Abstract
Polybrominated diphenyl ethers (PBDEs) and polybrominated bipyrroles are natural products that bioaccumulate in the marine food chain. PBDEs have attracted widespread attention due to their persistence in the environment and potential toxicity to humans. However, the natural origins of PBDE biosynthesis are not known. Here we report marine bacteria as producers of PBDEs and establish a genetic and molecular foundation for their production that unifies paradigms for the elaboration of bromophenols and bromopyrroles abundant in marine biota. We provide biochemical evidence of marine brominase enzymes revealing decarboxylative-halogenation enzymology previously unknown among halogenating enzymes. Biosynthetic motifs discovered in our study were used to mine sequence databases to discover unrealized marine bacterial producers of organobromine compounds.
Halogenation, particularly bromination, is a diagnostic feature of marine natural products. While bromide is present in only trace amounts terrestrially, its greater natural abundance in seawater provides a substrate reservoir for halogenating enzymes evolved to selectively activate and transfer bromide to organic molecules. Marine organisms, including bacteria, algae and invertebrates, are prolific sources of organobromine compounds that number over 2200 reported molecules with a wide range of biological properties1–2. These brominated natural products range from simple volatile polybromoalkanes3 to highly complex alkaloids4. Yet, our understanding of the biosynthesis of brominated natural products has lagged far behind their discovery largely because few biosynthetic gene loci have been identified and characterized. Herein we report a pervasive marine bacterial pathway to polybrominated aromatic compounds that accounts for a diverse suite of common polybrominated pyrrole and phenol-based natural products.
In their hydroxylated (OH-BDE) and methoxylated (MeO-BDE) forms, PBDEs are abundant across all trophic levels of marine life ranging from marine plants5, algae6–8 and invertebrates9–12, to marine mammals at the apex of the food chain13–15. OH-BDEs and MeO-BDEs detected in marine biota were once thought to be derived from chemical transformation of anthropogenically produced polybrominated flame retardants chemicals of similar structure16. However, derivatives such as 2’-MeO-BDE-47, a metabolite widely detected in the marine metabolome (Fig. 1a), were shown through Δ14C measurements to be of natural origin15. Despite the mounting evidence for the biosynthesis of OH-BDEs in the natural metabolome, no natural producer of these potential toxins that target mammalian nuclear hormone mediated signaling pathways17 has been confirmed.
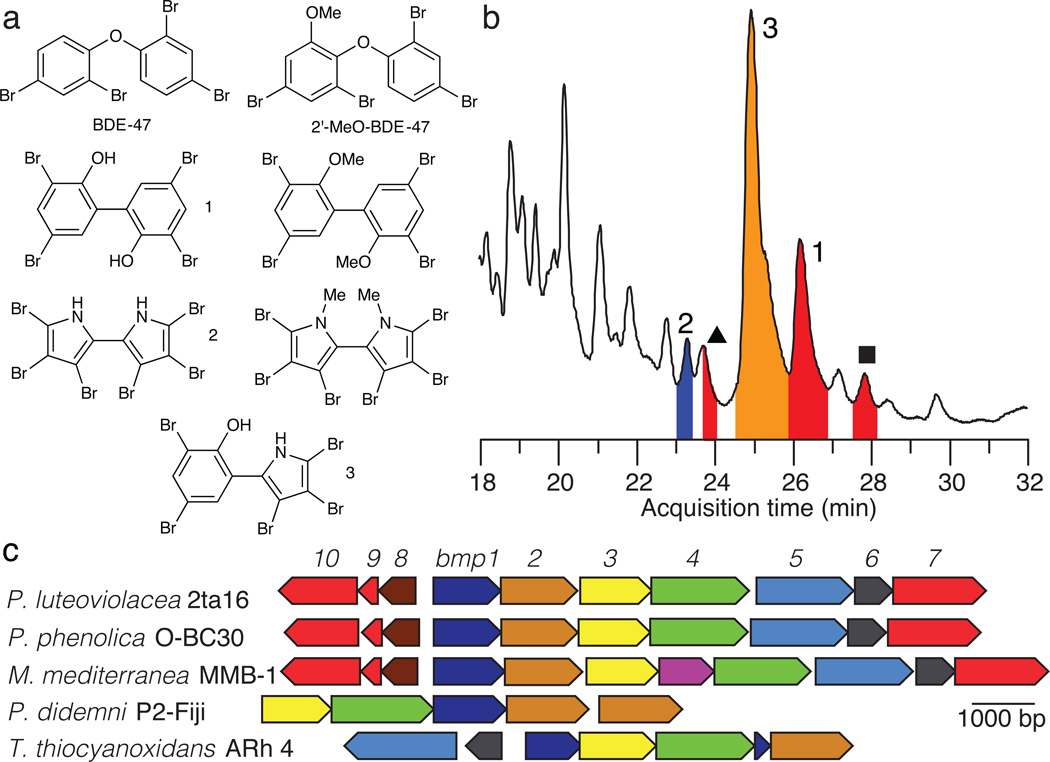
Figure 1. Structures and sources of marine polybrominated natural products.
(a) Representative structures of polybrominated molecules produced anthropogenically (BDE-47), or microbially (1–3), and their derivatives detected in the environment. (b) Total ion chromatogram (TIC) (black curve) from LC-MS/MS analysis of an organic extract of P. luteoviolacea 2ta16 demonstrating production of the 1–3. Peaks labeled ● and ▲ were later correlated to OH-BDEs 11 and 12, respectively (vide infra). (c) Organization of bmp gene loci in marine bacteria. Note that M. mediterranea MMB-1 encodes a putative permease between bmp3 and bmp4.
An abundance of PBDEs at all trophic levels of marine Eukarya suggests lower trophic level sources for their genesis. This hypothesis is supported by the production of chemically similar polybrominated molecules, such as 3,3’,5,5’-tetrabromo-2,2’-biphenyldiol (1)18, hexabromo-2,2’-bipyrrole (2)19, and the hybrid bromophenol-bromopyrrole pentabromopseudilin (3)19–20 (Fig. 1a), by Pseudoalteromonas spp., marine γ-proteobacteria often associated with eukaryotic hosts21. While 1–2 are not industrially synthesized, their methylated analogs are extensively detected in marine mammals14,22 (Fig. 1a). Notably, in contrast to the microbial biosynthesis of polybrominated biphenyls and bipyrroles, production of PBDEs has not been confirmed from marine bacterial sources.
Herein we report the discovery of a conserved biosynthetic gene cluster in marine bacteria responsible for the synthesis of widespread polybrominated aromatic compounds and describe two flavin-dependent brominases involved in the synthesis of universal polybromophenol- and polybromopyrrole-based metabolites. For the first time, our findings establish marine bacteria as sources of OH-BDEs. Moreover, we provide the first report and biochemical characterization of a flavin-dependent decarboxylative-brominase enzyme employing an inferred enzyme architecture not previously realized among halogenases.
RESULTS
A genetic basis for bromination in marine bacteria
In our search for marine prokaryotic sources of polybrominated molecules, we focused on the cosmopolitan marine genus Pseudoalteromonas spp. previously shown to produce 1–3. Specifically, we queried the small molecule natural products synthesized by coral-associated P. luteoviolacea 2ta16 isolated in the Florida Keys23, and by planktonic P. phenolica O-BC30 isolated off the coast of Japan18. We analyzed organic extracts of liquid cultures of these marine bacteria grown in the presence of bromide by high performance liquid chromatography-tandem mass spectrometry (LC-MS/MS). Among numerous polybrominated molecules present in culture extract of both bacteria, we confirmed the production of 1–3 by mass spectrometry and NMR spectroscopy of isolated compounds (Figs. 1b, Supplementary Results, Supplementary Figs. 1–2 and Supplementary Table 1). Additionally, we confirmed the production of a large series of bromophenol and bromopyrrole monomeric molecules, including 2,4-dibromophenol (4), 2,4,6-tribromophenol (5), 2,3,4-tribromopyrrole (6) and 2,3,4,5-tetrabromopyrrole (7) on the basis of their MS spectra and comparison to synthetic standards (Supplementary Fig. 2 and Supplementary Table 1).
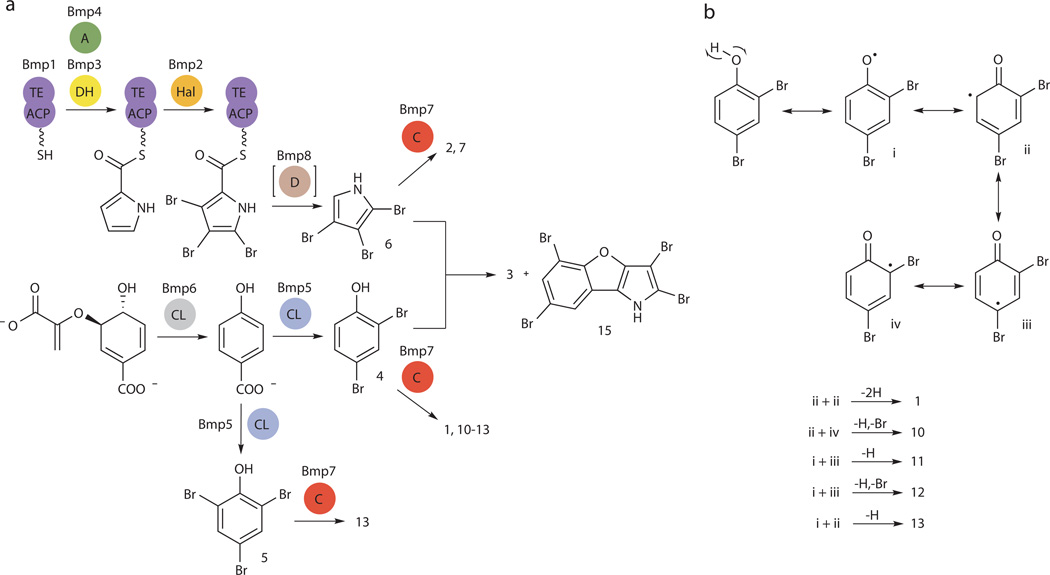
Having established experimentally tractable marine bacterial sources for the production of 1–7, we next sought to determine the genetic basis for the production of these polybrominated small molecule marine natural products. Towards this aim, we sequenced the genomes of P. luteoviolacea 2ta16 and P. phenolica O-BC30, resulting in draft 6.36 Mb and 4.76 Mb genomes, respectively. We hypothesized that the bromopyrroles, and by extension the bromopyrrole moieties of heterodimeric molecules such as 3, are derived in a manner analogous to the biosynthesis of the dichlorinated pyrrole moiety of the antifungal natural product pyoluteorin24, consistent with prior isotope feeding studies tracing the source of the pyrrole moiety of 3 to L-proline25 (Supplementary Fig. 3). Hence, we queried the genomes of P. luteoviolacea 2ta16 and P. phenolica O-BC30 for the proline dehydrogenase (PltE) and the pyrrole halogenase (PltA) enzymes from pyoluteorin biosynthesis. This analysis successfully led to the identification of the brominated marine pyrroles/phenols (bmp) biosynthetic gene locus in both bacteria. Homologs of pltE and pltA (bmp3 and bmp2, respectively, Fig. 1c) were found clustered together with genes putatively encoding an acyl carrier protein (ACP)-thioesterase (TE) di-domain protein (bmp1), proline adenyltransferase (bmp4), flavin-dependent oxygenase (bmp5), chorismate lyase (bmp6), cytochrome P450 (CYP450, bmp7), ferredoxin (bmp9) and ferredoxin reductase (bmp10), and a carboxymuconolactone decarboxylase homolog (bmp8). Primary sequence similarity of Bmp ORFs to pyoluteorin biosynthetic enzymes is provided in Supplementary Table 2.
In order to confirm the contribution of the bmp gene cluster to the biosynthesis of 1–7, we constructed an expression vector and heterologously expressed P. luteoviolacea 2ta16 bmp1–8 genes in Escherichia coli (Supplementary Fig. 4 and Supplementary Tables 3 and 5). In the presence of bromide, we observed the heterologous production of 1–7 (Fig. 2a,b). Note that the complexity of the culture extract precludes a concomitant visual representation of all polybrominated molecules within a single TIC. We next explored the individual functions of the bmp1–8 genes by expressing the bmp cluster with individual gene deletions (Supplementary Table 5). As postulated by homology to the pyoluteorin pyrrole dichlorinase enzyme PltA and consistent with its assignment as a pyrrole brominase, deletion of bmp2 abolished the production of bromopyrroles 2, 3, 6 and 7 (Fig. 2a). On the other hand, deletion of bmp5 eliminated 1, 3, and all other bromophenol-containing species, leading to the proposed role of Bmp5 as a phenol brominase. We further identified bmp7 as a coupling enzyme as its deletion led to the abolishment of all dimers 1–3, while monomers 4–6 could still be detected, with the notable exception of 7 (Supplementary Fig. 5). These experiments established the first genetic link to the production of polybrominated aromatic molecules in nature.
Figure 2. Genetic basis of the Bmp pathway.
(a) Total ion chromatograms (TICs) for organic extracts of E. coli expressing the P. luteoviolacea 2ta16 bmp gene cluster and selected gene deletions. Molecule 2 is only present in high abundance in the absence of bmp5 as E. coli expressing both bmp5 and bmp7 preferentially produce 1 and other coupled bromophenols. Retention times of 5 and 7 are nearly identical; hence, for the sake of clarity, the more abundant species is highlighted in chromatograms where both are present (i.e., bmp1–8, and Δbmp7). In addition to 1–7, the bmp1–8 extract contains other polybrominated phenol species. Peaks labeled ● and ▲ were later correlated to OH-BDEs 11 and 12 (vide infra). TIC for an organic extract of E. coli expressing bmp1–8 cultured in the absence of bromide is shown as a negative control. (b) Chemical structures of bromophenol and bromopyrrole monomers 4–7.
We next analyzed genomic datasets for the presence of orphan gene loci related to the bmp gene cluster. Indeed, we identified a highly conserved gene cluster in the melanogenic marine bacterium Marinomonas mediterranea MMB-126, with an insertion of a putative permease between bmp3 and bmp4 (Fig. 1c). As expected, we confirmed the production of brominated molecules 1–3 by M. mediterranea MMB-1 (Supplementary Fig. 1), providing the first report of halogenated natural products from this genus that commonly associates with sea grass. Indeed, the methylated variant of 7 (2,3,4,5-tetrabromo-1-methylpyrrole) and MeO-BDEs such as 2’-MeO-BDE-68 (vide infra) have been detected in the sea grass Halophila ovalis5 and obligate marine herbivores such as dugongs13. We also identified a truncated gene cluster in the Lissoclinum patella tunicate metagenomic dataset for the marine cyanobacterial symbiont Prochloron didemni P-2 Fiji27 (Fig. 1c). Additionally, we found a highly similar gene locus in the sulfur-oxidizing γ-proteobacterium Thioalkalivibrio thiocyanoxidans ARh 428, notably with the omission of CYP450 bmp7 and CYP450 electron transfer partners bmp9 and bmp10 (Fig. 1c).
Brominase Bmp2 catalyzes pyrrole tribromination
We next sought biochemical confirmation for individual enzymatic transformations in the Bmp pathway, suggested by proline-based halopyrrole biosynthetic logic and gene deletion experiments. As characterized for the biosynthesis of pyrrole moieties in numerous alkaloid natural products29, the proline adenyltransferase Bmp4, together with flavin-dependent dehydrogenase Bmp3 catalyzed the near stoichiometric conversion of holo-Bmp1 to pyrrolyl-S-Bmp1 (Supplementary Figs. 6–8, see Supplementary Note 1 for detailed analytical protocols). Enzymatic conversion was abolished in the absence of either adenosine triphosphate (ATP) or L-proline, while reduced conversion was observed in the absence of Mg2+. These observations were consistent with the proposed reaction scheme for adenyltransferases proceeding via an adenylated amino acid intermediate that is subsequently transferred to the phosphopantetheine sulfhydryl of the ACP with the concomitant release of adenosine monophosphate (AMP).
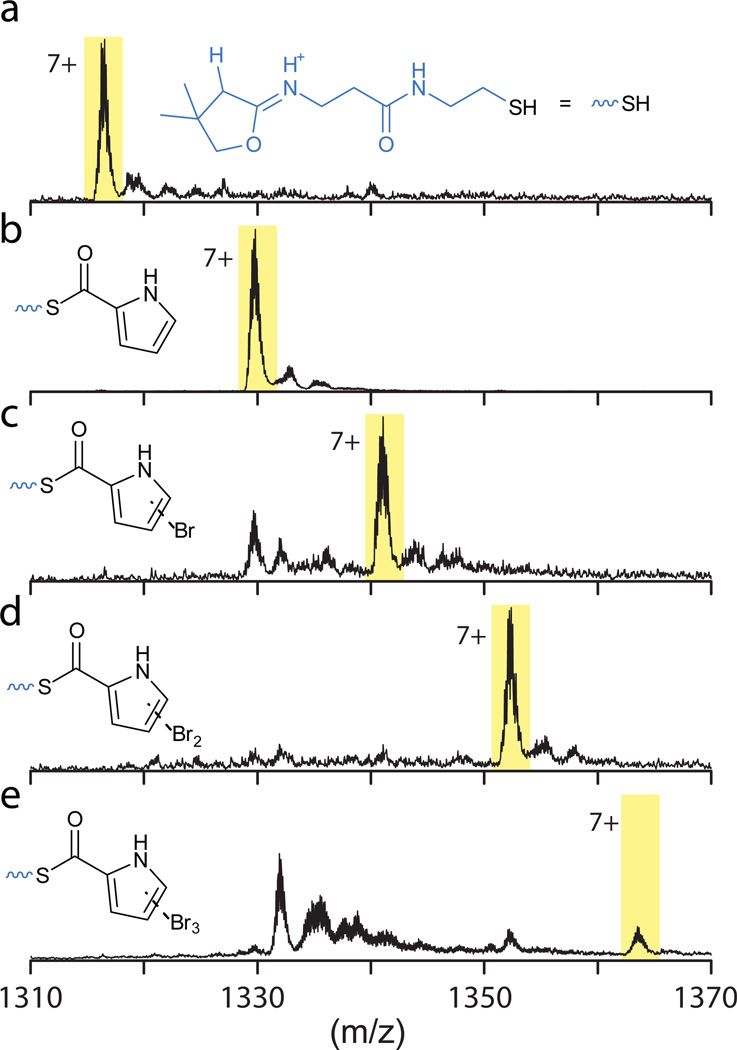
Bmp2 is homologous to canonical flavin-dependent halogenases that utilize ACP-tethered substrates30 (Supplementary Fig. 30). Flavin-dependent halogenase enzymes require an exogenous supply of reduced flavin co-factor (FADH2), typically provided in situ by a flavin reductase that catalyzes the reduction of the flavin cofactor with the concomitant oxidation of NAD(P)H24,31. In the presence of E. coli flavin reductase SsuE32, NADPH and bromide, Bmp2 catalyzed the bromination of pyrrolyl-S-Bmp1(ACP) to mono-, di- and tribromopyrrolyl-S-Bmp1 (Fig. 3). This result establishes the in vitro reconstitution of the first flavin-dependent halogenase with physiological bromination activity involved in natural product biosynthesis. Assignment of Bmp2 as a brominase is further supported by the observation that no incorporation of chloride was detected33. While numerous bromopyrrole containing marine natural products have been identified4,34, 2–3 bear a distinct chemical signature in that all three available positions on the pyrrole ring are brominated, rather than mono- or di- bromination that is more commonly observed in brominated alkaloids with the 3-position of the pyrrole being universally unmodified.
Figure 3. In vitro reconstitution of activity for brominase Bmp2.
Assay for the activity of Bmp2 relies on the detection of MS1 ions for tryptic peptide fragments of Bmp1 corresponding to (a) holo-S-Bmp1, (b) pyrrolyl-S-Bmp1, (c) monobromopyrrolyl-S-Bmp1 (d) dibromopyrrolyl-S-Bmp1 and (e) tribromopyrrolyl-S-Bmp1. All MS1 ions (a–e, shaded in yellow) bear a +7 charge, as identified by their isotopic distributions. MS/MS fragmentation of the shaded MS1 ions generates distinct diagnostic MS2 ions that are shown by their corresponding chemical structures. Note that the dehydrated cyclopantetheinyl MS2 ion for panel a is abbreviated for clarity in panels b–e. Upon transfer of pyrrolyl-S-CoA (16) to apo-S-Bmp1(ACP) by the B. subtilis phosphopantetheinyl transferase Sfp (described in the Online Methods section), pyrrolyl-S-Bmp1 is generated, that is mono-, di- and tribrominated by Bmp2 in the presence of NADPH, KBr and E. coli flavin-reductase SsuE. Calculation for MS1 peptide masses and peptide identification protocol, as inspired by prior reports24,50 is described in detail in the Supplementary Note 1. Note that the expression of Bmp1 in E. coli generates a mixture of apo-Bmp1 and holo-S-Bmp1. Of these, only apo-Bmp1 participates in the Bmp2 assay due to its amenability for acylation catalyzed by the Sfp transferase. Holo-S-Bmp1 remains unmodified as shown in panel a.
Chemical logic for the biosynthesis of 2–3 using enzyme functionalities present within the bmp gene locus dictates that an ACP-bound tribromopyrrole intermediate is released by the TE domain of Bmp1 and decarboxylated by Bmp8 to give 6. While the esterase activity of Bmp1 TE domain could indeed be confirmed by the hydrolysis of the model esterase substrate p-nitrophenylacetate (Supplementary Fig. 10), biochemical investigations into the formation of 6 by Bmp8 were precluded by our inability to generate soluble recombinant Bmp8 protein. However, the expression of bmp1–7 in E. coli excluding bmp8 resulted in altered levels of bromopyrroles, thus supporting a role of Bmp8 in bromopyrrole biosynthesis (Fig. 2a and Supplementary Fig. 11).
Bmp5, a decarboxylating phenol brominase
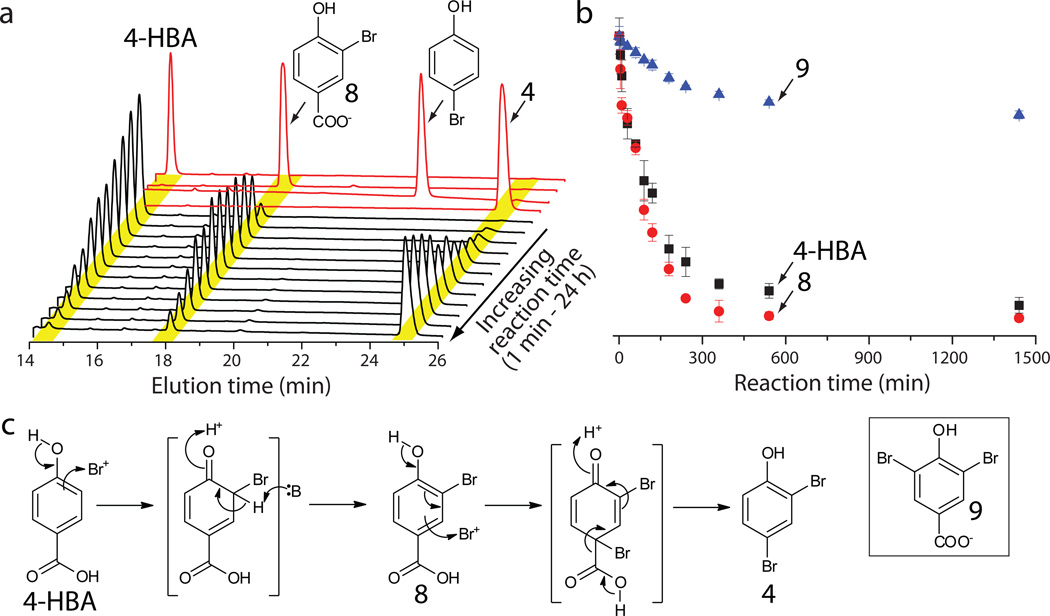
In contrast to assembly of bromopyrroles, the biosynthetic logic for the assembly of bromophenols, and thereafter of PBDEs and polybrominated biphenyls could not be readily anticipated. Previous isotope feeding experiments and the presence of chorismate lyase (Bmp6) in the gene cluster suggested 4-hydroxybenzoic acid (4-HBA) as the precursor for the bromophenol moiety of 335. While the bmp5 deletion experiments postulated its role as the phenol brominase, lack of sequence similarity of the flavoenzyme Bmp5 to canonical flavin-dependent halogenases, which includes Bmp2, led to uncertainty in this assignment. Moreover, incorporation of 4-HBA into bromophenols necessitates decarboxylation, a transformation not previously associated with flavin-dependent halogenases. Hence, we undertook a detailed in vitro characterization of the activity of Bmp5. Upon incubation of 4-HBA with recombinant Bmp5 in the presence of bromide, NADPH, and FAD, we observed a time-dependent formation of one major product via a stable intermediate (Fig. 4a and Supplementary Fig. 16). The identity of the intermediate as 3-bromo-4-hydroxybenzoic acid (8) and of the product as 4 was confirmed by mass spectrometry and retention time comparison to authentic synthetic standards (Fig. 4a and Supplementary Fig. 17). Bmp5 could utilize 8 as a substrate for the synthesis of 4, further establishing 8 as the physiological intermediate for the reaction (Fig. 4b). Bmp5 also generated 5 when expressed in E. coli in the presence of bromide (Supplementary Fig. 17). While production of 5 could not be detected in vitro, this observation is consistent with the acceptance of 3,5-dibromo-4-hydroxybenzoic acid (9) as a poor substrate by Bmp5 as compared to 4-HBA and 8 (Fig. 4b). Bmp5 activity was abolished in the absence of either bromide or NADPH, while a partial reduction in activity was observed upon omission of FAD, consistent with the partial occupancy of FAD in Bmp5 as purified (Supplementary Fig. 18). Consistent with prior characterization of halide specificity of aromatic halogenases33, Bmp5 could utilize iodide in vivo leading to the formation of iodophenols while no chloride incorporation was observed (Supplementary Fig. 28).
Figure 4. Biosynthesis of bromophenols by flavin-dependent decarboxylase-brominase Bmp5.
(a) Bmp5 was incubated with 4-HBA, KBr, NADPH, and FAD at 30 °C in 20 mM Tris-HCl (pH 8.0) buffer. At indicated time points, 40 µL assay volume was withdrawn and quenched by the addition of 16 µL MeCN + 0.35% TFA. HPLC separation of the substrate, intermediate and products of the Bmp5 reaction are shown as black traces after the following reaction times: 1 min, 5 min, 10 min, 30 min, 60 min, 90 min, 120 min, 180 min, 240 min, 360 min, 540 min and 1440 min. HPLC traces corresponding to synthetic standards analyzed under identical chromatographic conditions are shown in red. (b) Time-dependent conversion of 8 to 4, and of 9 to 5 was monitored in an identical fashion, and the area under the substrate peaks was integrated and plotted against reaction time to compare the relative rates for the consumption of 4-HBA, 8 and 9 by Bmp5. Experiments were conducted in triplicate; data represent mean values ± s.d. (c) A proposed reaction scheme for the conversion of 4-HBA to 4 catalyzed by Bmp5.
In contrast to the flavin-dependent halogenases described to date, Bmp5 did not require the addition of a flavin reductase to regenerate FADH2 in situ. This finding is consistent with sequence homology of Bmp5 to single-component flavin-dependent oxygenases (Supplementary Fig. 30). Consistent with the formation of electrophilic peroxo intermediates by flavin-dependent oxygenases and halonium intermediates by flavindependent halogenases36, a putative reaction scheme catalyzed by Bmp5 can be discerned (Fig. 4c). To the best of our knowledge, Bmp5 represents the first example of a decarboxylating flavin-dependent halogenase. Stability of benzoic acids distinguishes the Bmp5 decarboxylation activity from the extensively sampled marine haloperoxidases37 that participate in the decarboxylative degradation of labile halogenated derivatives of 3-oxo carboxylic acids and acyl homoserine lactones38–39.
Bmp7 catalyzed coupling of bromophenols and bromopyrroles
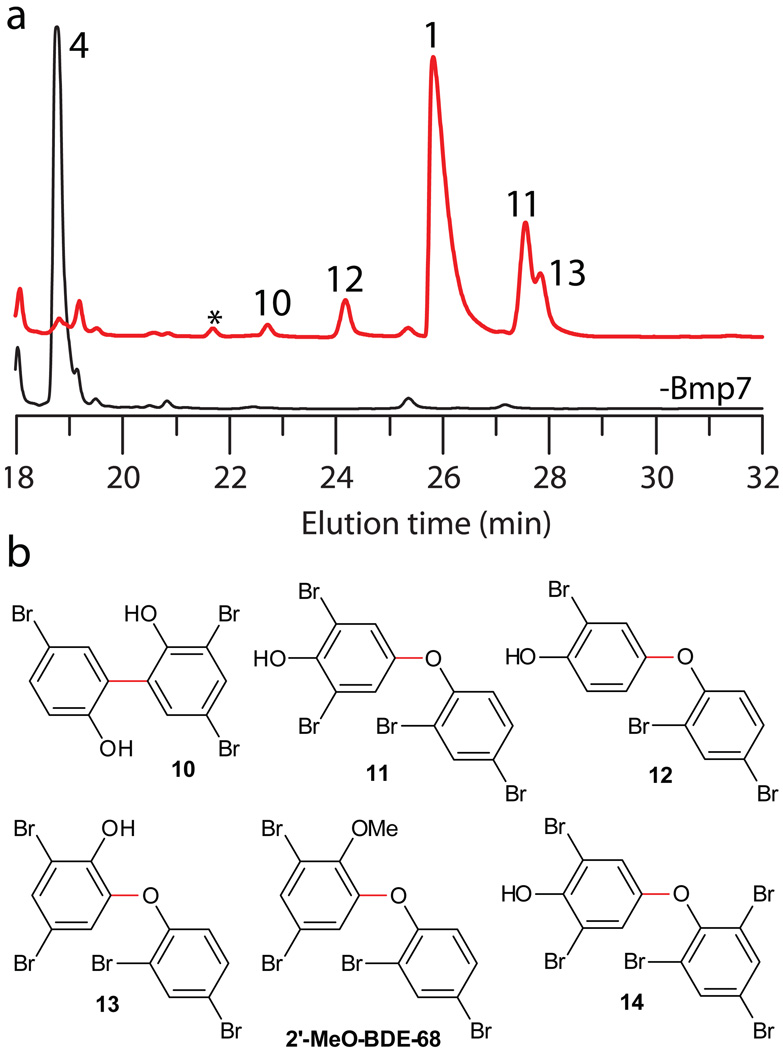
We next investigated whether the bromophenol products of Bmp5 could be accepted as substrates by CYP450 Bmp7. In the presence of ferredoxin (Bmp9), ferredoxin reductase (Bmp10), and NADH, Bmp7 catalyzed the coupling of 4 to at least six distinct products that could be identified by LC-MS/MS (Fig. 5a). Characterization of Bmp7 products provided by MS/MS, NMR and comparison to authentic standards is detailed in the Supplementary Note 2. We confirmed polybrominated biphenyls previously isolated from Pseudoalteromonas spp., 118 and 3,5,5’-tribromo-2,2’-biphenyldiol (10)40 as Bmp7 products (Fig. 5 and Supplementary Figs. 19–20). Additionally, a tribromo-biphenyldiol isomer of 10 (denoted by * in Fig. 5a) was detected as a Bmp7 product, but could not be isolated in sufficient quantities for comprehensive structure elucidation.
Figure 5. Enzymatic synthesis of polybrominated biphenyls and OH-BDEs.
(a) CYP450 Bmp7 dimerizes two molecules of 4 to generate biphenyls 1 and 10, and OH-BDEs 11–13 (red curve). The curve in black represents the negative control in the absence of the enzyme. (b) Chemical structures of polybrominated biphenyls and OH-BDEs generated by the bmp pathway as identified in this study. Coupling bonds generated by Bmp7 are shown in red. Also shown is 2’-MeO-BDE-68, the methoxylated derivative of 13.
Guided by differential MS/MS fragmentation of polybrominated biphenyls and OH-BDEs41, we further characterized three OH-BDE products, namely 2,6-dibromo-4-(2,4-dibromophenoxy)phenol (11), 2-bromo-4-(2,4-dibromophenoxy)phenol (12), and 4,6-dibromo-2-(2,4-dibromophenoxy)phenol (2’-OH-BDE68, 13) (Supplementary Note 2, Fig. 5 and Supplementary Figs. 20–22). The OH-BDE products possess two distinct chemical signatures, namely, para- (11–12) and ortho-coupling (13) with respect to the hydroxyl group (Fig. 5b). Guided by our in vitro identification of OH-BDEs, we analyzed organic extracts of P. luteoviolacea 2ta16, as well as E. coli expressing bmp1–8, and confirmed the in vivo production of 11–12 (denoted by ● and ▲, respectively, in Fig. 1b and Fig. 2a). This observation provides the first confirmation for marine bacterial biosynthesis of OH-BDEs. 11 and 13 are isomeric with 1, and have very similar retention time to each other. Moreover, in contrast to the diagnostic MS2 patterns for polybrominated biphenyls and para-OH-BDEs (see Supplemental Note 2), structural characterization of 13 relied on comparison to an authentic synthetic standard (Supplementary Fig. 22). These findings highlight some of the technical challenges inherent with the characterization of isomeric polybrominated biphenyl and OH-BDE products from whole-cell culture extracts.
Bmp7 could also dimerize two molecules of 5, albeit with reduced catalytic efficiency, to generate the OH-BDE product 2,6-dibromo-4-(2,4,6-tribromophenoxy)phenol (14) (Fig. 5b and Supplementary Figs. 23–24). Bmp7 converted chlorinated phenolic substrates as well, catalyzing the dimerization of 2,4-dichlorophenol to three isomeric products, corresponding to chlorinated counterparts to the polybrominated products 1, 11 and 13 generated with 4 (Supplementary Fig. 25). The mass spectrometry-based prediction of the polychlorinated products is described in detail in Supplementary Note 2.
Proceeding based on the biosynthetic logic for the hydrolytic offloading of tribromopyrrole carboxylic acid by Bmp1 TE domain and its decarboxylation by Bmp8, we propose 6 as the physiological substrate for Bmp7, which was supported by a typical substrate-induced UV-Vis absorbance shift (Supplementary Fig. 12). In vitro activity assays of Bmp7 in the presence of redox partners Bmp9–10 and NADH with 6 as a substrate generated 2 and 7 as major products (Supplementary Figs. 13–15). The identity of reaction products was confirmed by comparison to authentic standards. A final demonstration for the biosynthetic versatility for Bmp7 was provided by an in vitro reaction involving the heterodimerization of 4 and 6. We confirmed the enzymatic synthesis of 3 and 2,3,5,7-tetrabromobenzofuro[3,2-b]pyrrole (15)40 by comparison to authentic standards isolated from P. luteoviolacea 2ta16 (Fig. 6a and Supplementary Figs. 26–27). Additionally, we detected an isomer of 3 as a minor product generated by Bmp7 during the in vitro coupling of 4 and 6, which could also be detected in the culture extracts of the three producer strains. Though a comprehensive structural characterization by NMR spectroscopy could not be realized, a characteristic hydroxytribromopyrrole MS/MS product ion suggests a polybrominated phenol-pyrrole ether, a class of marine natural product not previously described (Supplementary Fig. 29).
Figure 6. Bi-modular scheme for the biosynthesis of polybrominated marine natural products by the bmp pathway.
(a) Chorismate, the precursor for bromophenols, is converted to 4-HBA by chorismate lyase (CL) Bmp6, and then to 4–5 by flavin-dependent halogenase (Hal) Bmp5. The CYP450 coupling (C) enzyme Bmp7 generates a suite of diverse polybrominated biphenyls (1 and 10) and OH-BDEs (11–14) from 4 and 5. The electron transfer partners for Bmp7, Bmp9 and Bmp10, are omitted for clarity. The bromopyrroles are derived from L-proline. Acylation of L-proline to the ACP domain of Bmp1 by the proline adenyltransferase (A) Bmp4 initiates its oxidation by the flavin-dependent dehydrogenase (DH) Bmp3 and tribromination by the flavin-dependent halogenase (Hal) Bmp2. The TE domain of Bmp1 likely catalyzes the offloading of a carboxylic acid intermediate that is decarboxylated (D) by carboxymuconolactone decarboxylase homolog Bmp8 to 6. 6 can be dimerized by Bmp7 to generate 2, or with 4 to generate heterodimers such as 3 and 15. During the homodimerization of 6, we also observed the formation of 7. The coloring scheme is consistent with Fig. 1c. (b) Proposed steps for radical generation (i), rearragement (ii–iv) and coupling of 4 by Bmp7 to generate biphenyls and OH-BDEs.
DISCUSSION
While several Bmp pathway products identified in this study were previously isolated from marine bacteria (1–318–20, 719, 10 and 1540) with 3 being characterized nearly half a century ago, the genetic and molecular bases of their biosyntheses have remained elusive until now. In this study, we identified a conserved gene locus in Gram-negative marine bacteria encoding the biosynthesis of at least fifteen polybrominated aromatic marine natural products containing ubiquitous bromopyrrole and bromophenol building blocks. A near complete in vitro reconstitution of all enzymatic steps within the Bmp pathway provides the basis for a bi-modular biosynthetic scheme as shown in Fig. 6a.
The bromopyrrole and bromophenol biosynthetic modules function independently and are united through the action of CYP450 Bmp7. CYP450-catalyzed phenol coupling reactions have extensive biochemical precedents, and the formation of polybrominated biphenyls 1 and 10, and OH-BDEs 11–14 are expected to follow accepted bi-radical mechanisms (Fig. 6b). A similar mechanism can be envisaged for coupling of bromopyrroles, consistent with CYP450-catalyzed coupling in indolocarbazole antibiotics42. The modularity of the Bmp pathway is reflected in the preservation of only the bromopyrrole biosynthetic module in the marine symbiotic cyanobacterium P. didemni P2-Fiji27 (Fig. 1c). As the bromophenol module is absent, it is likely that P. didemni P2-Fiji produces only halogenated pyrroles. Furthermore, a homologous gene locus identified in T. thiocyanoxidans ARh 4 lacks a gene coding for a coupling CYP450 (Fig. 1c). It is conceivable that an even wider applicability of the chemical logic described here for the biosynthesis of marine bromophenols and bromopyrroles is limited only by the availability of genomic data.
The discovery of flavoenzyme Bmp5 as a phenol brominase was unexpected. Bmp5 is homologous to flavin-dependent oxygenases43, rather than flavindependent halogenases. Primary sequence motifs for binding both the flavin cofactor, as well as the nicotinamide electron donor can be readily discerned within the primary sequence of Bmp5 (Supplementary Fig. 30). Indeed, bmp5 homologs in the biosynthetic gene clusters identified in M. mediterranea MMB-1 and T. thiocyanoxidans ARh 4 are annotated as flavin-dependent oxygenases (Fig. 1c). While decarboxylation was until now an unrealized activity within the catalytic repertoire of flavin-dependent halogenases, ortho-hydroxylation of 4-HBA44 as well as para-decarboxylative-hydroxylation of 4-HBA45 have been reported for flavin-dependent oxygenases, leading to a proposal for a two-step reaction scheme employed by Bmp5 (Fig. 4c). The collective action of an electron-donating phenol hydroxyl and electron-withdrawing carboxyl group would first direct electrophilic bromonium addition at the ortho position, and subsequent proton abstraction by a catalytic base would result in the formation of 8. This intermediate would then undergo a second bromonium addition para to the hydroxyl, followed by decarboxylation to generate 4. The Bmp5 reaction is regiospecific, in that ortho bromination is the first half reaction, followed by decarboxylative-bromination at the para position. This assertion for the strictly directed two-step Bmp5 reaction scheme is supported by 4-bromophenol not being detected as an intermediate, or as a product of the Bmp5 reaction (Fig. 4a). Furthermore, the Bmp5 active site likely sterically occludes bromonium addition at one of the ortho positions of the aryl ring, thus biasing the second half reaction towards decarboxylative-bromination at the para position, rather than a second ortho-bromination to yield 9 as a second intermediate. This postulate is supported by the observation that under identical experimental conditions, rates of decrease of 4-HBA and 8 in an in vitro reaction with Bmp5 are nearly identical, while 9 is a poor substrate for Bmp5 (Fig. 4b). Consequently, 5 is a minor product generated by Bmp5 that could not be detected in vitro, but is only detected to be produced in vivo when Bmp5 is heterologously expressed in E. coli in presence of bromide in the culture media (Supplementary Fig. 17). Additionally, Bmp5 could utilize iodide, but not chloride in vivo (Supplementary Fig. 28).
It is intriguing that neither bromophenol monomers nor OH-BDEs had previously been isolated from marine bacterial sources. Bromophenols and OH-BDEs identified in this study (and their methylated derivatives) have instead been extensively detected in marine eukaryotes, ranging from autotrophs and invertebrates, to apex predators5–15,46. While sponge-associated symbiotic marine cyanobacteria were hinted as producers of OH-BDEs10, a definite genetic basis for OH-BDE biosynthesis had not been established. Furthermore, in the absence of definitive genetic evidence for algal biosynthesis of OH-BDEs, a contribution of associated bacteria towards the production of OH-BDEs sourced from marine algae cannot be discounted8,12. It is thus noteworthy that all three marine bacteria harboring the Bmp pathway as identified in this study also belong to genera commonly associated with marine eukaryotes. Identification of the bmp gene locus should henceforth aid the computational mining of marine metagenomic datasets for the identification of bacterial, as well as potential eukaryotic OH-BDE producers utilizing similar motifs. Together with 1 (Fig. 1a), the methoxylated derivative for 13 (2’-MeO-BDE-68, Fig. 5b), is extensively detected in marine mammals and even humans14–15, suggesting that bromophenols and OH-BDEs identified in this study bioaccumulate in the marine food web and are transferred to humans via trophic connections. Furthermore, methylation of OH-BDEs, and by extension of polybrominated biphenyls and bipyrroles (Fig. 1a), appears to be a facile biochemical transformation consistent with a positive correlation between the existence of OH-BDEs and MeO-BDEs in marine environmental matrices16,47.
OH-BDEs, such as those characterized in this study, commonly feature two distinct chemical signatures in which the hydroxyl group is positioned either ortho or para to the ether linkage. Of the two regioisomers, para-OH-BDEs are potent inhibitors of thyroid hormone signaling48, with 14 having a higher affinity for binding to the thyroid hormone transport protein than its physiological substrates49. Hence, the detection of para-OH-BDEs, such as 11–12, and specifically 14, puts into perspective the toxic potential of the metabolites generated by the Bmp pathway. As the synthesis of biphenyls and diphenyl ethers could also be confirmed for Bmp7 using polychlorinated and polyiodinated phenolic substrates (Supplementary Figs. 25, 28), other naturally occurring polyhalogenated aryl compounds may also be derived from analogous enzymatic pathways with differing halogen specificities of the halogenases. As the industrial production of polybrominated flame retardant chemicals continues to be phased out in recognition of their persistence and toxicity, we anticipate that naturally produced marine bacterial PBDEs will take a center stage in informing our small molecule-mediated interactions with the environment.
ONLINE METHODS
Sources of polyhalogenated molecules described in this study
Analytical standards of compounds 1 and 10 were obtained by purifying an impure standard of 1 obtained from Sigma-Aldrich (S842753) as described in detail in Supplementary Fig. 19. Compounds 2, 3 and 15 were purified from culture extract of P. luteoviolacea 2ta16 as described in detail below and in Supplementary Figs. 14, 26 and 27. Compounds 4 (catalog number 258164, 95% purity), 5 (137715, 99% purity),7 (L165042, 99% purity), 8 (699926, 97% purity) and 17 (105953). 99% purity) were purchased from Sigma-Aldrich. 651 was synthesized according to established protocols and purified by HPLC. 1H and 13C NMR spectra for synthetic 6 matched that of values reported in literature. Compound 9 was purchased from Fisher Scientific (AC24363, 99% purity). Compound 13 was purchased from AccuStandard (HBDE-4006S–CN, 99% purity). Compound 16 was synthesized according to the protocol described below. Compounds 11, 12 and 14 were isolated from preparative scale reaction of Bmp7 with 4 or 5 and characterized by NMR and MS/MS as described in Supplementary Note 2 and Supplementary Figs 23–24.
Analytical scale extractions of bacterial cultures
P. luteoviolacea 2ta1623 was kindly provided by Professor Farooq Azam of Scripps Institution of Oceanography, University of California at San Diego. M. mediterranea MMB-126 was obtained from the American Type Culture Collection (ATCC 700492). P. phenolica O-BC30 was obtained from the Japan Collection of Microorganisms, Riken BRC (JCM 21460). Strains were cultured in Difco 2216 Marine Broth (BD) supplemented with 1 g/L KBr. 5 mL cultures of P. luteoviolacea 2ta16, P. phenolica O-BC30, or M. mediterranea MMB-1 were incubated for 48–72 h at 30 °C with shaking at 200 rpm. Cultures were extracted with two volumes of ethyl acetate. The organic layer was collected and solvent was removed in vacuo. The residue was dissolved in 100 µl MeOH. Extracts were analyzed by LC-MS/MS in negative polarity as described in Supplementary Fig. 1. Identical LC-MS/MS conditions are used throughout the study for analysis of all bacterial extracts, Bmp7 reaction products (vide infra), and synthetic standards.
Isolation of 2–3 and 15
Molecules 2–3 and 15 were isolated from extracts of P. luteoviolacea 2ta16. 10×100 mL cultures were grown as before and incubated for 24 h at 30 °C with shaking at 200 rpm. Cultures were pooled and extracted with an equal volume of ethyl acetate. The organic layer was dried with anhydrous MgSO4, filtered and solvent was removed in vacuo. The residue was dissolved in 5 mL MeOH and filtered through a 0.2 µM vacuum filter. 2–3, and 15 were purified by semi-preparative scale HPLC using a reverse phase C18 column (Phenomenex, 250 mm). Solvents used were water + 0.1% TFA (A) and MeCN + 0.1% TFA (B). An identical elution profile as described before for analytical scale experiments but at an increased flow rate of 2 mL/min was used for all semi-preparative purification experiments in this study. Guided by LC-MS/MS, fractions containing 2–3, and 15 were lyophilized. 500 µg of each product was dissolved in 50 µL CDCl3. 1H NMR, and 13C NMR spectra for 2 were obtained using a 600 MHz Varian NMR microprobe. Spectra of 3 and 15 were verified with literature values40.
Genome sequencing
Genomic DNA was isolated from 5 mL overnight cultures of P. luteoviolacea 2ta16 grown as before by phenol:chloroform extraction using a phase lock gel tube (5 Prime Inc.). Genomic DNA was extracted from a 5 mL overnight culture of P. phenolica O-BC30 grown in A1 media at 30 °C with shaking at 200 rpm using a DNeasy DNA Extraction Kit (Qiagen). The P. luteoviolacea 2ta16 genome was sequenced at the J. Craig Venter Institute using 454 Titanium chemistry on the Genome Sequencer FLX platform (Roche). 555,393 reads with an average length of 404 nucleotides were assembled using the CAMERA Meta-Assembler workflow (http://camera.calit2.net). The high quality draft genome of 6.36 Mb (137 contigs; N50, 166764 nt) contains 5380 protein coding sequences. Gene prediction and functional annotation was performed by the JGI Integrated Microbial Genomes-Expert Review pipeline 52. A sequencing library was constructed from genomic DNA of P. phenolica O-BC30 using an Ion Xpress™ Plus Fragment Library Kit (Life Technologies). An Ion PGM™ Template OT2 400bp Kit (Life Technologies) was used to generate sequencing templates for loading onto the sequencing chip. Sequencing was performed using an Ion Torrent Personal Genome Machine (Life Technologies) with an Ion PGM™ 400 bp Sequencing Kit (Life Technologies) on a 318v2 sequencing chip (Life Technologies). De novo genome assembly was performed using CLC Genomics Workbench software (CLC bio). The sequence of bmp cluster from P. phenolica O-BC30 was verified by Sanger sequencing (SeqXcel, La Jolla, CA) using forward primers based on the initial sequence identified from the draft genome assembly.
Assembly of pGAL -bmp1–7 yeast/ E. coli shuttle vector
All antibiotics in subsequent sections are used at the following final concentrations in media: carbenicillin 100 µg/mL, kanamycin 50 µg/mL, apramycin 50 µg/mL, and chloramphenicol 25 µg/mL. A modified protocol from Larinov and Kouprina53 was used to assemble bmp1–7 by recombination in Saccharomyces cerevisiae BJ5465 (ATCC 208289). A yeast/E. coli shuttle vector pGAL-bmp1–7 was constructed from PCR amplified fragments encompassing the vector backbone of the pGAL-MF yeast expression vector (DualSystems) and bmp1–7 (Supplementary Fig. 4). Primer pairs were designed to amplify a total of five ~2.5 kb regions incorporating ~35 bp flanking regions (Supplementary Table 3). The first two fragments were derived from digested pGAL-MF yeast expression vector, including an E. coli antibiotic resistance marker (AmpR), E. coli origin of replication (pBScri), yeast auxotrophic selective marker (URA3), and yeast origin of replication (2micron). The remaining three fragments encompassing bmp1–7 were amplified from P. luteoviolacea 2ta16 gDNA. SpeI and SacI restriction sites were incorporated at the cluster boundaries for integration into the compatible XbaI and SacI restriction sites of the pETDuet-1 Vector (Novagen). Fragments were amplified by PCR using the PrimeStar HS DNA polymerase standard protocol (Clontech). PCR amplified fragments were purified using a QIAquick PCR Purification Kit (Qiagen) combined, concentrated, and transformed into electrocompetent yeast S. cerevisiae BJ5465. Transformed S. cerevisiae BJ5465 was plated onto yeast minimal media deficient in uracil and incubated for 3 days at 30 °C. Yeast colonies were screened for cluster fragments by colony PCR. Yeast colonies positive for all three fragments of bmp1–7 were inoculated in 1.5 mL yeast minimal media and incubated overnight with shaking at 30 °C. Candidate pGAL-bmp1–7 constructs were extracted using a Zymoprep Yeast Plasmid Miniprep I extraction Kit (Zymo Research). Extracted plasmids were transformed into One Shot TOP10 Electrocomp E. coli (Invitrogen) and plated on LB-Agar supplemented with carbenicillin. Plasmids were extracted from E. coli using a QIAprep Spin MiniPrep Plasmid Extraction Kit (Qiagen). Candidate pGAL-bmp1–7 plasmids were sequenced by Sanger sequencing (SeqXcel, La Jolla, CA).
Ligation of bmp1–7 into E. coli expression plasmid
The fragment containing bmp1–7 was excised from pGAL-bmp1–7 by double digestion with SacI and SpeI restriction enzymes (NEB) and ligated using T4 DNA Ligase (NEB) into XbaI and SacI double digested pETDuet-1 Vector (Novagen) to yield the E. coli expression construct pETDuet-bmp1–7.
Deletions of bmp2, bmp5, and bmp7
Individual deletions of bmp2 and bmp5 were performed by PCR mutagenesis using PrimeStar Max DNA Polymerase standard protocol (Clontech) with pETDuet-bmp1–7 as the template. Primers were designed with the motif 5′-[20 nucleotide overlap] [35 nucleotide primer region]-3′ (Supplementary Table 4). The PCR product was purified using a QIAquick PCR Purification Kit (Qiagen). Purified products were treated with DpnI exonuclease (NEB) to digest methylated pETDuet-bmp1–7 template. Reactions were purified using the QIAquick PCR Purification Kit (Qiagen), concentrated, and transformed into One Shot TOP10 Electrocomp E. coli (Invitrogen), plated on LB-Agar with carbenicillin, and incubated overnight at 37 °C. Candidate colonies were inoculated into 5 mL LB containing carbenicillin and grown overnight at 37 °C with shaking. Plasmids were extracted using QIAprep Spin MiniPrep Plasmid Extraction Kit (Qiagen). Constructs were screened by digestion with HindIII exonuclease (NEB) followed by DNA gel electrophoresis. Sequences of candidate constructs were checked by Sanger Sequencing (SeqXcel, La Jolla, CA).
A bmp7 scar-less deletion was performed by λRED-mediated PCR targeted recombination using a protocol modified from Gust et al. 54. The extended apramycin disruption cassette aac(3)IV was amplified from digested vector pIJ773 using forward and reverse primers incorporating regions flanking bmp7 on pETDuet-bmp1–7 and terminating in XmaI restriction sites (Supplementary Table 4). Target plasmid pETDuet-bmp1–7 was transformed into electrocompetent E. coli BW25513/pIJ790 (containing the gene encoding the λRED recombinase). Transformants were plated on LB-Agar containing carbenicillin and chloramphenicol, and incubated overnight at 30 °C to sustain temperature-sensitive pIJ790. Five mL LB containing carbenicillin and chloramphenicol was inoculated with a single colony and grown overnight at 30°C with shaking. To prepare electrocompetent E. coli BW25513/pIJ790/pETDuet-bmp1–7, 50 mL LB containing carbenicillin and chloramphenicol were inoculated with a 1% inoculum of overnight culture. L-arabinose was added to a final concentration 10 mM to induce expression of the λRED recombinase. The culture was incubated at 30 °C with shaking for 3 hours, then pelleted by centrifugation and washed three times with 10% glycerol. The apramycin disruption cassette was transformed into electrocompetent E. coli BW25513/pIJ790/pETDuet-bmp1–7 and the transformants were plated on LB-Agar containing carbenicillin and apramycin and incubated overnight at 37 °C. Colonies were screened by PCR, and candidate colonies were grown in 5 mL LB containing carbenicillin and apramycin, incubated overnight at 37 °C with shaking. Plasmids were extracted and re-transformed in One Shot TOP10 Electrocomp E. coli (Invitrogen) plated on LB-Agar containing carbenicillin and apramycin, and incubated overnight at 37 °C. Single colonies were inoculated in 5 mL LB containing carbenicillin and apramycin, and incubated overnight at 37 °C with shaking, and plasmids were extracted. Replacement of bmp7 with aac(3)IV was confirmed by restriction analysis. The plasmid was digested with XmaI exonuclease (NEB) to excise the apramycin resistance cassette. Linearized plasmid was gel purified and self-ligated using T4 DNA ligase (NEB). The ligated plasmid was transformed into One Shot T10 Electrocomp E. coli (Invitrogen) and plated on LB-Agar containing carbenicillin and incubated overnight at 37 °C. Single colonies were inoculated in 5 mL LB containing carbenicillin, and incubated overnight at 37 °C with shaking. Plasmids were extracted and checked by restriction digestion with HindIII exonuclease followed by DNA gel electrophoresis.
Cloning of bmp8 and ppt from P. luteoviolacea 2ta16
Phosphopantetheinyl transferase (ppt, locus tag: Plt2ta16_02049; 57% similarity and 33% identity to B. subtilis Sfp) and bmp8 were amplified from P. luteoviolacea 2ta16 genomic DNA by PCR using appropriately designed primers. Ppt was cloned into MCS-1 of pCOLADuet-1 Vector (Novagen) to generate pCOLADuet-ppt. Bmp8 was cloned into MCS-2 of pCOLADuet to generate pCOLADuet-bmp8. Ppt was further cloned into MCS-2 of pCOLADuet-bmp8 to generate pCOLADuet-bmp8-ppt.
Heterologous expression, and LC-MS/MS analysis
Cluster expression constructs were transformed into electrocompetent E. coli BL21(DE3) (Lucigen) (Supplementary Table 5). 5 mL LB cultures containing carbenicillin and kanamycin were inoculated with the respective E. coli strains and incubated at 37 °C for 3 h with shaking. Fresh 5 mL cultures (100 µg/mL carbenicillin, 50 µg/mL kanamycin, 0.01% w/v FeSO4, 1 g/L 4-hydroxybenzoic acid, 1 g/L L-proline, 1 g/L KBr) were inoculated with a 2% inoculum of pre-culture at 30 °C for 3 h with shaking, induced with 2.5 µL of 100 mM IPTG, and incubated for an additional 16 h at 30 °C with shaking. E. coli expressing bmp1–8 and ppt grown in LB media not supplemented with KBr was used as a negative control. Sample preparation and LC-MS/MS analysis was performed as previously described for wild type bacterial strains.
Cloning, protein expression and purification
Based on primary sequence, Bmp1(ACP) domain was mapped to residues 1 through 77. ORFs were amplified by PCR, and cloned using appropriate restriction sites into pET28a(+) expression vector, with the exception of Sfp, which was cloned into a pET24a(+) vector, and introduced into competent E. coli BL21Gold(DE3) for recombinant protein expression. Terrific broth cultures were grown in the presence of 50 µg/mL kanamycin. Cultures for Bmp2 and Bmp9 were spiked with 10 mg of riboflavin (Fisher Scientific, AC13235-0250), Bmp10 with 50 mg Fe(II)SO4, and Bmp7 with 0.15 mM 5-aminolevulinic acid (Sigma-Aldrich A3785-1G). Cultures were grown at 30 °C till the optical density of 0.5 was obtained, and then shifted to 16 °C. After 1 h, protein expression was induced by the addition of 0.15 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Further growth was allowed at 16 °C for 24 h. For Bmp7, the culture was spiked with an additional 0.15 mM 5-aminolevulinic acid after 24 h, and growth allowed for an additional 24 h.
Cells were harvested by centrifugation and resuspended in 20 mM Tris-HCl (pH 8.0) 500 mM NaCl 10% glycerol buffer, and lysed by sonication. The lysate was centrifuged at 16,000g for 45 min, and the supernatant applied to 5 mL bed volume Ni-NTA column equilibrated in the harvest buffer. The column was extensively washed with 20 mM Tris-HCl (pH 8.0) 1 M NaCl 30 mM imidazole buffer, and the protein eluted with a linear gradient to 250 mM imidazole. With the exception of Bmp10 and SsuE, pure protein fractions were pooled, and extensively dialyzed in 20 mM Tris (pH 8.9) 50 mM NaCl buffer at 4 °C in the presence of 1 unit/mg thrombin protease (MP Biomedicals, 154163). No thrombin was added to Sfp and Bmp1(ACP).
After dialysis, all proteins were further purified by anion exchange chromatography using a Q-FF column. The proteins were then desalted to 100 mM potassium phosphate (pH 7.6) 10% glycerol buffer using PD-10 columns, and stored at −80 °C in small aliquots. N-His6-Bmp10 was directly desalted to 100 mM potassium phosphate (pH 7.6) 10% glycerol buffer using PD-10 column after affinity chromatography and stored at 4 °C. Thrombin was then added. Bmp10 was used for assays within 24 h of its purification. 10% glycerol was added to N-His6-SsuE protein after elution from affinity chromatography column, and used without further concentration. The plasmid containing an engineered variant of phosphite dehydrogenase N-His6-PtdH-17X–A176R was kindly provided by the laboratory of Prof. Satish K. Nair and expressed and purified in an identical manner.
Coexpression of Bmp1(ACP) with Bmp3
As shown in Supplementary Fig. 6, M. mediterranea MMB-1 bmp1 was ligated into RSFDuet-1 vector MCS-1 digested with BamHI and HindIII restriction enzymes. This construct encodes a N-His6 tag fused to Bmp1. In MCS-2 of the same vector, the ORF corresponding to an ACP synthase (GenBank: NC_015276.1) was inserted using NdeI and XhoI restriction sites. RSFDuet-1 vector utilizes a kanamycin resistance marker. M. mediterranea MMB-1 bmp3 was ligated into CDFDuet-1 vector MCS-2 digested with NdeI and XhoI restriction enzymes. CDFDuet-1 vector utilizes a streptomycin resistance marker. Also shown in Supplementary Fig. 6 is the di-domain architecture of Bmp1. As identified by sequence homology, the N-terminal 77 residues constitute the ACP domain, with Ser35 being the site for phosphopantetheinylation. Residues 78 to the C terminus constitute the TE domain, with Ser202 being the catalytic nucleophilic residue of a typical serine protease/esterase Ser-His-Asp/Glu triad. To generate Bmp1(ACP), the codon corresponding to residue 78 of bmp1 was mutated to a stop codon by site directed mutagenesis. The two vectors were then co-transformed into E. coli BL21Gold(DE3) competent cells. Protein expression cultures were spiked with 0.1 mM D-pantothenic acid (Sigma-Aldrich, P2250-5G), and 10 mg riboflavin. Bmp1(ACP) was then purified by affinity chromatography.
Assay for halogenation of pyrrolyl-S-Bmp1 by Bmp2
16 was synthesized guided by established protocols55. Briefly, commercially available pyrrole-2-carboxylic acid (Sigma-Aldrich, P73609-1G, 20 mg, 0.18 mmol) was dissolved in 500 µL DMF. To this solution PyBOP (Sigma-Aldrich, 377848-5G, 112 mg, 0.22 mmol), DIEA (Sigma-Aldrich, 387649-100ML, 38 µL, 22 mmol) and thiophenol (Alfa Aesar, 50–120–7468, 37 µL, 0.36 mmol) was added and the reaction mixture was stirred for 30 min at room temperature. The reaction was quenched by the addition of saturated brine and extracted twice with ethyl acetate. The combined organic phases were dried over MgSO4, concentrated in vacuo and purified on a silica gel column using hexanes/ethyl acetate 40:1 as solvent. S-phenyl-pyrrole-2-carbothioate (10 mg, 0.49 mmol, 27%) was obtained. S-phenyl-pyrrole-2-carbothioate (4 mg, 0.02 mmol) was dissolved in 300 µL sodium phosphate-buffer (50 mM, pH 8.5) and 200 µL THF. To this solution coenzyme A sodium-salt (Sigma-Aldrich, C4780-100MG, 14 mg, 0.018 mmol) was added and the reaction mixture was stirred at room temperature for 24 h. The product was purified by semi-preparative HPLC on a reversed phase C18 column (Phenomenex Luna 5 µm, 250 mm x 10 mm, 100 Å) using a gradient of 5–100% MeCN+0.1% TFA in H2O+0.1% TFA over 30 min. Fractions containing the product were lyophilized and pure S-coenzyme A-pyrrole-2-carbothioate (15) (1.2 mg, 0.0014 mmol, 7%) was obtained. 1H NMR spectra for 16 is reported in Supplementary Fig. 9. 16: 1H NMR (600 MHz, D2O) δ 8.67 – 8.63 (m, 1H), 8.33 (s, 1H), 7.16 – 7.12 (m, 1H), 7.15 (d, J = 7 Hz, 1H), 6.29 – 6.26 (m, 1H), 6.18 (d, J = 10 Hz 1H), 4.63-4.62 (m, 1H), 4.30 – 4.23 (m, 2H), 4.04 (s, 1H), 3.88 – 3.84 (m, 2H), 3.61 – 3.57 (m, 2H), 3.48 – 3.46 (m, 1H), 3.45-3.43 (m 1H) 3.41 (m, 1H), 3.23 (s, 1H), 3.12 (m, 1H), 2.44 (t, J = 10 Hz, 2H), 0.94 (s, 3H), 0.80 (s, 3H).
0.2 mM purified N-His6-Bmp1(ACP) was incubated with 1 mM 16 and 4 µM Sfp in a 100 µL assay volume containing 15 mM MgCl2 and 5 mM dithiothreitol (DTT) (Sigma-Aldrich, 43819-5G) in 50 mM Tris-HCl (pH 8.0) buffer at 30 °C for 1.5 h. To this reaction, 0.1 mM flavin adenine dinucleotide (FAD) (Sigma-Aldrich, F6625-250MG), 5 mM freshly prepared reduced nicotinamide adenine 2′-phosphate dinucleotide phosphate (NADPH) (Sigma-Aldrich, N5130-25MG), 100 mM KBr, 20 µM Bmp2 and 6 µM SsuE were added. The assay was incubated at 30 °C, and refreshed by addition of 5 mM NADPH after every hour. After every hour, 33 µL of the assay mixture was withdrawn, and diluted to 100 µL with 50 mM Tris-HCl (pH 8.0) buffer. The assay was then digested with 1 µg of proteomics grade trypsin protease (Sigma-Aldrich, T7575-1KT) for 45 min at 30 °C, and stored at −80 °C until further analysis.
Cloning, expression, and purification of Bmp5
The ORF encoding Bmp5 was amplified by PCR and ligated into pET28a(+) vector. N-His6-Bmp5 was expressed as previously described for other recombinant proteins in this study. The culture was harvested by centrifugation, and the cells were resuspended in 25 mL buffer A (50 mM Tris-HCl, pH 8.4, 400 mM NaCl, 0.1% Triton X-100, 20% glycerol) and lysed by sonication. The cell lysate was centrifuged at 11,000 rpm for 45 min. The supernatant was collected and imidazole was added to a final concentration of 10 mM. The supernatant was loaded onto a 5 mL column packed with 1.2 mL (wet volume) Ni-NTA Agarose (Qiagen). The loaded column was rinsed with 10 mL each of buffer A containing 10 mM and 50 mM imidazole. Majority of the recombinant Bmp5 did not bind to the Ni-NTA column and was obtained in the flow through and wash fractions as analyzed by SDS-PAGE. Column bound protein was eluted with 5 mL each of buffer A containing 75 mM, 100 mM, and 200 mM imidazole. Purified N-His6-Bmp5 was analyzed by SDS-PAGE (Supplementary Fig. 16a). The protein was then desalted to 20 mM Tris-HCl (pH 8.0) 20% glycerol buffer using Amicon Ultra centrifugal filters, and immediately used for assays. Bmp5 was found to be very prone to aggregation and precipitation, and care was taken to minimize the time the enzyme was stored prior to assays.
Enzyme assays for Bmp5
Bmp5 was incubated with 0.5 mM 4-HBA (Sigma-Aldrich, 240141-50G) in a 0.5 mL reaction, along with 5 mM NADPH, 0.1 mM FAD and 10 mM KBr in 20 mM Tris-HCl (pH 8.0) buffer. After the indicated time points, 40 µL of reaction aliquots were withdrawn and quenched by the addition of 16 µL of MeCN + 0.35% TFA, heated at 50 °C for 30 min, and centrifuged at 13,000 rpm for 10 min to remove precipitated protein. 25 µL of the quenched reaction was injected on a Phenomenex Luna C18 5 µm (4.6 × 100 mm) analytical HPLC column operating on an Agilent 1260 analytical HPLC setup at room temperature. Water + 0.1% TFA was used as buffer A, and MeCN + 0.1% TFA was used as buffer B. The elution profile was as follows: 5% buffer B for 5 min, linear gradient to 95% buffer B across 25 min, step increase to 100% buffer B, 100% buffer B for 3 min, linear decrease to 5% buffer B across 2 min, 5% buffer B for 5 min. Absorbance was monitored at 214 nm wavelength. Bmp5 reactions using 8–9 as substrates were conducted in an identical manner. For negative control reactions, Bmp5 was assayed using 4-HBA as the substrate, but with the omission of either the enzyme, NADPH, KBr or FAD. After 90 min, the reaction was quenched and analyzed by HPLC.
Enzyme assays for Bmp7
Purified Bmp7, together with Bmp9–10 was incubated with 0.2 mM 4, 2 mM NADH, and 10 µM FAD in a 1 mL reaction in 100 mM potassium phosphate (pH 7.6) buffer. After 12 h incubation at 30 °C, the reaction was extracted twice with ethyl acetate. The organic layer was collected and dried in vacuo. The precipitate was resuspended in 100 µL MeOH, and analyzed by LC-MS/MS as guided by literature41. Absorbance was monitored at 214 nm wavelength. Large scale reactions were performed in 100 mL reaction volume, with the substrate concentrations increased to 0.5 mM. Regeneration of NADH was achieved by commercially available glucose-6-phosphate dehydrogenase (Sigma-Aldrich, G2921-1KU) in the presence of glucose-6-phosphate (G6P) (Sigma-Aldrich, G7879-1G), or by PtdH-17X–A176R in the presence of sodium phosphite (Sigma-Aldrich, 04283-250G). Large scale reactions were refreshed with 2 mM G6P, or 2 mM sodium phosphite after 6 hours of reaction time. Reactions were extracted twice with ethyl acetate and the products separated by semi-preparative HPLC as described before. Detailed characterization of Bmp7 reaction products is described in the Supplementary Note 2.
Supplementary Material
Acknowledgments
We thank our colleagues E. Frick for preliminary in vitro studies with Bmp6, B. M. Duggan for assistance in NMR data collection, P. A. Jordan for NMR data analysis, Y. Su for mass spectrometry data collection and L. I. Aluwihare for useful discussions. This work was jointly supported by the NSF (OCE-1313747) and NIEHS (P01-ES021921) through the Oceans and Human Health program, the Gordon and Betty Moore Foundation Marine Microbial Sequencing Project, Helen Hay Whitney Foundation postdoctoral fellowship to V.A., NIH Marine Biotechnology Training Grant pre-doctoral fellowship to A.A.E. (T32-GM067550), and an NIH instrument grant (S10-RR031562).
Footnotes
Author Contributions
V.A., A.A.E., E.E.A. and B.S.M. designed research, V.A., A.A.E. and K.Y. performed genetic experiments, V.A., A.A.E. and R.D.K performed in vitro experiments, M.S. and E.E.A. generated sequencing data, D.P. contributed new analytical reagents, V.A., A.A.E., E.E.A. and B.S.M. analyzed data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Supplementary information is available in the online version of the paper. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Gribble GW. The natural production of organobromine compounds. Environ Sci Pollut Res Int. 2000;7:37–47. doi: 10.1065/espr199910.002. [DOI] [PubMed] [Google Scholar]
- 2.Gribble GW. Naturally Occurring Organohalogen Compounds - A Comprehensive Update. Vol. 91. Springer Vienna; 2010. [Google Scholar]
- 3.Liu YN, et al. Spatial and temporal distributions of bromoform and dibromomethane in the Atlantic Ocean and their relationship with photosynthetic biomass. J Geophys Res-Oceans. 2013;118:3950–3965. [Google Scholar]
- 4.Al-Mourabit A, Zancanella MA, Tilvi S, Romo D. Biosynthesis, asymmetric synthesis, and pharmacology, including cellular targets, of the pyrrole-2-aminoimidazole marine alkaloids. Nat Prod Rep. 2011;28:1229–1260. doi: 10.1039/c0np00013b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaul S, et al. Identification of the natural product 2,3,4,5-tetrabromo-1-methylpyrrole in Pacific biota, passive samplers and seagrass from Queensland, Australia. Mar Pollut Bull. 2011;62:2463–2468. doi: 10.1016/j.marpolbul.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Kitamura M, Koyama T, Nakano Y, Uemura D. Corallinafuran and Corallinaether, novel toxic compounds from crustose coralline red algae. Chemistry Letters. 2005;34:1272–1273. [Google Scholar]
- 7.Kuniyoshi M, Yamada K, Higa T. A biologically-active diphenyl ether from the green-alga Cladophora fascicularis. Experientia. 1985;41:523–524. [Google Scholar]
- 8.Malmvarn A, Zebuhr Y, Kautsky L, Bergman K, Asplund L. Hydroxylated and methoxylated polybrominated diphenyl ethers and polybrominated dibenzo-p-dioxins in red alga and cyanobacteria living in the Baltic Sea. Chemosphere. 2008;72:910–916. doi: 10.1016/j.chemosphere.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 9.King GM, Giray C, Kornfield I. Biogeographical, biochemical and genetic differentiation among North-American Saccoglossids (Hemichordata, Enteropneusta, Harrimaniidae) Mar Biol. 1995;123:369–377. [Google Scholar]
- 10.Unson MD, Holland ND, Faulkner DJ. A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar Biol. 1994;119:1–11. [Google Scholar]
- 11.Calcul L, et al. NMR strategy for unraveling structures of bioactive sponge-derived oxy-polyhalogenated diphenyl ethers. J Nat Prod. 2009;72:443–449. doi: 10.1021/np800737z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lofstrand K, et al. Brominated phenols, anisoles, and dioxins present in blue mussels from the Swedish coastline. Environ Sci Pollut Res Int. 2010;17:1460–1468. doi: 10.1007/s11356-010-0331-1. [DOI] [PubMed] [Google Scholar]
- 13.Vetter W, Scholz E, Gaus C, Muller JF, Haynes D. Anthropogenic and natural organohalogen compounds in blubber of dolphins and dugongs (Dugong dugon) from northeastern Australia. Arch Environ Contam Toxicol. 2001;41:221–231. doi: 10.1007/s002440010241. [DOI] [PubMed] [Google Scholar]
- 14.Marsh G, et al. Identification, quantification, and synthesis of a novel dimethoxylated polybrominated biphenyl in marine mammals caught off the coast of Japan. Environ Sci Technol. 2005;39:8684–8690. doi: 10.1021/es051153v. [DOI] [PubMed] [Google Scholar]
- 15.Teuten EL, Xu L, Reddy CM. Two abundant bioaccumulated halogenated compounds are natural products. Science. 2005;307:917–920. doi: 10.1126/science.1106882. [DOI] [PubMed] [Google Scholar]
- 16.Wan Y, et al. Origin of hydroxylated brominated diphenyl ethers: natural compounds or man-made flame retardants. Environ Sci Technol. 2009;43:7536–7542. doi: 10.1021/es901357u. [DOI] [PubMed] [Google Scholar]
- 17.Ren XM, Guo LH. Molecular toxicology of polybrominated diphenyl ethers: nuclear hormone receptor mediated pathways. Environ Sci Process Impacts. 2013;15:702–708. doi: 10.1039/c3em00023k. [DOI] [PubMed] [Google Scholar]
- 18.Isnansetyo A, Kamei Y. MC21-A a bactericidal antibiotic produced by a new marine bacterium Pseudoalteromonas phenolica sp nov O-BC30(T), against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:480–488. doi: 10.1128/AAC.47.2.480-488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen RJ, Wolfe MS, Faulkner DJ. Autotoxic antibiotic production by a marine chromobacterium. Mar Biol. 1974;27:281–285. [Google Scholar]
- 20.Burkholder PR, Pfister RM, Leitz FH. Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol. 1966;14:649–653. doi: 10.1128/am.14.4.649-653.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmstrom C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 22.Vetter W. Polyhalogenated alkaloids in environmental and food samples. Alkaloids Chem Biol. 2012;71:211–276. doi: 10.1016/b978-0-12-398282-7.00003-5. [DOI] [PubMed] [Google Scholar]
- 23.Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 24.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci U S A. 2005;102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peschke JD, Hanefeld U, Laatsch H. Biosynthesis of the marine antibiotic pentabromopseudilin 2 The pyrrole ring. Biosci Biotech Bioch. 2005;69:628–630. doi: 10.1271/bbb.69.628. [DOI] [PubMed] [Google Scholar]
- 26.Lucas-Elio P, et al. Complete genome sequence of the melanogenic marine bacterium Marinomonas mediterranea type strain (MMB-1(T)) Stand Genomic Sci. 2012;6:63–73. doi: 10.4056/sigs.2545743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donia MS, Fricke WF, Ravel J, Schmidt EW. Variation in tropical reef symbiont metagenomes defined by secondary metabolism. PLoS One. 2011;6:e17897. doi: 10.1371/journal.pone.0017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sorokin DY, Tourova TP, Lysenko AM, Mityushina LL, Kuenen JG. Thioalkalivibrio thiocyanoxidans sp nov Thioalkalivibrio paradoxus sp nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria capable of growth on thiocyanate, from soda lakes. Int J Syst Evol Microbiol. 2002;52:657–664. doi: 10.1099/00207713-52-2-657. [DOI] [PubMed] [Google Scholar]
- 29.Walsh CT, Garneau-Tsodikova S, Howard-Jones AR. Biological formation of pyrroles: nature’s logic and enzymatic machinery. Nat Prod Rep. 2006;23:517–531. doi: 10.1039/b605245m. [DOI] [PubMed] [Google Scholar]
- 30.Buedenbender S, Rachid S, Muller R, Schulz GE. Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases. J Mol Biol. 2009;385:520–530. doi: 10.1016/j.jmb.2008.10.057. [DOI] [PubMed] [Google Scholar]
- 31.van Pee KH. Enzymatic chlorination and bromination. Methods Enzymol. 2012;516:237–257. doi: 10.1016/B978-0-12-394291-3.00004-6. [DOI] [PubMed] [Google Scholar]
- 32.Eichhorn E, van der Ploeg JR, Leisinger T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 33.Blasiak LC, Drennan CL. Structural perspective on enzymatic halogenation. Acc Chem Res. 2009;42:147–155. doi: 10.1021/ar800088r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gribble GW. Occurrence of halogenated alkaloids. Alkaloids Chem Biol. 2012;71:1–165. doi: 10.1016/b978-0-12-398282-7.00001-1. [DOI] [PubMed] [Google Scholar]
- 35.Hanefeld U, Floss HG, Laatsch H. Biosynthesis of the marine antibiotic pentabromopseudilin .1. The benzene-ring. Journal of Organic Chemistry. 1994;59:3604–3608. [Google Scholar]
- 36.Walsh CT, Wencewicz TA. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat Prod Rep. 2013;30:175–200. doi: 10.1039/c2np20069d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewson WD, Hager LP. Bromoperoxidases and halogenated lipids in marine-algae. J Phycol. 1980;16:340–345. [Google Scholar]
- 38.Borchardt SA, et al. Reaction of acylated homoserine lactone bacterial signaling molecules with oxidized halogen antimicrobials. Appl Environ Microbiol. 2001;67:3174–3179. doi: 10.1128/AEM.67.7.3174-3179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beissner RS, Guilford WJ, Coates RM, Hager LP. Synthesis of brominated heptanones and bromoform by a bromoperoxidase of marine origin. Biochemistry. 1981;20:3724–3731. doi: 10.1021/bi00516a009. [DOI] [PubMed] [Google Scholar]
- 40.Feher D, Barlow R, McAtee J, Hemscheidt TK. Highly brominated antimicrobial metabolites from a marine Pseudoalteromonas sp. J Nat Prod. 2010;73:1963–1966. doi: 10.1021/np100506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mas S, et al. Comprehensive liquid chromatography-ion-spray tandem mass spectrometry method for the identification and quantification of eight hydroxylated brominated diphenyl ethers in environmental matrices. J Mass Spectrom. 2007;42:890–899. doi: 10.1002/jms.1224. [DOI] [PubMed] [Google Scholar]
- 42.Makino M, et al. Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton. Proc Natl Acad Sci U S A. 2007;104:11591–11596. doi: 10.1073/pnas.0702946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballou DP, Entsch B, Cole LJ. Dynamics involved in catalysis by single-component and two-component flavin-dependent aromatic hydroxylases. Biochem Biophys Res Commun. 2005;338:590–598. doi: 10.1016/j.bbrc.2005.09.081. [DOI] [PubMed] [Google Scholar]
- 44.Eppink MH, Cammaart E, Van Wassenaar D, Middelhoven WJ, van Berkel WJ. Purification and properties of hydroquinone hydroxylase, a FAD-dependent monooxygenase involved in the catabolism of 4-hydroxybenzoate in Candida parapsilosis CBS604. Eur J Biochem. 2000;267:6832–6840. doi: 10.1046/j.1432-1033.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 45.Eppink MH, Boeren SA, Vervoort J, van Berkel WJ. Purification and properties of 4-hydroxybenzoate 1-hydroxylase (decarboxylating), a novel flavin adenine dinucleotide-dependent monooxygenase from Candida parapsilosis CBS604. J Bacteriol. 1997;179:6680–6687. doi: 10.1128/jb.179.21.6680-6687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. Marine natural products. Nat Prod Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- 47.Wiseman SB, et al. Polybrominated diphenyl ethers and their hydroxylated/methoxylated analogs: environmental sources, metabolic relationships, and relative toxicities. Mar Pollut Bull. 2011;63:179–188. doi: 10.1016/j.marpolbul.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 48.Ucan-Marin F, Arukwe A, Mortensen AS, Gabrielsen GW, Letcher RJ. Recombinant albumin and transthyretin transport proteins from two gull species and human: chlorinated and brominated contaminant binding and thyroid hormones. Environ Sci Technol. 2010;44:497–504. doi: 10.1021/es902691u. [DOI] [PubMed] [Google Scholar]
- 49.Meerts IA, et al. Potent competitive interactions of some brominated flame retardants and related compounds with human transthyretin in vitro. Toxicol Sci. 2000;56:95–104. doi: 10.1093/toxsci/56.1.95. [DOI] [PubMed] [Google Scholar]
- 50.Dorrestein PC, Kelleher NL. Dissecting non-ribosomal and polyketide biosynthetic machineries using electrospray ionization Fourier-Transform mass spectrometry. Nat Prod Rep. 2006;23:893–918. doi: 10.1039/b511400b. [DOI] [PubMed] [Google Scholar]
- 51.John EA, Pollet P, Gelbaum L, Kubanek J. Regioselective syntheses of 2,3,4-tribromopyrrole and 2,3,5-tribromopyrrole. J Nat Prod. 2004;67:1929–1931. doi: 10.1021/np0498399. [DOI] [PubMed] [Google Scholar]
- 52.Markowitz VM, et al. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 53.Kouprina N, Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- 54.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li DBChapter 19, et al. In vitro studies of phenol coupling enzymes involved in vancomycin biosynthesis. Methods Enzymol. 2009;458:487–509. doi: 10.1016/S0076-6879(09)04819-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.