Abstract
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) combines the sensitivity and selectivity of mass spectrometry with spatial analysis to provide a new dimension for histological analyses to provide unbiased visualization of the arrangement of biomolecules in tissue. As such, MALDI IMS has the capability to become a powerful new molecular technology for the biological and clinical sciences. In this review, we briefly describe several applications of MALDI IMS covering a range of molecular weights, from drugs to proteins. Current limitations and challenges are discussed along with recent developments to address these issues.
The field of proteomics has matured to include a number of powerful analytical tools to help elucidate molecular mechanisms fundamental to understanding health and disease. Mass spectrometry has achieved a pivotal role in proteomics owing to the sensitivity and molecular specificity of this technology. For example, it is now possible to measure the proteome of yeast cells with near comprehensive coverage [1–3] in a single experiment. Although these experiments are technologically impressive, these achievements represent only a starting point in the understanding of the many processes occurring within the cell and further, to understand these processes in multicellular organisms. To determine the role of a protein in human health and disease, one must study the protein in the context of its natural environment, the tissue. Currently, many of the most widely used proteomics approaches are unable to measure proteins while maintaining critical spatial information that is necessary for an in depth understanding of the biological system being studied. Traditionally, spatial information concerning the distributions of biomolecules in tissue has been obtained by using techniques such as immunohistochemistry, which require prior knowledge of target analytes. While effective in some cases, these methods are not adequate for the detection of biologically important protein processing steps such as post-translational modifications or endogenous proteolysis.
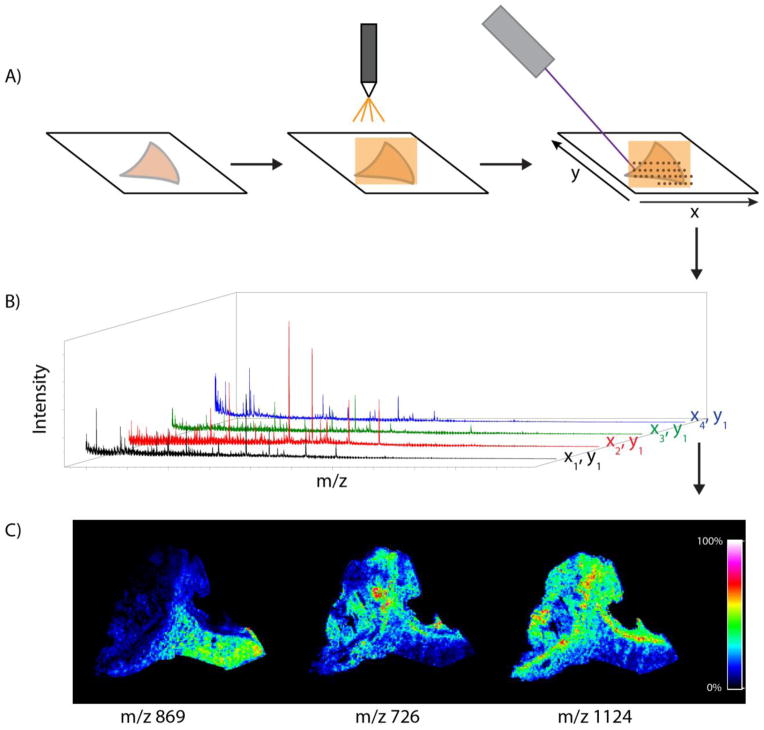
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) combines the tremendous sensitivity and selectivity of mass spectrometry with the spatial analysis provided by traditional histology, offering unbiased visualization of the spatial arrangement of biomolecules in tissue. An overview of a typical MALDI IMS workflow is presented in Figure 1. In this example, flash frozen tissue is cut into thin tissue sections and flat mounted onto a target. The sections are coated with a MALDI matrix that assists in the desorption and ionization of the molecules in the tissue. During the experiment, specific regions of the tissues are irradiated by a laser in an array of discrete points and mass spectra are generated for each x,y coordinate. Ion intensities are then plotted on a coordinate system matching the relative location of each spectrum that was acquired on the tissue surface, creating images of the ion. For reference, IMS images may also be compared to a histological stained micrograph of the tissue to determine molecular patterns that correlate to specific anatomical features. This approach is useful for a wide variety of biological systems (e.g., different species and analytes) as evidenced by the increasing frequency of publications citing MALDI IMS in biology and medical research (Figure 2).
Figure 1.
Overview of the MALDI IMS workflow on a section of a human kidney biopsy. (A) Fresh frozen tissue is cut and mounted on a conductive surface. Matrix is applied by a robotic sprayer or is sublimated, and the section is irradiated by the laser in a raster array. (C) Mass spectra are acquired for each x,y coordinate. (D) Selected ions may be mapped on the tissue surface to create images.
Figure 2.
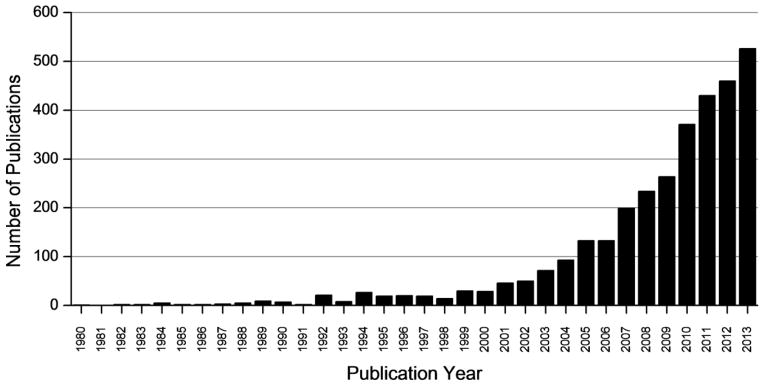
Publications reporting the development and application of IMS through 2013. Data was collected from a PubMed search of the following key words: (imaging mass spectrometry OR mass spectrometry imaging)) AND (“1980”[Date - Publication]: “2013”[Date - Publication])).
With a significant increase in the number and variety of applications employing this technology, there is ongoing interest in MALDI IMS and important work has been done to significantly improve both methods and instrumentation, in order to meet the needs of researchers. An ideal imaging MS experiment would offer high sensitivity for all analyte classes and submicron spatial resolution for analysis of sub-cellular structures, along with easy identification and quantification of analytes. Although these “wish list” capabilities have not yet been realized for routine IMS, the field has made great advances from its beginning nearly 20 years ago [4]. In this review, we describe recent advances in MALDI IMS that advance the field towards these performance metrics, including efforts to address the limitations of current instrumentation and challenges for future MALDI IMS research. This includes challenges in sample preparation, spatial resolution, analyte identification, and data processing. We also discuss the opportunities and special considerations regarding the use of MALDI IMS as a new molecular technology in the clinical laboratory, including the potential for a high throughput, quantitative, and unbiased informational approach for clinical analysis of patient samples
MALDI IMS as a robust bioanalytical tool
Below we outline four areas of ongoing interest and work in making IMS a robust bioanalytical tool. These include: (1) the development of standardized protocols for sample preparation, (2) improved spatial resolution of the images, (3) efficient protein and peptide identification and tools to increase the dynamic range of MALDI IMS, (4) and the development of next generation data processing algorithms that leverage the unique, highly dimensional data generated in these experiments.
1. Experiment-specific and standardized sample preparation methods
General MALDI IMS work flows have been described in a number of publications [5, 6]; however, the specifics of these protocols are often tailored for the analyte and tissue of interest. These protocol adjustments are commonly made in the sample preparation steps that precede MALDI IMS analysis. Sample preparation, including tissue section thickness, tissue washes, and matrix application are often optimized for each new type of experiment to maximize the sensitivity and provide sufficient spatial resolution for the experiment. For example, lipid analysis has been shown to benefit from an aqueous wash of tissue to remove salts [7], while organic washes are used for protein analysis, in order to fix tissue and remove lipids that suppress protein signal [8]. Other tissue treatments are often developed for analytes that are difficult to desorb and ionize. For example, a detailed sample preparation protocol for membrane proteins in retinal tissue was described that included a special tissue mounting technique, followed by pipetting deionized water 11 times over the tissue [9]. With slight modifications to this protocol, Nicklay and co-workers acquired images of the transmembrane myelin proteolypid protein (PLP) in rat brain tissue (Figure 3) [10]. Here, the authors added a final wash step that included 1% acetonitrile.
Figure 3.
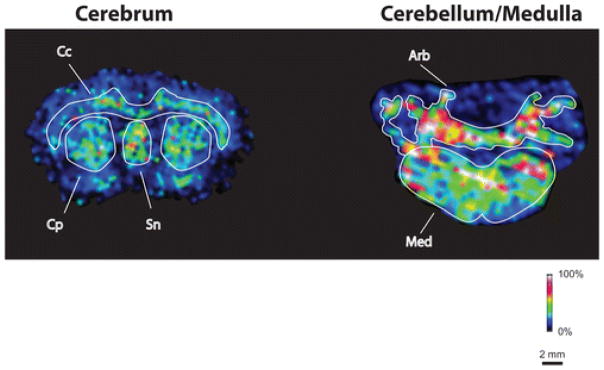
Images of the membrane protein myelin proteolipid protein (PLP, 30 kDa). Special sample preparation protocols were developed to enable imaging of membrane proteins [10]. The protein was imaged within cerebrum (left) and cerebellum/medulla (right). Exclusive localization to white matter regions of brain, particularly to the corpus callosum (Cc), caudate-putamen (Cp), and septal nucleus (Sn) of the cerebrum and the arbor vitae (Arb) of the cerebellum and the entire medulla (Med), is apparent.
The method of matrix application is important and should be optimized for each type of experiment. For example, robotic spotting of matrix on tissue allows for excellent analyte extraction, but low image resolution (200 μm). Spraying matrix solution onto the tissues surface, either by hand or with a robotic sprayer, allows for better resolution (10–20 μm), although care must be taken to keep the tissue surface from becoming too wet with solvent to avoid delocalization of analytes. Sublimation and rehydration offers excellent image resolution, but may result in comparably less effective extraction of analytes (i.e. lower sensitivity). Of course, care must be taken to ensure that the sublimation is homogeneous across the tissue and that the recommended rehydration step does not introduce delocalization [11]. In all of these methods, matrix thickness can affect ion intensities and different matrices are more sensitive for specific molecules. For example, α-cyano-4-hydroxycinnamic acid is best for peptides and small proteins, while sinapinic acid is better for larger proteins (> 20 kDa).
As this imaging technology is applied to new fields, the number and types of analytes and tissue types will increase, as will the details of various sample preparation protocols. With this in mind, inherent variation between protocols will require careful consideration of how novel sample preparations affect the accuracy and reproducibility of IMS data. Although sample preparation may vary between different experiment types, it will also be increasingly important to maintain carefully controlled and standardized sample preparation procedures, since even small alterations may produce variations in the MS data and the corresponding image. It is imperative that the MALDI IMS community develop guidelines and protocols to ensure consistent and accurate data acquisition [12].
2. Spatial resolution
Spatial resolution of the MALDI IMS experiment is a key parameter and must be chosen with care. High-resolution imaging (<20 μm raster) is necessary for certain applications, particularly applications designed to elucidate biological differences at the cellular level. IMS provides unique information to the study of cells because molecular components that define the cellular phenotype may be measured directly in each cell rather than having to infer their contribution from average measurements taken from cell populations.
Commercially available MALDI IMS instruments operate in microprobe mode, in which a raster by the laser is performed over an area of the tissue, generating mass spectra at each ablation point. In general, the minimum spatial resolution that can be achieved is limited by the diameter of the laser beam. Current commercial MALDI IMS instruments in microprobe mode can now achieve spatial resolutions at just below 20 μm. There is a considerable effort to increase the spatial resolution through the use of several instrumental approaches. One technique to increase resolution without instrument modification is oversampling. Here, the laser ablates overlapping areas between adjacent spots such that the signal originates from just part of the original spot size [13]. This method is useful but requires complete ablation of the sample at each spot and, therefore, longer data acquisition times. As an alternative, a simple, inexpensive modification to standard commercial instruments has been described to reduce the diameter of the laser beam using a 25 μm pinhole filter. The filter reduced the laser beam diameter from 20 μm to 5 μm on two MALDI TOF instruments [14].
Custom instrument configurations have also been developed that enable high spatial resolution IMS analysis. In one example, a Gaussian beam profile was created using optical single mode fibers, offering a small laser beam diameter [15]. In another case, a transmission geometry vacuum ion source was designed to allow sub-micron laser spot sizes [16, 17]. In this configuration, the laser beam irradiates the backside of the sample. The authors acquired sub-cellular images of synaptophysin (m/z 323) and somatostatin (m/z 532) in islet cells (Figure 4).
Figure 4.
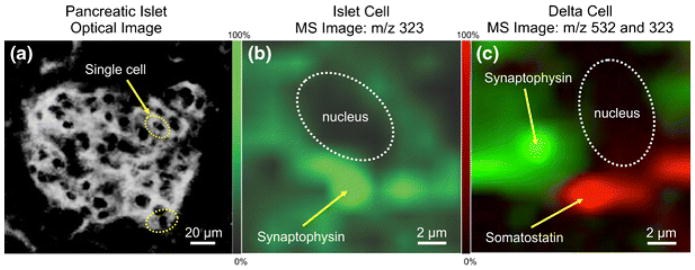
Target MALDI IMS analysis of synaptophysin (m/z 323) and somatostatin (m/z 532) in single islet cells. Images were obtained using a transmission geometry ion source for high-resolution imaging. (a) Optical image of islet cells. (b) MALDI IMS of an immunoreactive islet cell for synaptophysin. (c) Targeted IMS of a delta cell for synaptophysin (green) and somatostatin (red), showing different localization for each molecule within different cellular structures. [16]
In addition to advances in instrumentation, matrix application itself may limit the lateral resolution in a MALDI IMS experiment. With instruments capable of high resolution imaging, the size of the matrix crystals and the density of the applied matrix can be a limiting factor in MALDI IMS resolution. Sublimation of matrix onto tissue is currently the best method for high resolution analysis [18]. However, this type of solvent-free application limits sensitivity and must be followed by rehydration of the sample to extract proteins and peptides from the surface of the tissue into the matrix [11].
Despite its clear advantages, the capability for high spatial resolution imaging has some disadvantages. The large data sets that are generated result in long data acquisition and processing times and issues of data storage may arise. Most importantly, high resolution imaging often results in decreased sensitivity, since less material is analyzed using smaller ablation areas. Thus, high resolution imaging is most effective with metabolites, drugs, lipids, and peptides that are more easily extracted and ionized. Finally, the large number of laser shots and spectra acquired in a high resolution IMS experiment contributes significant wear and tear on MALDI IMS instruments, such that costly instrument parts, including lasers and detectors, must be replaced more frequently. Taken together, it is important to carefully evaluate the spatial resolution that is required to answer a given biological question, and employ the resolution needed to answer the question at hand. For cases that require high resolution imaging, increasing the speed of analysis and improving data storage and processing capabilities are active areas of instrument and are discussed further below.
Speed of analysis
As the capabilities for high spatial resolution are advanced, the need for faster data acquisition becomes increasingly apparent. Thus, a large effort in instrumentation development has been devoted to increasing the speed of analysis. At the time that this technology was first introduced, 1 Hz N2 lasers were common in MALDI MS instruments and imaging experiments took several hours or in some cases days to acquire images. As IMS has become more mainstream, instrumentation developers have worked to meet the needs of IMS research. For example, commercial instruments now incorporate lasers with the ability to operate at pulse repetition rates of ~1 kHz or more using frequency tripled solid-state lasers, although researchers are working to increase these rates further. For example, investigators have recently presented data using a 20 kHz Nd:YVO4 laser [19], with the best results obtained when operating at repetition rates between 5–10 kHz.
Another approach for decreasing sample acquisition time is to use continuous laser raster sampling. Typically, pixels are acquired by sampling at discrete spots over the tissue, with the laser turned off as the stage moves from spot to spot. With continuous sampling, the laser is fired continuously as the sample stage moves non-stop in a straight line. Using a 5kHz Nd:YLF laser with continuous laser raster sampling, a sagittal rat brain tissue section was analyzed in less than 10 min at a spatial resolution of 100 μm [20]. In another example, images of drugs in rat sections were acquired with a 1 kHz laser with a rastering speed of 18 mm/s and was completed in less than 15 minutes [21]. Lastly, it is worth noting that instruments operating in mass microscope mode would also offer the opportunity for faster analysis. The desorption/ionization beam in this configuration is 200–300 μm, so fewer measurement points are required for a given area, even for high resolution images [22]. The disadvantage to such an approach is the extremely limited m/z range and sensitivity of these instruments relative to more conventional mass analyzers.
Data storage and processing
Data file sizes obtained by MALDI IMS can be large even for routine, low resolution imaging. Individual images can generate thousands of spectra, which greatly impact data processing times, computation costs, and data storage [23]. Currently, no data reduction tools for MALDI IMS data are commercially available. However, as MALDI IMS researchers develop methods for increased spatial and mass resolution, high throughput analysis, and specialized applications such as 3-D imaging [24], the need for data reduction will become increasingly important.
3. Protein identification in MALDI IMS
MALDI IMS analysis is highly sensitive and allows researchers to map the spatial distribution of a large number of analytes simultaneously. However, identification of species within an MS image can be labor intensive in some cases. Targeted in situ MALDI MS/MS analysis for the identification of lipids and small molecules is generally routine using current technology, but identification of proteins and peptides pose greater challenges [25].
There are generally two approaches for identification of proteins from tissue. The first is imaging of intact proteins using a linear TOF mass spectrometer, which is necessary to accommodate the larger masses of these molecules. Signals from images of intact proteins are compared to MALDI MS data from fractionated tissue homogenates. Using imaging results as a guide, the protein fraction containing the signals of interest is determined and bottom up or top-down LC MS/MS experiments are performed to identify the species. A significant drawback to this method is that TOF mass analyzers have limited mass resolution and cannot resolve the multiple ion species that may lie within a single peak in the spectrum. Moreover, analysis of large proteins requires greater ionization energy and, therefore, imaging with TOF mass spectrometers using routine methods is practically limited to species under ~ 50 kDa, although examples of detection of large (e.g. 200 kDa) proteins have been reported [26].
A second identification strategy is to perform an on-tissue tryptic digest [27, 28]. Here, trypsin is deposited directly on tissue, whether by robotic spotters [29] or automated sprayers, and hydrolysis occurs on the tissue surface. Following matrix application, MS/MS analysis is performed directly from tissue, typically by collision induced dissociation. The issue of mass resolution remains with TOF instruments, as several overlapping signals are often present in the complex MS spectra generated from direct tissue analysis. Unresolved features in the mass spectra pose difficulties for the interpretation of MS/MS results and may obscure important details in MS images of near isobars with distinct spatial distributions. In order to avoid these issues, high mass resolution IMS using FTICR [30, 31] or Orbitrap [28, 32] mass analyzers are extremely valuable in MALDI IMS experiments. These instruments offer high mass resolution as well as high mass accuracy, giving increasing confidence for analyte identification. One drawback of Fourier transform based analyzers is that they are relatively slow, typically requiring long transients for each pixel. In addition, high mass resolution data generates large data files which, in turn, increase data processing times.
In some cases, ion mobility spectrometry [33] can be effectively incorporated into the IMS experiment. In ion mobility spectrometry, ions are passed through a drift cell containing a neutral buffer gas, under the influence of an electric field. Ions are separated by their collision cross sections; ions with small collision cross sections move through the drift cell more quickly than ions with large collisions cross sections. The additional separation by ion mobility increases the dynamic range of the analysis and has allowed for imaging of near isobars of tryptic fragments from actin and ubiquitin in rat brain tissue. By selecting ions with different mobilities, separate localization could be found for these peptides (Figure 5) [34]. Without ion mobility separation, this localization would not have been apparent and the image for this signal would have been an overlay of the images for each peptide.
Figure 5.
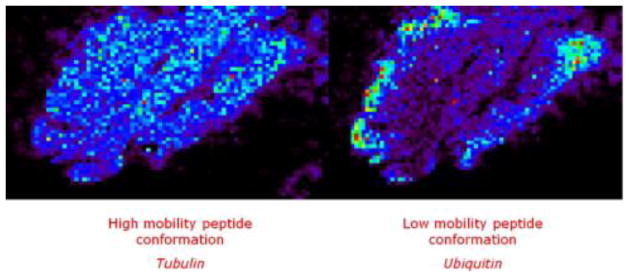
Images of separated and identified isobaric ions of tubulin and ubiquitin tryptic peptide fragments (1039 m/z) in rat brain were imaged using ion mobility spectrometry coupled with imaging mass spectrometry. For these images, two different drift times have been selected: 100–127 and 133–148 bins to reconstruct the respective images. Without any ion mobility, one image is obtained corresponding to the superposition of these two images. [34]
Several challenges for MS/MS in imaging MS experiments should be noted. For example, MS/MS data acquisition is not automated and fragmentation data must be collected manually, one parent mass at a time. This greatly increases data acquisition time, especially if there are multiple signals of interest. Moreover, as data is acquired for MALDI MS/MS, matrix is ablated from the tissue surface such that a given spot on the tissue surface may only be analyzed a limited number of times. Identification of multiple species, therefore, may require multiple samples. Multiplexed MS/MS methods, in which multiple MS/MS products are analyzed simultaneously, would greatly increase the efficiency of this approach. Finally, the z = +1 charge states that arise from the MALDI process greatly affect the efficiency of peptide fragmentation. Recently, new MALDI matrices that give rise to multiply charged MALDI ions have been introduced and may help resolve this issue [35].
4. Processing and Analysis of IMS Data
Data Processing and analysis are essential parts of an imaging mass spectrometry experiment, yet this is still a young and developing field. The complicated nature of IMS data has given rise to a number of different preprocessing and analysis methods, and requires that experimentalists understand correct usage of these methods for accurate results [23]. Moreover, many of these methods are not available commercially. Unfortunately, this lack of integration with commercial software packages results in a barrier for the use of these algorithms by investigators that don’t have the ability to write custom programs to manipulate the data. In these cases, investigators are encouraged to consult with those who have expertise in relevant bioinformatics fields to assess confidence of scientific findings and to correctly apply these computational tools.
After image acquisition, proper data processing and analysis are required for accurate interpretation of IMS results. In general, the data can be processed both at the spectral level and at the image level. Preprocessing of spectral data may include smoothing, background subtraction, and normalization to account for instrumental and chemical noise in IMS data [36]. Spectra in IMS often contain high-frequency noise and unresolved chemical background, such that smoothing and background subtraction are important for accurate representation of molecular localization in MS images. Most MALDI IMS acquisition and visualization software include automated algorithms for these preprocessing methods.
Normalization of spectral intensity is used to account for some variations in ionization due to suppression and matrix effects. Normalization to the total ion count (TIC) is most commonly used in MALDI IMS to account for global effects across the tissue. However, normalizing to the TIC is problematic if substantial differences in the TIC between pixels exist [23, 37]. In this case, normalization can significantly change the image [37]. Ultimately, there is unlikely a single best preprocessing method for all IMS data sets. Thus, the investigator should assess the appropriateness of these methods for a given experiment, to ensure the most accurate representation of the data.
In addition to normal mass spectral noise, image noise can arise due to the stochastic nature of the analysis. Recently, commercial IMS data analysis software packages have incorporated averaging and smoothing algorithms to reduce the impact of noise within an MS image. In addition to preprocessing, commercial software also offers additional tools for image manipulation, including the ability to select data from regions of interest within the tissue, co-registration of optical images (e.g. stained serial sections) with the MALDI MS image, and easy export of MS spectra from individual pixels to other software programs for further analysis, including statistical analysis and data base searches.
Finally, picking peaks for further analysis can be complicated in IMS, due to the large number of peaks in the data set. One option is to perform peak picking from the mean spectrum of the image, although this may not be sensitive, especially for signals that are only present in small areas of the tissue. Another problem encountered with peak picking in IMS is that m/z values can drift slightly between pixels due to instrumental drift during the long acquisition time and/or variability in the tissue surface [38, 39]. Peak alignment tools have been developed to account for this variation [36], but are not yet commercially available.
MALDI IMS in the clinical laboratory
One of reasons for increased interest in MALDI MS in biology is the potential of the technology to solve clinical problems. For example, the application of MALDI profiling approaches to the classification of microbes made significant impact on clinical microbiology [40–42]. The molecular specificity, high throughput, and accuracy of classification of this technology provide a means for clinical laboratories to save time and money while maintaining or even increasing the quality of the clinical results. The key challenge of MALDI IMS for routine clinical application is to develop automated protocols to accommodate a heavy workload and must maintain a high level of quality control. In addition to the developments described earlier, other factors are also important when considering the potential of this technology for routine pathological analysis in the clinical laboratory. These include the development of high-throughput MALDI IMS, the ability to quantitate MALDI IMS results, and finally bioinformatic tools for data analysis of patient samples.
1. Sample Throughput
Sample throughput is essential to establish MALDI IMS as a robust analytical technology for clinical applications. As discussed above, developments in instrumentation for fast data acquisition will greatly help towards this goal. Nevertheless, there are other places for improvement within the IMS workflow, which has several time-intensive steps, including tissue sectioning, matrix application, and setting up the image. As such, researchers are also targeting steps within this workflow in order to further increase sample throughput. For example, the MALDI slides pre-coated with matrix have been developed that could be prepared in advanced and would bypass the matrix application step after sectioning [43, 44]. These slides offer similar image quality to post coating approaches (Figure 6) and have a shelf life of more than 6 months, when kept in a desiccator.
Figure 6.
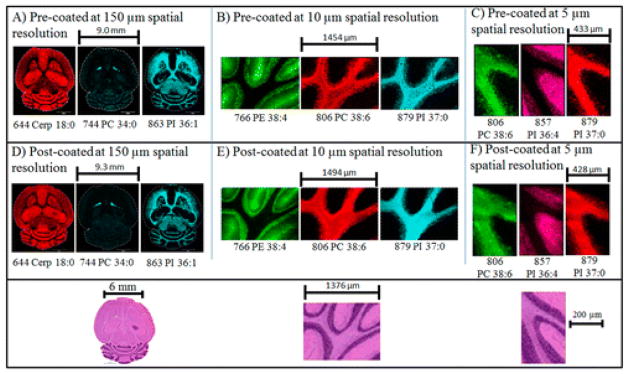
Comparison of imaging results of mouse brain serial sections with matrix pre- and post-coated targets at different spatial resolutions. Ion images were collected in negative ion mode. Tentative identification was based on previous MS/MS analyses of lipids at these masses in mouse brain. Below: H&E optical scanned images from a serial section are shown at the bottom of the figure.
Automated sample loading systems have been reported for continuous instrument operation [45]. The loading system includes a controlled environment storage chamber and sample-loading robot and was demonstrated to be reproducible for clinical applications. Such advances are essential, as high sample throughput is necessary to make the technology viable for the clinical laboratory.
2. Quantitation in IMS
Relative quantitation of analytes from the direct analysis of tissue requires special consideration when designing an experiment. Ion suppression, analyte extraction efficiency, ionization efficiency, and ion stability may affect results [46, 47]. Ion suppression results from endogenous species that compete for ionization, such as lipids or salts, resulting in decreased signal intensities for some analytes. These effects are not unique and are common in many types of MS analyses of complex biological mixtures [37]. In IMS, however, the chemical microenvironment of adjacent areas within the same tissue section may be composed of different cell types and morphologies [48, 49]. This tissue heterogeneity may, in some cases, result in varying desorption/ionization effects across a single tissue section. In addition, uneven matrix application may introduce variations in ion signal [11]. Even if matrix is homogenously applied, variations in tissue composition can affect matrix crystallization, producing different crystal sizes and morphologies. Control experiments should be conducted that are designed to account for different extraction efficiencies of analytes with the preparation protocol being used.
Several groups have introduced methods to obtain absolute quantitative information from IMS data. Typically, these methods incorporate an internal standard to correct for signal variability. One common approach used for pharmaceuticals is to spot a dilution range of a target analyte on a control tissue section and analyze the control tissue and dosed tissue in parallel [50–53]. The drug concentration in the dosed tissue is estimated by comparing the ion intensity in the image to the standard curve from the dilution range. This method is fast and relatively simple, although it may not always accurately represent the extraction efficiency of the compound. To overcome this, investigators have applied an internal standard directly on the MALDI target, before tissue mounting [54]. This method offered improvements in signal reproducibility, MS images, and calibration cure linearity [51, 54]. Furthermore, it has been shown to account for tissue-specific variations when monitoring the analyte to internal standard ratio across several tissues types [49]. Recently, a promising mimetic tissue method, in which tissue homogenates spiked with internal standard of varying concentrations were frozen into a polymer mold [55] was described. The frozen homogenates were sectioned, mounted, and imaged in parallel with dosed tissue. This method mimics both the analyte extraction and ion suppression effects of the dosed tissue. In one example, the authors demonstrate that this method is comparable to similar analysis by LC MS/MS for rat liver from a rat dosed with the drug nevirapine (Figure 7).
Figure 7.
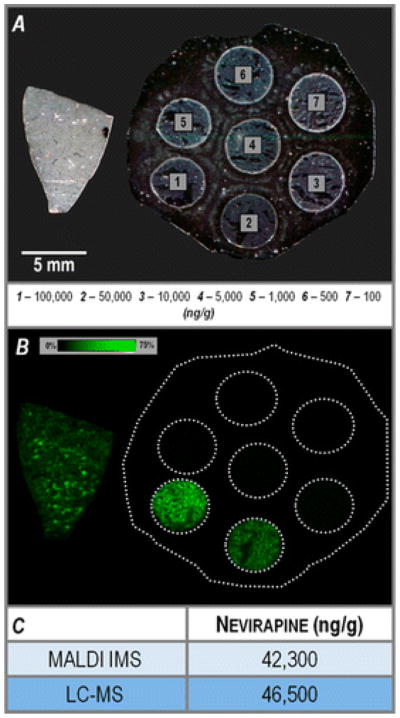
Quantitative analysis of a rat liver section dosed with the drug nevirapine (m/z 267.124) using a mimetic tissue model. (A) Optical scan of sections from the nevirapine-dosed rat liver (left) and the mimetic tissue model (right), before matrix application. (B) Ion image for nevirapine, m/z 267.124, at 75% maximum intensity threshold. (C) Comparison of nevirapine quantification results from the MALDI IMS and LC-MS analyses.
Even in cases when robust methods for quantitation are established, additional challenges will certainly arise for special cases such as whole body imaging [48, 49, 56–59], where diverse tissues types are sampled simultaneously and 3D imaging [60–62], in which the signal intensities and variations in sample preparation must be normalized throughout several tissue sections. Finally, it should be noted that the development of quantitative methods in MALDI IMS is primarily driven by an interest to quantitate small molecule pharmaceuticals in tissue and therefore most quantitation methods are developed for analysis of exogenous compounds in tissue from dosed animals. Still, it is possible to envision a future in which quantitation of endogenous molecules may be possible, using a similar approach.
3. Bioinformatic analysis
Statistical evaluation of IMS data is necessary for clinical research and biomarker discovery. Well-established multivariate analyses, such as principle component analysis and classification algorithms are used in both unsupervised data mining and supervised classification of IMS results. These methods are used to identify patterns in the localization of molecules in a histology-directed or histology-independent manner and are typically employed by treating each pixel as an independent sample. There are several excellent reviews describing various methods for these analyses [23, 63]. Commercial software from instrument vendors and open-source software are available for some statistical analyses. However, these platforms tend to be vendor-specific, and the details of the computational approach are not always well understood by the practitioners. As a result, many groups still rely on in-house solutions to preprocessing and statistical analysis to meet the needs specific to their research. In order to overcome these limitations, more development is needed done to add transparency to the statistical treatment of such data, and the results that are returned must be provided in a form that can be clearly interpreted by the clinician. Moreover, clinical applications of MALDI IMS will benefit greatly when standardized approaches to the analysis and classification of these data are available.
Conclusions and perspectives
Since the inception of MALDI IMS, improvements to instrumentation and the experimental workflow have increased the spatial resolution, speed, and sensitivity of analysis. Developments in sample preparation have allowed this technology to be applied to measurements in a diverse set of biological systems. Advances in instrumentation have allowed for increasing spatial resolution in IMS images, although this has introduced new challenges with data acquisition times and data storage and processing. New strategies to increase the dynamic range of analysis, including Fourier transform-based methods and the incorporation of ion mobility, have been shown to be effective for broadening the application of IMS to include many new analytes. Data processing and analysis are areas within the MALDI IMS workflow that are currently being rigorously pursued. Standardized preprocessing methods must be developed for complex IMS data, as well as validations for effective, cross-platform applications. Other advances, such as the integration of IMS data with other imaging modalities, such as microscopy and MRI would greatly enhance the potential for new enriched data sets to help unravel the complexity of living systems.
Overall, MALDI IMS is rapidly maturing to become a robust and routine bioanalytical technology for basic biological research and clinical analyses. MALDI IMS has already been applied in an increasing number of basic biological studies although inclusion into the clinical research arena has not yet begun. The utilization of this technology in clinical analysis is highly likely thanks to the convergence of technological development for advanced IMS instrumentation and emerging clinical problems that require high molecular specificity and sensitivity, in addition to detailed histopathological information. Automated MALDI MS analysis has already been found to have great utility in the clinical microbiology lab, providing a technological basis for the further development of automated MALDI IMS as part of a standard clinical approach in the molecular age of medicine.
Research Highlights.
MALDI IMS has the capability to become a powerful new molecular technology for the biological and clinical sciences.
We describe applications of MALDI IMS covering a range of molecular weights, from drugs to proteins.
Current limitations and challenges are discussed, as well as recent developments that address these issues.
Acknowledgments
The authors wish to acknowledge the support of grants from the National Center for Research Resources (5 P41 RR031461) and the National Institute of General Medical Sciences (8 P41GM103391) from the National Institutes of Health. MMG is supported by a Ruth L. Kirschstein National Research Service Award from the National Institute of Diabetes, Digestive and Kidney Diseases (1F32DK097875-01A1).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Froehlich F, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–U60. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 2.Lamond AI, Uhlen M, Horning S, Makarov A, Robinson CV, Serrano L, et al. Advancing Cell Biology Through Proteomics in Space and Time. Molecular & Cellular Proteomics. 2012:11. doi: 10.1074/mcp.O112.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hebert AS, Richards AL, Bailey DJ, Ulbrich A, Coughlin EE, Westphall MS, et al. The One Hour Yeast Proteome. Molecular & Cellular Proteomics. 2014;13:339–47. doi: 10.1074/mcp.M113.034769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: Localization of peptides and proteins using MALDI-TOF MS. Analytical Chemistry. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 5.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nature Medicine. 2001;7:493–6. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- 6.Angel PM, Caprioli RM. Matrix-Assisted Laser Desorption Ionization Imaging Mass Spectrometry: In Situ Molecular Mapping. Biochemistry. 2013;52:3818–28. doi: 10.1021/bi301519p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angel PM, Spraggins JM, Baldwin HS, Caprioli R. Enhanced Sensitivity for High Spatial Resolution Lipid Analysis by Negative Ion Mode Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry. Analytical Chemistry. 2012;84:1557–64. doi: 10.1021/ac202383m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. Journal of the American Society for Mass Spectrometry. 2008;19:1069–77. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grey AC, Chaurand P, Caprioli RM, Schey KL. MALDI Imaging Mass Spectrometry of Integral Membrane Proteins from Ocular Lens and Retinal Tissue. Journal of Proteome Research. 2009;8:3278–83. doi: 10.1021/pr800956y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicklay JJ, Harris GA, Schey KL, Caprioli RM. MALDI Imaging and in Situ Identification of Integral Membrane Proteins from Rat Brain Tissue Sections. Analytical Chemistry. 2013;85:7191–6. doi: 10.1021/ac400902h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Caprioli RM. Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Analytical Chemistry. 2011;83:5728–34. doi: 10.1021/ac200998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonnell LA, Heeren RMA, Andren PE, Stoeckli M, Corthals GL. Going forward: Increasing the accessibility of imaging mass spectrometry. Journal of Proteomics. 2012;75:5113–21. doi: 10.1016/j.jprot.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. Journal of the American Society for Mass Spectrometry. 2005;16:1654–9. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Zavalin A, Yang J, Caprioli R. Laser Beam Filtration for High Spatial Resolution MALDI Imaging Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2013;24:1153–6. doi: 10.1007/s13361-013-0638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiao H, Spicer V, Ens W. The effect of laser profile, fluence, and spot size on sensitivity in orthogonal-injection matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22:2779–90. doi: 10.1002/rcm.3675. [DOI] [PubMed] [Google Scholar]
- 16.Thiery-Lavenant G, Zavalin AI, Caprioli RM. Targeted Multiplex Imaging Mass Spectrometry in Transmission Geometry for Subcellular Spatial Resolution. Journal of the American Society for Mass Spectrometry. 2013;24:609–14. doi: 10.1007/s13361-012-0563-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM. Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS. Journal of Mass Spectrometry. 2012;47:1473–81. doi: 10.1002/jms.3108. [DOI] [PubMed] [Google Scholar]
- 18.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. Journal of the American Society for Mass Spectrometry. 2007;18:1646–52. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trim PJ, Djidja MC, Atkinson SJ, Oakes K, Cole LM, Anderson DMG, et al. Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadrupole time-of-flight mass spectrometer for MALDI-MS imaging. Analytical and Bioanalytical Chemistry. 2010;397:3409–19. doi: 10.1007/s00216-010-3874-6. [DOI] [PubMed] [Google Scholar]
- 20.Spraggins JM, Caprioli RM. High-Speed MALDI-TOF Imaging Mass Spectrometry: Rapid Ion Image Acquisition and Considerations for Next Generation Instrumentation. Journal of the American Society for Mass Spectrometry. 2011;22:1022–31. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopfgartner G, Varesio E, Stoeckli M. Matrix-assisted laser desorption/ionization mass spectrometric imaging of complete rat sections using a triple quadrupole linear ion trap. Rapid Communications in Mass Spectrometry. 2009;23:733–6. doi: 10.1002/rcm.3934. [DOI] [PubMed] [Google Scholar]
- 22.Klerk LA, Altelaar AFM, Froesch M, McDonnell LA, Heeren RMA. Fast and automated large-area imaging MALDI mass spectrometry in microprobe and microscope mode. International Journal of Mass Spectrometry. 2009;285:19–25. [Google Scholar]
- 23.Jones EA, Deininger S-O, Hogendoorn PCW, Deelder AM, McDonnell LA. Imaging mass spectrometry statistical analysis. Journal of Proteomics. 2012;75:4962–89. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Xiong X, Xu W, Eberlin LS, Wiseman JM, Fang X, Jiang Y, et al. Data Processing for 3D Mass Spectrometry Imaging. Journal of the American Society for Mass Spectrometry. 2012;23:1147–56. doi: 10.1007/s13361-012-0361-7. [DOI] [PubMed] [Google Scholar]
- 25.Mascini NE, Heeren RMA. Protein identification in mass-spectrometry imaging. Trac-Trends in Analytical Chemistry. 2012;40:28–37. [Google Scholar]
- 26.Chaurand P. Imaging mass spectrometry of thin tissue sections: A decade of collective efforts. Journal of Proteomics. 2012;75:4883–92. doi: 10.1016/j.jprot.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. Journal of Mass Spectrometry. 2007;42:254–62. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- 28.Schober Y, Guenther S, Spengler B, Roempp A. High-resolution matrix-assisted laser desorption/ionization imaging of tryptic peptides from tissue. Rapid Communications in Mass Spectrometry. 2012;26:1141–6. doi: 10.1002/rcm.6192. [DOI] [PubMed] [Google Scholar]
- 29.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Analytical Chemistry. 2006;78:827–34. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 30.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Analytical Chemistry. 2008;80:5648–53. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taban IM, Altelaar AFM, van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, et al. Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. Journal of the American Society for Mass Spectrometry. 2007;18:145–51. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Makarov A, Denisov E, Lange O, Horning S. Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. Journal of the American Society for Mass Spectrometry. 2006;17:977–82. doi: 10.1016/j.jasms.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Bohrer BC, Mererbloom SI, Koeniger SL, Hilderbrand AE, Clemmer DE. Biomolecule Analysis by Ion Mobility Spectrometry. Annual Review of Analytical Chemistry. 2008:293–327. doi: 10.1146/annurev.anchem.1.031207.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stauber J, MacAleese L, Franck J, Claude E, Snel M, Kaletas BK, et al. On-Tissue Protein Identification and Imaging by MALDI-Ion Mobility Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2010;21:338–47. doi: 10.1016/j.jasms.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Inutan ED, Wang B, Lietz CB, Green DR, Manly CD, et al. Matrix Assisted Ionization: New Aromatic and Nonaromatic Matrix Compounds Producing Multiply Charged Lipid, Peptide, and Protein Ions in the Positive and Negative Mode Observed Directly from Surfaces. Journal of the American Society for Mass Spectrometry. 2012;23:1625–43. doi: 10.1007/s13361-012-0413-z. [DOI] [PubMed] [Google Scholar]
- 36.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, et al. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. International Journal of Mass Spectrometry. 2007;260:212–21. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deininger S-O, Cornett DS, Paape R, Becker M, Pineau C, Rauser S, et al. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Analytical and Bioanalytical Chemistry. 2011;401:167–81. doi: 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonnell LA, Mize TH, Luxembourg SL, Koster S, Eijkel GB, Verpoorte E, et al. Using matrix peaks to map topography: Increased mass resolution and enhanced sensitivity in chemical imaging. Analytical Chemistry. 2003;75:4373–81. doi: 10.1021/ac034401j. [DOI] [PubMed] [Google Scholar]
- 39.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Analytical Chemistry. 2005;77:6118–24. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 40.Patel R. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in Clinical Microbiology. Clinical Infectious Diseases. 2013;57:564–72. doi: 10.1093/cid/cit247. [DOI] [PubMed] [Google Scholar]
- 41.Patel R. MALDI-TOF Mass Spectrometry: Transformative Proteomics for Clinical Microbiology. Clinical Chemistry. 2013;59:340–2. doi: 10.1373/clinchem.2012.183558. [DOI] [PubMed] [Google Scholar]
- 42.Clark AE, Kaleta EJ, Arora A, Wolk DM. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry: a Fundamental Shift in the Routine Practice of Clinical Microbiology. Clinical Microbiology Reviews. 2013;26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grove KJ, Frappier SL, Caprioli RM. Matrix Pre-Coated MALDI MS Targets for Small Molecule Imaging in Tissues. Journal of the American Society for Mass Spectrometry. 2011;22:192–5. doi: 10.1007/s13361-010-0013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Caprioli RM. Matrix Precoated Targets for Direct Lipid Analysis and Imaging of Tissue. Analytical Chemistry. 2013;85:2907–12. doi: 10.1021/ac303554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McDonnell LA, van Remoortere A, van Zeijl RJM, Dalebout H, Bladergroen MR, Deelder AM. Automated imaging MS: Toward high throughput imaging mass spectrometry. Journal of Proteomics. 2010;73:1279–82. doi: 10.1016/j.jprot.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 46.Prideaux B, Stoeckli M. Mass spectrometry imaging for drug distribution studies. Journal of Proteomics. 2012;75:4999–5013. doi: 10.1016/j.jprot.2012.07.028. [DOI] [PubMed] [Google Scholar]
- 47.Reich RF, Cudzilo K, Levisky JA, Yost RA. Quantitative MALDI-MSn Analysis of Cocaine in the Autopsied Brain of a Human Cocaine User Employing a Wide Isolation Window and Internal Standards. Journal of the American Society for Mass Spectrometry. 2010;21:564–71. doi: 10.1016/j.jasms.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. International Journal of Mass Spectrometry. 2007;260:195–202. [Google Scholar]
- 49.Pirman DA, Kiss A, Heeren RMA, Yost RA. Identifying Tissue-Specific Signal Variation in MALDI Mass Spectrometric Imaging by Use of an Internal Standard. Analytical Chemistry. 2013;85:1090–6. doi: 10.1021/ac3029618. [DOI] [PubMed] [Google Scholar]
- 50.Reyzer ML, Hsieh YS, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. Journal of Mass Spectrometry. 2003;38:1081–92. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 51.Pirman DA, Reich RF, Kiss A, Heeren RMA, Yost RA. Quantitative MALDI Tandem Mass Spectrometric Imaging of Cocaine from Brain Tissue with a Deuterated Internal Standard. Analytical Chemistry. 2013;85:1081–9. doi: 10.1021/ac302960j. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh Y, Casale R, Fukuda E, Chen JW, Knemeyer I, Wingate J, et al. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Communications in Mass Spectrometry. 2006;20:965–72. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 53.Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos J-M, Bouzom F, et al. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. Journal of Proteomics. 2012;75:4952–61. doi: 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 54.Pirman DA, Yost RA. Quantitative Tandem Mass Spectrometric Imaging of Endogenous Acetyl-L-carnitine from Piglet Brain Tissue Using an Internal Standard. Analytical Chemistry. 2011;83:8575–81. doi: 10.1021/ac201949b. [DOI] [PubMed] [Google Scholar]
- 55.Groseclose MR, Castellino S. A Mimetic Tissue Model for the Quantification of Drug Distributions by MALDI Imaging Mass Spectrometry. Analytical Chemistry. 2013;85:10099–106. doi: 10.1021/ac400892z. [DOI] [PubMed] [Google Scholar]
- 56.Trim PJ, Henson CM, Avery JL, McEwen A, Snel MF, Claude E, et al. Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. Analytical Chemistry. 2008;80:8628–34. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 57.Shahidi-Latham SK, Dutta SM, Conaway MCP, Rudewicz PJ. Evaluation of an Accurate Mass Approach for the Simultaneous Detection of Drug and Metabolite Distributions via Whole-Body Mass Spectrometric Imaging. Analytical Chemistry. 2012;84:7158–65. doi: 10.1021/ac3015142. [DOI] [PubMed] [Google Scholar]
- 58.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Analytical Chemistry. 2006;78:6448–56. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 59.Drexler DM, Tannehill-Gregg SH, Wang L, Brock BJ. Utility of quantitative whole-body autoradiography (QWBA) and imaging mass spectrometry (IMS) by matrix-assisted laser desorption/ionization (MALDI) in the assessment of ocular distribution of drugs. Journal of Pharmacological and Toxicological Methods. 2011;63:205–8. doi: 10.1016/j.vascn.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 60.Attia AS, Schroeder KA, Seeley EH, Wilson KJ, Hammer ND, Colvin DC, et al. Monitoring the Inflammatory Response to Infection through the Integration of MALDI IMS and MRI. Cell Host & Microbe. 2012;11:664–73. doi: 10.1016/j.chom.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen R, Hui L, Sturm RM, Li L. Three Dimensional Mapping of Neuropeptides and Lipids in Crustacean Brain by Mass Spectral Imaging. Journal of the American Society for Mass Spectrometry. 2009;20:1068–77. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nature Methods. 2008;5:101–8. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 63.Alexandrov T. MALDI imaging mass spectrometry: statistical data analysis and current computational challenges. Bmc Bioinformatics. 2012:13. doi: 10.1186/1471-2105-13-S16-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]