Abstract
Prostate cancer (PCa) has become to have the highest incidence and the second mortality rate in western countries, affecting men's health to a large extent. Although prostate-specific antigen (PSA) was discovered to help diagnose the cancer in an early stage for decades, its specificity is relative low, resulting in unnecessary biopsy for healthy people and over-treatment for patients. Thus, it is imperative to identify more and more effective biomarkers for early diagnosis of PCa in order to distinguish patients from healthy populations, which helps guide an early treatment to lower disease-related mortality by noninvasive or minimal invasive approaches. This review generally describes the current early diagnostic biomarkers of PCa in addition to PSA and summarizes the advantages and disadvantages of these biomarkers.
Keywords: early diagnostic biomarkers, MALAT1-derived miniRNA, PCA3, prostate cancer, PSA
INTRODUCTION
Prostate cancer (PCa) plagues male population and has become a major public health problem in most western countries. It is reported to be the first most frequently diagnosed cancer and the second leading cause of cancer death in the United States in 2013, accounting for 28% (238 590) of the total new cases and 10% (29 720) of the total cancer death.1 With the vast increase of PCa population, early detection becomes one of the major approaches to solve the problem, which is supposed to be the key to an early intervention to reduce disease-related mortality.
Due to the clinical application of prostate-specific antigen (PSA) since 20 years ago, it has shown great of value in PCa detection, staging, and monitoring with high sensitivity. However, many other factors affect the usefulness of PSA as an early diagnostic biomarker. Researchers have found that benign prostate hyperplasia (BPH) and prostatitis may cause an elevation in PSA and there is no evidence to prove that BPH and prostatitis will develop into PCa. Further, it is confirmed that only 30% of patients with unusual PSA value (above 4 ng ml−1) were finally diagnosed with PCa, leading to the over-treatment of low-risk patients,2 unnecessary biopsies and nonessential radical prostatectomies.3,4 In contrast, the prevention of such over-diagnosis and over-treatment may cause the progression of potential aggressive, life-threatening PCa.
Thus, novel early diagnostic biomarkers of PCa are urgently needed to be excavated and evaluated to distinguish patients with cancers from healthy population. The review aims to generally summarize the current diagnostic biomarkers of PCa, their status and future prospective.
PROSTATIC ACID PHOSPHATASE AND PROSTATE SPECIFIC ANTIGEN
Prostate acid phosphatase (PAP) was first reported to be elevated in the serum of patients with PCa metastasized to bone and could be regarded as a serum diagnostic biomarker in 1930s.5 Unfortunately, PAP was proved to be not sensitive to detect localized lesion of PCa6 and totally replaced by PSA that was discovered in 1970s. Recently, PAP was studied in deep because it was found that high levels of PAP expression were detected in high Gleason score PCa,7 offering a new and interesting functional aspect of differentiating indolent and aggressive PCa. PAP can be also used as a target antigen for PCa therapy. A novel Food and Drug Administration (FDA)-approved therapy termed Provenge (Sipuleucel-T) aims at 95% PAP expressed PCa,8 and a phase III clinical trial showed 4.1 months improvement in median overall survival compared with the control group.9
Prostate specific antigen, also known as kallikrein-3 (KLK3), is a glycoprotein enzyme encoded in humans by the KLK3 gene. It was first discovered in late 1970s from prostate extracts.10 PSA was later found increased in serum of patients with PCa resulting from the disruption of basal cell layer of prostate. It was initially used as a screening biomarker and officially approved for PCa screening by FDA in 1994. However, with the low specificity of PSA in indicating PCa, which may cause relative high false-positive rate in screening, the United States Prevention Services Task Force doesn’t recommend PSA screening for early diagnosis and early treatment due to asymptomatic status of PCa for life and risks of complications originated by over-treatment. European randomized study of screening for PCa evaluated the effect of PSA screening and suggested that PSA-based screening could reduce the rate of death from PCa by 20%, but was associated with a high-risk of over-diagnosis.2 Thus, scientists are focusing on the ways to improve PSA test in order to better distinguish patients with cancers from people with benign conditions. These advanced PSA tests include: (a) free PSA (fPSA) (unbound to other proteins in the serum), is usually used as the form of ratio of fPSA/total PSA (tPSA) which can exclude the suspect of having PCa if the ratio level is lower than normal when PSA is 2.6–10 ng ml−1.11,12 (b) PSA velocity (rate of change in a man's PSA level per year), is reported that people with PSA velocity >2 ng ml−1 year−1 may have a relative high-risk of death from PCa before diagnosis, demonstrating its role in predicting cancerous state that will lead to death. Moreover, FDA approved the availability of prostate health index (PHI) as a newly developed test combination in 2012. It composes of the blood measurements of %fPSA (fPSA/tPSA), PSA and proPSA, to calculate a score. A recent meta-analysis has proved PHI's diagnostic ability of increasing the sensitivity to 90% and specificity to 32% particularly in the group of patients with PSA between 2 ng ml−1 and 10 ng ml−1.13 Further, Perdona et al.14 tested PHI score in 160 suspected patients followed by biopsies and found that PHI score was significantly higher in PCa group than PCa-negative group. These results reconfirmed that PHI could be used as a diagnostic tool in suspected patients before the first time biopsy.
Current diagnostic biomarkers
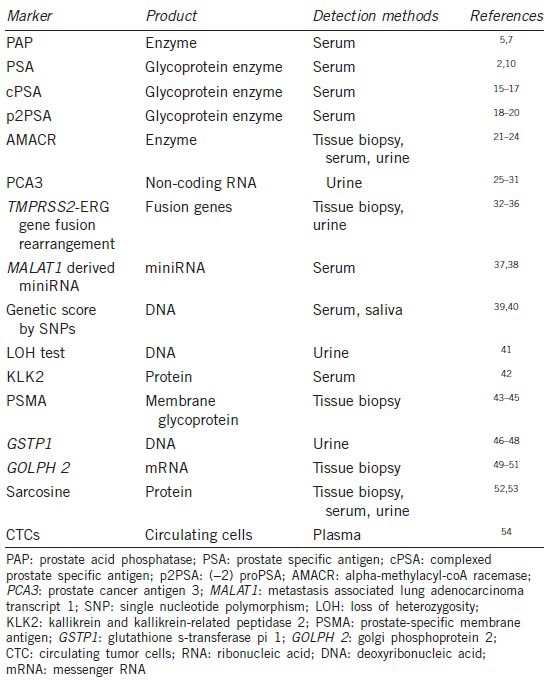
Based on the low specificity of PSA in detecting PCa, numerous novel diagnostic biomarkers found to be valuable in researches are currently applied or going to be applied in clinical uses to detect patients with a disease or abnormal condition. In general, these biomarkers can be roughly divided into three groups including deoxyribonucleic acid-based biomarkers, ribonucleic acid (RNA)-based biomarkers and protein biomarkers. Further, based on approaches to find the biomarkers, they can be divided into tissue, serum, urine, and semen diagnostic biomarkers (Table 1).
Table 1.
Current early diagnostic biomarkers of prostate cancer
Prostate specific antigen isoforms
Serum PSA originates from proPSA synthesized in prostate cells. On an average 70%–90% of serum PSA is complexed with serum protease inhibitors and is termed complexed PSA (cPSA), a kind of PSA isoform. In the process of proPSA degradation, different forms of proPSA are developed, including (−2), (−4), and (−5) proPSA, of which (−2) proPSA (p2PSA) plays the most important role and accounts for the most concentration in PCa.15
Complexed PSA is the sum of immunodetectable forms of PSA with serum protease inhibitors. Several studies with a large testing population have proved cPSA to have a better diagnostic efficiency, especially in specificity with a larger area under the curve (AUC) value than tPSA alone.16,17,18
(−2) proPSA, also known as p2PSA, is tested in its derivatives, %p2PSA (p2PSA/tPSA) which may improve both discrimination between male patients with PCa-positive and those with PCa-negative.19,20 In a recent study, Lazzeri et al.21 testified clinical performance of serum p2PSA and its derivatives, %p2PSA in 1026 men with a family history of PCa, demonstrating that both p2PSA value and %p2PSA were significantly higher in patients than healthy men, and indicating that a better threshold of %p2PSA as 1.66 with sensitivity and specificity of 70.4% and 70.1%, respectively. The results showed great improvement of p2PSA as an isoform of PSA in early detection of PCa. In addition to the diagnostic value, p2PSA also presents characteristics in the prediction of pathologic outcomes before operation,22 which needs more researches to confirm in the future.
Alpha-methylacyl-coa racemase
Alpha-methylacyl-coa racemase (AMACR) belongs to the family of isomerase, which specifies racemases and epimerases acting on other compounds. It has been shown to be associated with human cancers including neuroendocrine neoplasms of the stomach,23 hepatocellular carcinoma,24 and colorectal adenomas25 with higher expression in cancerous tissues than normal. In prostate, AMACR was proved to be overexpressed in cancer epithelium, hence becoming a potential diagnostic biomarker for cancer cells within PCa.26 In addition, epidemiologics, genetics, and laboratory studies have all pointed out the importance of AMACR in PCa.27 Although, it has been recognized that AMACR may play a key role in PCa genesis, detection approaches in the prostate tissue still perplex whether to have a biopsy for early diagnosis because of potential severe complications brought by invasive examinations. Sreekumar et al.28 screened sera for AMACR in both patients with PCa and controls. They found AMACR immunoreactivity to be statistically significantly higher in the sera from cancer case subjects than that from control subjects. In urine samples, Rogers et al.29 detected AMACR elevation in 100% patients with adenocarcinoma of prostate confirmed by biopsy. All these results show the potential application of AMACR as an early diagnostic biomarker, replacing invasive biopsy. However, AMACR still has some limitations that affect its improvement. It can cause humoral responses and production of endogenous antibody, which might lower the serum AMACR detection rate.30 Nevertheless, more testing is under way to evaluate the possibility of using AMACR as an early diagnostic biomarker for PCa
Prostate cancer antigen 3
Prostate cancer antigen 3 (PCA3 or DD3) is a non-coding RNA, which is only detected in human prostate tissue and highly overexpressed in PCa.31 PCA3 can be detected in urine and has a relative lower sensitivity, but much higher specificity in diagnosing PCa when compared with PSA.32 In the first two clinical trials from Canada33 and Austria,34 PCA3 was evaluated for the potential diagnostic value in urine. More than 700 men undergoing prostate biopsy donated urine after digital rectal examination in order to make sure that there were sufficient prostate epithelial materials in the urine to be detected. The sensitivity and specificity of PCA3 were 66%–82% and 76%–89%, respectively. The PCA3 score has been considered as a new genetic test that determines whether products of genes associated with PCa are present in the urine, which can be calculated by the ratio of PCA3 to PSA messenger RNA (mRNA). It represents the expression of PCA3 corrected for the background of normal or BPH epithelial cells present in the specimen. A strong correlation was observed between PCA3 score and the probability of PCa, ranging from 14% when PCA3 score lower than 5 to 69% when higher than 100. The authors also concluded that PCA3 score is useful in patients with a previous negative biopsy or without any biopsy.35,36 It can also be used to predict tumor aggressiveness and treatment options.37 In addition, a recent prospective study suggested that urinary PCA3 testing with increased PSA value can decrease the number of unnecessary prostate biopsy.38 However, the limitation of PCA3 includes lacking most appropriate cutoff level of PCA3 score and false negative results that some PCa, especially aggressive tumors, may present with a low PCA3 score, affecting the determination.
TMPRSS2-ERG gene fusion rearrangement
Gene fusion between TMPRSS2 (21q22) and ERG (21q22), ETV1 (7q21), ETV4 (17q21) or ETV5 (3q27) are commonly identified in PCa.39 Among them, TMPRSS2-ERG fusions-positive is detected in approximately 50% of Caucasian with PCa.40 TMPRSS2 refers to transmembrane protease serine two which is androgen responsive. ERG encodes for a protein that functions as a transcriptional regulator, the overexpression of which may contribute to development of androgen-independence in PCa. Essentially, the fusion between TMPRSS2 and ERG may disrupt the ability of cells to differentiate into proper and normal prostate cells, resulting in forming unorganized tissue.41 The fusion can be detected in patients’ urine without any invasive approaches, and it was shown that the fusion could be detected in 42% of patients with localized PCa, indicating the ability for noninvasive early diagnosis.42 Moreover, with the deepening of studies, researchers found that TMPRSS2-ERG gene fusion was associated with disease progression. A cohort tracking study was performed by Pettersson et al.43 for 12.6 years after 1180 men being treated with radical prostatectomy, and a meta-analysis including 5074 men, both found the relationship between TMPRSS2-ERG gene fusion and the stage of diagnosis. Hessels et al.44 intriguingly, discovered the combination of TMPRSS2-ERG and PCA3 could increase the sensitivity from 37% to 73% and give a better indication for patients to have repeat biopsies. In the latest research, combining urinary detection of TMPRSS2-ERG and PCA3 with serum PSA can greatly improve the prediction (AUC = 0.88; specificity = 90% at 80% sensitivity).45 However, the limitation of using TMPRSS2-ERG gene fusion rearrangement as the early diagnostic biomarker lies in its inconsistence between different populations, as we found the detection rate of TMPRSS2-ERG was only 18.5% in Chinese population, which was obviously lower than that in Caucasians.46
Metastasis associated lung adenocarcinoma transcript 1 derived mini ribonucleic acid
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) was first discovered to be a long noncoding RNA that can predict metastasis and survival in early-stage non-small cell lung cancer in 2003.47 Further studies have confirmed that the overexpression of MALAT1 is related to human hepatocellular carcinoma, breast cancer, pancreatic cancer, colon cancer, and PCa.48 We performed an RNA-sequencing in 14 paired Chinese PCa samples and identified significant MALAT1 overexpression in PCa tissue.46 Further, to decide whether MALAT1 can be applied to early diagnosis of PCa in serum, we discovered that MALAT1 had the form of miniRNA fragment in the sera of patients with PCa and thus examined the diagnostic ability in 87 cancer, 82 BHP and 23 healthy controls sera samples. We found that MALAT1-derived miniRNA was more effective to distinguish PCa from non-PCa than PSA.49 The results showed a great potential of MALAT1-derived miniRNA applied in PCa early diagnosis in the future.
Genetic score based on single nucleotide polymorphisms
The fact that PCa is linked to genetic factors was verified by twin studies which suggested that over 42% of etiologic factors result from genes.50 Scientists have discovered more than 50 risk single nucleotide polymorphisms (SNPs) associated with the tumorigenesis of PCa in Caucasian population,51 and more than twenty ones in Chinese population, including two novel ones in our previous findings.52 Genetic score is a recently proposed measurement of inherited risk for PCa, calculated by genotypes of the panel of PCa-related risk SNPs that are weighted by their relative risk (RR) to PCa.53 In the REduction by DUtasteride of PCa events study, genetic score was presented to increase PCa risk prediction in patients with PSA > 2.5 ng ml−1 who needed rebiopsy operation.54 We selected 33 PCa-related risk SNPs as a panel to be tested in Chinese population and found the score was significantly higher in patients with cancer than in controls. This confirmed the improved prediction ability of PCa by genetic score.55 However, genetic score has the plateau effect on discriminating PCa. We evaluated the performance of 29 risk SNPs related to PCa and compared with the previous 24 ones. Consequently, genetic scores and AUC were both similar between them, leading to the indication that PCa-related risk SNPs with small RR was unlikely to further improve the predictive effect.56
Loss of heterozygosity test
Loss of heterozygosity (LOH) occurs in PCa as a form of copy number changes, which locates in 7q, 8p, 10q, 12p, 13p, 16p, 17p, and 18q, respectively.57 Urinary LOH test was performed and discriminated between patients with PCa and healthy ones, obtaining sensitivity as high as 87% through a PCR based assay. However, the cases for analysis were limited to 19 specimens.57 Although LOH test is widely used in detecting bladder cancer, it has little been applied to PCa detection.
Tissue kallikrein and kallikrein-related peptidase 2
Kallikrein comprise a family of 15 homologous secreted trypsin-or chymotrypsin-like serine protease.58 Kallikrein-related peptidase 2 (KLK2) has the most organ restricted expression in the luminal epithelium of human prostate with KLK3 PSA. Unlike PSA, most of KLK2 have an unbound form in patients’ sera. The ratio of KLK2 to PSA was reported to increase PCa detection rate in patients with tPSA ranging from 2 to 10 ng ml−1.59 Further studies are required to evaluate the diagnostic effect of KLK2.
Prostate-specific membrane antigen
Prostate-specific membrane antigen (PSMA), also known as glutamate carboxypeptidase II, is a kind of membrane glycoprotein that is strongly expressed in epithelia cells of prostate, encoded by folate hydrolase 1 gene.60 PSMA was proved to be overexpressed 8- to 12-fold in PCa tissues rather than in noncancerous tissues.61 Although PSMA has been regarded as a targeted biomarker for therapy and prognosis prediction for a long time due to its high concentration in PCa,62,63 it showed much greater effect on PCa detection when combined with PET imaging recently.64 Moreover, researchers in University of California have developed a test by using modified phage viruses to detect PSMA for PCa early diagnosis, which is supposed to be operated at home.65
Glutathione s-transferase pi 1
Glutathione s-transferase pi 1 (GSTP1) is a polymorphic gene encoding active, functionally different GSTP1 variant proteins that are thought to function in xenobiotic metabolism and play a role in susceptibility to cancer, and other diseases.66 Many studies have detected and confirmed that GSTP1 methylation in urine can help enhance the sensitivity and specificity of PCa diagnosis in a noninvasive manner than PSA.67,68,69 Due to several specific characteristics, including its prevalent status in the cancer condition and noninvasive detection through urine, GSTP1 is regarded as a good and promising diagnostic biomarker.
Golgi phosphoprotein 2
Golgi phosphoprotein 2 (GOLPH 2) mRNA was first found to have upregulated expression in PCa tissue in 2002.70 Further study confirmed the overexpression of GOLPH 2 mRNA in PCa tissues rather than normal and adjacent tissues, and 91.4% of PCa cases showed an upregulation of GOLPH, which could serve as a novel diagnostic biomarker in the condition of AMACR-negative tissues.71 In addition to detecting GOLPH 2 in PCa tissue, GOLPH 2 mRNA was described to be one of the complex biomarker panels in urine to diagnose PCa even regardless of serum PSA.72 All these results greatly manifest the extraordinary talent of GOLPH 2 in early diagnosis of PCa, especially in AMACR-negative ones.
Sarcosine
Sarcosine is known as an intermediate and byproduct in glycine synthesis and degradation. Sreekumar et al.73 profiled more than 1126 metabolites across 262 clinical samples including tissue, serum and urine and found that sarcosine was highly increased in urine in group of PCa, indicating its vital role in prostate tumorigenesis. Another study investigated serum samples from 290 patients with PCa and 310 healthy men, and concluded that serum sarcosine could increase the accuracy of PCa detection, especially in patients with tPSA <4.0 ng ml−1.74 However, several studies discovered that sarcosine was not good enough to be detected and considered it as a promising diagnostic and prognostic prediction biomarker in urine and serum.75,76 More detailed investigations are required to decide whether or not sarcosine can be a diagnostic biomarker of PCa.
Circulating tumor cells
Circulating tumor cells (CTCs) are cells that shed from the origin part of the cancer and circulate into the bloodstream, forming seeds for cancer growth of additional cancers in vital distant organs and triggering a mechanism responsible for cancer-related deaths.77 CTCs were first observed in 1869 by Thomas Ashworth and validated the existence in various cancers. In Schulich School of Medicine and Dentistry, Dr. Hong Leong ran a blood test of CTCs in 50 noncancer and cancer patients. The results showed 90% accuracy in predicting patients with actual PCa and no false positives (no data shown). Since the application of single-cell sequencing is widespread recently,78 the test of CTCs will be easily carried out and the effectiveness of cancer related events will be largely improved, especially in prognostic prediction of overall survival in patients with metastatic PCa,79 and in therapy target decision in personalized medicine era.80 Taking more efforts, such as generation of enormous data and comprehensive studies, are necessary and promising to contribute to the development of CTCs applied to PCa diagnosis and treatment.
CONCLUSIONS AND PROSPECTIVE
Prostate cancer is a kind of heterogeneous cancer with different phenotypes and outcomes.81 Most PCa present to be indolent with mild and atypical clinical manifestations, which may confuse and mislead clinicians and patients at the point of disease diagnosis. The screening of PCa by PSA in the last century has greatly promoted early diagnosis and intervention, thus lowering the disease related mortality. However, PSA has the shortage of low specificity, which leads to over-diagnosis and over-treatment for the patients. Thus, extensive efforts have been made to identify better biomarkers in order to guide early diagnosis and prevent the disease from progression. Numerous emerging biomarkers for PCa have been discovered and been applied to clinical uses recently, bringing new insight of PCa to researchers and clinicians as well as producing plenty of novel screening tests for potential patients. However, it is a challenge to substitute for PSA because of its minimally invasive characteristic and relatively low cost, accompanying its high sensitivity.
High-throughput technology methods and advances in molecular biology are helping and accelerating the exploration to useful biomarkers. Next generation sequencing (NGS), as a newly developed technology, has been widely applied to detect biomarkers in the area of PCa and other diseases, as it produces thousands or millions of sequences concurrently with lower cost.82 The methods of NGS include single-molecule real-time sequencing, ion semiconductor, pyrosequencing, sequencing by synthesis and sequencing by ligation with different mechanisms in genetic sequence mensuration.
Although an increasing number of biomarkers are discovered to contribute to the early diagnosis of PCa, the consideration of both their sensitivity and specificity is still challenging. The analysis of a panel of multiple biomarkers may better indicate the presence and progression of the disease. Besides, testing methods such as via patients’ serum, or urine, or biopsy is critical to the application of a diagnostic biomarker for it should follow the rules that noninvasive testing is the most suitable approach to the patients without any traumatic complications.
In summary, as a significant public health threat worldwide, especially in countries where patients can have life expectancies long enough to a clinical manifestation of the disease, the early detection of PCa is critical and necessary to reduce the burden of men's health problem. In the future, more studies are needed to reconfirm the features of the existing biomarkers and further discover novel potential ones to better predict the presence of the disease.
AUTHOR CONTRIBUTIONS
YHS and SCR conceived the idea and made up the structure. MQ drafted and revised the manuscript.
COMPETING INTERESTS
The authors declare no competing interests.
ACKNOWLEDGMENT
This study was supported by the National Basic Research Program of China (No. 2012CB518306), the National Natural Science Foundation of China (No. 81101946), the Prostate Cancer Foundation Young Investigator Award, and the Shanghai Pujiang Program (No. 12PJD008).
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, et al. Screening and prostate-cancer mortality in a randomized European study. The New England journal of medicine. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Moyer VA. Screening for prostate cancer: U.S. Preventive Services Task Force recommendation statement. Annals of internal medicine. 2012;157:120–34. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 4.Schroder FH. Stratifying risk-the U.S. Preventive Services Task Force and prostate-cancer screening. The New England journal of medicine. 2011;365:1953–5. doi: 10.1056/NEJMp1112140. [DOI] [PubMed] [Google Scholar]
- 5.Gutman AB, Gutman EB. An “Acid” Phosphatase Occurring in the Serum of Patients with Metastasizing Carcinoma of the Prostate Gland. The Journal of clinical investigation. 1938;17:473–8. doi: 10.1172/JCI100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez J, Thompson IM. Prostate-specific antigen: a review of the validation of the most commonly used cancer biomarker. Cancer. 2004;101:894–904. doi: 10.1002/cncr.20480. [DOI] [PubMed] [Google Scholar]
- 7.Gunia S, Koch S, May M, Dietel M, Erbersdobler A. Expression of prostatic acid phosphatase (PSAP) in transurethral resection specimens of the prostate is predictive of histopathologic tumor stage in subsequent radical prostatectomies. Virchows Archiv : an international journal of pathology. 2009;454:573–9. doi: 10.1007/s00428-009-0759-1. [DOI] [PubMed] [Google Scholar]
- 8.Drake CG. Re: interdisciplinary critique of sipuleucel-T as immunotherapy in castration-resistant prostate cancer. Journal of the National Cancer Institute. 2012;104:1422–3. doi: 10.1093/jnci/djs340. author reply. [DOI] [PubMed] [Google Scholar]
- 9.Gerritsen WR. The evolving role of immunotherapy in prostate cancer. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2012;23(Suppl 8):viii22–7. doi: 10.1093/annonc/mds259. [DOI] [PubMed] [Google Scholar]
- 10.Wang MC, Valenzuela LA, Murphy GP, Chu TM. Purification of a human prostate specific antigen. Investigative urology. 1979;17:159–63. [PubMed] [Google Scholar]
- 11.Catalona WJ, Smith DS, Ornstein DK. Prostate cancer detection in men with serum PSA concentrations of 2.6 to 4.0 ng/mL and benign prostate examination. Enhancement of specificity with free PSA measurements. JAMA : the journal of the American Medical Association. 1997;277:1452–5. [PubMed] [Google Scholar]
- 12.Ito K, Yamamoto T, Ohi M, Kurokawa K, Suzuki K, et al. Free/total PSA ratio is a powerful predictor of future prostate cancer morbidity in men with initial PSA levels of 4.1 to 10.0 ng/mL. Urology. 2003;61:740–4. doi: 10.1016/s0090-4295(02)02427-5. [DOI] [PubMed] [Google Scholar]
- 13.Filella X, Gimenez N. Evaluation of [-2] proPSA and Prostate Health Index (phi) for the detection of prostate cancer: a systematic review and meta-analysis. Clinical chemistry and laboratory medicine : CCLM / FESCC. 2013;51:729–39. doi: 10.1515/cclm-2012-0410. [DOI] [PubMed] [Google Scholar]
- 14.Perdona S, Bruzzese D, Ferro M, Autorino R, Marino A, et al. Prostate health index (phi) and prostate cancer antigen 3 (PCA3) significantly improve diagnostic accuracy in patients undergoing prostate biopsy. The Prostate. 2013;73:227–35. doi: 10.1002/pros.22561. [DOI] [PubMed] [Google Scholar]
- 15.Ozen H, Sozen S. PSA isoforms in prostate cancer detection. Eur Urol Suppl. 2006;5:495–9. [Google Scholar]
- 16.Tanguay S, Begin LR, Elhilali MM, Behlouli H, Karakiewicz PI, et al. Comparative evaluation of total PSA, free/total PSA, and complexed PSA in prostate cancer detection. Urology. 2002;59:261–5. doi: 10.1016/s0090-4295(01)01497-2. [DOI] [PubMed] [Google Scholar]
- 17.Sozen S, Eskicorapci S, Kupeli B, Irkilata L, Altinel M, et al. Complexed prostate specific antigen density is better than the other PSA derivatives for detection of prostate cancer in men with total PSA between 2.5 and 20 ng/ml: results of a prospective multicenter study. European urology. 2005;47:302–7. doi: 10.1016/j.eururo.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Partin AW, Brawer MK, Bartsch G, Horninger W, Taneja SS, et al. Complexed prostate specific antigen improves specificity for prostate cancer detection: results of a prospective multicenter clinical trial. The Journal of urology. 2003;170:1787–91. doi: 10.1097/01.ju.0000092695.55705.dd. [DOI] [PubMed] [Google Scholar]
- 19.Guazzoni G, Nava L, Lazzeri M, Scattoni V, Lughezzani G, et al. Prostate-specific antigen (PSA) isoform p2PSA significantly improves the prediction of prostate cancer at initial extended prostate biopsies in patients with total PSA between 2.0 and 10 ng/ml: results of a prospective study in a clinical setting. European urology. 2011;60:214–22. doi: 10.1016/j.eururo.2011.03.052. [DOI] [PubMed] [Google Scholar]
- 20.Catalona WJ, Partin AW, Sanda MG, Wei JT, Klee GG, et al. A multicenter study of [-2]pro-prostate specific antigen combined with prostate specific antigen and free prostate specific antigen for prostate cancer detection in the 2.0 to 10.0 ng/ ml prostate specific antigen range. The Journal of urology. 2011;185:1650–5. doi: 10.1016/j.juro.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzeri M, Haese A, Abrate A, de la Taille A, Redorta JP, et al. Clinical performance of serum prostate-specific antigen isoform [-2]proPSA (p2PSA) and its derivatives, %p2PSA and the prostate health index (PHI), in men with a family history of prostate cancer: results from a multicentre European study, the PROMEtheuS project. BJU international. 2013;112:313–21. doi: 10.1111/bju.12217. [DOI] [PubMed] [Google Scholar]
- 22.Guazzoni G, Lazzeri M, Nava L, Lughezzani G, Larcher A, et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer. European urology. 2012;61:455–66. doi: 10.1016/j.eururo.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 23.Annenkov A, Nishikura K, Domori K, Ajioka Y. Alpha-methylacyl-coenzyme A racemase expression in neuroendocrine neoplasms of the stomach. Virchows Archiv : an international journal of pathology. 2012;461:169–75. doi: 10.1007/s00428-012-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helal Tel A, Radwan NA, Abdel Kader Z, Helmy NA, Hammad SY. Role of alpha methylacyl-coenzyme A racemase in differentiating hepatocellular carcinoma from dysplastic and nondysplastic liver cell lesions. Annals of diagnostic pathology. 2012;16:330–4. doi: 10.1016/j.anndiagpath.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Lakis S, Papamitsou T, Panagiotopoulou C, Kotakidou R, Kotoula V. AMACR is associated with advanced pathologic risk factors in sporadic colorectal adenomas. World journal of gastroenterology : WJG. 2010;16:2476–83. doi: 10.3748/wjg.v16.i20.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Z, Woda BA, Rock KL, Xu Y, Savas L, et al. P504S: a new molecular marker for the detection of prostate carcinoma. The American journal of surgical pathology. 2001;25:1397–404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Carter HB, Isaacs WB. Improved biomarkers for prostate cancer: a definite need. Journal of the National Cancer Institute. 2004;96:813–5. doi: 10.1093/jnci/djh174. [DOI] [PubMed] [Google Scholar]
- 28.Sreekumar A, Laxman B, Rhodes DR, Bhagavathula S, Harwood J, et al. Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. Journal of the National Cancer Institute. 2004;96:834–43. doi: 10.1093/jnci/djh145. [DOI] [PubMed] [Google Scholar]
- 29.Rogers CG, Yan G, Zha S, Gonzalgo ML, Isaacs WB, et al. Prostate cancer detection on urinalysis for alpha methylacyl coenzyme a racemase protein. The Journal of urology. 2004;172:1501–3. doi: 10.1097/01.ju.0000137659.53129.14. [DOI] [PubMed] [Google Scholar]
- 30.Tricoli JV, Schoenfeldt M, Conley BA. Detection of prostate cancer and predicting progression: current and future diagnostic markers. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:3943–53. doi: 10.1158/1078-0432.CCR-03-0200. [DOI] [PubMed] [Google Scholar]
- 31.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer research. 1999;59:5975–9. [PubMed] [Google Scholar]
- 32.Vlaeminck-Guillem V, Ruffion A, Andre J. Value of urinary PCA3 test for prostate cancer diagnosis. Progres en urologie : journal de l’Association francaise d’urologie et de la Societe francaise d’urologie et de la Societe francaise d’urologie. 2008;18:259–65. doi: 10.1016/j.purol.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 33.Fradet Y, Saad F, Aprikian A, Dessureault J, Elhilali M, et al. uPM3, a new molecular urine test for the detection of prostate cancer. Urology. 2004;64:311. doi: 10.1016/j.urology.2004.03.052. [DOI] [PubMed] [Google Scholar]
- 34.Tinzl M, Marberger M, Horvath S, Chypre C. DD3PCA3 RNA analysis in urine--a new perspective for detecting prostate cancer. European urology. 2004;46:182–6. doi: 10.1016/j.eururo.2004.06.004. discussion 7. [DOI] [PubMed] [Google Scholar]
- 35.Deras IL, Aubin SM, Blase A, Day JR, Koo S, et al. PCA3: a molecular urine assay for predicting prostate biopsy outcome. The Journal of urology. 2008;179:1587–92. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 36.Haese A, de la Taille A, van Poppel H, Marberger M, Stenzl A, et al. Clinical utility of the PCA3 urine assay in European men scheduled for repeat biopsy. European urology. 2008;54:1081–8. doi: 10.1016/j.eururo.2008.06.071. [DOI] [PubMed] [Google Scholar]
- 37.Nakanishi H, Groskopf J, Fritsche HA, Bhadkamkar V, Blase A, et al. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. The Journal of urology. 2008;179:1804. doi: 10.1016/j.juro.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Crawford ED, Rove KO, Trabulsi EJ, Qian J, Drewnowska KP, et al. Diagnostic performance of PCA3 to detect prostate cancer in men with increased prostate specific antigen: a prospective study of 1,962 cases. The Journal of urology. 2012;188:1726–31. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 40.Esgueva R, Perner S, JL C, Scheble V, Stephan C, et al. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23:539–46. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer cell. 2010;17:443–54. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, et al. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8:885–8. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersson A, Graff RE, Bauer SR, Pitt MJ, Lis RT, et al. The TMPRSS2:ERG rearrangement, ERG expression, and prostate cancer outcomes: a cohort study and meta-analysis. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessels D, Smit FP, Verhaegh GW, Witjes JA, Cornel EB, et al. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:5103–8. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 45.Salami SS, Schmidt F, Laxman B, Regan MM, Rickman DS, et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urologic oncology. 2013;31:566–71. doi: 10.1016/j.urolonc.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren S, Peng Z, Mao JH, Yu Y, Yin C, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell research. 2012;22:806–21. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji P, Diederichs S, Wang W, Boing S, Metzger R, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 48.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 49.Ren S, Wang F, Shen J, Sun Y, Xu W, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–59. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 50.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England journal of medicine. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 51.Nordstrom T, Aly M, Eklund M, Egevad L, Gronberg H. A Genetic Score Can Identify Men at High Risk for Prostate Cancer Among Men With Prostate-Specific Antigen of 1-3 ng/ml. European urology. 2013 doi: 10.1016/j.eururo.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Mo Z, Ye D, Wang M, Liu F, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nature genetics. 2012;44:1231–5. doi: 10.1038/ng.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun J, Na R, Hsu FC, Zheng SL, Wiklund F, et al. Genetic score is an objective and better measurement of inherited risk of prostate cancer than family history. European urology. 2013;63:585–7. doi: 10.1016/j.eururo.2012.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kader AK, Sun J, Reck BH, Newcombe PJ, Kim ST, et al. Potential impact of adding genetic markers to clinical parameters in predicting prostate biopsy outcomes in men following an initial negative biopsy: findings from the REDUCE trial. European urology. 2012;62:953–61. doi: 10.1016/j.eururo.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng J, Liu F, Lin X, Wang X, Ding Q, et al. Predictive performance of prostate cancer risk in Chinese men using 33 reported prostate cancer risk-associated SNPs. The Prostate. 2012;72:577–83. doi: 10.1002/pros.21462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren S, Xu J, Zhou T, Jiang H, Chen H, et al. Plateau effect of prostate cancer risk-associated SNPs in discriminating prostate biopsy outcomes. The Prostate. 2013 doi: 10.1002/pros.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thuret R, Chantrel-Groussard K, Azzouzi AR, Villette JM, Guimard S, et al. Clinical relevance of genetic instability in prostatic cells obtained by prostatic massage in early prostate cancer. British journal of cancer. 2005;92:236–40. doi: 10.1038/sj.bjc.6602311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundwall A, Band V, Blaber M, Clements JA, Courty Y, et al. A comprehensive nomenclature for serine proteases with homology to tissue kallikreins. Biological chemistry. 2006;387:637–41. doi: 10.1515/BC.2006.082. [DOI] [PubMed] [Google Scholar]
- 59.Magklara A, Scorilas A, Catalona WJ, Diamandis EP. The combination of human glandular kallikrein and free prostate-specific antigen (PSA) enhances discrimination between prostate cancer and benign prostatic hyperplasia in patients with moderately increased total PSA. Clinical chemistry. 1999;45:1960–6. [PubMed] [Google Scholar]
- 60.O’Keefe DS, Su SL, Bacich DJ, Horiguchi Y, Luo Y, et al. Mapping, genomic organization and promoter analysis of the human prostate-specific membrane antigen gene. Biochimica et biophysica acta. 1998;1443:113–27. doi: 10.1016/s0167-4781(98)00200-0. [DOI] [PubMed] [Google Scholar]
- 61.O’Keefe DS, Bacich DJ, Heston WD. Comparative analysis of prostate-specific membrane antigen (PSMA) versus a prostate-specific membrane antigen-like gene. The Prostate. 2004;58:200–10. doi: 10.1002/pros.10319. [DOI] [PubMed] [Google Scholar]
- 62.Wang X, Yin L, Rao P, Stein R, Harsch KM, et al. Targeted treatment of prostate cancer. Journal of cellular biochemistry. 2007;102:571–9. doi: 10.1002/jcb.21491. [DOI] [PubMed] [Google Scholar]
- 63.Perner S, Hofer MD, Kim R, Shah RB, Li H, et al. Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Human pathology. 2007;38:696–701. doi: 10.1016/j.humpath.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 64.Afshar-Oromieh A, Malcher A, Eder M, Eisenhut M, Linhart HG, et al. PET imaging with a [68Ga]gallium-labelled PSMA ligand for the diagnosis of prostate cancer: biodistribution in humans and first evaluation of tumour lesions. European journal of nuclear medicine and molecular imaging. 2013;40:486–95. doi: 10.1007/s00259-012-2298-2. [DOI] [PubMed] [Google Scholar]
- 65.Home tests for prostate-specific membrane antigen being developed for prostate cancer diagnosis. Expert review of molecular diagnostics. 2013;13:523–5. doi: 10.1586/14737159.2013.825130. [DOI] [PubMed] [Google Scholar]
- 66.Smith CM, Bora PS, Bora NS, Jones C, Gerhard DS. Genetic and radiation-reduced somatic cell hybrid sublocalization of the human GSTP1 gene. Cytogenetics and cell genetics. 1995;71:235–9. doi: 10.1159/000134117. [DOI] [PubMed] [Google Scholar]
- 67.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clinical cancer research : an official journal of the American Association for Cancer Research. 2003;9:2673–7. [PubMed] [Google Scholar]
- 68.Cairns P, Esteller M, Herman JG, Schoenberg M, Jeronimo C, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:2727–30. [PubMed] [Google Scholar]
- 69.Woodson K, O’Reilly KJ, Hanson JC, Nelson D, Walk EL, et al. The usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancer. The Journal of urology. 2008;179:508. doi: 10.1016/j.juro.2007.09.073. [DOI] [PubMed] [Google Scholar]
- 70.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, et al. Gene expression analysis of prostate cancers. Molecular carcinogenesis. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 71.Kristiansen G, Fritzsche FR, Wassermann K, Jager C, Tolls A, et al. GOLPH2 protein expression as a novel tissue biomarker for prostate cancer: implications for tissue-based diagnostics. British journal of cancer. 2008;99:939–48. doi: 10.1038/sj.bjc.6604614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer research. 2008;68:645–9. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 74.Lucarelli G, Fanelli M, Larocca AM, Germinario CA, Rutigliano M, et al. Serum sarcosine increases the accuracy of prostate cancer detection in patients with total serum PSA less than 4.0 ng/ml. The Prostate. 2012;72:1611–21. doi: 10.1002/pros.22514. [DOI] [PubMed] [Google Scholar]
- 75.Jentzmik F, Stephan C, Miller K, Schrader M, Erbersdobler A, et al. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumours. European urology. 2010;58:12–8. doi: 10.1016/j.eururo.2010.01.035. discussion 20-1. [DOI] [PubMed] [Google Scholar]
- 76.Struys EA, Heijboer AC, van Moorselaar J, Jakobs C, Blankenstein MA. Serum sarcosine is not a marker for prostate cancer. Annals of clinical biochemistry. 2010;47:282. doi: 10.1258/acb.2010.009270. [DOI] [PubMed] [Google Scholar]
- 77.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nature reviews. Genetics. 2013;14:618–30. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 79.Thalgott M, Rack B, Maurer T, Souvatzoglou M, Eiber M, et al. Detection of circulating tumor cells in different stages of prostate cancer. Journal of cancer research and clinical oncology. 2013;139:755–63. doi: 10.1007/s00432-013-1377-5. [DOI] [PubMed] [Google Scholar]
- 80.Gorges TM, Pantel K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer immunology, immunotherapy : CII. 2013;62:931–9. doi: 10.1007/s00262-012-1387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren SC, Qu M, Sun YH. Investigating intratumour heterogeneity by single-cell sequencing. Asian journal of andrology. 2013;15:729–34. doi: 10.1038/aja.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–25. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]


