Abstract
Studies on biflagellated algae Chlamydomonas reinhardtii mutants have resulted in significant contributions to our understanding of the functions of cilia/flagella components. However, visual inspection conducted under a microscope to screen and classify Chlamydomonas swimming requires considerable time, effort, and experience. In addition, it is likely that identification of mutants by this screening is biased toward individual cells with severe swimming defects, and mutants that swim slightly more slowly than wild-type cells may be missed by these screening methods. To systematically screen Chlamydomonas swimming mutants, we have here developed the cell-locating-with-nanoscale-accuracy (CLONA) method to identify the cell position to within 10-nm precision through the analysis of high-speed video images. Instead of analyzing the shape of the flagella, which is not always visible in images, we determine the position of Chlamydomonas cell bodies by determining the cross-correlation between a reference image and the image of the cell. From these positions, various parameters related to swimming, such as velocity and beat frequency, can be accurately estimated for each beat cycle. In the examination of wild-type and seven dynein arm mutants of Chlamydomonas, we found characteristic clustering on scatter plots of beat frequency versus swimming velocity. Using the CLONA method, we have screened 38 Chlamydomonas strains and detected believed-novel motility-deficient mutants that would be missed by visual screening. This CLONA method can automate the screening for mutants of Chlamydomonas and contribute to the elucidation of the functions of motility-associated proteins.
Introduction
Cilia and flagella are cellular organelles that beat to move the surrounding fluid or to propel the cell itself. These movements play a fundamental role in various physiological functions, such as embryonic development (1), clearing mucus from airways (2), the immune system (3), and the reproductive system (4). Malfunctions of cilia/flagella cause various ciliopathies (5): hydrocephalus (6), juvenile myoclonic epilepsy (7), retinal pigmentosa (8), situs inversus (a left-right asymmetry defect) (1), and polycystic kidney disease (9). Therefore, understanding the mechanisms of cilia and flagella function are important for advancing our knowledge in biology as well as making advances in medicine.
At the core of the cilia and flagella is a cytoskeletal structure called an axoneme, which consists of nine doublet microtubules cross-bridged with dynein motors and a central pair of microtubules (9+2 structure). The bending of the axoneme is driven by dynein motors, which convert chemical energy derived from ATP hydrolysis to sliding movements between doublet microtubules. Despite its compact appearance, the axoneme is a highly complex structure composed of hundreds of proteins (10–12). Despite numerous studies that have been published on these organelles, functions of many of the flagella components remain unknown.
The eukaryotic unicellular algae Chlamydomonas reinhardtii has been one of the most widely used model organisms to elucidate the function of flagella components for several reasons:
-
1.
Flagella mutants can be easily screened by visual selection of slow swimming cells (13),
-
2.
A wide variety of genetic tools are available, and
-
3.
Biochemical and structural studies can be performed on the flagella.
However, identifying new flagella mutants is becoming difficult, partly because the genes involved in slow swimming mutants are saturated. Therefore, new methods for screening more subtle changes in swimming patterns are required.
In this study, we developed a method that can localize the position of each cell to within 10-nm precision from high-speed video images (1200 fps). The position of the cell in each frame was determined by the cross-correlation of the cell image in each frame and the reference image. By using this cell-locating-with-nanoscale-accuracy (CLONA) method, we obtained detailed information on the parameters related to swimming for wild-type Chlamydomonas, as well as for seven dynein mutants: four mutants lacking parts of the outer dynein arm, and three mutants lacking parts of the inner dynein arm. To show the effectiveness of the CLONA method in screening for motility-deficient mutants, we analyzed 38 Chlamydomonas strains and detected believed-novel mutants that showed subtle deficiency in the motility. These results demonstrate that precise analysis of seemingly simple movies of moving objects, such as flagella-driven cells, provides valuable information about the objects.
Materials and Methods
Chlamydomonas mutants
Chlamydomonas flagella mutants oda1, oda11, suppf1, suppf2, ida1, ida5, and ida9 were kindly provided by Dr. R. Kamiya (The University of Tokyo and Gakushuin University). The general and specific locations of the mutations and the affected genes in each mutant are listed in Table 1. Each Chlamydomonas strain was cultured in 250 mL Tris-acetate-phosphate medium with aeration at 25°C (14). To reduce the variations caused by different phases of the cell cycles, the circadian phases of cells were synchronized in a light-controlled room under a 12-h light: 12-h dark cycle and all the observations were carried out between 8 and 10 a.m.
Table 1.
Chlamydomonas flagella mutants used in this study
| Type | Strain | Affected components | References |
|---|---|---|---|
| Outer dynein arm mutants | oda1 | Outer arm, ODA-DC (deleted) | (27–29) |
| oda11 | αHC+LC5+LIS1 (deleted) | (30,31) | |
| suppf1 | βHC (truncated) | (32,33) | |
| suppf2 | γHC (truncated) | (32,34) | |
| Inner dynein arm mutants | ida1 | f (deleted) | (35) |
| ida5 | a, c, d, e (deleted) | (36,37) | |
| ida9 | c (deleted) | (38) |
The 38 Chlamydomonas strains used for the screening trial were prepared in the previous study (15). Each strain was cultured in a six-well plate using a shaking incubator at 25°C, under a 12-h light/12-h dark cycle. All videos in this study were recorded at 25°C.
Video microscopy
High-speed cameras, EX-F1 (Casio, Tokyo, Japan) or EoSens MC1362 (Mikrotron, Unterschleissheim, Germany), were connected to a dark-field light microscope (BX51; Olympus, Tokyo, Japan) equipped with a 20× objective lens (UPLFLN 20×; Olympus, Tokyo, Japan) and a mercury lamp (BH2-RFL-T3; Olympus). For each video, a 30-μL quantity from a cell suspension was pipetted in an chamber closed on either side by a glass slide (S1214; Matsunami, Osaka, Japan) and a 18 × 18 mm cover glass (C218181; Matsunami), and sealed with Vaseline (Unilever, London, UK). The distance between the glass slide and cover glass was estimated to be ∼180 μm (30 μL volume/154 mm2 area). To avoid overlaps between cell images and cell-cell collisions, the concentration of the cells was adjusted to <105 cells/mL. Videos were recorded at 1200 fps with 336 × 96 pixel field size or at 600 fps with 432 × 192 pixel field size. The pixel size is 0.56 μm at the specimen. The recorded videos were transferred to an iMac computer (Apple, Cupertino, CA), and cells were tracked using a multiple-particle tracker plug-in (MTrack2) for the software ImageJ (16). The cells in each frame was detected, and then tracked by determining which cells in the successive frames were the closest. The location for each cell was roughly calculated using the centroid of the binary image. Further analysis was done using custom Ruby-Helix scripts (17). The software R (18) was used for plotting the results and statistical analysis. Statistical significances between different strains (see Fig. 4, Fig. 5 C, and those mutants categorized into the third group) were tested using the F test of Wilks’s lambda.
Figure 4.
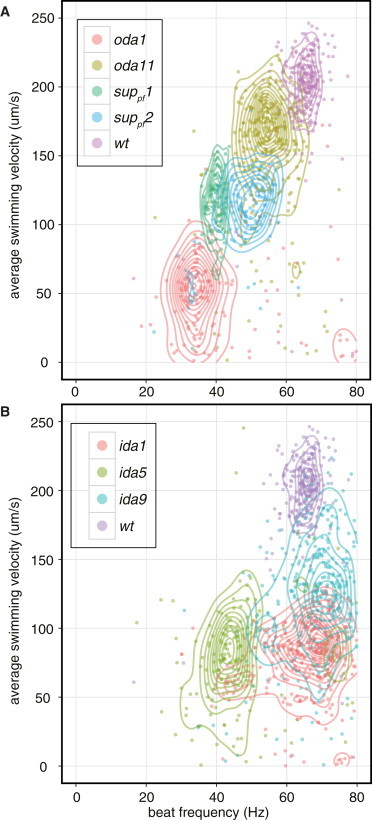
Phenotyping Chlamydomonas mutants based on the CLONA method. Scatter plots of beat frequency f versus average swimming velocity of the beat cycles, generated using the software R (18). Each dot represents one beat cycle, and mutant and wild-type strains are shown with distinct colors. Descriptions of the flagella mutants used here are shown in Table 1. The contour lines were generated to aid with interpreting the distribution of the dots using STAT_DENSITY2D of the GGPLOT2 software package (24), which estimates two-dimensional kernel densities. The difference between the strains are statistically significant (p < 0.001). (A) Scatter plot of beat cycles of four outer dynein arm mutants (oda1, oda11, suppf1, and suppf2) and wild-type. The beat cycles of wild-type and mutants formed a cluster around each characteristic area in the scatter plot. (B) Scatter plot of the beat cycles of three inner dynein arm mutants (ida1, ida5, and ida9) and the beat cycles of wild-type.
Figure 5.
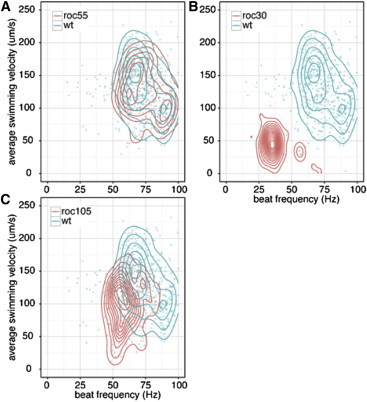
Screening of 38 Chlamydomonas strains for motility-deficient mutants using the CLONA method. Scatter plots of beat frequency f versus average swimming velocity of the beat cycles. (Blue contour lines) The characteristic cluster of the control. The difference between roc30 and wild-type and the difference between roc105 and wild-type are statistically significant (assuming Wilks’s lambda distribution, p < 0.001). (Each dot) One beat cycle. Focus index > 80. (A) A scatter plot of roc55, a representative strain showing no deficiency in motility. (B) A scatter plot of roc30 (ODA4), a representative strain showing severe defects in motility. (C) A scatter plot of roc105, a representative strain of newly detected motility deficient mutants, showing mild deficiency in average swimming speed and beat frequency.
Selecting target cells
High-speed video of swimming Chlamydomonas cells was recorded using a dark-field light microscope and the recorded videos were analyzed to select cells that were in focus and swimming parallel to the glass slide (Fig. 1 A).
Figure 1.
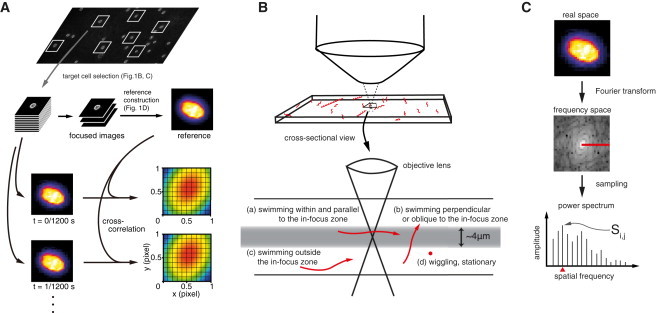
Processes of locating cellular size objects at high spatial and temporal resolution. (A) Overview of the localizing method. The recorded videos were first analyzed to identify target cells. The position of the cell in each frame was determined by cross-correlation between the cell image in each frame and the reference image. (B) Schematic representation of the four types of swimming paths. (Red solid lines) The possible swimming paths of Chlamydomonas cells: (a) swimming within and parallel to the in-focus zone, (b) swimming perpendicular or oblique to the XY plane, (c) swimming outside of the in-focus zone, and (d) wiggling within the trajectory or stationary. Cells that swim parallel to the glass slide and that have an in-focus image were selected for further analysis. (C) The definition of the score Si,j. The image of cell i in frame j was transformed into frequency space, and the values at the horizontal axis were sampled to produce a power spectrum. The amplitude of the frequency component at 32/3 pixel ≈ 6 μm gives Si,j.
The cells were classified based on their swimming path (Fig. 1 B). We selected cells that swam long distance in the XY plane (Fig. 1 B, a and c) and excluded the cells that swam in the perpendicular direction, wiggled, or were stationary (Fig. 1 B, b and d, respectively). Next, cells that were in the in-focus zone, which has a depth of ∼4 μm (Fig. 1 B, a), were identified based on the focus index, F, which reflects high-frequency components in the image (Fig. 1 C).
We introduced the index to measure the quality of focus of each image. The index was calculated as follows: from each frame, a 32 × 32 pixel region centered on the centroid of the cell binary image was selected. The selected image was transformed to a frequency space by fast Fourier transform (19). For each image, a score Si,j (score of frame j of cell i; see Fig. 1 C) was calculated based on the amplitude of high frequency components in the Fourier transform of the image. In-focus images have higher scores, due to the amplitude of the high frequency component being relatively higher than that of poorly focused images (20). This scoring was applied to all frames, and the focus index of cell i or (Fi) was determined as
| (1) |
where t is a given time window and Si,j is the score of frame j of cell i. Here, the theory of Minimax was used to guarantee a certain level of focusing within the given time window t (t = 50 frames in this study). Selecting image sequences with a threshold of F > 90 excludes cells that swam outside the in-focus zone (Fig. 1, B and C).
Procedures for position determination
The position of the cell in each frame was determined by maximizing cross-correlation between the image of cell in each frame and the reference image constructed from the cell itself (Fig. 1 A). The procedure consists of two steps:
-
1.
Construction of the reference image (see Fig. S1 in the Supporting Material), and
-
2.
Taking cross-correlation between the image of the cell in each frame and the reference (lower-half of Fig. 1 A).
To construct a reference, the best 30 in-focus images were selected. Each image was recentered using a two-dimensional Gaussian fit (21), and these recentered images were averaged using subpixel location and rotation angle based on bicubic interpolation (22) to produce the initial reference image (see Fig. S1). Taking this initial reference image as the reference, each cropped image was again translationally and rotationally aligned using cross-correlation (19). The final reference image was obtained by iterating this procedure two-to-three times using the preceding reference image as a reference.
The cross-correlation between the reference image and the cell image in each frame was calculated using a fast-Fourier transform-based procedure, and the position and rotation angle that give the maximum correlation coefficient were obtained (lower-half of Fig. 1 A).
Results
Precision of cell localization
To estimate the precision of localizing the cell, we applied the CLONA method to stationary cells. Wild-type Chlamydomonas cells were fixed using 5% formaldehyde and mounted to the slide. Video of these nonmotile cells was recorded at 1200 fps with a pixel size of 0.56 μm, and the position of each cell was calculated by the CLONA method. Fig. 2 A shows a scatter plot with marginal histograms of the stationary cell over 2000 frames. The red solid line is a fitted normal distribution, with σx = 6.5 nm and σy = 7.4 nm. The mean ± standard deviation of the positions of stationary cells was 8.0 nm (n = 16). This standard deviation of the obtained coordinate values indicates that the location can be obtained with precision of ≈10 nm using this method.
Figure 2.
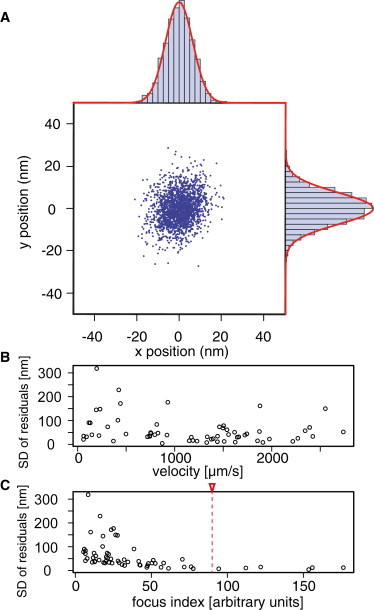
Precision of determining the cell position using the CLONA method. (A) Position of stationary cells for 2000 frames calculated using the CLONA method. The average position of the 2000 frames was used as the origin of coordinate system. (Red solid line) Fitted normal distribution, with σx = 6.5 and σy = 7.4 nm. Pixel size of the original images: 560 nm. (B) The standard deviation of residuals plotted against the velocity of moving cells. The precision varies among cells but does not depend on velocity. (C) The standard deviation of residuals plotted against the focus index. The two variables show a clear inverse relation. For a high focus index, all the targets were located in high-precision region, independent of the velocity. The precision of the CLONA method can be <10 nm, independent of the velocity if cells have a focus index >90 (red dotted line).
We next evaluated the precision of the CLONA method when applied to moving cells. Because the images of moving cells can be blurred, we were concerned that the precision would be lowered. To test the effect of this movement on image clarity, wild-type Chlamydomonas cells were formalin-fixed and placed in a flow chamber (well width, 2 mm; well depth, 0.080 mm). We assumed that the velocity of the cells was constant over 5 ms, and the residuals between the linear regression and the position determined by the CLONA method were used to evaluate the precision. The standard deviation of the residuals, which indicates the precision, was plotted against the velocity of the cells (Fig. 2 B). The precision was found to vary among cells but did not depend on the velocity. On the other hand, plotting the standard deviation of the residuals against the focus index showed a clear inverse relation (Fig. 2 C): when the focus index is low, the standard deviation of the residuals varies greatly, showing that some cells were localized with high precision whereas others were not. When the focus index was high, all the cells were localized with high precision. These results show that the precision of the CLONA method is dependent mostly on whether the images are well focused. Therefore, the CLONA method can locate the cells within 10-nm precision, irrespective of the velocity, if cells with a focus index >90 are selected (Fig. 2 C). As shown below, the maximum velocity of Chlamydomonas swimming is ∼1000 μm/s and, thus, error associated with image blur can be neglected for analysis of the swimming of Chlamydomonas.
Application of the CLONA method to wild-type Chlamydomonas cells
High-speed video images of swimming wild-type Chlamydomonas cells, CC125+, were analyzed using the CLONA method. Fig. 3, A and B, shows the typical swimming paths of a wild-type cell (121 frames, each time step is 1/1200 s ≈ 0.83 ms; see Movie S1 in the Supporting Material) determined by the CLONA method (Fig. 3 A), and by the conventional binary-centroid method (Fig. 3 B). Compared to the path determined by binary-centroid method (Fig. 3 B), the smooth curve in Fig. 3 A indicates the accuracy of the CLONA method in tracing the swimming path of the Chlamydomonas cell.
Figure 3.
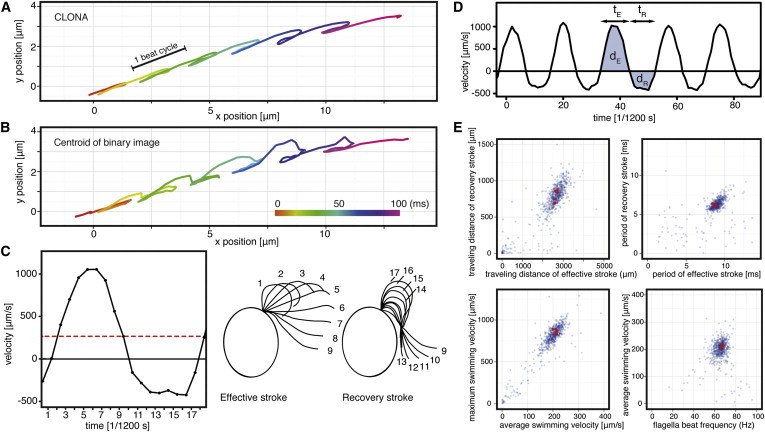
Application of the CLONA method to high-speed video images of wild-type Chlamydomonas cells (see Movie S1 in the Supporting Material). Swimming paths of a typical wild-type Chlamydomonas cell determined by (A) the CLONA methods and (B) conventional binary-centroid method. The position in each time frame is indicated by a dot and the frame interval is 1/1200 s ≈ 0.83 ms. The smooth curve indicates the better accuracy of the CLONA method in tracing the swimming path of Chlamydomonas cells. (C) The relation between the instantaneous swimming velocity parallel to the swimming direction and the flagellar beat stage obtained from Movie S2. The curve was obtained from a single beat without applying any filters. The mean swimming velocity is indicated by the red dotted line. (D) A typical plot of instantaneous swimming velocity. The following swimming parameters obtained from each beat cycle are shown: period of the effective and recovery strokes (tE and tR, respectively); travel distance of each stroke (dE and dR, respectively). (E) Scatter plots of wild-type beat cycles. Each point represents one beat cycle in each two-dimensional space. (Red points) Beat cycles of a single typical wild-type cell.
Using the accurate positions of the cells, we determined the swimming velocity based on the movement between two frames (Fig. 3, C and D). The shape of the swimming velocity curves was surprisingly constant among strokes (Fig. 3 D) and among cells (Fig. 3 E). Note that the velocity curves were obtained from a single beat without applying any filters. Although movements perpendicular to the XY plane were not determined, we estimated the contribution of perpendicular movements to be <10% because we selected cells that swam parallel to the XY plane using the focus index criterion (see Fig. S2). The average time and distance traveled through the in-focus zone was 58 ms and 12 μm.
Fig. 3 C illustrates the relation between the swimming velocity parallel to the swimming direction and the position of the flagellum (the red dotted line represents the mean swimming velocity; see also Movie S2). The Chlamydomonas cell accelerates as the flagellum bends at the base and the distal part straightens (Fig. 3 C, index = 1–5). The velocity reaches a maximum (1.05 × 103 μm/s) when the flagellum straightens vertically in the swimming direction (Fig. 3 C, index = 6). The cell gradually decelerates as the bending of the flagellum slows down (Fig. 3 C, index 7–10). When the flagellum reverses the bending direction at the base (Fig. 3 C, index = 10–13), the cell went backward despite that the distal half of the flagellum was still moving backward. The velocity curve of the recovery stroke has a dull phase (index = 10–17); that is, when the base of the flagellum starts to bend forward while the distal half is still moving backward, the forward propulsive force is canceled.
To quantitatively describe the characteristics of the swimming velocity curve, i.e., the period of the effective stroke and the recovery stroke (tE and tR, respectively) and the distance through which each stroke travels (dE and dR, respectively), we calculated the mean swimming velocity,
and flagella beat frequency,
for each beat cycle (Fig. 3 D). Scatter plots that were generated using these parameters (665 strokes, 167 wild-type cells, 600 fps) showed a cluster in each plot (Fig. 3 E). Each point represents one beat cycle in each two-dimensional space. The red points represent the beat cycles of a single typical wild-type cell. The spread of the clusters, which show the variance of the swimming parameters, is due to inter- and intracell variances. Intercell variance is the variance among different cells, whereas intracell variance is the variance among different beat cycles by the same cell. As shown in Fig. 3 E, the cluster of the red points was more dense than the actual black clouds in the plot, indicating that variances among cells contribute greatly to the overall variance. Regarding beat frequency, f, the variance among cells (intercell variance) was 8.9%, whereas that within cells for different beat cycles (intracell variance) was 2.9%. Brokaw and Luck (23) showed the variance of the beat frequency of the entire population to be 15.4% (65 ± 10 Hz, 20°C), whereas our study showed that beat cycles are fairly uniform for individual cells and that the variance among cells dominates the overall variance.
Application of the CLONA method to flagella mutants
To demonstrate the ability of CLONA method to distinguish motility-deficient mutants, we have analyzed high-speed video images of seven flagella mutants (Table 1). The scatter plots of beat frequency, f, versus mean swimming velocity, , of the beat cycles of four outer dynein arm mutants (oda1, oda11, suppf1, and suppf2) are shown in Fig. 4 A, and those for the inner dynein arm mutants (ida1, ida5, and ida9) are shown in Fig. 4 B (24). The beat cycles of each strain formed a characteristic cluster in the scatter plot and the difference between the strains are statistically significant (assuming Wilks’s lambda distribution, p > 0.001). The swimming speeds of these outer dynein arm mutants are proportional to the beat frequencies, consistent with the idea that the waveform of flagella is similar to that of wild-type (25,26). On the other hand, ida1 and ida9 cells swam slower, although the beat frequencies are in the same range as those of wild-type cells. This was also consistent with the idea that the beat frequencies of the inner dynein arm mutants are at almost normal value, but with reduced bend angle (25,26). Note that the swimming patterns of oda11 (missing α-heavy chain of outer dynein arm) are known to be similar to that of wild-type and difficult to distinguish by visual screening. Using the CLONA method, these mutants are distinguished in the scatter plot, and subtle changes in swimming behavior can be detected. Details about the mutant strains in the table are described in the literature (27–38).
Application of the CLONA method to flagella mutant screening
To test the effectiveness of the CLONA method in screening for motility-deficient mutants, we have examined 38 Chlamydomonas strains that had been previously isolated as circadian mutants (15). These mutants are insertional mutants with a CBR genetic background and screened by defective circadian rhythms. The responsible gene for each strain has been identified, but their swimming phenotypes had not been examined. For each strain, 150–200 s of high-speed video was taken and its swimming was phenotyped using the CLONA method (Fig. 5, and see Fig. S3). Note that the swimming phenotype of CBR+ (39) is different from CC125+ in Figs. 3 and 4. Roughly, the swimming phenotypes of these strains were divided into three groups:
-
1.
The first group shows no or little deficiency in motility (Fig. 5 A).
-
2.
This group (roc30, roc34, roc54, roc63, and roc106) shows severe deficiency in motility (Fig. 5 B).
-
3.
This group (roc23, roc77, roc78, roc88, roc105, and roc112) shows mild deficiency in motility (Fig. 5 C).
See also Movie S3 and Movie S4 for high-speed videos of the control strain and a representative strain of the third group. All the responsible genes of the second group have been previously reported to be flagella-related, whereas those of the third group were newly detected as motility-deficient. We also applied statistical analysis to the third group and showed that the difference between these mutant strains and wild-type were statistically significant (p < 0.001).
Discussion
Our CLONA method can determine the position of cell-sized objects (diameter, 10 μm; pixel size, 0.56 μm) with an accuracy of ∼10 nm, and this method provides valuable information about flagella motility. By applying the CLONA method to high-speed videos (600 or 1200 fps) of Chlamydomonas cells exhibiting swimming behavior, parameters related to swimming can be obtained with high spatial and temporal resolution for each individual cell. This application provides new ways to quantify aspects of Chlamydomonas motility that were difficult to observe without high spatiotemporal analysis: maximum swimming velocity, ratio of mean and maximum swimming velocity, variance among cells, and variance among beat cycles of the same cell. Analysis of seven Chlamydomonas flagella strains demonstrated that subtle changes in motility can be detected using the CLONA method, and the screening of 38 Chlamydomonas strains demonstrated that selection of mutants can be quickly and easily accomplished.
Precision of cell localization
For superresolution methods, such as stochastic optical reconstruction microscopy (40), one of the key factors in achieving high spatial resolution is to collect enough photons from the target object. Our CLONA method collects many photons diffracted from large objects, and achieves both high temporal (1 ms) and spatial (∼10 nm) resolution. One of the main factors for achieving temporal resolution is to collect many photons diffracted by a large object. According to our estimate, >105 photons were collected from a Chlamydomonas cell in 1 ms and were used in the localization of the cell. Therefore, if the same number of photons were collected from a molecule, precision of a few nanometers can be achieved according to the following equation (21),
| (2) |
where s is the standard deviation of the point spread function, a is the pixel size of the detector, N is the number of total collected photons, and b is background noise. We assumed s = 300 nm, a = 1000 nm, and N = 105. This equation is often used to estimate the resolution of superresolution methods, such as fluorescence imaging with 1-nm accuracy (FIONA) (41) or stochastic optical reconstruction microscopy, and based on the assumption that the point-spread function can be approximated by the Gaussian distribution and that the photon shot noise is the limiting factor for determining precision.
The image of a Chlamydomonas cell, of course, cannot be approximated by Gaussian distribution, because the cell is larger than the point-spread function of the microscope. To overcome this problem, another key technique of our CLONA method is that the reference images used for cross-correlation are generated from experimental images by averaging well-focused images of the target cell. For the determination of the position of the cell in Fig. 2 A using the Gaussian distribution as a reference, the standard deviation is 2.5 times larger than for the CLONA method, showing the effectiveness of using the averaged image. Although Chlamydomonas cells rotate while swimming, the shape of the cells seems unchanged probably because the cell shape is symmetric around the swimming axis. Therefore, the rotation of the cell would not largely affect the accuracy of the CLONA method. It is noteworthy that the standard deviation of the cell position doubles when the image in the neighboring frame is used as the reference, indicating that having a high signal/noise of the reference image is important. In conclusion, the use of large cell images and having a high signal/noise image on average are the two key factors for the success of the CLONA method.
Analysis of propulsive force generated by free-swimming Chlamydomonas cells
Because we determined the velocity of swimming cells to a high degree of accuracy, we can now translate the velocity into the drag force acting on the cell body, which balances the propulsive force produced by the two flagella. The viscous force on the cell body can be estimated from the formulas describing the motion of an ellipsoid in Stokes’ flow. Chlamydomonas cells can be considered to be well described by Stokes’ Law because the Reynolds number (Re) is very small (Re ≈ 10−3 to 10−2 (42)). Chwang and Wu (43) presented the relationship between velocity and viscous drag force on a prolate spheroid moving in the direction of the major axis with a very low Reynolds numbers to be
| (3) |
| (4) |
| (5) |
where F is the drag force, μ is the viscosity, e is the eccentricity, a is the major axis, and b is the minor axis. From the equations above, the average of maximum instantaneous propulsive force of Chlamydomonas cells was estimated to be 46.7 ± 4.8 pN (assuming that v = 802.0 ± 93.0 μm/s, μ = 0.893 × 10 −3 Pa s, a = 4 μm, b = 3 μm).
There are several ways to estimate the force generated by Chlamydomonas flagella. One way is to use velocity and the Stokes’ formula as described above. Minoura and Kamiya (44) have calculated the propulsive force of a free-swimming Chlamydomonas, using the mean swimming velocity, to be 10–12 pN. The values are consistent with our results, because the propulsive force estimated from the mean velocity determined from our measurement was 11.8 pN. Our results show that the instantaneous maximum swimming velocity can be approximately four times the mean velocity, which is consistent with results obtained from measuring flow field around the swimming cell (45). Therefore, the propulsive force estimated from the instantaneous maximum swimming velocity in this study was 40–48 pN. Racey et al. (46) also examined the velocity of Chlamydomonas cells using quasielastic light scattering and cinematographical techniques at 100–200 fps. Because the temporal resolution is limited to 10 ms in their analysis, they obtained the mean forward velocity of 379 ± 108 μm/s, which is approximately one-half that of the maximum velocity found in our study.
The force generated by flagella can be also be measured by an optical trap. McCord et al. (47) determined that a force of 26.5 ± 10.4 pN was required for Chlamydomonas to escape from the optical trap. Although the value is smaller than the maximum propulsive force determined in our study (46.7 ± 4.8 pN), it seems reasonable because the force required to escape the optical trap needs to be sustained for several milliseconds for a successful escape. Note that the force exerted by optical traps was determined by applying Stokes’ equation to dead trapped cells in a medium with a constant flow rate (47). Therefore, our method is a more direct measurement of the propulsion force than these trapping methods.
New approach toward the elucidation of flagella mechanisms
Our CLONA method provides a quantitative approach for screening flagella mutants. As described by Kamiya (13), motility-deficient Chlamydomonas cells have been identified by visual inspection under a microscope, which requires considerable time and effort. This visual inspection method may be biased toward selecting mutants that show substantial decreases in mean swimming velocity and many mutants that exhibit only small changes in swimming velocity might be missed. The CLONA method can select mutants that have subtle changes in mean velocity, or changes in other parameters, such as maximum velocity or the proportion of effective stroke time and recovery stroke time. Selection of mutants using the CLONA method could provide expanded screening capabilities and represents a labor-saving method because of its accurate velocity analysis and automated procedures.
Although several methods have been developed for the detailed analysis of flagella movement, they are not suitable for the screening of new flagella mutants. For example, Brokaw and Luck (23) established waveform analysis using the uni1 mutant strain. The uni1 mutant is suitable for conducting extensive waveform analysis because the strain has only one trans-flagellum (48) and rotates in place. Recently, a computed flagella waveform analysis was developed for uni1 cells (49) and used for the high-resolution estimation of the force generated by the flagellum (50). However, this waveform analysis requires selection of double mutant strains, which carry both uni1 mutation and a target flagella mutation. Therefore, this uni1-based waveform analysis is not suitable for screening for new mutants. Micropipettes have been used to hold Chlamydomonas cells and observe the detailed waveform (51,52). Because the micropipette analysis acquires the precise waveform by preventing movement and rotation, it can reveal the complex dynamics exhibited by flagella (53,54). However, capturing cells by micropipette requires a certain amount of time, making it unsuitable for screening a large number of mutants.
One of the important findings of our study is that the position and velocity of Chlamydomonas cells accurately reflect the flagella movement. Therefore, even though the shape of the flagella is not always visible in the images due to the rotation of cell, the CLONA method is able to analyze the flagellar phenotypes of the free swimming cells in their natural, unrestrained state.
In the screening using 38 Chlamydomonas strains, CLONA detected motility-deficient mutants that would have been difficult to detect previously, just by taking a 150–200 s high-speed video for each strain. We have chosen a set of circadian mutants for this screening because the mutated genes include previously reported flagella genes, which could be used as a positive control (ODA4, PF9, DLC7a, and FLA14; see Table S1 in the Supporting Material). CLONA detected all of these mutants in the screening, ensuring the validity of the screening. Within the 38 strains, we have actually detected six mutants that were not known to play a role in motility (roc23, roc77, roc78, roc88, roc105, and roc112; see Table S1 and Fig. S3).
One of these mutants, roc78 is a kinase, which may modify both circadian rhythm and motility via the signaling pathway. In fact, several studies have shown that there are multiple phosphorylation sites on outer and inner dynein arms (55,56), which suggests that kinases such as roc78 could modify the activity of dynein and thus affect motility. It is interesting that all of the mutants that showed severe deficiency in motility were previously characterized mutants, whereas all of the mutant that showed mild defects were genes that were not known to play a role in motility. This may suggest that more unidentified motility related gene can be found by screening and examining mutants that show mild defects in motility. Although future studies are required to find how the newly detected genes caused the motility defects, this screening test have demonstrated the strength of CLONA as a mutant screening tool.
Conclusions
This study demonstrated an observational method, CLONA, which localizes cells and similarly sized objects with high spatial (<10 nm) and temporal (millisecond scale) resolution. The features of CLONA that distinguish it from other superresolution methods:
-
1.
CLONA generates the reference image from experimental images of the target itself.
-
2.
CLONA can locate target whose size is significantly larger than the precision.
-
3.
It does not require special instruments.
One suitable application of CLONA is analyzing the fluid flow driven by bacterial flagella: fluid flows are often estimated by monitoring the movements of small plastic beads (57). The positions of beads can be determined more accurately by CLONA than by commonly-used binary-centroid procedures. Similarly, the CLONA method may contribute analyzing behavior analysis of model organisms including sperms and evaluation of cilia movement in fallopian tubes or ventricles of brain (58). Hence, the CLONA method can be applicable also to various systems that require high spatiotemporal resolution.
Acknowledgments
We thank all of the members of the Kikkawa Lab, especially Dr. T. Yagi, for his valuable suggestions and critical reading of the manuscript.
This work was supported by the Funding Program for Next Generation World-Leading Researchers (grant No. LS027), the Uehara Memorial Foundation, and the Takeda Science Foundation (all to M.K.).
Supporting Material
Supporting Citations
Ref. (59) appears in the Supporting Material.
References
- 1.Nonaka S., Tanaka Y., Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 2.Leigh M.W., Pittman J.E., Zariwala M.A. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet. Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finetti F., Baldari C.T. Compartmentalization of signaling by vesicular trafficking: a shared building design for the immune synapse and the primary cilium. Immunol. Rev. 2013;251:97–112. doi: 10.1111/imr.12018. [DOI] [PubMed] [Google Scholar]
- 4.Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 5.Badano J.L., Mitsuma N., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 6.Ibañez-Tallon I., Pagenstecher A., Omran H. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Hum. Mol. Genet. 2004;13:2133–2141. doi: 10.1093/hmg/ddh219. [DOI] [PubMed] [Google Scholar]
- 7.Ikeda T., Ikeda K., Kamiya R. The mouse ortholog of EFHC1 implicated in juvenile myoclonic epilepsy is an axonemal protein widely conserved among organisms with motile cilia and flagella. FEBS Lett. 2005;579:819–822. doi: 10.1016/j.febslet.2004.12.070. [DOI] [PubMed] [Google Scholar]
- 8.Marszalek J.R., Liu X., Goldstein L.S. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 9.Pazour G.J., Dickert B.L., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avidor-Reiss T., Maer A.M., Zuker C.S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 11.Li J.B., Gerdes J.M., Dutcher S.K. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 12.Merchant S.S., Prochnik S.E., Grossman A.R. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya R. Selection of Chlamydomonas dynein mutants. Methods Enzymol. 1991;196:348–355. doi: 10.1016/0076-6879(91)96031-l. [DOI] [PubMed] [Google Scholar]
- 14.Gorman D.S., Levine R.P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuo T., Okamoto K., Ishiura M. A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 2008;22:918–930. doi: 10.1101/gad.1650408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klopfenstein D.R., Vale R.D. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell. 2004;15:3729–3739. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metlagel Z., Kikkawa Y.S., Kikkawa M. Ruby-Helix: an implementation of helical image processing based on object-oriented scripting language. J. Struct. Biol. 2007;157:95–105. doi: 10.1016/j.jsb.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: a Language and Environment for Statistical Computing. [Google Scholar]
- 19.Russ J.C. 6th Ed. CRC Press; Boca Raton, FL: 2011. The Image Processing Handbook. [Google Scholar]
- 20.Groen F.C., Young I.T., Ligthart G. A comparison of different focus functions for use in autofocus algorithms. Cytometry. 1985;6:81–91. doi: 10.1002/cyto.990060202. [DOI] [PubMed] [Google Scholar]
- 21.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Debella-Gilo M., Kaab A. Sub-pixel precision image matching for measuring surface displacements on mass movements using normalized cross-correlation. Remote Sens. Environ. 2011;115:130–142. [Google Scholar]
- 23.Brokaw C.J., Luck D.J. Bending patterns of Chlamydomonas flagella I. Wild-type bending patterns. Cell Motil. 1983;3:131–150. doi: 10.1002/cm.970030204. [DOI] [PubMed] [Google Scholar]
- 24.Wickham H. Springer; New York: 2009. GGPLOT2 Elegant Graphics for Data Analysis. [Google Scholar]
- 25.Brokaw C.J., Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- 26.Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- 27.Kamiya R. Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J. Cell Biol. 1988;107:2253–2258. doi: 10.1083/jcb.107.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada S., Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from α-β and γ subunits: requirement of a third factor. J. Cell Biol. 1994;126:737–745. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takada S., Wilkerson C.G., Witman G.B. The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell. 2002;13:1015–1029. doi: 10.1091/mbc.01-04-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakakibara H., Mitchell D.R., Kamiya R. A Chlamydomonas outer arm dynein mutant missing the α-heavy chain. J. Cell Biol. 1991;113:615–622. doi: 10.1083/jcb.113.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell D.R., Brown K.S. Sequence analysis of the Chlamydomonas reinhardtii flagellar α-dynein gene. Cell Motil. Cytoskeleton. 1997;37:120–126. doi: 10.1002/(SICI)1097-0169(1997)37:2<120::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 32.Huang B., Ramanis Z., Luck D.J. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 33.Porter M.E., Knott J.A., Dutcher S.K. Mutations in the SUP-PF-1 locus of Chlamydomonas reinhardtii identify a regulatory domain in the β-dynein heavy chain. J. Cell Biol. 1994;126:1495–1507. doi: 10.1083/jcb.126.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupp G., O’Toole E., Porter M.E. The sup-pf-2 mutations of Chlamydomonas alter the activity of the outer dynein arms by modification of the γ-dynein heavy chain. J. Cell Biol. 1996;135:1853–1865. doi: 10.1083/jcb.135.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamiya R., Kurimoto E., Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J. Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato T., Kagami O., Kamiya R. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct. Funct. 1993;18:371–377. doi: 10.1247/csf.18.371. [DOI] [PubMed] [Google Scholar]
- 37.Kato-Minoura T., Hirono M., Kamiya R. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J. Cell Biol. 1997;137:649–656. doi: 10.1083/jcb.137.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yagi T., Minoura I., Kamiya R. An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J. Biol. Chem. 2005;280:41412–41420. doi: 10.1074/jbc.M509072200. [DOI] [PubMed] [Google Scholar]
- 39.Niwa Y., Matsuo T., Ishiura M. Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 2013;110:13666–13671. doi: 10.1073/pnas.1220004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildiz A., Selvin P.R. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 42.Tam D., Hosoi A.E. Optimal feeding and swimming gaits of biflagellated organisms. Proc. Natl. Acad. Sci. USA. 2011;108:1001–1006. doi: 10.1073/pnas.1011185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chwang A.T., Wu T.Y.T. Hydromechanics of low Reynolds number flows. Part 2. Singularity method for Stokes flows. J. Fluid Mech. 1975;67:787–815. [Google Scholar]
- 44.Minoura I., Kamiya R. Strikingly different propulsive forces generated by different dynein-deficient mutants in viscous media. Cell Motil. Cytoskeleton. 1995;31:130–139. doi: 10.1002/cm.970310205. [DOI] [PubMed] [Google Scholar]
- 45.Guasto J.S., Johnson K.A., Gollub J.P. Oscillatory flows induced by microorganisms swimming in two dimensions. Phys. Rev. Lett. 2010;105:168102. doi: 10.1103/PhysRevLett.105.168102. [DOI] [PubMed] [Google Scholar]
- 46.Racey T.J., Hallett R., Nickel B. A quasi-elastic light scattering and cinematographic investigation of motile Chlamydomonas reinhardtii. Biophys. J. 1981;35:557–571. doi: 10.1016/S0006-3495(81)84812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCord R.P., Yukich J.N., Bernd K.K. Analysis of force generation during flagellar assembly through optical trapping of free-swimming Chlamydomonas reinhardtii. Cell Motil. Cytoskeleton. 2005;61:137–144. doi: 10.1002/cm.20071. [DOI] [PubMed] [Google Scholar]
- 48.Huang B., Ramanis Z., Luck D.J. Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell. 1982;29:745–753. doi: 10.1016/0092-8674(82)90436-6. [DOI] [PubMed] [Google Scholar]
- 49.Bayly P.V., Lewis B.L., Dutcher S.K. Efficient spatiotemporal analysis of the flagellar waveform of Chlamydomonas reinhardtii. Cytoskeleton (Hoboken) 2010;67:56–69. doi: 10.1002/cm.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bayly P.V., Lewis B.L., Dutcher S.K. Propulsive forces on the flagellum during locomotion of Chlamydomonas reinhardtii. Biophys. J. 2011;100:2716–2725. doi: 10.1016/j.bpj.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoops H.J., Wright R.L., Witman G.B. Flagellar waveform and rotational orientation in a Chlamydomonas mutant lacking normal striated fibers. J. Cell Biol. 1984;98:818–824. doi: 10.1083/jcb.98.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruffer U., Nultch W. Comparison of the beating of cis- and trans-flagella of Chlamydomonas cells held on micropipettes. Cytoskeleton. 1987;7:87–93. doi: 10.1002/(SICI)1097-0169(1998)41:4<297::AID-CM3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 53.Polin M., Tuval I., Goldstein R.E. Chlamydomonas swims with two “gears” in a eukaryotic version of run-and-tumble locomotion. Science. 2009;325:487–490. doi: 10.1126/science.1172667. [DOI] [PubMed] [Google Scholar]
- 54.Wan K.Y., Leptos K.C., Goldstein R.E. Lag, lock, sync, slip: the many ‘phases’ of coupled flagella. J. R. Soc. Interface. 2014;11:20131160. doi: 10.1098/rsif.2013.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith J.H., Denhardt D.T. Evidence for two pathways of protein kinase C induction of 2ar expression: correlation with mitogenesis. J. Cell. Physiol. 1989;139:189–195. doi: 10.1002/jcp.1041390126. [DOI] [PubMed] [Google Scholar]
- 56.King S.M., Witman G.B. Multiple sites of phosphorylation within the α-heavy chain of Chlamydomonas outer arm dynein. J. Biol. Chem. 1994;269:5452–5457. [PubMed] [Google Scholar]
- 57.Darnton N., Turner L., Berg H.C. Moving fluid with bacterial carpets. Biophys. J. 2004;86:1863–1870. doi: 10.1016/S0006-3495(04)74253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lechtreck K.F., Sanderson M.J., Witman G.B. High-speed digital imaging of ependymal cilia in the murine brain. Methods Cell Biol. 2009;91:255–264. doi: 10.1016/S0091-679X(08)91013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foster K.W., Smyth R.D. Light antennas in phototactic algae. Microbiol. Rev. 1980;44:572–630. doi: 10.1128/mr.44.4.572-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


