Abstract
Over the past decade, the integrase enzyme from phage phiC31 has proven to be a useful genome engineering tool in a wide variety of species, including mammalian cells. The enzyme efficiently mediates recombination between two distinct sequences, attP and attB, producing recombinant product sites, attL and attR. The reaction proceeds exclusively in a unidirectional manner, because integrase is unable to synapse attL and attR. To date, use of phiC31 integrase has been limited to attP x attB recombination. The factor needed for the reverse reaction – the excisionase or recombination directionality factor (RDF) – was identified recently and shown to function in vitro and in bacterial cells. To determine whether the phiC31 RDF could also function in mammalian cells, we cloned and tested several vectors that permit assessment of phiC31 RDF activity in mammalian environments. In the human and mouse cell lines tested (HeLa, HEK293, NIH3T3), we observed robust RDF activity, using plasmid and/or genomic assays. This work is the first to demonstrate attL-attR serine integrase activity in mammalian cells and validates phiC31 RDF as a new tool that will enable future studies to take advantage of phiC31 integrase recombination in the forward or reverse direction.
Keywords: cassette exchange, genome engineering, phage integrase, recombination directionality factor, site-specific integration
INTRODUCTION
The Streptomyces phage phiC31 uses a large serine recombinase, phiC31 integrase (Int), to merge its genome with that of its bacterial host [1, 2]. Its two substrates – attP and attB – are distinct (only ~40% identical, Supporting Information, Table S1) [2]. Recombination of attP and attB (PxB) produces the attL and attR sites, which, without assistance, are not synapsed by the Int enzyme [3]. The phiC31 prophage is excised when an additional protein, the recombination directionality factor (RDF), is coexpressed with Int [4]. RDF binding to Int imparts it with a reversed directionality, where attL-attR recombination (LxR) is mediated, but PxB is not [4].
Despite its phage origin, Int has been shown to function efficiently in a variety of microbial, plant, and animal species [5]. In these organisms, it is primarily used for cassette exchange into the genome or for plasmid integration. A unidirectional recombinase like Int is particularly well-suited for these applications, compared to bidirectional recombinases such as Cre.
Since the identity of the phiC31 RDF remained unknown until recently [4], all use of Int in eukaryotes over the past decade has been limited to the PxB reaction. With the isolation and characterization of the phiC31 RDF in bacteria and purified extracts [4], reversal of the Int PxB reaction in eukaryotes became possible in theory. To assess whether RDF could actually function in mammalian cells, we constructed and tested several vectors that permitted detection of LxR recombination on extrachromosomal and integrated substrates. In this study, we establish that the phiC31 RDF functions efficiently in several mammalian cell lines on a plasmid substrate. We also show that the RDF:Int complex is capable of driving efficient genomic excision. This demonstration of phiC31 RDF functionality in mammalian cells now enables studies that utilize LxR recombination in eukaryotes, thus adding a new dimension to the utility of the phiC31 integrase system.
MATERIAL AND METHODS
Details on plasmid construction, the plasmid inversion assay, the genomic excision assay, junction sequencing, and pseudo site mapping are provided in Supporting Information, Materials and Methods.
RESULTS
The phiC31 RDF functions in mammalian cells
Details of the RDF sequence employed in this study are provided in Supporting Information, Results. For RDF:Int and Int-only expression, we built the pCS-kRI and pCS-kI plasmids, respectively (Fig. 1A and 1B). In pCS-kRI, RDF and Int are present on the same CMV-driven transcript, but the translated proteins are kept separate by inclusion of a ribosome-skipping 2A-peptide sequence from porcine teschovirus [6]. pCS-kI is an Int-only expression derivative of pCS-kRI, and is similar to our previously described pCMVInt vector [7].
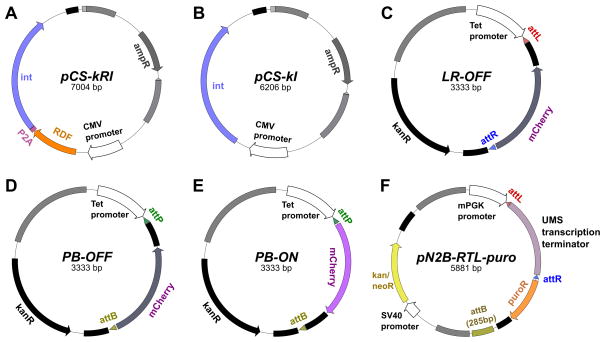
Figure 1.
Schematic diagram of plasmids used in this study. (A) pCS-kRI, used for polycistronic expression of phiC31 RDF and integrase (Int) in mammalian cells. (B) pCS-kI, for Int-only expression. (C) LR-OFF, used to detect attL-attR recombination (LxR). (D) PB-OFF, for detection of attP-attB activity. (E) PB-ON, a positive-control vector used for normalization of recombination activity. (F) pN2B-RTL-puro, for construction of cell lines that permit assessment of genomic LxR activity. Unless otherwise indicated, all cloned att-sites are 50 bases in length.
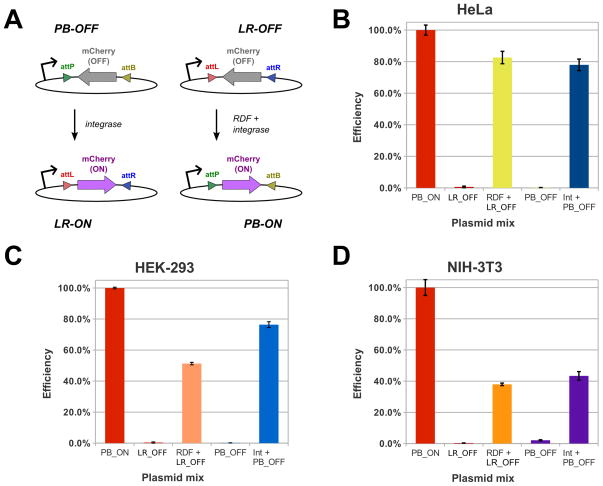
To permit quick measurement of RDF and phiC31 Int activity in mammalian cells, we constructed two inversion reporter plasmids, LR-OFF and PB-OFF, respectively (Fig. 1C and 1D). These vectors carry an inducible tetracycline promoter that has been placed upstream of an inverted, att-site flanked, mCherry ORF (Fig. 2A). Recombination flips the mCherry sequence, which permits its expression, visible as red fluorescence. To normalize our data, we also constructed the positive control PB-ON plasmid, which is the LxR recombination product of LR-OFF (Fig 2).
Figure 2. Plasmid inversion assays.
(A) Simplified diagram of detection schemes for PxB and LxR inversion. In the un-flipped state, mCherry is not expressed, as its ORF is inverted relative to the upstream promoter. When acted upon by Int or RDF:Int (for PB-OFF and LR-OFF, respectively), the mCherry ORF is flipped, which leads to its expression, detectable via fluorescence. Assays with these plasmids were conducted in HeLa (B), HEK-293 (C) and NIH-3T3 (D) cells. The percentage of cells expressing mCherry was measured in triplicate for each plasmid combination and then normalized to the positive control, the PB-ON transfection efficiency of same cell type. Representative data from one of three independent trials is shown here. ‘RDF’ and ‘Int’ refer to the pCS-kRI and pCS-kI plasmids, respectively. The error bars indicate the standard error of the mean.
To look for evidence of LxR recombination, we transiently transfected our mCherry reporter and RDF:Int expression plasmids, along with PB-CA-rtTA to activate the Tet promoter, into three mammalian cell lines, HeLa, HEK293, NIH 3T3. In all of these experiments, we observed moderate to high percentages of red-fluorescing cells (35–80%, normalized to PB-ON transfection), a result that is consistent with efficient LxR activity in these lines (Fig. 2, B–D). For an additional negative control, we also co-transfected our Int-only expression plasmid pCS-kRI with LR-OFF, and observed no fluorescence above background (data not shown). To confirm that the expected LxR recombinations took place, we sequenced the product junctions (attP and attB; Supporting Information, Table S3) and found them to be correct in all cases.
RDF:Int can mediate LxR chromosomal excision
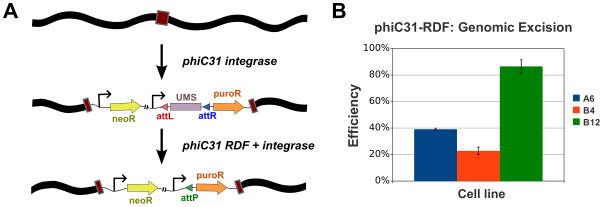
To extend these results from extrachromosomal substrates to chromosomally-integrated substrates, we next looked for evidence of RDF activity on a genomic substrate by using an excision assay. We focused on three HeLa-derived cell lines that carry an integrated copy of the pN2B-RTL-puro plasmid (Fig. 1F), each at at a distinct phiC31 pseudo site locus (Fig. 3A, Supporting Information, Table S4). LxR recombination imparts these cells with puromycin resistance, as it triggers expression of the PAC gene (puroR) via excision of an upstream stuffer (Fig. 3A). In all three lines, we observed strong evidence of chromosomal RDF activity. Puromycin-resistant colonies were obtained from cells transfected with the RDF:Int expression vector (Fig. 3B; ~20–80% efficiency), but not from those only transfected with a control (pCS; data not shown). In addition, all sequences of the PCR-amplified genomic attP remnant were as expected, which confirmed that the expected excision occurred in these lines.
Figure 3. Genomic excision experiment.
(A) Simplified diagram of the selection scheme for LxR excision. Stable lines are made with the pN2B-RTL-puro plasmid by phiC31 pseudo site integration, then isolated through G418 selection. Next, each line was transfected with the pCS-kRI expression vector, which will impart it with resistance to puromycin if the RDF:Int complex can act at the integrated locus, due to excision of the UMS transcription terminator. (B) Normalized excision efficiency. The HeLa pN2B-RTL-puro lines A6, B4 and B12 were transfected in triplicate with pCS-kRI, and puromycin-resistant colonies were obtained in every case. Puromycin-resistant colonies were never obtained from control-vector transformations. Colony counts were normalized to the transfection efficiency of pEGFP-C1 for each line. Representative data from one of two independent trials is shown here. The mean efficiency of each line was plotted here with error bars that indicate the standard error of the mean.
DISCUSSION
This study is the first demonstration of LxR recombination mediated by a serine recombinase in a eukaryotic environment. While the ability of the phiC31 integrase to function in mammalian cells is well-established, whether RDF would function in mammalian cells was unknown. For example, factors known to bind Int [8, 9] might have blocked RDF:Int complex formation or nuclear import. After cloning the RDF, we were quickly able to confirm its activity in mammalian cell lines with plasmid inversion assays. Our finding that phiC31 RDF functions in mammalian cells strongly predicts that it will function in other eukaryotic environments as well. The nature of the reaction and its precision were verified by DNA sequencing. We observed considerable variation in efficiency across cell lines (~35–75%; Fig 2, B–D). This result was expected, since Int activity is known to vary significantly across species and cell types [10–13].
Because a popular application of the phiC31 RDF is likely to be chromosomal excision, we also assessed genomic LxR activity in HeLa cells. For this study, we focused on three cell lines that each had a single puro-reporter construct integrated by phiC31 at a distinct location and that possessed the typical micro-deletions in the pseudo attL/R sites [14, 15], rendering them unlikely to be recombined by the RDF:Int complex, to minimize the possibility of whole-plasmid excision. For all three lines, we observed clear evidence of genomic LxR activity. Puro-resistant colonies with sequence-verified junctions were obtained from cells transfected with the RDF-Int plasmid, but not from those that received a control vector (pCS; Fig 3B and data not shown). Similar to observations from genomic integration assays [14, 16], the chromosomal LxR activity range varied widely among the three lines (~20–80%), suggesting that the efficiency of the genomic LxR reaction is also likely to be affected by chromosomal context. Because most integration events at pseudo attP sites create small deletions and other mutations in the integration site [15], the RDF-Int complex is unlikely to reverse these events efficiently.
Concluding remarks
We have demonstrated that the RDF from phage phiC31 can be used in conjunction with integrase to reverse the attB x attP recombination in eukaryotic cells. This finding makes it possible to employ the phiC31 reaction in either the forward or reverse direction. phiC31 RDF is thus likely to take its place alongside phiC31 integrase as a valuable tool for genome engineering and synthetic biology in a wide variety of species and applications.
Supplementary Material
Acknowledgments
This work was supported by grant RL1-00634-1 from the California Institute for Regenerative Medicine to M.P.C.
Abbreviations
- attB
bacterial attachment site
- attP
phage attachment site
- attL
left attachment site
- attR
right attachment site
- Int
integrase
- LxR
recombination of attL and attR
- PxB
recombination of attP and attB
- RDF
recombination directionality factor
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
Author contributions: A.P.F.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; C.L.C.: conception and design, collection and assembly of data, analysis and interpretation; C.L.M.: collection and assembly of data, data analysis and interpretation; M.P.C.: conception and design, funding of project, manuscript writing.
References
- 1.Kuhstoss S, Rao RN. Analysis of the Integration Function of the Streptomycete Bacteriophage phiC31. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 2.Thorpe HM, Smith MCM. In Vitro Site-specific Integration of Bacteriophage DNA Catalyzed by a Recombinase of the Resolvase/invertase Family. Proc Natl Acad Sci U S A. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorpe HM, Wilson SE, Smith MCM. Control of Directionality in the Site-specific Recombination System of the Streptomyces Phage phiC31. Mol Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 4.Khaleel T, Younger E, McEwan AR, Varghese AS, et al. A Phage Protein That Binds phiC31 Integrase to Switch Its Directionality. Molecular Microbiology. 2011;80:1450–1463. doi: 10.1111/j.1365-2958.2011.07696.x. [DOI] [PubMed] [Google Scholar]
- 5.Smith MCM, Brown WRA, McEwan AR, Rowley PA. Site-specific Recombination by φC31 Integrase and Other Large Serine Recombinases. Biochem Soc Trans. 2010;38:388. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly MLL, Luke G, Mehrotra A, Li X, et al. Analysis of the Aphthovirus 2A/2B Polyprotein “cleavage” Mechanism Indicates Not a Proteolytic Reaction, but a Novel Translational Effect: A Putative Ribosomal “skip”. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 7.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A Phage Integrase Directs Efficient Site-specific Integration in Human Cells. Proc Natl Acad Sci U S A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y, Li Z, Wang JJ, Xu G, et al. Sp100 Interacts with Phage ΦC31 Integrase to Inhibit Its Recombination Activity. Acta Biochim Pol. 2011;58:67–73. [PubMed] [Google Scholar]
- 9.Chen J, Ji C, Xu G, Pang R, et al. DAXX Interacts with Phage PhiC31 Integrase and Inhibits Recombination. Nucleic Acids Res. 2006;34:6298–6304. doi: 10.1093/nar/gkl890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maucksch C, Aneja MK, Hennen E, Bohla A, et al. Cell Type Differences in Activity of the Streptomyces Bacteriophage phiC31 Integrase. Nucleic Acids Res. 2008;36:5462–5471. doi: 10.1093/nar/gkn532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liesner R, Zhang W, Noske N, Ehrhardt A. Critical Amino Acid Residues Within the PhiC31 Integrase DNA Binding Domain Affect Recombination Activities in Mammalian Cells. Hum Gene Ther. 2010;21:1104–1118. doi: 10.1089/hum.2010.034. [DOI] [PubMed] [Google Scholar]
- 12.Andreas S, Schwenk F, Kuter-Luks B, Faust N, et al. Enhanced Efficiency Through Nuclear Localization Signal Fusion on Phage phiC31-integrase: Activity Comparison with Cre and FLPe Recombinase in Mammalian Cells. Nucleic Acids Res. 2002;30:2299–2306. doi: 10.1093/nar/30.11.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific Cassette Exchange and Germline Transmission with Mouse ES Cells Expressing phiC31 Integrase. Nat Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 14.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, et al. Site-Specific Genomic Integration in Mammalian Cells Mediated by Phage phiC31 Integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, et al. Integration Specificity of Phage phiC31 Integrase in the Human Genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 16.Monetti C, Nishino K, Biechele S, Zhang P, et al. PhiC31 Integrase Facilitates Genetic Approaches Combining Multiple Recombinases. Methods. 2011;53:380–385. doi: 10.1016/j.ymeth.2010.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.