Abstract
The significant diversity among HIV-1 variants poses serious challenges for vaccine development and for developing sensitive assays for screening, surveillance, diagnosis and clinical management. Recognizing a need to develop a panel of HIV representing the current genetic and geographic diversity NIH/NIAID contracted the External Quality Assurance Program Oversight Laboratory (EQAPOL) to isolate, characterize and establish panels of HIV-1 strains representing global diverse subtypes and circulating recombinant forms (CRFs), and to make them available to the research community. HIV-positive plasma specimens and previously established isolates were collected through a variety of collaborations with a preference for samples from acutely/recently infected persons diagnosed since 2009. Source specimens were cultured to high-titer/high-volume using well-characterized cryopreserved PBMCs from healthy donors. Panel samples were stored as neat culture supernatant or diluted into defibrinated plasma. Characterization for the final expanded virus stocks included viral load, p24 antigen, infectivity (TCID), sterility, coreceptor usage, and near full-length genome sequencing. Viruses are made available to approved, interested laboratories using an online ordering application. The current EQAPOL Viral Diversity panel includes over 101 viral specimens representing 6 subtypes (A, B, C, D, F, and G), 2 sub-subtypes (F1 and F2), 7 CRFs (01, 02, 04, 14, 22, 24, and 47), 19 URFs and 3 group O viruses from 22 countries. The EQAPOL Viral Diversity panel is an invaluable collection of well-characterized reagents that are available to the scientific community, including researchers, epidemiologists, and commercial manufacturers of diagnostics and pharmaceuticals to support HIV research and diagnostic and vaccine development.
Keywords: HIV, EQAPOL, Subtype Panel, Recombinant
1. Introduction
At the end of 2011, there were an estimated 34 million people living with human immunodeficiency virus type 1 (HIV-1) (UNAIDS, 2012). Extensive efforts to prevent transmission, diagnose infection, monitor disease progression and treat infected individuals are ongoing (WHO, 2011; O'Connell et al., 2012; Piot and Quinn, 2013). Standardized reagents and assays are critical to support these efforts as well as for evaluating immune responses to vaccine candidates.
The rapid evolution of HIV has given rise to multiple subtypes and recombinant strains of HIV-1 and HIV-2. Presently, HIV-1, the major source of the global pandemic, is divided into four groups: M (Major), O (Outlier), N (Non-M, Non-O) and P (Charneau et al., 1994; Simon et al., 1998; Corbet et al., 2000; Plantier et al., 2009). HIV-1 M group, which accounts for the vast majority of HIV-1 infections, has been further subdivided into nine subtypes (A–D, F–H, J, and K), 58 circulating recombinant forms (CRFs) and many unique recombinant forms (URFs) (Los Alamos HIV Sequence Database; URL: http://www.hiv.lanl.gov). The significant diversity observed within HIV poses serious challenges for vaccine development and for maintaining sensitive assays for screening, surveillance, diagnosis and clinical management.
Qualitative detection of HIV-1 RNA is employed in blood screening and for diagnosis of acute infections. Quantification of HIV-1 RNA viral load (VL) in plasma is essential for monitoring response to antiretroviral therapy, estimating risk of disease progression and to establish diagnosis when a negative or indeterminate HIV-1 antibody test is present (Coombs et al., 1996; Mellors et al., 1996; O'Brien et al., 1996; Welles et al., 1996). The assays to detect and quantify HIV RNA have been developed and validated mainly using B subtype samples whereas non-B subtypes represent more than 89% of strains that are circulating with the proportion of CRFs and URF increasing (Hemelaar et al., 2011). Furthermore, assays are targeted to different genes in the viral genome using sequence-specific probes necessitating thorough evaluation of different viral load platforms across an array of subtypes. Differences between commercially available qualitative and quantitative viral load assays for non-B subtype strains have been noted (Antunes et al., 2003; Gottesman et al., 2006; Kim et al., 2007; Xu et al., 2010; Amendola et al., 2011; Chudy et al., 2012). In addition, differences between subtypes in serological diagnostic and screening assays have been reported (Ly et al., 2007; Ly et al., 2012).
In addition to posing challenges for screening and HIV quantification, subtype-specific differences have been implicated in disparate rates of disease progression, development of drug resistance mutations and responses to antiretroviral therapies (Adazu et al., 2005). Recognizing the challenges that HIV diversity poses, HIV diversity panels have been previously assembled to support the development of HIV detection, treatment and vaccination (Huang et al., 2003; Brown et al., 2005). However, they were generated from source samples collected more than a decade ago and, therefore, do not adequately represent the current HIV-1 diversity. Furthermore, many of these samples have not been characterized with current molecular assays. Recognizing a need to develop a panel which encompasses the genetic and geographic diversity currently present worldwide, the National Institute of Health / National Institute of Allergy and Infectious Diseases (NIH/NIAID) established a pilot program to develop a HIV subtype panel in collaboration with the Food and Drug Administration (FDA), Walter Reed Army Institute of Research (WRAIR), academia, and industry representatives (Manak et al., 2012). Through this pilot program, 30 viral isolates representing subtypes B, C, B/F, CRF04_cpx, and CFR02_AG were obtained from plasma samples. The viral strains in the pilot panel were well-characterized, and all of the data was compiled to be made available to researchers interested in those samples.
To expand on the important work initiated in the pilot panel program, the NIH/NIAID-sponsored External Quality Assurance Program Oversight Laboratory (EQAPOL) program was funded and includes as a major goal the continued development of contemporaneous HIV global diversity panels. The EQAPOL panel focused on identifying virus source specimens mainly from recently diagnosed individuals with acute/early stages of infection. Plasma samples were obtained through collaborations with blood banks worldwide taking advantage of their HIV nucleic acid technology (NAT) screening programs whereby HIV RNA positive/antibody-negative donors were detected and deferred. Additional samples were acquired from previously collected plasma from individuals who are part of recently HIV infected cohorts. In some cases, it is difficult to export plasma/Peripheral Blood Mononuclear Cell (PBMC) samples from countries due to regulatory restrictions. Therefore, previously established viral isolates from these countries were also obtained to expand the overall diversity of the panel. Since the EQAPOL program's inception in 2011, over 100 viral samples have been propagated to high-titer/ high-volume and fully characterized.
2. Materials and Methods
2.1. Viral Source Specimen Collection and Sources
2.1.1. NAT-Yield Plasma Samples
Blood Donor Sample Collection
Acquisition of samples from acutely infected blood donors for use in this study was coordinated by the Blood System Research Institute (BSRI). Samples were acquired through regular blood donations identified by individual or pooled screening as NAT-yield positive (RNA positive, antibody negative), and collected as whole blood donations processed into red blood cells and plasma components. All donations had been collected with ethical review and approval of the collection protocol and informed consent as previously established for the donor organization's collection activities.
Sample Sources
Plasma samples were obtained from six countries on four continents; South Africa (South African National Blood Service (n=148)), the U.S. (American Red Cross (n=52)), France (Institut National de la Transfusion Sanguine (n=5)), Brazil (Department of Infectious Disease/University of Sao Paulo, (n=3)), Spain (Banc de Sang i Teixits (n=13)), and Japan (Central Blood Institute, Blood Service Headquarters, Japanese Red Cross Society (n=11)). All samples except those from Brazil and Spain were confirmed as NAT-yield (RNA positive, antibody negative) based on documented testing criteria at the collecting organizations. The HIV positive samples from Brazil (Fiebig staging: 2 × V stage and 1 × VI stage) and Spain were from recently infected donors as defined by very recent seroconversion, indeterminate Western blots or application of HIV recency assays with results indicating the donations were given with 1–2 months of acquisition of infection (Busch et al., 2010).
Sample Characterization
South African NAT-yield (RNA positive, antibody negative) donors included in this study were identified from 2009 through 2011 by individual donation (ID) NAT screening using Ultrio followed by HIV discriminatory transcription mediated amplification (TMA) (Gen Probe, San Diego, CA). All were Abbott HIV-1/2 ChLIA negative (Abbott Laboratories, Abbott Park, IL). Based on sequence analysis, the majority of these samples were clade C. U.S. NAT-yield donors were identified from 2001 through 2009 by min pool (MP) NAT screening in pools of sixteen using Procleix followed by HIV discriminatory TMA (Novartis Diagnostics, Emeryville, USA). All were Abbott-HIV-1/2 EIA negative. The majority were clade B based on sequence analysis. (Stramer et al., 2000; Stramer et al., 2004; Zou et al., 2010). French NAT-yield donors were identified from 2000 through 2011 by ID or MP NAT screening. All were Genscreen Ultra Ag-AB negative (Genscreen Pty Ltd, Australia) as well as Innotest p24 antigen negative (Innogenetics NV, Gent, Belgium). Descriptive information is not available for the Brazilian, Spanish, and Japanese NAT-yield and recently infected specimens.
2.1.2. Additional Plasma and PBMCs Samples
Plasma samples from 55 donors previously collected as part of the International AIDS Vaccine Initiative (IAVI) Protocol C were kindly provided by Pat Fast, Pauli Amornkul and Matt Price for inclusion in the EQAPOL program. The samples received by EQAPOL were collected at seven clinical research centers between 2009 to 2011: Kigali, Rwanda (Etienne Karita, Susan Allen and Eric Hunter); Lusaka and Copperbelt, Zambia (William Kilembe, Susan Allen, Mubiana Inambao, and Eric Hunter); Nairobi and Kilifi, Kenya (Eduard Sanders, Omu Anzala and Gaudensia Mutua); Entebbe and Masaka, Uganda (Anatoli Kamali and Eugene Ruzagira) (Price et al., 2011). At-risk volunteers were followed quarterly or month in large incidence cohorts, and it is estimated that samples were collected within 6 months of infection.
Additionally, Peripheral Blood Mononuclear Cells (PBMCs) and plasma from four donors in Russia were kindly provided by Hagen von Brisen at the Fraunhofer Institut fϋr Biomedizinisch Technik (IBMT). Samples were collected in 2008 within an estimated 1–6 months of infection.
2.1.3. Viral Culture Supernatants and Infectious Molecular Clones (IMCs)
Previously cultured viral supernatants from ten donors from China were kindly provided by Hong Shang, Wenqing Geng, Hualu Cui, and Hong Sun at the First Affiliated Hospital of China Medical University. Donor samples were collected between 2009 and 2011, and included a mix of supernatants obtained from recently infected (Fiebig IV and V) and chronically infected individuals as defined by the contributing laboratory. Thirty-three viral supernatants were obtained from the Centre for AIDS Reagents, NIBSC HPA UK, supported by the EC FP6/7 Europrise Network of Excellence, and NGIN consortia and the Bill and Melinda Gates GHRCCAVD Project and were donated by Lucia Pérez Alvarez (Instituto de Salud Carlos III) (Fernandez-Garcia et al., 2009; Cuevas et al., 2010; Fernandez-Garcia et al., 2010; Delgado et al., 2012; Thomson et al., 2012; Diez-Fuertes et al., 2013). Twenty-three culture supernatants from donors in Cameroon, Spain, USA and Germany were kindly provided by Indira Hewlett (CBER, FDA) (Vallejo et al., 2003; Ragupathy et al., 2011; Zhao et al., 2012). Additionally, EQAPOL received 12 Infectious Molecular Clones (IMCs) made and contributed by the Beatrice Hahn and George Shaw Laboratories (University of Pennsylvania) from plasma collected from acute donors (four Fiebig I, two Fiebig II, one Fiebig III, and four Fiebig IV) in Uganda and Rwanda by IAVI and the Uganda Virus Research Institute-British Medical Research Council program on HIV/AIDS (UVRI/MRC) (Price et al., 2011; Tang et al., 2011; Price et al., 2012; Baalwa et al., 2013).
2.2. Feeder Cell Acquisition and Permissivity Testing
2.2.1. Feeder Cell Acquisition
PBMCs used as feeder cells for EQAPOL virus culture were collected from healthy donors by leukapheresis. All donors were negative for HIV-1/2 Antibody, Hepatitis B Surface Antigen and/or Core, and cytomegalovirus (CMV). All donors were properly consented according to Duke University IRB, Federal and State regulations. Using the leukapheresis material, PBMCs were separated on Ficoll-Hypaque gradients and cryopreserved in a solution of 10% DMSO (Sigma-Aldrich, St. Louis, MO) in heat-inactivated fetal bovine serum (FBS) (Gemini Bio-products, West Sacramento, CA). PBMCs were slowly frozen using a computerized control rate freezer system and then quickly transferred into −180°C LN2 freezers. All procedures were performed under Good Clinical Laboratory Practices (GCLP).
2.2.2. Donor PBMC Permissivity
Donor PBMC permissivity was tested to select donors to pool for viral culture. Briefly, donor PBMCs were infected with a panel of six different viruses representing a variety of subtypes, groups and coreceptors of HIV. Tissue Culture Infectivity Dose (TCID) in TZM-bl cells was measured on each of these donor/virus conditions 2, 4 or 6 days after infection (see below for TCID measurement). Five donors were selected to pool for all viral cultures based on the permissivity observed. Donors in the pool were replaced as needed.
2.3. Virus isolation and expansion
All work was carried out under BSL-3 guidelines in a BSL-3 environment at the Regional Biocontainment Laboratory at Duke University Medical Center using GCLP practices. Viruses were cultured in a two-step procedure termed the small and large-scale cultures. The protocol utilized from culture was adapted from previously published methods (Albert et al., 1987; Falk et al., 1987; Costa et al., 1996; Aguado-Martinez et al., 2009). Briefly, cryopreserved PBMCs pooled from five donors, termed feeder cells, were activated with α-CD28 at 100ng/mL (BD Biosciences, San Jose, CA) and α-CD3 at 50ng/mL (eBioscience, San Diego, CA) three days prior to the start of culture. On the culture start date, PBMCs were CD8-depleted using CD8 Dynabeads® (Invitrogen, Grand Island, NY) according the manufacturer's instructions as removal of CD8+ T cells has been reported to increase HIV replication (Walker et al., 1986). When the source viral specimen was plasma, virus was first enriched by a 20% sucrose density gradient centrifuged at 40,000 × g for 2 hours. Virus isolated from plasma or virus culture supernatant (0.1–3mL) was added to 2 × 107 CD3/CD28-stimulated, CD8-depleted PBMCs, centrifuged at 1,200 × g for 2 hours to enhance infection (“spinoculation” see (O'Doherty et al., 2000)), and cultured at 37°C and 5%CO2 in a total volume of 20mL in culture media (RPMI 1640 w/L glutamine, 20% FBS, 1% Pen Strep and rh IL-2 at 2ng/mL).
Cells were cultured for two days, and then additional (2 × 107) CD8-depleted, activated feeder cells were added to the culture. Cell culture supernatant was removed, filtered, and frozen at −80°C on day 2 (prior to addition of supplementary cells), and then every two to three days through the life of the culture (usually less than 14 days). Spinoculation was performed at each harvest day. Representative aliquots from each harvest day were checked for viral load using the COBAS AmpliPrep/COBAS Taqman 48 HIV-1 test (Roche, Pleasanton, CA), and the culture was terminated after the viral load peaked. The culture supernatant isolated from the peak culture day (approximately 30mL) was thawed and aliquoted into LDMS-labeled vials at 0.5mL/vial and stored at −80°C. This product was termed the “master lot” and used to start the subsequent large-scale culture.
For the large-scale culture, 0.5mL of the master lot and 1 × 108 activated, CD8-depleted pooled PBMCs were spinoculated and cultured in a total volume of 100mL. Similar to the small-scale culture, on day 2 the culture was further expanded by adding 1.75 × 108 activated, CD8-depleted pooled PBMCs and bringing the total culture volume to 250mL. The process of expanding the culture was repeated every 2–3 days with the viral load closely monitored. When the culture reached high titer (> 1 × 109 cp/mL), neat culture supernatant was filtered then stored in 1mL aliquots without any freeze/thaws prior to aliquoting. Virus was also stored by spiking defibrinated human AB plasma (Gemini Bio-Products, West Sacramento, CA) with culture supernatants to yield a variety of viral load concentrations.
2.4. Sample characterization
Source material (plasma, culture supernatants), intermediate culture supernatant products and final culture products (culture supernatant and HIV-spiked plasma) were characterized using a variety of assays. Testing included commercially available FDA-approved nucleic acid and antibody assays and a Research Use Only (RUO) p24 antigen assay. The Roche COBAS AmpliPrep/TaqMan HIV-1 Version 2.0 viral load assay was used to measure viral RNA in source material, intermediate culture supernatant products and final culture products.
Anti-HIV reactivity of source plasma material was tested by the Bio-Rad Genetic Systems Anti-HIV-1/HIV-2 Plus O EIA and the Bio-Rad Genetic Systems HIV-1 Western blot methods (Hercules, CA). The PerkinElmer HIV-1 p24 Antigen Capture Assay (Waltham, MA) was used to measure supernatant antigen levels in final culture supernatant products. All assays were performed under GCLP and were performed according to manufacturer's instructions. Source viral plasma specimens were classified based on Fiebig stage as determined by the presence of viral RNA and antibody by EIA and specific bands on Western blot according to published methods (Fiebig et al., 2003). Testing of p24 antigen was not done to distinguish Fiebig stage I versus II.
Infectivity was determined using a TZM-bl-based TCID assay as previously described (Li et al., 2005). Briefly, TZM-bl cells (10,000 per well) were incubated with a 5-fold serial dilution of viral supernatant stock (1:10 to 1:97,656,250) with four replicates per dilution. Cells were cultured for two days, and then infectivity was measured using Britelite Plus reagent (Perkin Elmer, Waltham, MA). Wells were considered positive for infection if the RLU was 2.5-fold higher than non-infected cells. Infectivity was calculated as the dilution at which half of the four wells per sample dilutions were positive. The assay was repeated as two independent replicate infectious titration studies, and the average TCID was reported.
2.4.1. Sterility Testing
Selected viral source specimen and all final viral culture supernatant products were tested to ensure they were free of adventitious agents, including bacterial contamination and mycoplasmas. Sterility testing included direct inoculation to detect bacterial contamination, a Mycoplasma test, and a qualitative Limulus amebocyte lysate (LAL) assay to detect endotoxin. Briefly, 0.5mL of culture supernatant was added to 8mL prepared BBL™ Fluid Thioglycollate Medium (BD, Franklin Lakes, NJ) or Trypticase™ Soy Broth with 6.5% sodium chloride (BD, Franklin Lakes, NJ) and incubated at 35± 2°C or 22.5± 2.5°C, respectively. Samples were visually inspected for turbidity on days 3, 4, 7, and 14 after inoculation and were considered negative if no turbidity was observed during the 14 day culture period. Mycoplasma testing was done using a MycoAlert® Detection Kit (Lonza, Cologne, Germany) according to the manufacturer's instructions, and samples were reported as either positive or negative. Samples were tested for endotoxin using a qualitative gel clot LAL assay (Lonza, Cologne, Germany) according to the manufacturer's instructions. Prior to testing, samples were diluted 1:10 in culture medium to prevent inhibition of the clot reaction. If no clot was observed, then the sample was reported as containing less than 1.25EU/mL endotoxin.
2.4.2. Genotyping based on near full-length HIV-1 genome sequences
Viral RNA was extracted from 100μL of culture supernatant using the Qiagen EZ1 Virus mini kit on the BioRobot EZ1 (Qiagen, Valencia, CA) and used for cDNA synthesis using Superscript III Reverse Transcriptase (Life Technologies; Carlsbad, CA) and primers 1R3.B3R (5'-ACTACTTGAAGCACTCAAGGCAAGCTTTATTG-3' nt 9611–9642) and 07Rev9 (5'-CTTCCTGCCATAGGAGATGCCTAA-3' nt 5957–5980) for 3'- and 5'-half HIV-1 genome, respectively. Single genome amplification (SGA) was performed to obtain two overlapping half genomes for group M viruses as we previously described (Salazar-Gonzalez et al., 2009). For amplification of the 5'-half genome, the primers for the first round PCR were 1U5.B1F (5'-CCTTGAGTGCTTCAAGTAGTGTGTGCCCGTCTGT-3' nt 538–571), 07Rev8 (5'-CCTARTGGGATGTGTACTTCTGAACTT-3' nt 5193–5219), and the primers for the second round PCR were UP1A (5'-AGTGGCGCCCGAACAGG-3' nt 634–650) and Rev11 (5'-ATCATCACCTGCCATCTGTTTTCCAT-3' nt 5041–5066). For amplification of the 3'-half genome, the primers for the 1st round PCR were 07For7 (5'-CAAATTAYAAAAAATTCAAAATTTTCGGGTTTATTACAG-3'; nt 4875–4912) and 2.R3.B6R (5'-TGAAGCACTCAAGGCAAGCTTTATTGAGGC-3'; nt 9607–9636), and the primers for the 2nd round PCR were VIF1 (5'-GGGTTTATTACAGGGACAGCAGAG-3'; nt4900–4923) and Low2C (5'-TGAGGCTTAAGCAGTGGGTTCC-3'; nt 9591–9612). The same primers were used for amplification of both 5' and 3' halves of group O viral genomes, except the second round primers for the 5' half genome; O-LOW2 (5'-GAACCCGCTGCTTAACGCTCAATA-3' nt 507–530 and O-REV12 (5'-TGTCTGCTGTTTCAGGGTCAATCTGTGTT-3' nt 5325–5353). Eight PCR reactions were performed using the cDNA dilution that gave a PCR positive rate of 25% or less to obtain at least one 5' or 3' half SGA for each sample. The PCR amplification was performed using the Platinum Taq DNA polymerase High Fidelity (Life Technologies, Carlsbad, CA). The PCR thermocycling conditions were as follows: one cycle at 94°C for 2 minutes; 35 cycles of a denaturing step at 94°C for 15 seconds, an annealing step at 58°C for 30 second, an extension step at 68°C for 5.5 minutes; and one cycle of an additional extension at 68°C for 10 minutes.
SGA amplicons were sequenced directly by cycle sequencing and dye terminator methods using an ABI 3730xl genetic analyzer (Applied Biosystems, Foster City, CA). Individual sequences were assembled and edited using the Sequencher program 4.10 (Gene Codes, Ann Arbor, MI). Phylogenetic relationships of the newly characterized near full-length genome sequences were estimated from sequence comparisons with previously reported HIV-1 subtype, circulating recombinant form (CRFs) and group O virus reference sequences from the Los Alamos database. Sequences were added to an existing master alignment using the profile alignment option of CLUSTAL W and adjusted manually when necessary using SEAVIEW. Sites where there was a gap in any of the sequences, as well as areas of uncertain alignment, were excluded. Pairwise evolutionary distances were estimated by using the Kimura two-parameter model (Kimura, 1980). Phylogenetic trees were constructed by the neighbor-joining method (Saitou and Nei, 1987), and the reliability of topologies was estimated by performing bootstrap analysis with 1000 replicates. Recombination patterns were determined using the REGA Subtyping tool version 2 (http://jose.med.kuleuven.ac.be/genotypetool/html/subtypinghiv.html) and the jumping profile Hidden Markov Model (jpHMM) at GOBICS website (http://jphmm.gobics.de/) (Schultz et al., 2006; Schultz et al., 2009).
2.4.3. Coreceptor usage using cell culture
Coreceptor usage was determined on NP-2 cells which express CD4 along with CCR5 or CXCR4 as previously described (Shimizu et al., 2009; Jiang et al., 2011). Briefly, NP-2 cells (5 × 104) were seeded in 48-well plates the day before infection. Each virus (25μl cell culture supernatant) was added to the well in triplicates. After absorption at 37°C for 3 hours, cells were washed twice and maintained in complete medium at 37°C with 5% CO2. Virus replication was determined by measuring the p24 Antigen concentrations in culture supernatants collected at day 0 and day 7 with the Alliance HIV-1 p24 Antigen ELISA kit.
3. Results
3.1. Overview of EQAPOL Viral Diversity Project Process
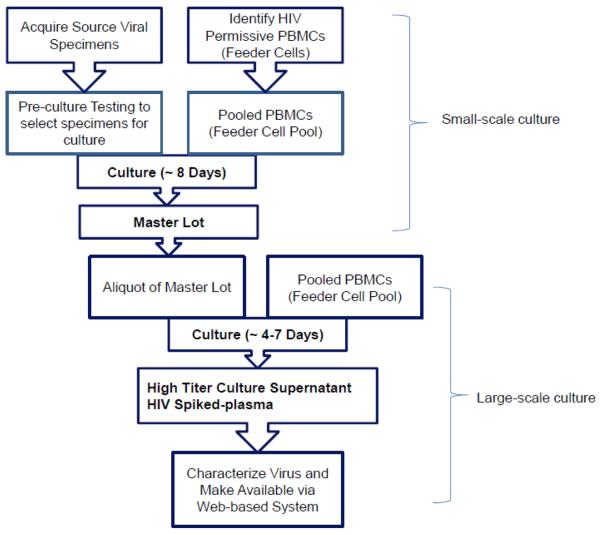
For the EQAPOL Viral Diversity Program, 50 HIV are cultured per year using a well-documented workflow for the culture and characterization process as depicted in Figure 1. Briefly, source viral specimen samples were acquired from a variety of collaborators (see Methods and Acknowledgements sections) and tested to select samples with characteristics desirable for culture. Preference was given for samples from acute infection (Fiebig Stage V or earlier) and samples that would expand the geographic or genetic diversity of the panel.
Figure 1. Flow Chart of EQAPOL Viral Diversity Project.
Viral Source Specimens were acquired and cultured in a two-step process to yield high-titer/high volume HIV culture supernatant. Virus is stored as neat culture supernatant and HIV spiked plasma at a variety of concentrations. Cultured viruses were extensively characterized and made available to approved interested sites using a secure web-based system.
Separately, cryopreserved PBMCs were screened for HIV permissivity to a variety of HIV subtypes/coreceptors as different donors have different susceptibilities to HIV (Williams and Cloyd, 1991; Spira and Ho, 1995; Greco et al., 1998). PBMCs from five donors with the best permissivity were selected to be part of a Feeder Cell Pool to be used in all cultures. As needed new donor PBMCs were added to the Feeder Cell Pool following HIV permissivity test.
Source viral specimen material was cultured with PBMCs in a two-step culture process to generate high-titer/high-volume culture stock, which was saved as neat culture supernatant and spiked into defibrinated plasma. Spiked plasma is made at approximately 1 ×107, 5 ×107 and 1 × 104 cp/mL. Viral stocks were then extensively characterized and made available for order to approved laboratories via a web-based application (https://eqapolapp.dhvi.duke.edu). All products produced for distribution include a Certificate of Analysis (COA) signed by the EQAPOL Central Quality Assurance Unit, which monitors GLCP compliance in the EQAPOL program (see Todd et al. in this issue).
All EQAPOL samples were named using a standardized naming convention based on a previously generated standard (Korber et al., 1994). The name provides information about the clade/group, country of origin, year of collection, and sample type (i.e. Culture supernatant, HIV-spiked plasma). As an example, in the name DEMB10US001.S1, DE indicates “Duke EQAPOL”, MB indicates Group M, Subtype B, 10 indicates the year the sample was collected, US is the two letter country code of origin, 001 indicates the sample number for that country and .S1 indicates that the sample type is culture supernatant..
3.2. Viral Source Specimens and Initial Characterization
Since the start of the EQAPOL Viral Diversity program in September 2010, 370 viral source specimens have been acquired as summarized in Table 1. Samples include 288 plasma specimens obtained through collaborations with BSRI and IAVI. To increase both genetic and geographic diversity, we have also collected previously-cultured viruses and IMCs for inclusion in the panel. Source specimens underwent an initial characterization to help select samples for culture. Testing included viral load, Fiebig staging, sterility testing, and full-length sequencing. Viral load was used to select plasma samples that would likely grow in culture; greater than 1 × 104 cp/mL was preferred as lower concentrations have a much lower success rate in culture (Manak et al., 2012). Previously cultured virus supernatants were tested for sterility to ensure they were free of bacterial and mycoplasma contamination to assure contamination was not introduced into the BSL-3 culture space and to allow the material to be used for the development of unique culture-based bioassays. Near full-length genome sequences were obtained by population sequencing for all source specimens prior to culture to determine the HIV subtype and if they contained more than one genotypes. All of this characterization data was compiled and used to select which samples would contribute to the overall genetic and geographic diversity of the panel.
Table 1.
Summary of Viral Source Specimens for EQAPOL Viral Diversity Program.
| Country of Origin | # of Samples |
|---|---|
| Brazil | 3 |
| Cameroon | 20 |
| China | 10 |
| France | 5 |
| Germany | 1 |
| Japan | 11 |
| Kenya | 8 |
| Russia | 4 |
| Rwanda | 4 |
| South Africa | 148 |
| Spain | 48 |
| Uganda | 34 |
| USA | 53 |
| Zambia | 21 |
|
| |
| Total | 370 |
Listing of countries and number of received from EQAPOL collaborators at the following institutions: Blood Systems Research Institute (BSRI), International AID Vaccine Initiative (IAVI), Instituto de Salud Carlos III, First Affiliated Hospital, China Medical University, SeraCare (collected as part of pilot panel), University of Pennsylvania, Food and Drug Administration, and Fraunhofer Institut für Biomedizinisch Technik (IBMT). Source specimens include plasma, PBMCs, previously cultured HIV and infectious molecular clones.
3.3. Virus Isolation and Expansion to High-titer/High-volume
Viruses were cultured in a two-step culture process (see Figure 1). In the first culture, virus isolated from plasma or culture supernatant from a previous HIV culture were mixed with activated, CD8-depleted, cryopreserved PBMCs pooled. This culture is termed the small-scale culture because the total culture volume is about 40mL. Viral propagation was monitored by testing the culture supernatant for viral load every two to three days during the culture with culture supernatant collected and frozen at each timepoint. Once the culture viral load peaks, the culture was terminated, and the culture supernatant harvested from the peak was aliquoted. The product of the first culture was termed the “master lot”.
Of the 370 viral source specimens, an initial high-titer (small-scale) culture has been started with 148 source specimens, and 122 have been successfully cultured to high-titer (82.4% success rate). The success rate for isolation of the viruses from plasma samples was 69.7% (53 out of 76). The success rate with plasma is similar to rates previously reported for plasma samples with greater than 1 × 104 cp/mL (Manak et al., 2012). The average culture time for the small-scale culture was 8.87 days, and the average peak VL was 8.9 ×109cp/mL. Figure 2a shows example growth curves for four viruses: DEMB09CN002 (Supernatant source, peaked at Day 4), DEURF11UG004 (Plasma source, peaked at Day 7), DEURF09UG002 (Plasma source, peaked at Day 11), and DEMB09US002 (Plasma source, peaked at Day 11). Master lot samples were all named according to the sample naming convention described.
Figure 2. Example Growth Curves for Small-scale and Large-scale Cultures.
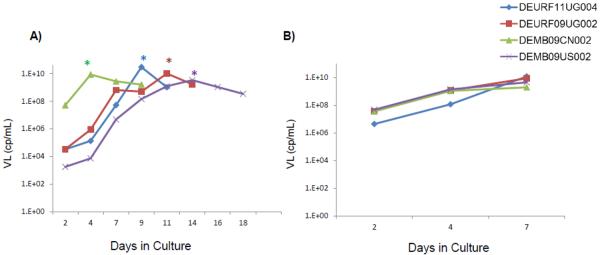
A) Example growth curves from four small-scale cultures as measured by viral load (VL). Culture DEMB09CN002 (Green) was initiated with previously cultured virus, whereas cultures DEURF11UG004 (blue), DEURF09UG002 (Red) and DEMB09US002 (Purple) were started with plasma. For cultures started with plasma, the VL of the source plasma is indicated as Day 0. Cultures were terminated following their peak (marked as * on graph) and an aliquot of virus from the peak day was used to start the large-scale culture. B) Example growth curves from four large-scale cultures as measured by viral load.
The second step of the culture process is termed the large-scale culture, which is initiated using an aliquot of the master lot. For the large-scale culture the total culture volume was 250mL. Once the culture was at high-titer, the culture was terminated and stored as 1mL aliquots. Some culture supernatant was reserved to spike into defibrinated plasma. All final products were aliquoted without any freeze/thaw cycles to preserve the infectivity of the virus. Since the inception of the EQPAOL program, a total of 101 viruses have been taken through both the small-scale and large-scale culture procedures, and the average culture time for the large-scale culture was 6.7 days.
3.4. Characterization of Expanded Viruses
As viruses were cultured to high-titer/high-volume, they were fully characterized including final viral load, p24, TCID, sterility testing, full length sequencing, and coreceptor analysis. A summary of the current EQAPOL panel is provided in Table 2. Viral load was quantified using the Roche COBAS Taqman Version 2.0 assay, which can quantify a broad range of subtypes, including Group O (De Bel et al., 2010). The average culture supernatant final product concentration was 4.20 × 109cp/mL, and the average p24 was 206.59 ng/mL (Table 3). Infectivity was measured using a TZM-bl-based TCID assay, and the average TCID for the panel was 2.22 × 106 (Table 3). Sterility testing was performed on culture supernatant samples to ensure that they are free of bacterial any mycoplasma contamination. Thus far all sterility testing has been negative, except for one culture where the source mycoplasma contamination was traced to the original stock culture supernatant.
Table 2.
Characterization of expanded viruses in EQAPOL Viral Diversity Panel. Fiebig staging data provided by collaborating sites or when plasma was available performed by EQAPOL laboratory as described in the Materials and Methods. HIV subtype is based on full-length sequencing, which was submitted to Genbank. Both Source specimen Genbank and Expanded virues Genbank numbers are provided. Coreceptor usage evaluated using a NP-2-cell-based assay (see Materials and Methods).
| Sample Name | Fiebig Stage | Donation Year | Country of Origin | Source GenBank | HIV Subtype | Coreceptor Usage | GenBank |
|---|---|---|---|---|---|---|---|
| DEMA106ES002 | ND | 2006 | Spaina | FJ670523 | A1 | CCR5 | JX140651 |
| DEMA105TZ001 | ND (CHI) | 2005 | Tanzaniaa | FJ670519 | A1 | CXCR4 | JX140650 |
| DEMA07UG005 | IV | 2007 | Ugandaf | JX236676 | A1 | CCR5 | KC596070 |
| DEMA03RW001 | III | 2003 | Rwandaf | JX236678 | A1 | CCR5 | Pending |
| DEMA03UG001 | IV | 2003 | Ugandaf | JX236671 | A1 | Pending | Pending |
| DEMA03UG002 | IV | 2003 | Ugandaf | JX236669 | A1 | CCR5 | Pending |
| DEMA11KE001 | IV | 2011 | Kenyag | KC018749 | A1 | CCR5 | Pending |
| DEMA110UG001 | VI | 2010 | Ugandag | KC019075 | A1 | CCR5 | Pending |
| DEMB09BO001 | ND (AHI) | 2009 | Boliviaa |
KC112996,
KC112997 |
B | CCR5 | JX140656 |
| DEMB10CN002 | ND (CHI) | 2010 | Chinac | None | B | CCR5 | JX140658 |
| DEMB05FR001 | II/III | 2005 | Franced | None | B | CCR5 | JX140652 |
| DEMB08FR002 | II/III | 2008 | Franceb | None | B | CCR5 | JX140654 |
| DEMBXXDE001 | I/II | UNK | Germanyd | None | B | CCR5 | KC596067 |
| DEMB03JP004 | I/I | 2003 | Japanb | None | B | CCR5 | KC473846 |
| DEMBXXPL001 | II | UNK | Polandd |
JN687774,
JN687691 |
B | CCR5 | KC596069 |
| DEMB08ES001 | ND (AHI) | 2008 | Spaina | FJ670531 | B | CCR5 | JX140653 |
| DEMB08UY001 | ND (AHI) | 2008 | Uruguaya | GU362886 | B | CCR5 | JX140655 |
| DEMB09US002 | I/II | 2009 | USAb | None | B | CCR5 | JX140657 |
| DEMB10VE001 | ND (AHI) | 2010 | Venezuelaa |
KC113011,
KC113010 |
B | CCR5 | JX140659 |
| DEMB10ES002 | ND (AHI) | 2010 | Spaina | KC113004 | B | CCR5 | KC473842 |
| DEMB09CN002 | ND (CHI) | 2009 | Chinac | None | B | CCR5 | KC596066 |
| DEMB10US001 | I/II | 2010 | USAb | None | B | CCR5 | KC473825 |
| DEMB11US006 | I/II | 2011 | USAb | None | B | CCR5 | KC473833 |
| DEMB10US007 | I/II | 2010 | USAb | None | B | CCR5 | KC473828 |
| DEMB10US011 | VI | 2010 | USAb | None | B | CCR5 | KC473830 |
| DEMB11US002 | VI | 2011 | USAb | None | B | CCR5 | KC473831 |
| DEMB10ES003 | ND (AHI) | 2010 | Spaina | KC113005 | B | CCR5 | KC473843 |
| DEMB09ES007 | ND (AHI) | 2009 | Spaina |
KC112998,
KC112999 |
B | CCR5 | KC473841 |
| DEMB10US003 | VI | 2010 | USAb | None | B | CCR5 | KC473826 |
| DEMB11US015 | VI | 2011 | USAb | None | B | CCR5 | KC473835 |
| DEMB10US004 | VI | 2010 | USAb | None | B | CCR5 | KC473827 |
| DEMB11US004 | VI | 2011 | USAb | None | B | CCR5 | KC473832 |
| DEMB11US011 | V | 2011 | USAb | None | B | CCR5 | KC473834 |
| DEMB10US009 | VI | 2010 | USAb | None | B | CCR5 | KC473829 |
| DEMB09US003 | I/II | 2009 | USAb | None | B | CCR5 | KC473824 |
| DEMB11FR001 | I/II | 2011 | Franceb | None | B | CCR5 | Pending |
| DEMC07AO001 | ND | 2007 | Angolaa | EU884500 | C | CCR5 | JX140662 |
| DEMC07BR003 | VI | 2007 | Brazild |
JN687737,
JN687655 |
C | CCR5 | JX140663 |
| DEMC08NG001 | ND | 2008 | Nigeriaa | EU786681 | C | CCR5 | JX140665 |
| DEMC07ZA011 | II | 2007 | South Africad |
JN687618–
JN687627, JN687694– JN687703, JN687778– JN687798 |
C | CCR5 | JX140664 |
| DEMC08ZA011 | II | 2008 | South Africad |
JN687724,
JN687641 |
C | CCR5 | JX140666 |
| DEMC09ZA008 | IND | 2009 | South Africab | None | C | CCR5 | JX140667 |
| DEMC09ZA009 | I/II | 2009 | South Africab | None | C | CCR5 | JX140668 |
| DEMC10ZA001 | I/II | 2010 | South Africab | None | C | CCR5 | JX140669 |
| DEMC06ES003 | ND (AHI) | 2006 | Spaina | EU786673 | C | CCR5 | KC473844 |
| DE00210CM013 | ND | 2010 | Cameroone | JN864054 | CRF_02 | CCR5 | Pending |
| DE00109CN003 | IV | 2009 | Chinac | JX960615 | CRF01_AE | CCR5 | KC596061 |
| DE00109CN004 | IV | 2009 | Chinac | JX960618 | CRF01_AE | CCR5 | KC596062 |
| DE00110CN001 | IV | 2010 | Chinac | JX960610 | CRF01_AE | CCR5 | KC596063 |
| DE00111CN003 | ND (CHI) | 2011 | Chinac | JX960600 | CRF01_AE | CCR5 | KC596065 |
| DE00111CN002 | IV | 2011 | Chinac | None | CRF01_AE | CCR5 | KC596064 |
| DE00206AO001 | ND (RHI) | 2006 | Angolaa | EU884501 | CRF02_AG | CCR5 | JX140645 |
| DE00208CM004 | VI | 2008 | Cameroone | None | CRF02_AG | CCR5 | JX140647 |
| DE00208CM001 | VI | 2008 | Cameroone | None | CRF02_AG | CCR5 | JX140646 |
| DE00400GR002 | ND | 2000 | Greeced |
JN687744,
JN687745, JN687660, JN687661 |
CRF04_CPX | CCR5/CXCR4 | JX140648 |
| DE01405BR001 | ND | 2005 | Brazila | FJ670522 | CRF14_BG | CCR5/CXCR4 | JX140649 |
| DE01405ES002 | ND (CHI) | 2005 | Spaina | FJ670528 | CRF14_BG | CCR5/CXCR4 | KC473837 |
| DE02210CM011 | ND | 2010 | Cameroone | JN864051 | CRF22 | CCR5 | Pending |
| DE02210CM012 | ND | 2010 | Cameroone | JN864058 | CRF22 | CCR5/CXCR4 | Pending |
| DE02210CM014 | ND | 2010 | Cameroone | JN864059 | CRF22 | CCR5 | Pending |
| DE02210CM010 | ND | 2010 | Cameroone | JN864050 | CRF22_01A1 | CCR5 | Pending |
| DE02408ES002 | ND (RHI) | 2008 | Spaina | FJ670526 | CRF24_BG | CCR5 | KC473838 |
| DE04708ES004 | ND (AHI) | 2008 | Spaina | FJ670529 | CRF47_BF | CCR5 | KC473840 |
| DE04708ES003 | ND (AHI) | 2008 | Spaina | GQ372987 | CRF47_BF | CCR5/CXCR4 | KC473839 |
| DEMD10CM009 | VI | 2010 | Cameroone | None | D | CCR5/CXCR4 | JX140670 |
| DEMD07UG002 | II | 2007 | Ugandaf | JX236670 | D | CCR5 | KC596071 |
| DEMD08UG001 | I | 2008 | Ugandaf | JX236672 | D | CCR5 | KC596072 |
| DEMD07UG007 | I | 2007 | Ugandaf | JX236673 | D | CCR5 | Pending |
| DEMD07UG001 | IV | 2007 | Ugandaf | JX236679 | D | CCR5 | Pending |
| DEMD05UG001 | II | 2005 | Ugandaf | JX236668 | D | CCR5 | Pending |
| DEMD11KE003 | IV | 2011 | Kenyag | KC018957 | D | Pending | Pending |
| DEMD10UG004 | VI | 2010 | Ugandag | KC018740 | D | CCR5/CXCR4 | Pending |
| DEMD11UG003 | VI | 2011 | Ugandag | KC018603 | D | CCR5/CXCR4 | Pending |
| DEMF110ES001 | ND (RHI) | 2010 | Spaina |
JN010222,
KC113009 |
F1 | CCR5 | JX140671 |
| DEMF210CM001 | VI | 2010 | Cameroone | None | F2 | CCR5 | JX140672 |
| DEMF210CM007 | VI | 2010 | Cameroone | None | F2 | CCR5 | JX140673 |
| DEMG10CM008 | VI | 2010 | Cameroone | None | G | CCR5 | JX140676 |
| DEMG05ES001 | ND (RHI) | 2005 | Spaina | EU786670 | G | CCR5 | JX140674 |
| DEMG09ES002 | ND (AHI) | 2009 | Spaina | GU362882 | G | CCR5 | JX140675 |
| DEOXXDE004 | ND | UNK | Germanye | None | Group O | CCR5 | Pending |
| DEOXXES001 | ND | UNK | Spaine | None | Group O | CCR5 | Pending |
| DEOXXUS001 | ND | UNK | USAe | None | Group O | CCR5 | Pending |
| DEURF09ES005 | ND | 2009 | Spaina | KC113007 | URF_A1B | CCR5 | JX140678 |
| DEURF10US008 | I/II | 2010 | USAb | None | URF_A1B | CCR5 | KC473836 |
| DEURF07UG006 | I | 2007 | Ugandaf | JX236675 | URF_A1D | CCR5 | KC596073 |
| DEURF08UG002 | I | 2008 | Ugandaf | JX236674 | URF_A1D | CCR5 | Pending |
| DEURF10UG011 | VI | 2010 | Ugandag | KC019010 | URF_A1D | CCR5 | Pending |
| DEURF11UG004 | VI | 2011 | Ugandag | KC019104 | URF_A1D | CCR5 | Pending |
| DEURF09KE002 | VI | 2009 | Kenyag |
KC018751,
KC018658, KC018750 |
URF_A1D | CCR5 | Pending |
| DEURF10UG005 | VI | 2010 | Ugandag | KC018940 | URF_A1D | CCR5 | Pending |
| DEURF09UG002 | VI | 2009 | Ugandag | KC018893 | URF_A1D | CCR5 | Pending |
| DEURF08CM005 | ND | 2008 | Cameroone | JN864047 | URF_A1G | CCR5 | Pending |
| DEURF09GQ001 | ND | 2009 | Equatorial Guineaa |
KC113000,
KC113001 |
URF_ADG | CCR5 | KC473845 |
| DEURF07BR002 | V | 2007 | Brazild | None | URF_BC | CCR5 | KC596068 |
| DEMBF09ES003 | ND | 2009 | Spaina | KC113006 | URF_BF | CCR5 | JX140660 |
| DEMBF09ES006 | ND (AHI) | 2009 | Spaina | KC113008 | URF_BF | CCR5 | JX140661 |
| DEURF07ES002 | ND (AHI) | 2007 | Spaina | GQ372986 | URF_CRF02_AG/A3 | CCR5 | JX140677 |
| DEURF10DZ001 | ND | 2010 | Algeriaa |
KC113003,
KC113002 |
URF_CRF02_AG/CRF06_cpx | CCR5 | JX140679 |
| DEURF11RW001 | VI | 2011 | Rwandag | KC018537 | URF_DC | CCR5 | Pending |
| DEURF06CM001 | ND | 2006 | Cameroone | FJ014688 | URF_X | CCR5/CXCR4 | Pending |
| DEURF10KE002 | VI | 2010 | Kenyag | KC018842 | URF_X | CCR5 | Pending |
Alvarez, Instituto de Salud Carlos III
Blood Systems Research Institute
H. Shang, First Affiliated Hospital, China Medical University
SeraCare
I. Hewlett, Food and Drug Administration
B. Hahn/G. Shaw, University of Pennsylvania
International AIDS Vaccine Initiative
ND, not determined; IND, Indeterminate (Negative EIA, Indeterminate WB); AHI, Acute HIV Infection; RHI, Recent HIV Infection; CHI, Chronic HIV Infection; UNK, Unknown; URF_X, Unique recombinant form with greater than three subtypes present.
Table 3.
Viral Load, p24 and Infectivity of EQAPOL Viral Diversity Panel.
| Viral Load (cp/mL) | p24 (ng/mL) | TCID | |
|---|---|---|---|
| Average | 4.20E+09 | 206.59 | 2.22E+06 |
| Max | 1.75E+10 | 738.00 | 3.01E+07 |
| Minimum | 8.57E+07 | 25.00 | 2.80E+04 |
The average and range of viral load (Roche COBAS AmpliPrep/TaqMan HIV-1 v2.0), p24 (PerkinElmer HIV-1 p24 Antigen Capture Assay) and TCID (TZM-bl-Based Cell Assay) for the expanded viruses in the EQAPOL Viral Diversity Panel.
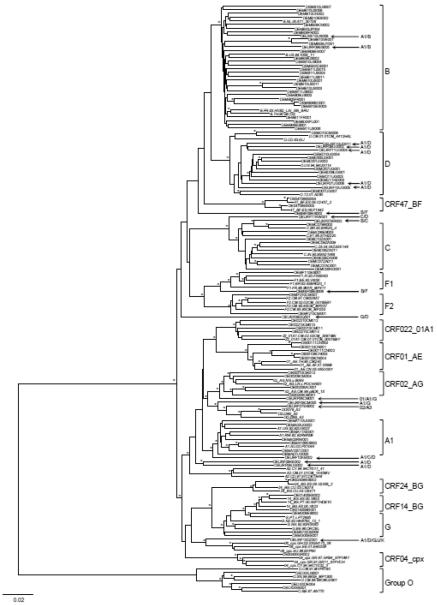
One near full-length genome was obtained by two overlapping half SGA amplicons for each isolate. The ambiguous bases were detected in 14 viruses, and the most of them contain only one ambiguous base. Thus, those rare ambiguous bases could not affect genetic subtyping of the viral isolates in the diversity panel. Although the diversity of the each expanded viral isolate was not investigated, one near full-length genome sequence from each isolate was sufficient to define the genotype of the isolate. Figure 3 shows a phylogenetic tree for the 101 samples present in the EQAPOL panel. Thus far the EQAPOL viral diversity panel includes 6 subtypes (A, B, C, D, F, G), 2 sub-subtypes (F1 and F2), 7 CRFs (01, 02, 04, 14, 22, 24 and 47), 19 URFs and group O viruses from 22 countries (Table 4). The panel continues to grow with a focus on identifying samples to expand the overall genetic and geographic diversity of the panel.
Figure 3. Phylogenetic analysis of newly characterized near full-length HIV-1 genome sequences.
The phylogenetic tree was constructed with near full-length HIV-1 genome nucleotide sequences, using the neighbor-joining method. The tree was midpoint rooted. Horizontal branch lengths are drawn to scale (the scale bar represents 0.02 nucleotide substitution per site). Vertical separation is for clarity only. Asterisks at the nodes indicate the bootstraps in which the cluster to the right was supported by 80% and higher. Brackets at the right denote the major subtypes, CRFs and group Os. The reference sequences are indicated by underline. The unique recombinant form (URF) sequences are indicated by arrow.
Table 4.
Genetic and Geographic Diversity Present in Viral Diversity Panel
| Country | Number of Samples | Genotype |
|---|---|---|
| Algeria | 1 | URF CRF02 AG/CRF06 cpx |
| Angola | 1 | C |
| 1 | CRF02_AG | |
| Bolivia | 1 | B |
| Brazil | 1 | C |
| 1 | CRF14 BG | |
| 1 | URF BC | |
| Cameroon | 3 | CRF02_AG |
| 4 | CRF22_01A1 | |
| 1 | D | |
| 2 | F2 | |
| 1 | G | |
| 1 | URF 01A1G | |
| 1 | URF A1G | |
| China | 2 | B |
| 5 | CRF01_AE | |
| Equatorial Guinea | 1 | URF ADG |
| France | 3 | B |
| Germany | 1 | B |
| 1 | Group O | |
| Greece | 1 | CRF04 CPX |
| Japan | 1 | B |
| Kenya | 1 | A1 |
| 1 | D | |
| 1 | URF A1CD | |
| 1 | URF_A1D | |
| Nigeria | 1 | C |
| Poland | 1 | B |
| Rwanda | 1 | A1 |
| 1 | URF DC | |
| South Africa | 5 | C |
| Spain | 1 | A1 |
| 4 | B | |
| 1 | C | |
| 1 | CRF14_BG | |
| 1 | CRF24_BG | |
| 2 | CRF47_BF | |
| 1 | F1 | |
| 2 | G | |
| 1 | Group O | |
| 1 | URF A1B | |
| 2 | URF_BF | |
| 1 | URF CRF02 AG/A3 | |
| Tanzania | 1 | A1 |
| Uganda | 3 | A1 |
| 7 | D | |
| 6 | URF A1D | |
| Uruguay | 1 | B |
| USA | 13 | B |
| 1 | Group O | |
| 1 | URF A1B | |
| Venezuela | 1 | B |
| Total | 100 |
Coreceptor usage was determined using a NP-2-cell-based assay (Shimizu et al., 2009; Jiang et al., 2011). Thus far 74 of the samples have been tested; 68 (91.9%) of the 74 samples had a CCR5 phenotype, five were dual tropic (CCR5/CXCR4, 6.8%) and one had a CXCR4 phenotype (1.3%).
3.5. System for Access to EQAPOL Samples
Fully characterized samples are made available to order on the EQAPOL web-based system. The web-based system captures characterization data for HIV source specimens, intermediate products and final products, as well as details about the culture process. Additionally the web-based system displays a COA for each virus, which includes all of the characterization data and source specimen data. External sites, which are approved by NIAID, are able to access the web-based system and search through an inventory of viruses available to order. The web-based system provides a real-time inventory of available vials, and allows users to search/view characterization data to select samples of interest for order. The ordering process is captured in the web-based system and allows for tracking of samples from order to receipt. Samples and panels are provided to qualified investigators, commercial assay and vaccine developers and regulators at no charge.
4. Discussion
For the EQAPOL Viral Diversity program, a comprehensive, well-controlled and standardized system to collect, propagate and characterize HIV was developed. Work by BSRI and other collaborators have yielded 370 viral specimens with an emphasis on samples from recently-acquired infections. Work is ongoing to acquire additional panel source specimens, in particular those from genetic subtypes and geographic locations not represented on the current panel, as well as from emerging recombinants. The goal is for the panel to represent the current trends observed in transmitted HIVs throughout world. As the panel expands, it may be able to identify viral diversity within geographic areas and be used to determine vaccine immunogens to include within candidate vaccines that may be required to confer protection.
One of the hallmarks for the EQAPOL program is the commitment to quality and standardization through the implementation of GCLP. This is of particular importance for the Viral Diversity Panel where products will be utilized in the development and assessment of a variety of assays. As discussed in Todd et al. in this issue, the EQAPOL Viral Diversity program adheres to GCLP when available, in particular through the culture process and much of the characterization. Work is currently underway to bring all aspects of the characterization work to GCLP. The inclusion of a COA with all products allows investigators to have documentation attesting to the quality and characteristics of each sample.
In support of EQAPOL, we have developed a user-friendly web-based system where approved laboratories can search through an inventory of viral samples to select items for order. Users can search for items using criteria such as viral load, subtype, country of origin and year of donation. The system provides a link to the GenBank sequence so that sites can have access to the full-length sequence of each product. Once a user orders items, the web-based system is used to ensure the site has the appropriate compliance documents to receive samples and to alert sites via email shipment tracking information. The web-based system, in combination with LDMS, allows for traceability of each vial from creation to shipment to a site.
Now that the panel has expanded to include a sizeable number of viruses, we are beginning to assemble the viruses into pre-made panels that will be made available to researchers for specific uses. Through collaboration with the Program for Appropriate Technology (PATH) we have created two panels using subtype B, C and D viruses to assist laboratories participating in the Gates Foundation Grand Challenges Point-of-Care Diagnostics program. One panel included a dilution series of HIV Spiked Plasma for the three subtypes. The other panel included a dilution series of isolated RNA for the three subtypes to be used by laboratories not equipped with the safety equipment required to work with live virus.
Globally diverse HIV strains may prove to be a challenge to current methods of genotyping and drug resistance testing. The Recipient Epidemiology and Donor Evaluation Study-III (REDS-III) program funded by the National Heart, Lung and Blood Institute was established to ensure safe and effective blood banking and transfusion medicine at domestic and international blood donation centers (https://reds-iii.rti.org/REDSProgram.aspx). The REDS program wanted to assess the molecular methods for accurately testing genotype and drug resistance in the international reference laboratories. A Genotype Diversity Panel was assembled from available viral isolates from the EQAPOL program; this contains 50 viral isolates representing globally diverse HIV genotypes and drug resistant phenotypes. It has been distributed to the international REDSIII sites located in South Africa, Brazil and China. Each of the laboratories will use standard procedures used for testing and this will determine accuracy or potential limitations in the current methods used. Both the PATH Panel and the Genetic Diversity Panel developed for the REDS program are available to interested laboratories. As the EQAPOL Viral Program continues to expand, additional panels will be constructed to meet the needs of the research community, with plans to develop diversity panels for evaluation of relative analytical sensitivity across HIV diagnostic assay platforms.
In summary, we have developed a HIV Subtype panel to capture the current genetic and geographic diversity observed as a continuation of a pilot viral diversity program (Manak et al., 2012). Combined with the pilot program, the Viral Subtype Diversity panel currently includes over unique 130 virus isolates and continues to grow each year. Laboratories interested in gaining access to the EQAPOL panel should contact the EQAPOL program via email (EQAPOL@duke.edu). Products are available free of charge to NIAID-approved laboratories. The EQAPOL Viral Diversity panel as an invaluable collection of well-characterized reagents are currently made available to the scientific community, including researchers, epidemiologists, and commercial manufacturers of diagnostics and pharmaceuticals to support HIV research.
Acknowledgments
This project has been funded with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract Nos. HHSN272201000045C and HHSN272200800014C.
We wish to thank all of our collaborators for providing HIV source material: Susan Stramer, American Red Cross; Marion Vermeulen, South African National Blood Service; Mark Manak, US Military HIV Research Program (MHRP), HJF; Michael Schmidt, German Red Cross Institute Baden-Württemberg – Hessen; Syria Laperche, Institut National de la Transfusion Sanguine; Ester Sabino, University of Sao Paulo; Kenji Tadokoro, Japanese Red Cross; Lucía Pérez-Álvarez, Elena Delgado, Michael Thomson, Aurora Fernández-García and Teresa Cuevas, Instituto de Salud Carlos III; Hong Shang, Wenqing Geng, Hualu Cui, and Hong Sun, First Affiliated Hospital of China Medical University; Indira Hewlett and Viswanath Ragupathy, Food and Drug Administration; Joshua Baalwa, University of Alabama, Birmingham; Beatrice Hahn and George Shaw, University of Pennsylvania; Hagen von Brisen, Fraunhofer Institut für Biomedizinisch Technik (IBMT), Pat Fast and Matt Price, International AIDS Vaccine Initiative (IAVI); Pauli Amornkul, University of California, San Francisco; Etienne Karita William Kilembe and Mubiana Inambao, Rwanda Zambia HIV Research Group; Susan Allen and Eric Hunter, Rwanda Zambia HIV Research Group and Emory University; Anatoli Kamali and Eugene Ruzagira, MRC/UVRI Uganda Research Unit on AIDS; Eduard Sanders, Kenya Medical Research Institute and University of Oxford; and Omu Anzala and Gaudensia Mutua, Kenya AIDS Vaccine Initiative.
We are grateful to Jim Lane for his leadership and guidance. We also thank Sara Brown, Linda Walker, Shelly Woody, Rachel Lovingood, Khalil Itani, Laura Racz, Kyle Liebl, and Holly Alley for their assistance with HIV culture work. Additionally, we are grateful for Marcella Sarzotti-Kelsoe and Chris Todd from the EQAPOL Central Quality Assurance Unit for monitoring GCLP compliance and for assistance developing the sterility testing program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. References
- Adazu K, Lindblade KA, Rosen DH, Odhiambo F, Ofware P, Kwach J, Van Eijk AM, Decock KM, Amornkul P, Karanja D, Vulule JM, Slutsker L. Health and demographic surveillance in rural western Kenya: a platform for evaluating interventions to reduce morbidity and mortality from infectious diseases. The American journal of tropical medicine and hygiene. 2005;73:1151–8. [PubMed] [Google Scholar]
- Aguado-Martinez A, Alvarez-Garcia G, Fernandez-Garcia A, Risco-Castillo V, Marugan-Hernandez V, Ortega-Mora LM. Failure of a vaccine using immunogenic recombinant proteins rNcSAG4 and rNcGRA7 against neosporosis in mice. Vaccine. 2009;27:7331–8. doi: 10.1016/j.vaccine.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Albert J, Gaines H, Sonnerborg A, Nystrom G, Pehrson PO, Chiodi F, von Sydow M, Moberg L, Lidman K, Christensson B, et al. Isolation of human immunodeficiency virus (HIV) from plasma during primary HIV infection. Journal of medical virology. 1987;23:67–73. doi: 10.1002/jmv.1890230108. [DOI] [PubMed] [Google Scholar]
- Amendola A, Coen S, Belladonna S, Pulvirenti FR, Clemens JM, Capobianchi MR. Improving clinical laboratory efficiency: a time-motion evaluation of the Abbott m2000 RealTime and Roche COBAS AmpliPrep/COBAS TaqMan PCR systems for the simultaneous quantitation of HIV-1 RNA and HCV RNA. Clinical chemistry and laboratory medicine : CCLM / FESCC. 2011;49:1283–8. doi: 10.1515/CCLM.2011.625. [DOI] [PubMed] [Google Scholar]
- Antunes R, Figueiredo S, Bartolo I, Pinheiro M, Rosado L, Soares I, Lourenco H, Taveira N. Evaluation of the clinical sensitivities of three viral load assays with plasma samples from a pediatric population predominantly infected with human immunodeficiency virus type 1 subtype G and BG recombinant forms. Journal of clinical microbiology. 2003;41:3361–7. doi: 10.1128/JCM.41.7.3361-3367.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baalwa J, Wang S, Parrish NF, Decker JM, Keele BF, Learn GH, Yue L, Ruzagira E, Ssemwanga D, Kamali A, Amornkul PN, Price MA, Kappes JC, Karita E, Kaleebu P, Sanders E, Gilmour J, Allen S, Hunter E, Montefiori DC, Haynes BF, Cormier E, Hahn BH, Shaw GM. Molecular identification, cloning and characterization of transmitted/founder HIV-1 subtype A, D and A/D infectious molecular clones. Virology. 2013;436:33–48. doi: 10.1016/j.virol.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BK, Darden JM, Tovanabutra S, Oblander T, Frost J, Sanders-Buell E, de Souza MS, Birx DL, McCutchan FE, Polonis VR. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. Journal of virology. 2005;79:6089–101. doi: 10.1128/JVI.79.10.6089-6101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch MP, Pilcher CD, Mastro TD, Kaldor J, Vercauteren G, Rodriguez W, Rousseau C, Rehle TM, Welte A, Averill MD, Garcia Calleja JM, Assays, W.H.O.W.G.o.H.I. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. Aids. 2010;24:2763–71. doi: 10.1097/QAD.0b013e32833f1142. [DOI] [PubMed] [Google Scholar]
- Charneau P, Borman AM, Quillent C, Guetard D, Chamaret S, Cohen J, Remy G, Montagnier L, Clavel F. Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology. 1994;205:247–53. doi: 10.1006/viro.1994.1640. [DOI] [PubMed] [Google Scholar]
- Chudy M, Weber-Schehl M, Pichl L, Jork C, Kress J, Heiden M, Funk MB, Nubling CM. Blood screening nucleic acid amplification tests for human immunodeficiency virus Type 1 may require two different amplification targets. Transfusion. 2012;52:431–9. doi: 10.1111/j.1537-2995.2011.03281.x. [DOI] [PubMed] [Google Scholar]
- Coombs RW, Welles SL, Hooper C, Reichelderfer PS, D'Aquila RT, Japour AJ, Johnson VA, Kuritzkes DR, Richman DD, Kwok S, Todd J, Jackson JB, DeGruttola V, Crumpacker CS, Kahn J. Association of plasma human immunodeficiency virus type 1 RNA level with risk of clinical progression in patients with advanced infection. AIDS Clinical Trials Group (ACTG) 116B/117 Study Team. ACTG Virology Committee Resistance and HIV-1 RNA Working Groups. The Journal of infectious diseases. 1996;174:704–12. doi: 10.1093/infdis/174.4.704. [DOI] [PubMed] [Google Scholar]
- Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. Journal of virology. 2000;74:529–34. doi: 10.1128/jvi.74.1.529-534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa CI, Morgado MG, Santos VG, Bongertz V. HIV-1 isolation from plasma specimens. HEC/FIOCRUZ AIDS Clinical Research Group. Memorias do Instituto Oswaldo Cruz. 1996;91:745–6. doi: 10.1590/s0074-02761996000600017. [DOI] [PubMed] [Google Scholar]
- Cuevas MT, Fernandez-Garcia A, Pinilla M, Garcia-Alvarez V, Thomson M, Delgado E, Gonzalez-Galeano M, Miralles C, Serrano-Bengoechea E, Ojea de Castro R, Lopez-Alvarez MJ, Lezaun MJ, Sanchez-Garcia AM, Sanchez-Martinez M, Munoz-Nieto M, Perez-Alvarez L. Short communication: Biological and genetic characterization of HIV type 1 subtype B and nonsubtype B transmitted viruses: usefulness for vaccine candidate assessment. AIDS research and human retroviruses. 2010;26:1019–25. doi: 10.1089/aid.2010.0018. [DOI] [PubMed] [Google Scholar]
- De Bel A, Marissens D, Debaisieux L, Liesnard C, Van den Wijngaert S, Lauwers S, Pierard D. Correction of underquantification of human immunodeficiency virus type 1 load with the second version of the Roche Cobas AmpliPrep/Cobas TaqMan assay. Journal of clinical microbiology. 2010;48:1337–42. doi: 10.1128/JCM.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado E, Fernandez-Garcia A, Vega Y, Cuevas T, Pinilla M, Garcia V, Sanchez M, Gonzalez M, Sanchez AM, Thomson MM, Perez-Alvarez L. Evaluation of genotypic tropism prediction tests compared with in vitro co-receptor usage in HIV-1 primary isolates of diverse subtypes. The Journal of antimicrobial chemotherapy. 2012;67:25–31. doi: 10.1093/jac/dkr438. [DOI] [PubMed] [Google Scholar]
- Diez-Fuertes F, Delgado E, Vega Y, Fernandez-Garcia A, Cuevas MT, Pinilla M, Garcia V, Perez-Alvarez L, Thomson MM. Improvement of HIV-1 coreceptor tropism prediction by employing selected nucleotide positions of the env gene in a Bayesian network classifier. The Journal of antimicrobial chemotherapy. 2013 doi: 10.1093/jac/dkt077. [DOI] [PubMed] [Google Scholar]
- Falk LA, Jr., Paul D, Landay A, Kessler H. HIV isolation from plasma of HIV-infected persons. The New England journal of medicine. 1987;316:1547–8. doi: 10.1056/NEJM198706113162413. [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia A, Cuevas MT, Vinogradova A, Rakhmanova A, Perez-Alvarez L, de Castro RO, Osmanov S, Thomson MM. Near full-length genome characterization of a newly identified HIV type 1 subtype F variant circulating in St. Petersburg, Russia. AIDS research and human retroviruses. 2009;25:1187–91. doi: 10.1089/aid.2009.0140. [DOI] [PubMed] [Google Scholar]
- Fernandez-Garcia A, Perez-Alvarez L, Cuevas MT, Delgado E, Munoz-Nieto M, Cilla G, Iribarren JA, Pinilla M, Ocampo A, Miralles C, Perez-Castro S, Gonzalez-Galeano M, de Castro RO, Trigo M, Garcia V, Sanchez AM, Thomson MM. Identification of a new HIV type 1 circulating BF intersubtype recombinant form (CRF47_BF) in Spain. AIDS research and human retroviruses. 2010;26:827–32. doi: 10.1089/aid.2009.0311. [DOI] [PubMed] [Google Scholar]
- Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. Aids. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- Gottesman BS, Grossman Z, Lorber M, Levi I, Shitrit P, Katzir M, Shahar E, Gottesman G, Chowers M. Comparative performance of the Amplicor HIV-1 Monitor Assay versus NucliSens EasyQ in HIV subtype C-infected patients. Journal of medical virology. 2006;78:883–7. doi: 10.1002/jmv.20653. [DOI] [PubMed] [Google Scholar]
- Greco G, Barker E, Levy JA. Differences in HIV replication in CD4+ lymphocytes are not related to beta-chemokine production. AIDS research and human retroviruses. 1998;14:1407–11. doi: 10.1089/aid.1998.14.1407. [DOI] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S, Isolation, W.-U.N.f.H. and Characterisation Global trends in molecular epidemiology of HIV-1 during 2000–2007. Aids. 2011;25:679–89. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DD, Giesler TA, Bremer JW. Sequence characterization of the protease and partial reverse transcriptase proteins of the NED panel, an international HIV type 1 subtype reference and standards panel. AIDS research and human retroviruses. 2003;19:321–8. doi: 10.1089/088922203764969528. [DOI] [PubMed] [Google Scholar]
- Jiang C, Parrish NF, Wilen CB, Li H, Chen Y, Pavlicek JW, Berg A, Lu X, Song H, Tilton JC, Pfaff JM, Henning EA, Decker JM, Moody MA, Drinker MS, Schutte R, Freel S, Tomaras GD, Nedellec R, Mosier DE, Haynes BF, Shaw GM, Hahn BH, Doms RW, Gao F. Primary Infection by a Human Immunodeficiency Virus with Atypical Coreceptor Tropism. Journal of virology. 2011;85:10669–81. doi: 10.1128/JVI.05249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Beckthold B, Chen Z, Mihowich J, Malloch L, Gill MJ. Short communication: identification of a novel HIV type 1 subtype H/J recombinant in Canada with discordant HIV viral load (RNA) values in three different commercial assays. AIDS research and human retroviruses. 2007;23:1309–13. doi: 10.1089/aid.2007.0080. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Korber BT, Osmanov S, Esparza J, Myers G. The World Health Organization Global Programme on AIDS proposal for standardization of HIV sequence nomenclature. WHO Network for HIV Isolation and Characterization. AIDS research and human retroviruses. 1994;10:1355–8. doi: 10.1089/aid.1994.10.1355. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. Journal of virology. 2005;79:10108–25. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly TD, Ebel A, Faucher V, Fihman V, Laperche S. Could the new HIV combined p24 antigen and antibody assays replace p24 antigen specific assays? Journal of virological methods. 2007;143:86–94. doi: 10.1016/j.jviromet.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Ly TD, Plantier JC, Leballais L, Gonzalo S, Lemee V, Laperche S. The variable sensitivity of HIV Ag/Ab combination assays in the detection of p24Ag according to genotype could compromise the diagnosis of early HIV infection. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2012;55:121–7. doi: 10.1016/j.jcv.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Manak M, Sina S, Anekella B, Hewlett I, Sanders-Buell E, Ragupathy V, Kim J, Vermeulen M, Stramer SL, Sabino E, Grabarczyk P, Michael N, Peel S, Garrett P, Tovanabutra S, Busch MP, Schito M. Pilot studies for development of an HIV subtype panel for surveillance of global diversity. AIDS research and human retroviruses. 2012;28:594–606. doi: 10.1089/aid.2011.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors JW, Rinaldo CR, Jr., Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–70. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- O'Brien TR, Blattner WA, Waters D, Eyster E, Hilgartner MW, Cohen AR, Luban N, Hatzakis A, Aledort LM, Rosenberg PS, Miley WJ, Kroner BL, Goedert JJ. Serum HIV-1 RNA levels and time to development of AIDS in the Multicenter Hemophilia Cohort Study. JAMA : the journal of the American Medical Association. 1996;276:105–10. [PubMed] [Google Scholar]
- O'Connell RJ, Kim JH, Corey L, Michael NL. Human immunodeficiency virus vaccine trials. Cold Spring Harbor perspectives in medicine. 2012;2:a007351. doi: 10.1101/cshperspect.a007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. Journal of virology. 2000;74:10074–80. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot P, Quinn TC. Response to the AIDS pandemic--a global health model. The New England journal of medicine. 2013;368:2210–8. doi: 10.1056/NEJMra1201533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, Damond F, Robertson DL, Simon F. A new human immunodeficiency virus derived from gorillas. Nature medicine. 2009;15:871–2. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- Price MA, Rida W, Mwangome M, Mutua G, Middelkoop K, Roux S, Okuku HS, Bekker LG, Anzala O, Ngugi E, Stevens G, Chetty P, Amornkul PN, Sanders EJ. Identifying at-risk populations in Kenya and South Africa: HIV incidence in cohorts of men who report sex with men, sex workers, and youth. Journal of acquired immune deficiency syndromes. 2012;59:185–93. doi: 10.1097/QAI.0b013e31823d8693. [DOI] [PubMed] [Google Scholar]
- Price MA, Wallis CL, Lakhi S, Karita E, Kamali A, Anzala O, Sanders EJ, Bekker LG, Twesigye R, Hunter E, Kaleebu P, Kayitenkore K, Allen S, Ruzagira E, Mwangome M, Mutua G, Amornkul PN, Stevens G, Pond SL, Schaefer M, Papathanasopoulos MA, Stevens W, Gilmour J, Group, I.E.I.C.S. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS research and human retroviruses. 2011;27:5–12. doi: 10.1089/aid.2010.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragupathy V, Zhao J, Wood O, Tang S, Lee S, Nyambi P, Hewlett I. Identification of new, emerging HIV-1 unique recombinant forms and drug resistant viruses circulating in Cameroon. Virology journal. 2011;8:185. doi: 10.1186/1743-422X-8-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–89. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz AK, Zhang M, Bulla I, Leitner T, Korber B, Morgenstern B, Stanke M. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic acids research. 2009;37:W647–51. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz AK, Zhang M, Leitner T, Kuiken C, Korber B, Morgenstern B, Stanke M. A jumping profile Hidden Markov Model and applications to recombination sites in HIV and HCV genomes. BMC Bioinformatics. 2006;7:265. doi: 10.1186/1471-2105-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Tanaka A, Oue A, Mori T, Ohtsuki T, Apichartpiyakul C, Uchiumi H, Nojima Y, Hoshino H. Broad usage spectrum of G protein-coupled receptors as coreceptors by primary isolates of HIV. AIDS. 2009;23:761–9. doi: 10.1097/QAD.0b013e328326cc0d. [DOI] [PubMed] [Google Scholar]
- Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Georges-Courbot MC, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nature medicine. 1998;4:1032–7. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- Spira AI, Ho DD. Effect of different donor cells on human immunodeficiency virus type 1 replication and selection in vitro. Journal of virology. 1995;69:422–9. doi: 10.1128/jvi.69.1.422-429.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramer SL, Caglioti S, Strong DM. NAT of the United States and Canadian blood supply. Transfusion. 2000;40:1165–1168. doi: 10.1046/j.1537-2995.2000.40101165.x. [DOI] [PubMed] [Google Scholar]
- Stramer SL, Glynn SA, Kleinman SH, Strong DM, Sally C, Wright DJ, Dodd RY, Busch MP. Detection of HIV-1 and HCV infections among antibody-negative blood donors by nucleic acid-amplification testing. N Engl J Med. 2004;351:760–8. doi: 10.1056/NEJMoa040085. [DOI] [PubMed] [Google Scholar]
- Tang J, Cormier E, Gilmour J, Price MA, Prentice HA, Song W, Kamali A, Karita E, Lakhi S, Sanders EJ, Anzala O, Amornkul PN, Allen S, Hunter E, Kaslow RA, Network, I.A.H.R. Human leukocyte antigen variants B*44 and B*57 are consistently favorable during two distinct phases of primary HIV-1 infection in sub-Saharan Africans with several viral subtypes. Journal of virology. 2011;85:8894–902. doi: 10.1128/JVI.00439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MM, Fernandez-Garcia A, Delgado E, Vega Y, Diez-Fuertes F, Sanchez-Martinez M, Pinilla M, Castro MA, Marino A, Ordonez P, Ocampo A, da Silva AR, Perez-Castro S, Lopez-Alvarez MJ, Trigo M, Perez-Alvarez L. Rapid expansion of a HIV-1 subtype F cluster of recent origin among men who have sex with men in Galicia, Spain. Journal of acquired immune deficiency syndromes. 2012;59:e49–51. doi: 10.1097/QAI.0b013e3182400fc4. [DOI] [PubMed] [Google Scholar]
- UNAIDS . Report on the Global AIDS Epidemic. 2012. [Google Scholar]
- Vallejo A, Gurtler L, Zekeng L, Hewlett IK. Nucleotide sequence analysis of the accessory genes of HIV-1 group O isolates. Virus research. 2003;91:189–93. doi: 10.1016/s0168-1702(02)00270-8. [DOI] [PubMed] [Google Scholar]
- Walker CM, Moody DJ, Stites DP, Levy JA. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–6. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- Welles SL, Jackson JB, Yen-Lieberman B, Demeter L, Japour AJ, Smeaton LM, Johnson VA, Kuritzkes DR, D'Aquila RT, Reichelderfer PA, Richman DD, Reichman R, Fischl M, Dolin R, Coombs RW, Kahn JO, McLaren C, Todd J, Kwok S, Crumpacker CS. Prognostic value of plasma human immunodeficiency virus type 1 (HIV-1) RNA levels in patients with advanced HIV-1 disease and with little or no prior zidovudine therapy. AIDS Clinical Trials Group Protocol 116A/116B/117 Team. The Journal of infectious diseases. 1996;174:696–703. doi: 10.1093/infdis/174.4.696. [DOI] [PubMed] [Google Scholar]
- WHO . Progress report 2011: Global HIV/AIDS response. 2011. [Google Scholar]
- Williams LM, Cloyd MW. Polymorphic human gene(s) determines differential susceptibility of CD4 lymphocytes to infection by certain HIV-1 isolates. Virology. 1991;184:723–8. doi: 10.1016/0042-6822(91)90442-e. [DOI] [PubMed] [Google Scholar]
- Xu S, Song A, Li X, Nie J, Zhang C, Wang Y. Performance of the automated COBAS AmpliPrep/COBAS TaqMan HIV-1 test on a genetically diverse panel of specimens from China: comparison to the COBAS amplicor HIV-1 monitor test, v1.5. Intervirology. 2010;53:221–8. doi: 10.1159/000299064. [DOI] [PubMed] [Google Scholar]
- Zhao J, Tang S, Ragupathy V, Gaddam D, Wang X, Zhang P, Nyambi PN, Hewlett I. CRF22_01A1 is involved in the emergence of new HIV-1 recombinants in Cameroon. Journal of acquired immune deficiency syndromes. 2012;60:344–50. doi: 10.1097/QAI.0b013e318258c7e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Musavi F, Notari EP, Stramer SL, Dodd RY. Prevalence, incidence, and residual risk of major blood-borne infections among apheresis collections to the American Red Cross Blood Services, 2004 through 2008. Transfusion. 2010;50:1487–94. doi: 10.1111/j.1537-2995.2010.02621.x. [DOI] [PubMed] [Google Scholar]