholar]
- 26.Karolin J. Fluorescence and absorption spectroscopic properties of dipyrrometheneboron difluoride (BODIPY) derivatives in liquids, lipid membranes, and proteins. J. Am. Chem. Soc. 1994;116:7801–7806. [Google Scholar]
- 27.Olofsson M., Kalinin S., Johansson L.B. Tryptophan-BODIPY: a versatile donor-acceptor pair for probing generic changes of intraprotein distances. Phys. Chem. Chem. Phys. 2006;8:3130–3140. doi: 10.1039/b601313a. [DOI] [PubMed] [Google Scholar]
- 28.Levenberg K. A method for the solution of certain non-linear problems in least squares. Q. J. Appl. Math. 1944;2:164–168. [Google Scholar]
- 29.Weber G., Teale F.W.J. Fluorescence excitation spectrum of organic compounds in solution.Part 1—Systems with quantum yield independent ot the exciting wavelength. Trans. Faraday Soc. 1958;54:640–648. [Google Scholar]
- 30.Zhou H.X. Polymer crowders and protein crowders act similarly on protein folding stability. FEBS Lett. 2013;587:394–397. doi: 10.1016/j.febslet.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marmé N., Knemeyer J.P., Wolfrum J. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconjug. Chem. 2003;14:1133–1139. doi: 10.1021/bc0341324. [DOI] [PubMed] [Google Scholar]
- 32.Tachiya M. Kinetics of quenching of luminescent probes in micellar systems. II. J. Chem. Phys. 1982;76:340–348. [Google Scholar]
- 33.Ranganathan R., Vautier-Giongo C., Bales B.L. Toward a hydrodynamic description of bimolecular collisions in micelles. An experimental test of the effect of the nature of the quencher on the fluorescence quenching of pyrene in SDS micelles and in bulk liquids. J. Phys. Chem. B. 2003;107:10312–10318. [Google Scholar]
- 34.Miklos A.C., Li C., Pielak G.J. Volume exclusion and soft interaction effects on protein stability under crowded conditions. Biochemistry. 2010;49:6984–6991. doi: 10.1021/bi100727y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benton L.A., Smith A.E., Pielak G.J. Unexpected effects of macromolecular crowding on protein stability. Biochemistry. 2012;51:9773–9775. doi: 10.1021/bi300909q. [DOI] [PubMed] [Google Scholar]
- 36.Christiansen A., Wittung-Stafshede P. Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin in vitro. Biophys. J. 2013;105:1689–1699. doi: 10.1016/j.bpj.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christiansen A., Wang Q., Wittung-Stafshede P. Factors defining effects of macromolecular crowding on protein stability: an in vitro/in silico case study using cytochrome c. Biochemistry. 2010;49:6519–6530. doi: 10.1021/bi100578x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


