Abstract
Metastatic prostate cancer is currently incurable. Metastasis is thought to result from changes in the expression of specific metastasis-driving genes in nonmetastatic prostate cancer tissue, leading to a cascade of activated downstream genes that set the metastatic process in motion. Such genes could potentially serve as effective therapeutic targets for improved management of the disease. They could be identified by comparative analysis of gene expression profiles of patient-derived metastatic and nonmetastatic prostate cancer tissues to pinpoint genes showing altered expression, followed by determining whether silencing of such genes can lead to inhibition of metastatic properties. Various hurdles encountered in this approach are discussed, including (i) the need for clinically relevant, nonmetastatic and metastatic prostate cancer tissues such as xenografts of patients’ prostate cancers developed via subrenal capsule grafting technology and (ii) limitations in the currently available methodology for identification of master regulatory genes.
BACKGROUND
Metastatic prostate cancer, as distinct from localized prostate cancer, is highly resistant to conventional therapy.1,2 The development of new, effective therapeutic strategies especially targeting metastatic prostate cancer is urgently needed for improved disease management.3,4 Metastasis is thought to result from changes in the expression of specific genes (e.g. upregulation) that lead to activation of multiple downstream genes mediating the metastatic process (Figure 1).5,6 Such metastasis-driving genes could serve as effective therapeutic targets for management of metastatic prostate cancer.
Figure 1.
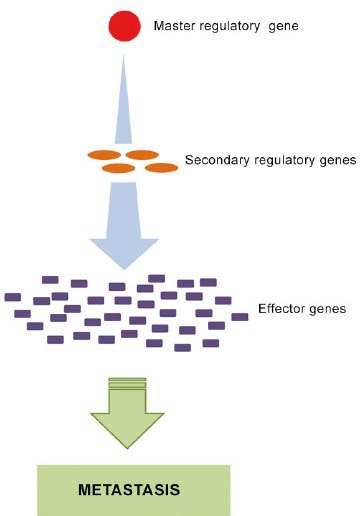
Metastasis is thought to result from changes in the expression of specific master regulatory genes (e.g. upregulation) that lead to activation of cascades of downstream genes that set the metastatic process in motion.
Metastasis-driving genes could be identified by comparing gene expression profiles of metastatic and nonmetastatic human prostate cancer cells in patients’ specimens to pinpoint genes showing altered expression, followed by determining whether silencing of such genes can lead to inhibition of metastatic properties. A major hurdle using this approach, however, is that primary prostate cancer samples, the usual source of nonmetastatic prostate cancer cells, do not consist of pure nonmetastatic cells, but also contain metastatic cells. To overcome this hurdle, we have developed transplantable, metastatic and nonmetastatic patient-derived prostate cancer tissue xenograft lines in NOD-SCID (nonobese diabetic/severe combined immunodeficient) mice. The subrenal capsule grafting technique used to develop such cancer tissue lines tends to retain important properties of the original cancers, including histopathology, chromosomal aberrations and gene expression profiles, resulting in xenograft lines that closely resemble the original cancers.7,8,9 Recently, we have developed paired metastatic (LTL-313H) and nonmetastatic (LTL-313B) prostate cancer sublines from one patient's specimen, that are suitable for investigating the molecular basis of metastasis.10
In the present study, comparative analysis of gene expression profiles of the LTL-313H and LTL-313B sublines led to identification of a number of novel, differentially expressed genes, including TIMELESS and DLX1, that have not previously been associated with prostate cancer metastasis and could have a metastasis-driving function.
STUDY DESIGN
Gene expression profiles of paired metastatic LTL-313H and nonmetastatic LTL-313B prostate cancer tissue xenografts were obtained using microarray technology. Raw gene expression data were filtered for improved quality prior to analysis of differential gene expression. Specifically, probes without corresponding gene annotations and probes without detectable expression levels (less than 3 in log2 scale) were removed. The MIAME-compliant gene expression data are accessible through GEO Series accession number GSE41193 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE41193).
Gene expression profiles of Memorial Sloan-Kettering Cancer Center (MSKCC) prostate tissues (normal tissues, primary and secondary prostatic adenocarcinomas) and associated clinical information11 were downloaded from the CBio Cancer Genomics Portal website.12 Correlations were sought between poor prognostic factors of the patients (MSKCC cohort) and the relative expression levels in their prostate cancer tissues of selected genes (identified in the xenografts).
To validate differential gene expressions found, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was applied using normal human prostate and prostate cancer specimens frozen in optimal cutting temperature compounds (OCT); RNA was isolated using the RNeasy FFPE kit (Qiagen Inc., Hilden, Germany) following the manufacturer's instructions. The specimens had been obtained from patients, with their informed written consent, following a protocol approved by a local ethics committee (Kelowna Hospital, British Columbia, Canada) and examination by a pathologist.13
Effects of gene silencing on cell proliferation, cell migration and tissue invasion were determined using; e.g. PC3M, PC3 and C4-2 human prostate cancer cells, as previously described.14
Statistical significance was established using the Student's t-test.
HIGHLIGHTED RESULTS
Elevated expression of TIMELESS and DLX1
Comparative analysis of microarray gene expression data (GSE41193), obtained from paired metastatic LTL-313H and nonmetastatic LTL-313B prostate cancer tissue xenografts, was used to identify genes in the metastatic tissue that showed elevated expression (>1.5-fold). For further studies, transcription factor-related genes (as defined in gene ontology) were selected as they are thought to play a critical role in metastasis.15,16 Subsequently, genes that had previously been associated with metastatic prostate cancer were excluded. Of the remaining genes, TIMELESS and DLX1, both showing elevated expression in the metastatic LTL-313H xenografts (validated by qRT-PCR; Table 1), were of particular interest, since they also showed elevated expression in secondary prostate cancer samples of the MSKCC cohort study.11 Thus, as shown in Figure 2a, the expression level of the TIMELESS gene was significantly higher in secondary prostate cancer samples in comparison to normal prostate and primary prostate cancer samples. As well, elevated TIMELESS expression was positively associated with poor patients’ prognostic factors, including increased lymph node involvement, increased Gleason score and elevated PSA levels (Figure 2b). In the case of DLX1, its expression level in the MSKCC cohort was significantly higher in both primary and secondary cancers when compared to normal prostate samples (Figure 2c). This differential expression pattern of the DLX1 gene was validated by qRT-PCR analysis in our laboratory using a smaller pool of patients’ samples (six normal prostate samples and five prostate cancer samples; (Figure 2d).
Table 1.
Elevated gene expression of TIMELESS and DLX1 in metastatic LTL-313H prostate cancer tissue xenografts compared to nonmetastatic LTL-313B counterparts
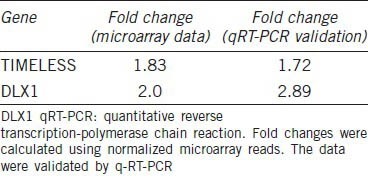
Figure 2.
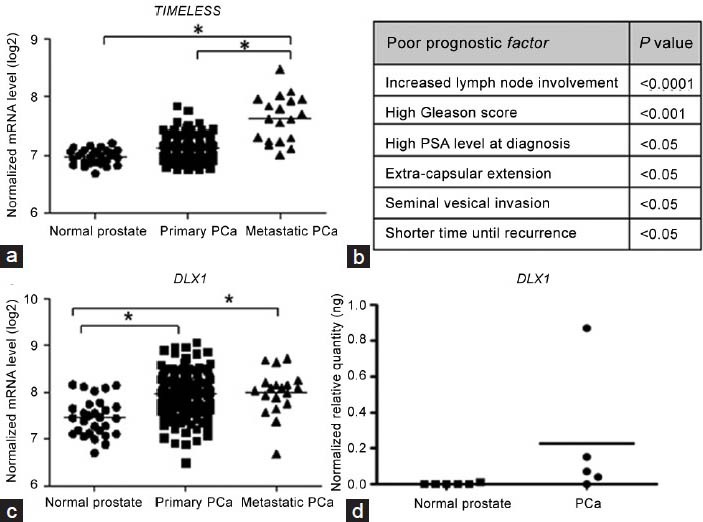
Elevated expression of TIMELESS and DLX1 associated with metastatic prostate cancer (PCa). (a) Elevated TIMELESS expression in metastatic PCa (n = 19) compared to primary PCa (n = 131) and normal prostate tissues (n = 28; microarray gene expression data from the MSKCC Prostate Oncogenome Project). (b) Elevated expression of TIMELESS associated with poor patient prognosis. (c) Elevated DLX1 expression in both primary and metastatic tumors compared to normal prostate samples (MSKCC cohort). (d) Elevated DLX1 expression in five prostate tumors compared to six normal prostate samples. Note that three out of six normal samples have undetectable DLX1 levels. Statistical significance was established using the Student's t-test; *P < 0.05. MSKCC: Memorial Sloan-Kettering Cancer Center.
Effect of TIMELESS and DLX1 silencing on proliferation, migration and tissue invasion of prostate cancer cell lines
siRNA-induced knockdown of TIMELESS significantly inhibited the migration of PC3M cells in a wound healing assay (Figure 3c) without affecting their proliferation rate (Figure 3b) or their cell cycle progression (data not shown). In contrast, siRNA-induced knockdown of DLX1 did not affect the migration or proliferation of PC3M (Figure 4) or C4-2 cells (data not shown).
Figure 3.
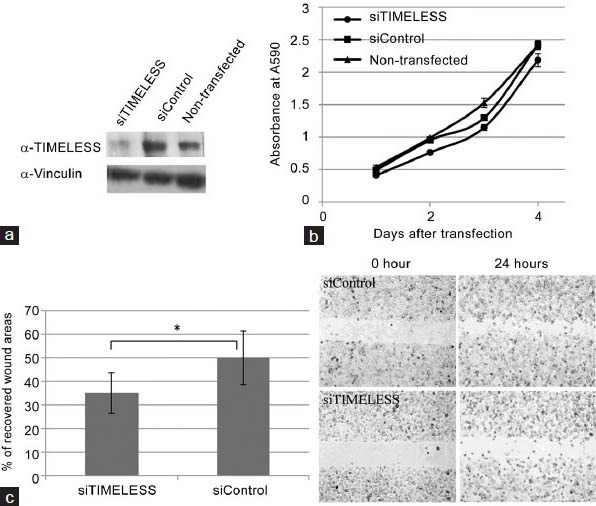
siRNA-induced down-regulation of TIMELESS in PC3M cells reduces cell migration. (a) Treatment of PC3M cells with siTIMELESS leads to a marked reduction in TIMELESS protein levels, (b) but have no effect on cell proliferation. (c) A monolayer of PC3M cells was scratched to examine the rate of cell migration into the wounded area. The bar graph represents the percentage of cell-recovered wound areas after 24 h of incubation (*P < 0.01). Representative images of the wound captured at different time points are shown (at right). Results are representative of three individual experiments (mean ± S.D.). S.D., standard deviation.
Figure 4.
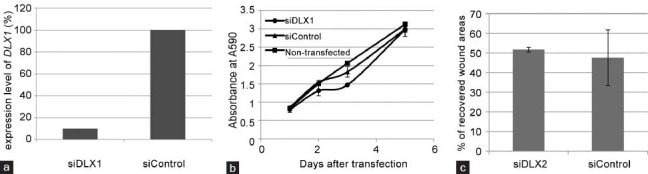
siRNA-induced down-regulation of DLX1 in PC3M cells has no effect on cell proliferation or migration. (a) Treatment with siDLX1 in PC3M cells leads to >90% reduction of DLX1 gene expression. Downregulated DLX1 does not affect (b) cell proliferation and (c) cell migration of PC3M cells. Results are representative of three individual experiments (mean ± s.d.).
DISCUSSION
Development of metastasis is generally thought to stem from changes in the expression of specific, metastasis-driving (master regulatory) genes that drive multiple downstream genes to set the metastatic process in motion (Figure 1). However, the exact molecular and cellular mechanisms involved are poorly understood. This is in part due to a lack of clinically relevant, experimental cancer metastasis models. Solid cancers typically consist of complex configurations of multiple, diverse and interacting cell types. While commonly used xenografts of cultured cancer cell lines can be useful as models for studying basic aspects of the metastatic process, they lack the molecular and biological complexity of the malignancies (e.g. tumor heterogeneity) and hence are not adequate for elucidating the multifaceted genetic architecture of metastatic cancers and the mechanisms underlying their development.17,18,19 The LTL-313H and LTL-313B prostate cancer xenograft sublines used in present study are based on grafting of cancer tissues and hence more closely resemble the original malignancy. As such, they appear to be more suitable for identification of prostate cancer metastasis-driving genes.
While altered expressions of TIMELESS and DLX1 have been reported for a variety of cancers,20,21,22,23,24 they have not been associated with prostate cancer metastasis. The elevated expression of TIMELESS found in our metastatic xenografts (Table 1), as well as in metastatic prostate cancer patients’ samples (MSKCC cohort; Figure 2a) and a strong correlation between elevated TIMELESS expression and poor clinical patients’ outcome (increased seminal vesicle invasion and lymph node involvement; Figure 2b), suggest that this gene has a role in prostate cancer metastasis and perhaps as a metastasis-driving gene. However, silencing of TIMELESS in PC3M cells only led to limited inhibition of cell migration (Figure 3c) and had no effect on the metastatic properties of C4-2 and PC3 cells, such as cell migration and tissue invasiveness (data not shown). Similarly, elevated expression of DLX1 was found to be clinically relevant (Figure 2c and 2d), but silencing of DLX1 did not show any inhibitory effect of metastatic properties of the above three prostate cancer cell lines in vitro (Figure 4b and 4c). These data suggest that TIMELESS and DLX1 are not metastasis-driving genes. On the other hand, the conventional in vitro single cells-based assays used might not be adequate (lacking biological complexity25) for evaluating the metastasis-driving functions of the genes, since it has been reported that metastasis can involve the participation of a variety of cells, e.g. certain immune cells.26 In view of the above, further studies are needed to confirm whether or not TIMELESS or DLX1 are prostate cancer metastasis-driving genes.
With regard to alternative approaches for identifying metastasis-driving genes, it has long been known that master regulatory gene networks often act as amplification cascades.27,28 In such a case, our initial approach, aimed at identifying metastasis-driving genes on the basis of relatively high gene expression changes, would not be effective, since changes in the expression of upstream master regulatory genes would be much smaller than those of their downstream target genes. Recently, a promising new approach has become available for identification of metastasis-driving genes in amplification cascades. It is possible to predict upstream master regulatory genes through integrative, software-based analysis of differential gene expression profiles coupled to knowledge of upstream regulatory genes (obtained from molecular studies). Various programs are available such as ARACNE (Algorithm for the Reconstruction of Accurate Cellular Networks; a novel algorithm, using microarray expression profiles) and an Upstream Regulator tool from Ingenuity Pathway Analysis.29,30,31,32,33 Such strategy was effective evidenced by our recent discovery of GATA2 gene as a potential metastasis-driving gene in prostate cancer.34
Other attempts to identify potential master regulatory genes include functional genomics approaches. For example, the Sleeping Beauty (SB) transposon system can be used, in which transposons can be randomly inserted into the DNA and subsequent development of metastasis can be linked to culprit genes.35
In conclusion, identification of metastasis-driving genes is a most challenging process. Amongst others, the question is raised whether only a transient change in the expression of these genes is required to activate and maintain the metastatic process, or whether their continuous expression is needed. In case of the latter, prostate cancer metastasis could be arrested by silencing of metastasis-driving genes, leading to improved management of the disease.
ACKNOWLEDGMENTS
We thank Drs Akira Watahiki and Fang Zhang for stimulating and helpful discussions. RNA quality control and microarray analysis were performed by the Laboratory for Advanced Genome Analysis at the Vancouver Prostate Centre, Vancouver, Canada. We gratefully thank Dr. Xiaoming Yang for anti-TIMELESS antibody. This study was supported by the Canadian Institutes of Health Research (YZW, CCC), BC Cancer Foundation (YZW), Prostate Cancer Canada (YZW) and Prostate Cancer Foundation BC (YTC).
COMPETING INTEREST
The authors declare that there is no conflict of interest.
REFERENCES
- 1.Stephenson AJ, Scardino PT, Kattan MW, Pisansky TM, Slawin KM, et al. Predicting the outcome of salvage radiation therapy for recurrent prostate cancer after radical prostatectomy. J Clin Oncol. 2007;25:2035–41. doi: 10.1200/JCO.2006.08.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Osanto S, Van Poppel H. Emerging novel therapies for advanced prostate cancer. Ther Adv Urol. 2012;4:3–12. doi: 10.1177/1756287211432777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukherji D, Eichholz A, De Bono JS. Management of metastatic castration-resistant prostate cancer: recent advances. Drugs. 2012;72:1011–28. doi: 10.2165/11633360-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–69. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Y, Wu H, Lei R, Chong RA, Wei Y, et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J Biol Chem. 2012;287:33533–44. doi: 10.1074/jbc.M112.392332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tung WL, Wang Y, Gout PW, Liu DM, Gleave M, et al. Use of irinotecan for treatment of small cell carcinoma of the prostate. Prostate. 2011;71:675–81. doi: 10.1002/pros.21283. [DOI] [PubMed] [Google Scholar]
- 8.Collins CC, Volik SV, Lapuk AV, Wang Y, Gout PW, et al. Next generation sequencing of prostate cancer from a patient identifies a deficiency of methylthioadenosine phosphorylase, an exploitable tumor target. Mol Cancer Ther. 2012;11:775–83. doi: 10.1158/1535-7163.MCT-11-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Xue H, Cutz JC, Bayani J, Mawji NR, et al. An orthotopic metastatic prostate cancer model in SCID mice via grafting of a transplantable human prostate tumor line. Lab Invest. 2005;85:1392–404. doi: 10.1038/labinvest.3700335. [DOI] [PubMed] [Google Scholar]
- 10.Watahiki A, Wang Y, Morris J, Dennis K, O’Dwyer HM, et al. MicroRNAs associated with metastatic prostate cancer. PLoS One. 2011;6:e24950. doi: 10.1371/journal.pone.0024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung PK, Woolcock B, Adomat H, Sutcliffe M, Bainbridge TC, et al. Protein profiling of microdissected prostate tissue links growth differentiation factor 15 to prostate carcinogenesis. Cancer Res. 2004;64:5929–33. doi: 10.1158/0008-5472.CAN-04-1216. [DOI] [PubMed] [Google Scholar]
- 14.Lin D, Watahiki A, Bayani J, Zhang F, Liu L, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–9. doi: 10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Ehsanian R, Waes CV. Nuclear transcription factors and signaling pathways in oral cancer metastasis. In: Myers J, editor. Oral Cancer Metastasis. New York: Springer; 2009. pp. 197–229. [Google Scholar]
- 16.Nebert DW. Transcription factors and cancer: an overview. Toxicology. 2002;181(2):131–41. doi: 10.1016/s0300-483x(02)00269-x. [DOI] [PubMed] [Google Scholar]
- 17.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–39. [PubMed] [Google Scholar]
- 18.Bott SR, Arya M, Shergill IS, Williamson M. Molecular changes in prostatic cancer. Surg Oncol. 2005;14:91–104. doi: 10.1016/j.suronc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–54. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- 20.Mazzoccoli G, Panza A, Valvano MR, Palumbo O, Carella M, et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–51. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 21.Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33:149–55. doi: 10.1007/s13277-011-0258-2. [DOI] [PubMed] [Google Scholar]
- 22.Fu A, Leaderer D, Zheng T, Hoffman AE, Stevens RG, et al. Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol Carcinog. 2012;51:923–9. doi: 10.1002/mc.20862. [DOI] [PubMed] [Google Scholar]
- 23.Starkova J, Gadgil S, Qiu YH, Zhang N, Hermanova I, et al. Up-regulation of homeodomain genes, DLX1 and DLX2, by FLT3 signaling. Haematologica. 2011;96:820–8. doi: 10.3324/haematol.2010.031179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba S, Takeshita K, Imai Y, Kumano K, Kurokawa M, et al. Homeoprotein DLX-1 interacts with Smad4 and blocks a signaling pathway from activin A in hematopoietic cells. Proc Natl Acad Sci U S A. 2003;100:15577–82. doi: 10.1073/pnas.2536757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer N, Walzl A, Unger C, Rosner M, Krupitza G, et al. In vitro cell migration and invasion assays. Mutat Res. 2013;752:10–24. doi: 10.1016/j.mrrev.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 27.Chan SS, Kyba M. What is a Master Regulator? J Stem Cell Res Ther. 2013 May 4;3 doi: 10.4172/2157-7633.1000e114. pii: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013;503:487–92. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–25. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pe’er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144:864–73. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Saito M, Bisikirska BC, Alvarez MJ, Lim WK, et al. Genome-wide identification of post-translational modulators of transcription factor activity in human B cells. Nat Biotechnol. 2009;27:829–39. doi: 10.1038/nbt.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koutsogiannouli E, Papavassiliou AG, Papanikolaou NA. Complexity in cancer biology: is systems biology the answer? Cancer Med. 2013;2:164–77. doi: 10.1002/cam4.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang YT, Wang K, Fazli L, Qi RZ, Gleave ME, et al. GATA2 as a potential metastasis-driving genes in prostate cancer. Oncotarget. 2014;5:451–61. doi: 10.18632/oncotarget.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–33. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]


