Abstract
The present study examined genetic and shared environment contributions to quantitatively-measured autism symptoms and categorically-defined ASD. Participants included 568 twins from the Interactive Autism Network. Autism symptoms were obtained using the Social Communication Questionnaire and Social Responsiveness Scale. Categorically-defined ASD was based on clinical diagnoses. DeFries-Fulker and liability threshold models examined etiologic influences. Very high heritability was observed for extreme autism symptom levels (h2g=.92-1.20). Extreme levels of social and repetitive behavior symptoms were strongly influenced by common genetic factors. Heritability of categorically-defined ASD diagnosis was comparatively low (.21, 95% CI=0.15-0.28). High heritability of extreme autism symptom levels confirms previous observations of strong genetic influences on autism. Future studies will require large, carefully ascertained family pedigrees and quantitative symptom measurements.
Keywords: autism, twins, genetic, heritability, environment, diagnosis
Introduction
Autism spectrum disorders (ASD) represent a diverse set of neurodevelopmental disorders with a wide range of presentations. DSM-5 criteria, supported by recent investigations (Frazier et al. 2010; Frazier et al. 2012; Mandy et al. 2012), define ASD as a broad category with two symptom dimensions – social communication/interaction (SCI) and restricted/repetitive behavior (RRB). Specific genetic variations contributing to ∼15-20% of ASD cases have been identified, primarily through relationships with genetic syndromes (Abrahams and Geschwind 2008), copy number variation (Levy et al. 2011; Sanders et al. 2011a; Sebat et al. 2007), and small-scale gene-disrupting variants (Iossifov et al. ; Jiang et al. ; Neale et al.; O'Roak et al. 2011; O'Roak et al. ; Sanders et al. 2012; Schaaf et al. 2011). It is possible that, as emerging bioinformatics technologies are applied and refined, a substantial proportion of idiopathic cases will harbor causative variation. However, at present, the etiology of the majority of ASD cases remains unknown. Epigenetic (Gregory et al. 2009; Hu et al. 2006) and environmental effects (Newschaffer et al. 2002) may also contribute to ASD. Epidemiological studies have identified a host of risk factors, including increased parental age (Grether et al. 2009; Reichenberg et al. 2006; Shelton et al. 2010), neonatal complications (Bilder et al. 2009; Gardener et al. 2011; Schmidt et al.), and environmental exposures (McCanlies et al. ; Windham et al. 2006). However, these findings have tended to result in small increases in the risk of ASD (Newschaffer et al. 2007; Newschaffer et al. 2002), are difficult to replicate (Croen et al. 2005; Holmes et al. 2003; Ip et al. 2004), or may operate via increased risk of germline mutation in offspring (Zhao et al. 2007). As a result, the presence, type, and magnitude of environmental influences on the development of ASD remain uncertain.
Clarifying etiologic factors is crucial for guiding future research. Twin studies have the potential to inform the relative contributions of genetic and environment factors to ASD etiology (Ronald and Hoekstra 2011). Early twin studies supported a strong genetic etiology to ASD (Bailey et al. 1995; Folstein and Rutter 1977; Ritvo et al. 1985; Steffenburg et al. 1989). More recent diagnostic concordance studies have confirmed high monozygotic concordance for ASD (>88%), but also higher than previously appreciated dizygotic (>30%) and sibling (>15%) concordance rates (Ozonoff et al. 2011; Rosenberg et al. 2009; Taniai et al. 2008). Observations of high dizygotic concordance have raised the prospect of non-trivial environmental contributions to ASD. A recent study by Hallmayer and colleagues (Hallmayer et al. 2011), using a large, carefully ascertained ASD-affected twin sample, identified a substantial environmental contribution to ASD diagnosis (∼58%), although other studies have found minimal shared environmental effects (Bailey et al. 1995; Lichtenstein et al. 2010). Population and community-based quantitative trait studies conducted over the last 12 years have generally supported very strong heritability (40-87%), with a modest shared environment component (0-32%) (Ronald and Hoekstra 2011). Literature discrepancies likely reflect measurement, statistical, and sampling differences across studies. For example, Hallmayer et al. examined a liability threshold model for categorically-defined ASD using an affected twin sample. In contrast, population and community studies have modeled quantitatively-assessed autism symptoms, and presumably included only a minority of ASD-affected twin pairs distributed at the extreme trait levels (Constantino and Todd 2000; Constantino and Todd 2003; Constantino and Todd 2005; Ronald et al. 2006a; Ronald et al. 2005; Ronald et al. 2006b; Skuse et al. 2005; Stilp et al. 2010). The present study clarifies these methodological factors by simultaneously evaluating both quantitatively-assessed symptoms and categorically-defined ASD, using DeFries-Fulker regression and liability threshold models, and focusing on a large ASD-affected twin sample that also includes non-ASD twin pairs.
The two predominant behavioral genetic approaches to examining ASD etiology - assessing autism symptoms in the population versus concordance of ASD diagnoses in clinical samples - mirror two distinct views of ASD. The first viewpoint proposes that autism symptoms are best represented dimensionally (Constantino 2009), with broad autism traits being intermediate between severe and typical symptom levels. In the dimensional model, differences between typical and ASD symptom levels are a matter of degree (i.e., no distinct ASD category is present). The majority of data from population studies of quantitatively-assessed autism traits support a dimensional conceptualization (Ronald and Hoekstra 2011). In these studies, heritability estimates are generally consistent across typical and extreme symptom levels (Lundstrom et al. 2012; Ronald et al. 2006a) and independent genetic effects influence each symptom domain (Robinson et al. 2012; Ronald et al. 2006a; Ronald et al. 2008).
The second viewpoint is that ASD represents a natural symptom category with related SCI and RRB sub-domains (Frazier et al. 2012; Mandy et al. 2012). In this model, ASD-affected and unaffected individuals show qualitative differences in symptom levels and relatives with broad phenotypic traits (Losh et al. 2009) could be considered sub-threshold for ASD. The categorical view of ASD has been supported by recent empirical investigations of autism symptoms (Frazier et al. 2010; Frazier et al. 2012) and a population study of toddler twin pairs that found higher heritability for a more extreme threshold (Stilp et al. 2010). Understanding the genetic and environmental architecture of ASD will be crucial to resolving these views and speeding the search for etiologic mechanisms.
The present study evaluated genetic and environmental influences using the largest ASD-affected twin sample ascertained to date. Specific aims were: 1) To characterize and compare the heritable and environmental components of quantitatively-assessed autism symptoms and categorically-defined ASD, 2) to determine whether the magnitude of heritability estimates is consistent between extreme (group heritability) and typical symptom levels (individual differences heritability), and 3) to estimate the magnitude of common and independent heritable influences on SCI and RRB symptom domains.
Methods
Participants
Twin pairs with an ASD-affected member (ASD twins) and pairs without an ASD-affected member (non-ASD twins) were selected from the Interactive Autism Network (IAN) registry (IAN Data Export ID: IAN_DATA_2011-08-01). Data from 568 twin pairs (1136 youth), including 471 ASD-affected and 97 non-ASD pairs were available. The proportion of twins in IAN (6.9%) is higher than population expectations (1.8-3.2%) (Martin et al. 2012), consistent with observations of increased rates of ASD in twins (Zachor and Ben Itzchak 2011). The proportion of monozygotic twins in IAN (22.5%) was similar to rates identified in the population (12-20%) (Bortolus et al. 1999) and the proportion of dizygotic opposite sex twins in IAN (42.2%) was only slightly lower than population expectation (∼50%). Twin zygosity was reported by caregivers and higher-order multiple births were excluded. ASD was defined by collapsing specific DSM-IV-TR diagnoses following current epidemiologic surveillance protocols maintained by the U.S. Centers for Disease Control (Rice 2009). Online Supplement 1 provides a detailed description of the IAN registry and clinical ASD diagnoses.
Informed consent was obtained from parents/guardians before entry into IAN. Use of IAN data for the present study was reviewed and approved by the institutional review board of the Cleveland Clinic.
ASD Measures
Autism symptom data were provided using Social Communication Questionnaire (SCQ) (Rutter et al. 2003) and the Social Responsiveness Scale (SRS) (Constantino and Gruber 2005). The SCQ is a dichotomously keyed rating scale tapping DSM-IV-TR symptoms. The SRS is a 65-item, ordinally-scaled (1=“not true” to 4=“almost always true”) questionnaire that provides a quantitative assessment of autism traits. Convergence of findings from these measure increases confidence that results are not simply due to the measurement scale or item content.
Categorical ASD was defined in three ways: 1) caregiver-reported clinical ASD diagnoses, 2) SCQ total raw score ≥ 15, and 3) SRS total t-score ≥ 70. Quantitative autism symptoms were assessed using total raw scores on the SCQ, total T-scores on the SRS, and SCI and RRB domain scores derived from each instrument. Total raw scores for the SCQ and T-scores for the SRS were computed based on the published scoring (Constantino and Gruber 2005; Rutter et al. 2003). SCI and RRB domain scores were computed following published latent structure modeling (Frazier et al. 2012) and guidance from proposed DSM-5 criteria (American Psychiatric Association - DSM-5 Development 2011). SCI and RRB domain scores are useful for determining whether these domains are driven by common or unique etiologic factors (Online Supplement 2).
Statistical Approach
Descriptive statistics and Pearson chi-square analyses characterized the sample and compared zygosity groups (MZ-monozygotic, DZSS-dizygotic same sex, DZOS-dizygotic opposite sex). Possible differences between the SCQ and SRS sub-samples and the full IAN twin sample were evaluated using independent samples t-tests comparing twin pairs with complete data (SCQ 333 pairs, SRS 179 pairs) versus pairs with one or both twins having missing data (SCQ 235 pairs, SRS 389 pairs).
To examine etiologic influences on quantitatively-assessed autism symptoms, DeFries-Fulker (DF) regression analyses (Cherney et al. 1992; DeFries and Fulker 1985) of the total and domain-specific scores were computed in extreme sub-samples (>97th and >99th population percentiles) and in ASD-diagnosed twin pairs (Online Supplement 3). Basic and augmented DF models estimated: twin similarity independent of zygosity (B1), group heritability (h2g; B2), shared environment (c2; B3), the difference between group heritability and individual differences heritability (h2g - h2; B4), and individual differences heritability (h2; B5). DF models were computed following procedures described by Stevenson (Stevenson 1992), using a zygosity-specific mean transformation (DeFries and Fulker 1988), consistent with recent studies (Robinson et al. 2011; Robinson et al. 2012). Using these transformed scores, it is theoretically possible to obtain heritability estimates greater than 1.0 if the difference in regression to the mean for MZ and DZ cotwins is very large. DF regression analyses were used rather than model fitting (Purcell and Sham 2003) for direct comparison to other studies using DF models (Lundstrom et al. 2012). Because the non-ASD group was small (k=49 pairs) and extreme DF analyses were under-powered, this group was not included in the above extremes analyses. Instead, we only included the non-ASD twin pairs when examining changes in group heritability across increasingly extreme scores using quantile regression DF analyses (Logan et al. 2012). These analyses have better power because quantile estimates are based on the full sample.
Common and unique genetic influences between SCI and RRB domains were estimated using a modified version of the basic DF model (Ronald et al. 2006b; Stevenson et al. 1993). In these analyses, the proband's SCI score was used to predict the co-twin's RRB score and vice versa. Bivariate and group heritability estimates were used to compute genetic correlations following Knopik et. al. (Knopik et al. 1997). Genetic correlations (rg) evaluate the proportion of heritable influences that are common across SCI and RRB domains.
To estimate etiologic influences on categorically-defined ASD, we first computed probandwise concordances and 95% confidence intervals (Davie 1979) for MZ and DZ twins using clinical diagnosis, SCQ≥15, and SRS≥70 cutoffs. Next, liability threshold model parameters were calculated using maximum likelihood estimation (Online Supplement 4). These models were highly similar to those of Hallmayer et. al. (Hallmayer et al. 2011) with two exceptions. First, ascertainment was assumed to be complete (Online Supplement 5); all ASD-affected twins were probands (Rosenberg et al. 2009). Second, only MZ and DZSS probandwise concordances estimated model parameters. Opposite sex dizygotic pairs were not used. Sex-limited models with combinations of additive genetic, shared environment, and unique environment/error (ACE models) were estimated. It has been argued that a dominant transmission pattern exists in at least a subset of ASD cases (Zhao et al. 2007). Therefore, we also fit a model with additive genetic, dominant genetic, and unique environment (ADE). The model with all four components (ACDE) is not identifiable and cannot be estimated from the data. Standard errors and 95% confidence interval were calculated using a bootstrap approach with 1000 re-samplings.
Results
Sample Description
Table 1 presents demographic and clinical characteristics separately by zygosity group. Consistent with the larger IAN registry, twins were disproportionately described as white/non-Hispanic with highly educated parents. MZ twin pairs tended to have slightly less educated parents (Cohen's d=.28), possibly reflecting greater research engagement in caregivers with MZ twins. There were no significant zygosity differences in the proportion of ASD-affected twin pairs, age, race, or parent age. Not surprisingly, male twins were more prevalent in the MZ and DZSS groups, consistent with autism sex ratios (Centers for Disease Control and Prevention 2012). SCQ and SRS completion rates were lower in the MZ group. This raises the possibility that some MZ pairs may have missing questionnaire data for one twin due to strong phenotypic similarity (Rosenberg et al. 2009) and may result in more conservative estimates of heritability. Levels of autism symptoms on the SCQ and SRS were congruent with previous reports in ASD-affected and non-ASD youth on these measures (Chandler et al. 2007; Constantino and Todd 2003) and reflect a broad range of autism severity. SCQ and SRS sub-samples were highly similar to the larger IAN twin sample and missing SCQ and SRS data did not significantly influence results (Online Supplement 3).
Table 1.
Sample characteristics of 568 twin pairs by zygosity.
| MZ | DZ-same sex | DZ-opposite sex | ||
|---|---|---|---|---|
| Twin Pairs | M (SD) | M (SD) | M (SD) | χ2 / F (p) |
|
|
||||
| K | 128 | 254 | 186 | |
| ASD in one or both twins (%) | 110 (86%) | 210 (83%) | 151 (81%) | 1.23 (.541) |
| Concordant pairs (% of ASD pairs) | 84 (76%) | 71 (34%) | 27 (18%) | 95.54 (<.001) |
| Age of child | 10.6 (4.6) | 9.9 (4.0) | 9.8 (4.0) | 1.42 (.243) |
| Race (% white/Non-Hispanic) | 111 (87%) | 226 (89%) | 164 (88%) | 0.42 (.812) |
| Age of parent | 38.9 (6.4) | 39.3 (6.0) | 39.4 (6.3) | 0.20 (.800) |
| Parent education | 3.5 (1.0) | 3.8 (1.1) | 3.7 (1.0) | 3.55 (.029) |
|
|
||||
| Individual Twins | M (SD) | M (SD) | M (SD) | χ2 / F (p) |
|
|
||||
| N | 256 | 508 | 372 | |
| Individuals with ASD (%) | 194 (76%) | 281 (55%) | 178 (48%) | 50.17 (<.001) |
| Male N (%) | 204 (80%) | 376 (74%) | 186 (50%) | 79.01 (<.001) |
| Completed SCQ (%) | 155 (61%) | 346 (68%) | 257 (69%) | 5.77 (.056) |
| SCQ total – full sample | 18.2 (10.8) | 14.3 (12.0) | 13.5 (13.3) | 6.40 (.002) |
| SCQ total – ASD-affected | 22.0 (8.7) | 20.6 (9.2) | 21.3 (11.6) | 0.58 (.562) |
| SCQ total – non-ASD | 7.9 (9.2) | 6.6 (10.6) | 6.2 (10.4) | 0.35 (.703) |
| Completed SRS (%) | 78 (30%) | 182 (36%) | 149 (40%) | 6.06 (.048) |
| SRS total T – full sample | 71.7 (23.0) | 66.4 (24.1) | 66.9 (23.7) | 1.18 (.308) |
| SRS total T – ASD-affected | 79.2 (19.2) | 77.7 (21.3) | 81.5 (19.9) | 0.64 (.530) |
| SRS total T – non-ASD | 51.9 (20.6) | 53.6 (20.4) | 53.9 (18.7) | 0.72 (.930) |
Note: MZ=monozygotic twin pairs, DZ-same sex=dizygotic same-sex twin pairs, DZ-opposite sex=dizygotic opposite-sex twin pairs. Parent education is coded 1=did not finish high school, 2=high school diploma or equivalent, 3=some college or associate's degree, 4=bachelor's degree, 5=master's degree, 6=doctoral or professional degree. SCQ total = Social Communication Questionnaire total raw score. SRS total = Social Responsiveness Scale total raw score. Concordant pairs is based on the clinical DSM diagnosis.
Quantitatively-Assessed Autism Symptoms
Table 2 presents SCQ and SRS raw scores, z-scores, and transformed scores for extreme groups. Inspection of the MZ and DZ co-twin means reveals substantial regression to the population mean in the DZ co-twins relative to MZ co-twins (Figure 1). Table 3 presents extreme group intra-class correlations and basic DF model results. Intra-class correlations were consistently 1.5 to 2 times higher in MZ relative to DZ twins, suggesting substantial genetic influences in the ASD and extreme scoring groups. Group heritability estimates were large and highly significant for both SCQ (B2=.92-1.07, p<.001) and SRS (B2=1.01-1.20, p<.001) when extreme sub-samples and ASD-affected twins were selected. The very large extreme group heritability estimates were plausible, with confidence intervals spanning 1.0. As expected, these values are almost exactly twice the difference of the transformed co-twin scores in Table 2. The reason for these large values is even more clearly seen in the large regression to the mean in DZ co-twins but not MZ co-twins (Figure 2) (DeFries and Fulker 1988). Removing DZOS pairs only slightly altered group heritability estimates (SCQ h2g=.71-.88; SRS h2g=.94-1.13). Further constraining heritability estimates to the MZ cotwin mean continued to produce very high heritability estimates (SCQ h2g=.52-.73; SRS h2g=.70-.97).
Table 2.
Untransformed and transformed MZ and DZ co-twin means and standard deviations for SCQ and SRS scores, across extreme group and non-ASD twins.
| Untransformed Means | Transformed Means | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Measure | Sub-sample | Twin Pairs | Cut Scores | MZ proband | MZ cotwin | DZ proband | DZ cotwin | MZ cotwin | DZ cotwin |
| MZ/DZ | M (z) | M (z) | M (z) | M (z) | M | M | |||
| SCQ (raw score) | |||||||||
| ASD | 52/232 | - | 23.3 (3.7) | 20.2 (3.0) | 22.4 (3.5) | 10.1 (1.0) | .83 | .29 | |
| SCQ 99th% | 45/168 | 21 | 26.8 (4.4) | 20.9 (3.2) | 26.2 (4.2) | 10.8 (1.2) | .73 | .27 | |
| SCQ 97th% | 50/217 | 15 | 25.1 (4.0) | 20.8 (3.2) | 23.8 (3.8) | 10.5 (1.1) | .78 | .29 | |
| SRS (T-score) | |||||||||
| ASD | 26/126 | - | 81.9 (3.2) | 77.3 (2.7) | 86.2 (3.6) | 59.3 (0.9) | .85 | .26 | |
| SRS ≥99th% | 17/88 | 81 | 95.0 (4.5) | 83.5 (3.3) | 95.7 (4.6) | 60.8 (1.1) | .74 | .24 | |
| SRS ≥97th% | 21/113 | 70 | 88.4 (3.8) | 82.9 (3.3) | 89.6 (4.0) | 61.6 (1.2) | .86 | .30 | |
Note. SCQ cutoffs are based on raw scores, while SRS cutoffs are based on T-scores. ASD and non-ASD groups are derived from clinical diagnoses. Untransformed means are presented in raw score metric and after computing z-scores for all twins using the population mean and standard deviation. Transformed means divide co-twin z-scores by proband z-scores. Thus, transformed proband means are 1.0 for both zygosities.
Figure 1.
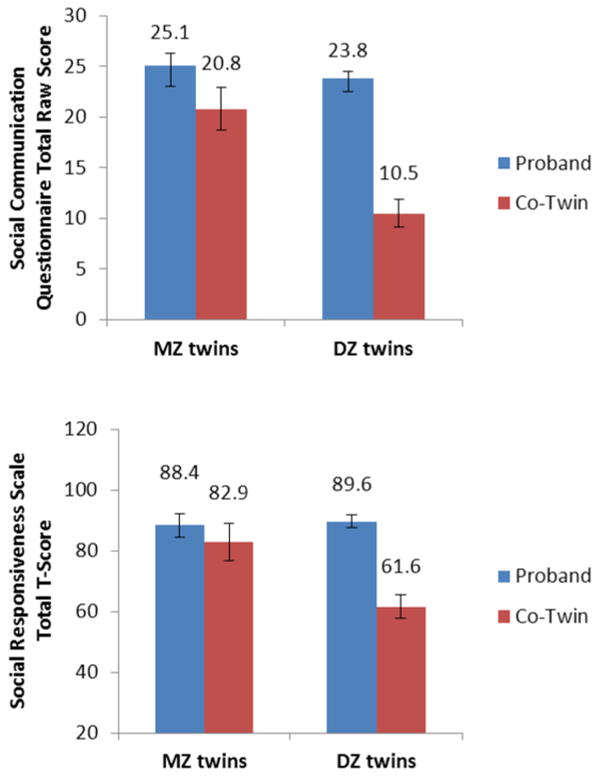
SCQ total raw and SRS total T-scores (M +/− 95% CIs) from MZ and DZ probands with extreme scores (≥97th percentile) and their co-twins.
Table 3.
Extreme group correlations, twin similarity, and group heritability for SCQ and SRS total scores.
| Extreme Group | Basic DF Model | |||||
|---|---|---|---|---|---|---|
| Intra-Class Correlations | ||||||
|
| ||||||
| Measure | Sub-sample | Twin Pairs | MZ twins | DZ twins | Twin similarity | Group heritability (h2g) |
| MZ/DZ | r | r | B1 (SE) | B2 (SE) | ||
| SCQ | ||||||
| ASD | 52/232 | .38 | .16 | .14 (−.04, .32) | 1.07 (.75, 1.39)* | |
| SCQ 99th% | 45/168 | .17 | .05 | .12 (−.22, .46) | .92 (.62, 1.22)* | |
| SCQ 97th% | 50/217 | .27 | .12 | .11 (−.13, .35) | .98 (.66, 1.30)* | |
| SRS | ||||||
| ASD | 26/126 | .64 | .23 | .31 (.09, .53)ˆ | 1.20 (.74, 1.66)* | |
| SRS ≥99th% | 17/88 | .26 | .08 | .18 (−.07, .43) | 1.01 (.53, 1.49)* | |
| SRS ≥97th% | 21/113 | .37 | .15 | <.01 (−.32, .33) | 1.12 (.64, 1.60)* | |
Note. SCQ=Social Communication Questionnaire. SRS=Social Responsiveness Scale, SCI=Social Communication and Interaction, RRB=Restricted, Repetitive Behavior. SE=corrected standard error.
p<.05
p<.001.
Figure 2.
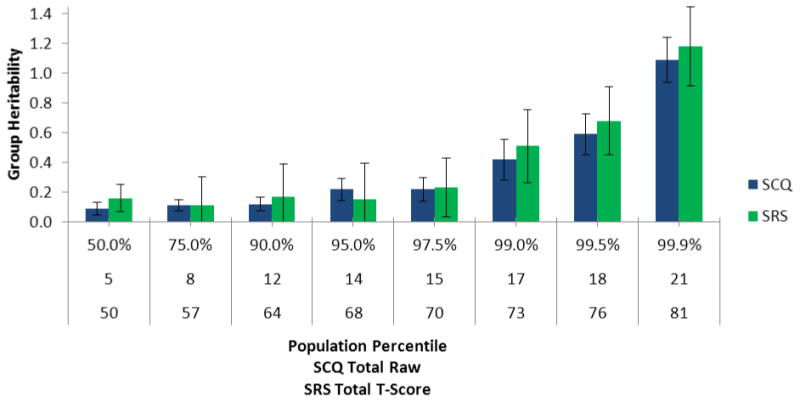
Increases in group heritability across levels of quantitatively-assessed autism symptoms.
To examine changes in heritability across the continuum of scores in this sample, quantile regression DF analyses were computed. These analyses were powered to detect moderate differences in heritability (≥.50) across score levels. Results indicated large increases in group heritability across levels of each quantitative symptom measure using quantile regression (Figure 2). Group heritability estimates increased dramatically in the range of recent ASD prevalence estimates (∼1%). For the augmented DF model, shared environment (B3) and individual differences heritability (B5) coefficients tended to be small and non-significant for most sub-samples (Online Supplement 6). Table 4 presents results for domain-specific DF models. The same pattern of findings emerged for SCI and RRB domains. Group heritability was very strong and statistically significant for extreme sub-samples. Individual differences heritability and shared environment were weaker and non-significant (Online Supplement 7).
Table 4.
Extreme group correlations, twin similarity, and group heritability for SCI and RRB symptom domains.
| Extreme Group | Basic DF Model | |||||
|---|---|---|---|---|---|---|
| Intra-Class Correlations | ||||||
|
| ||||||
| Measure | Sub-sample | Twin Pairs | MZ twins | DZ twins | Twin similarity | Group heritability (h2g) |
| MZ/DZ | r | r | B1 (95% CI) | B2 (95% CI) | ||
| SCQ – SCI | ||||||
| ASD | 52/232 | .40 | .23 | .16 (.02, .30)ˆ | .99 (.66, 1.32)* | |
| SCI ≥99th% | 45/162 | .18 | .09 | .29 (−.01, .58) | .77 (.47, 1.07)* | |
| SCI ≥97th% | 46/195 | .19 | .13 | .17 (−.05, .39) | .83 (.52, 1.14)* | |
| SCQ – RRB | ||||||
| ASD | 52/232 | .42 | .19 | .20 (.02, .38)ˆ | .99 (.69, 1.29)* | |
| RRB ≥99th% | 43/169 | .06 | .07 | .17 (−.23, .57) | .93 (.67, 1.19)* | |
| RRB ≥97th% | 47/212 | .16 | .12 | .09 (−.17, .35) | .96 (.67, 1.25)* | |
| SRS – SCI | ||||||
| ASD | 26/126 | .64 | .25 | .33 (.11, .55)ˆ | 1.21 (.73, 1.69)* | |
| SCI ≥99th% | 15/72 | .22 | .07 | .47 (−.16, 1.10) | .84 (.36, 1.32)* | |
| SCI ≥97th% | 20/98 | .35 | .13 | .23 (−.16, .62) | 1.04 (.58, 1.50)* | |
| SRS – RRB | ||||||
| ASD | 26/126 | .68 | .18 | .22 (.01, .43)ˆ | 1.18 (.75, 1.61)* | |
| RRB ≥99th% | 19/97 | .43 | .07 | .01 (−.36, .38) | 1.07 (.63, 1.51)* | |
| RRB ≥97th% | 23/115 | .57 | .12 | .08 (−.21, .37) | 1.14 (.70, 1.58)* | |
Note. SCQ=Social Communication Questionnaire. SRS=Social Responsiveness Scale, SCI=Social Communication and Interaction, RRB=Restricted, Repetitive Behavior. SE=corrected standard error.
p<.05
p<.001.
Table 5 presents cross-construct DF models. Bivariate genetic correlations were very high and statistically significant across each domain and sub-sample. In each case, these correlations approached group heritability estimates from basic DF analyses. Genetic correlations were very high (SCI-RRB rg=.84-.99).
Table 5.
Twin similarity and bivariate heritability for cross-construct analyses.
| Measure | Sub-sample | Twin Pairs | Twin similarity | Bivariate heritability (h2g) |
|---|---|---|---|---|
| MZ/DZ | B1 (95% CI) | B2 (95% CI) | ||
| SCQ – SCI predicts RRB | ||||
| ASD | 284 | −.01 (−.13, .11) | 1.00 (.70, 1.30)* | |
| ≥97th% | 241 | −.01 (−.22, .20) | .87 (.57, 1.17)* | |
| SCQ – RRB predicts SCI | ||||
| ASD | 284 | −.11 (−.30, .08) | 1.00 (.66, 1.34)* | |
| ≥97th% | 259 | −.30 (−.57, .03) | .81 (.50, 1.11)* | |
| SRS – SCI predicts RRB | ||||
| ASD | 152 | .23 (.03, .43)ˆ | 1.18 (.74, 1.62)* | |
| ≥97th% | 118 | .16 (−.24, .56) | 1.12 (.64, 1.60)* | |
| SRS – RRB predicts SCI | ||||
| ASD | 152 | .23 (−.01, .46) | 1.21 (.71, 1.71)* | |
| ≥97th% | 138 | .05 (−.24, .34) | .99 (.55, 1.43)* | |
p<.05
p<.001.
SCQ=Social Communication Questionnaire. SRS=Social Responsiveness Scale. SCI=Social communication/interaction. RRB=Restricted/Repetitive Behavior.
Categorically-Defined ASD
Online Supplement 8 presents probandwise concordances across the three definitions of categorical ASD. MZ concordances were substantial across definitions (range=.50-.88). DZSS and DZOS concordances were smaller and variable (DZSS range=.17-.54, DZOS range=.22-.48), similar to those seen in a smaller subset of IAN (Rosenberg et al. 2009). Table 6 presents ACE model estimates for additive genetic, shared environment, and individual environment/error influences on categorical ASD. Results indicated very high shared environment estimates. Small, but significant, additive genetic estimates (0.21, 95% CI=0.15-0.28) were seen for caregiver-reported ASD diagnosis. Additive genetic effects and the associated confidence intervals increased slightly for SCQ≥15 (0.26, 95% CI=0.16-0.39) and were larger still for SRS≥70 (0.35, 95% CI=0.20-0.56). Results for the ADE models (not shown) had implausible negative estimates of the dominance component, which is expected in twin models if a shared environmental component is present in the data.
Table 6.
ACE model results for caregiver-reported ASD diagnosis, SCQ≥15, and SRS≥70.
| A-female (SE) | A-male (SE) | C-female (SE) | C-male (SE) | LL | AIC | LR | p | ||
|---|---|---|---|---|---|---|---|---|---|
| Caregiver-reported ASD diagnosis | |||||||||
| ACE unequal | 0.31 (0.12) | 0.19 (0.04) | 0.68 (0.11) | 0.81 (0.04) | 0 | 8.00 | |||
| AE unequal | 0.99 (0.01) | 0.99 (0.01) | – | – | −72.07 | 148.13 | 144.13 | <.001 | |
| CE unequal | – | – | 0.91 (0.02) | 0.95 (0.01) | −26.48 | 56.97 | 52.97 | <.001 | |
| ACE equal | 0.21 (0.04) | 0.21 (0.04) | 0.78 (0.04) | 0.78 (0.04) | −1.23 | 6.46 | 2.46 | .293 | |
| AE equal | 0.99 (0.01) | 0.99 (0.01) | – | – | −72.34 | 146.69 | 144.69 | <.001 | |
| CE equal | – | – | 0.95 (0.01) | 0.95 (0.01) | −28.18 | 58.36 | 56.36 | <.001 | |
| E | – | – | – | – | −655.76 | 1311.52 | 1311.52 | ||
|
| |||||||||
| SCQ≥15 | |||||||||
| ACE unequal | 0.19 (0.14) | 0.28 (0.08) | 0.77 (0.13) | 0.70 (0.08) | 0.0 | 8.00 | |||
| AE unequal | 0.98 (0.02) | 0.98 (0.01) | – | –- | −28.55 | 61.10 | 57.10 | <.001 | |
| CE unequal | – | – | 0.89 (0.04) | 0.91 (0.02) | −9.12 | 22.24 | 18.24 | <.001 | |
| ACE equal | 0.26 (0.07) | 0.26 (0.07) | 0.71 (0.06) | 0.71 (0.06) | −0.22 | 4.43 | 0.43 | .805 | |
| AE equal | 0.98 (0.01) | 0.98 (0.01) | – | – | −28.60 | 59.20 | 57.20 | <.001 | |
| CE equal | – | – | 0.90 (0.02) | 0.90 (0.02) | −9.17 | 20.35 | 18.35 | <.001 | |
| E | – | – | – | – | −252.64 | 505.29 | 505.29 | ||
|
| |||||||||
| SRS≥70 | |||||||||
| ACE unequal | 0.50 (0.35) | 0.32 (0.12) | 0.40 (0.30) | 0.67 (0.12) | 0 | 8.00 | |||
| AE unequal | 0.93 (0.30) | 0.99 (0.01) | – | – | −10.35 | 24.69 | 20.69 | <.001 | |
| CE unequal | – | – | 0.75 (0.25) | 0.91 (0.02) | −7.77 | 19.54 | 15.54 | <.001 | |
| ACE equal | 0.35 (0.11) | 0.35 (0.11) | 0.64 (0.11) | 0.64 (0.11) | −1.49 | 6.97 | 2.97 | .226 | |
| AE equal | 0.99 (0.01) | 0.99 (0.01) | – | – | −11.28 | 24.57 | 22.57 | <.001 | |
| CE equal | – | – | 0.90 (0.03) | 0.90 (0.03) | −9.04 | 20.08 | 18.08 | <.001 | |
| E | – | – | – | – | −116.36 | 232.72 | 232.72 | ||
Note: LL=Log-Likelihood scaled so that the saturated model has a value of 0. AIC=Akaike Information Criterion. LR=Likelihood Ratio test. Bolded models are best fitting. SCQ≥15=Social Communication Questionnaire raw scores greater than or equal to 15. SRS≥70=Social Responsiveness Scale t-scores greater than or equal to 70. A-female = additive genetic variance for females. A-male=additive genetic variance for males. C-female =common environment or dominant genetic variance for females. C-male =common environment or dominant genetic variance for males. ACE=additive genetic, common environment, error/unique environment models. Error/unique environment estimates simply represent the remaining variance in liability threshold models. Unequal=sex-specific parameter estimates. Equal=parameter estimates constrained to equality across males and females.
Discussion
To our knowledge, this study represents the largest sample of ASD-affected twin pairs that simultaneously includes both quantitative and categorical approaches to autism measurement. Four major findings emerged: 1) Extreme levels of quantitatively-measured autism symptoms were strongly heritable with no significant shared environment. 2) Less extreme autism symptom levels showed lower heritability. 3) SCI and RRB symptoms had high genetic correlations, indicating that extreme scores on these domains are driven by common genetic sources. 4) Liability threshold model estimates of additive genetic effects tended to be much lower, but varied depending on model selection (i.e., ACE vs. AE vs. CE models). Each of these results has substantial implications for etiologic models of autism.
High extreme group heritability is consistent with previous quantitative symptom studies (Constantino and Todd 2005; Hoekstra et al. 2007; Ronald et al. 2006a; Ronald et al. 2005; Ronald et al. 2006b; Ronald et al. 2008; Skuse et al. 2005). However, heritability was smaller at less extreme symptom levels. While this contrasts with the majority of previous investigations (Lundstrom et al. 2012; Robinson et al. 2012), a recent study of autism in toddlers also identified stronger genetic effects at more extreme symptom levels (Stilp et al. 2010), and a population study of language ability reported greater heritability for impaired language levels (Spinath et al. 2004). There are several plausible explanations for the differences between the present study and the majority of population studies. The present study likely under-estimated individual differences heritability due to rater contrast. Caregivers of ASD-affected children may rate unaffected siblings as uniformly unimpaired. With this caveat in mind, it is important to note that the present study is methodologically quite different than previous population studies and therefore, complements them rather than contradicting. Quantile regression analyses in the current investigation were not estimating heritability of autism symptoms in the population, but rather in ASD-affected and non-ASD twin pairs, as discussed further in Online Supplement 5. Even using extreme cutoffs, previous population studies sampled mostly non-ASD twin pairs. For example, selecting scores ≤1%ile on a screening measure (Lundstrom et al. 2012; Robinson et al. 2011), less than half the twin pairs in this extreme sub-sample will have a member with categorically-defined ASD, even if all ASD-affected children score high. In the more likely scenario, where some ASD-affected twins have less extreme scores, the ability to detect differences in group heritability across increasingly more extreme scores is further diminished. This implies that previous population twin samples are more likely to identify continuity of heritability across score levels and may be under-powered to detect extreme group heritability explicitly attributable to ASD. In the present study, extreme symptom levels consisted exclusively of ASD-affected pairs.
Common genetic effects were the primary drivers of SCI and RRB symptoms in the present study. This also contrasts with previous population quantitative symptom studies, where genetic correlations tended to be more modest (Robinson et al. 2012; Ronald et al. 2006a). Again, this difference may reflect the presence of a large number of ASD-affected twin pairs in the present study. It is possible that SCI and RRB symptoms are separately influenced by a combination of polygenic, common environmental, and/or individual differences factors at typical population levels; but that powerful, pleiotropic effects drive extreme symptom levels. Findings of greater heritability at extreme symptom levels and common genetic effects across domains jointly point toward a categorical model of ASD. In this model, a qualitatively distinct symptom pattern is generated by strong, pleiotropic genetic influences. The categorical model has gained momentum from strong diagnostic stability across childhood (Chawarska et al. 2009; Lord et al. 2006; Moss et al. 2008) and empirically-driven symptom structure studies (Frazier et al. 2010; Frazier et al. 2012; Ingram et al. 2008). It is also consistent with recent findings of powerful genetic effects, such as CNVs (Glessner et al. 2009; Levy et al. 2011; Sanders et al. 2011b; Sebat et al. 2007) and rare gene-disrupting mutations (Iossifov et al. ; Jiang et al. ; O'Roak et al. 2011; Schaaf et al. 2011; Vaags et al. 2012), driving a non-trivial proportion of phenotypic variance in ASD. To date, these stronger genetic effects have been identified in a minority of ASD cases (10-25%). It is possible that, with emerging whole genome and bioinformatics technologies, a higher proportion of cases with mono- or oligogenic effects leading to autism will be identified. It is also conceivable that the remaining genetic effects are polygenic with a prominent phenotypic threshold effect or that the strong heritability observed in this study represents gene-environment interaction effects.
Intuitively, given the above findings for quantitatively-assessed autism symptoms, ACE model analyses of categorically-defined ASD would be expected to produce similarly large genetic effects. However, the opposite was observed. Categorically-defined ASD had low heritability and much higher estimates of shared environment. This finding was consistent with the recent Hallmayer study, which used similar corrections for ASD prevalence and proband ascertainment. However, there are several reasons to be very skeptical when interpreting results from liability threshold models of categorically-defined ASD. First, estimates from the liability threshold model are heavily influenced by small changes in categorical ASD classifications. For example, a subtle bias increasing ASD diagnosis would, in turn, increase DZ concordance, yielding smaller heritability and larger shared environment effects. Online Supplement 8 demonstrates the effect for only a handful of misclassifications using concordances obtained from Hallmayer and colleagues (Hallmayer et al. 2011). Second, higher shared environment estimates for categorical ASD may reflect correlated error within twin pairs. It is likely that clinical diagnoses are correlated within a twin pair for reasons beyond concordance. Diagnoses were often generated by the same evaluator/parent combination, and this may be true in other twin samples as well, even exerting influence when standardized instruments are administered. While correlated classification errors within twin pairs should theoretically be equally problematic for MZ and DZ pairs, in practice, the effect is likely to differentially increase DZ concordance and shared environment effects. This is because categorical ASD concordances are already quite high for MZ pairs - causing a ceiling effect, while DZ concordances have greater room to increase.
Third, in this data, liability threshold models produced unrealistically low estimates of unique environment and measurement error (often<3%). While it is possible to have a categorical phenotype strongly influenced by common environmental effects and not influenced by individual environment or error, a difference this large seems unlikely. This odd behavior of the threshold model is the result of assumptions about the underlying liability distribution. For example, it has been noted that fitting a model which assumes an underlying major gene effect to the data may yield entirely different results than the standard model, which assumes multivariate normality (Kidd and Cavalli-Sforza 1973). Similarly, it has been shown that numerous realistic scenarios for the distribution of the underlying liability can lead to large asymptotic biases, while small samples sizes may be biased even if the model assumptions are met (Benchek and Morris 2013). If, as we have argued, ASD is more of a categorical concept that is heavily influenced by highly penetrant alleles or a polygenic threshold, then there is no reason to trust that the classic ACE liability model will yield valid results because the liability distribution will differ strongly from multivariate normal. Furthermore, liability threshold results may be influenced by the combination of ascertainment and prevalence parameters in model estimation. Modest changes in these values can influence model results. On these grounds, ACE liability modeling studies of categorical ASD, including the present results, should be viewed cautiously.
Classical twin studies assume that common environments affects both monozygotic and dizygotic twins the same (Rijsdijk and Sham 2002). There is certainly reason to suspect that some of the known environmental risk factors for ASD may be differentially present in dizygotic twins. For example, there is evidence that maternal age (Sandin et al. 2012) and the use of assistive reproductive technology (Zachor and Ben Itzchak 2011) are risk factors for both dizygotic twinning and for ASD. Such factors would disproportionately increase the concordance rates among dizygotic twins leading to lower estimates of heritability. Perhaps more importantly, it is likely that many of these so called “environmental” risk factors act by increasing the mutational load. There is evidence that even monozygotic twins are not truly genetically “identical” (Bruder et al. 2008). Thus, even the relatively few cases of non-concordant monozygotic twins may be explained by genetic differences between the twins. It follows that a trait may be strongly genetically determined, but not particularly heritable.
Future behavioral genetic investigations of categorical ASD should consider the limitations of liability threshold models in the planning stages. Crucial factors will include consideration of ascertainment, prevalence, and recruitment of very large samples yielding greater precision (see Online Supplement 5 for additional discussion). These studies will also need to implement diagnostic procedures that accurately classify cases across the full autism spectrum and are administered by clinicians who are blinded to other family member's, reducing the potential for biased concordance. While this work will be labor-intensive and expensive, these methodological improvements are needed to advance our understanding of etiologic influences on categorical ASD.
Additional limitations of the present study included reliance on caregiver-reported ASD clinical diagnoses and zygosity, missing data for SCQ and SRS quantitative symptom measures, and the use of non-ASD twin pairs from ASD-affected families. Clinical ASD diagnoses are not as reliable and valid as diagnoses based on gold-standard semi-structured interviews or observational instruments. However, these gold-standard measures were often used as part of the diagnostic process and available data suggests that clinical diagnoses in IAN are quite accurate (Lee et al. 2010). Parent-reported zygosity is valid (Rietveld et al. 2000) and the proportions of MZ and DZ twins in this study were consistent with expectation and did not suggest a bias toward parents misclassifying DZ twins as MZ twins. Even if zygosity misclassifications existed, they should have decreased heritability estimates.
Missing questionnaire data were also potentially problematic. Fortunately, SCQ and SRS sub-samples did not markedly differ in terms of demographic and clinical features from the total sample and results were highly similar across the SCQ and SRS. This is comforting because the SCQ is derived from diagnostic criteria whereas the SRS is a quantitative trait instrument. Future work should include clinician ratings to exclude the possibility that findings are influenced by rater perspective. Lastly, the IAN registry does not include twins from families unaffected by ASD and higher SES families are over-represented. The latter factor may artificially inflate heritability estimates in DF models. Future work will need to include twin pairs from families without ASD and use samples more representative of SES in the broader population.
In spite of these limitations, the national scope and large number of ASD-affected twin pairs in the IAN registry provided a unique opportunity for evaluating etiologic influences on autism. Results supported strong heritability of extreme (clinical) levels of quantitatively-assessed autism symptoms, but also raise the possibility that extreme levels of symptoms may be substantially influenced by highly penetrant pleiotropic alleles or threshold effects rather than a graded polygenetic transmission. The current view of ASD genetics is a complex confluence of etiologies, including unique transmission patterns (de novo vs. inherited), different thresholds for males and females (Szatmari et al. 2012), and distinct mixtures of high and low penetrance genetic, epigenetic, and environmental influences. To assist in teasing out the relative importance of all these factors, large family studies using careful ascertainment methods are needed. Family pedigree designs measuring quantitative symptoms and categorical ASD can simultaneously evaluate additive genetic, dominant genetic, shared environment, and unique environment effects to clarify the relative importance of these distinct etiologic influences. Ultimately, behavioral genetics approaches will be most powerful when combined with comprehensive genomic studies that include sequence interrogation and gene expression.
Supplementary Material
Acknowledgments
The authors also wish to acknowledge the important contribution of the participants with autism, their siblings, and their families.
Funding/Support: This work was made possible by the Case Western Reserve University/Cleveland Clinic CTSA Grant Number UL1 RR024989 provided by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health. The IAN registry is supported by funding from Autism Speaks and the Kennedy Krieger Institute. The views and opinions expressed in this article are those of the authors and should not be construed to represent the views of any of the sponsoring organizations, agencies, or US government.
Footnotes
Author Contributions: All authors participated in framing the research question, designing the analysis plan, and/or interpreting results. Drs. Frazier and Thompson conducted the DeFries-Fulker analyses and Dr. Nathan Morris estimated liability threshold models. Dr. Frazier prepared the first draft of the manuscript. All authors participated in revising the manuscript after the first draft was prepared. Drs. Frazier and Morris take responsibility for the integrity of the data and accuracy of the data analysis.
Contributor Information
Thomas W. Frazier, Center for Autism and the Genomic Medicine Institute at the Cleveland Clinic
Lee Thompson, Department of Psychological Sciences at Case Western Reserve University.
Eric A. Youngstrom, Department of Psychology at the University of North Carolina at Chapel Hill
Paul Law, Department of Medical Informatics and the Interactive Autism Network at Kennedy Krieger Institute.
Antonio Y. Hardan, Department of Psychiatry and Behavioral Sciences at Stanford
Charis Eng, Genomic Medicine Institute at the Cleveland Clinic and Department of Genetics at Case Western Reserve University.
Nathan Morris, Department of Epidemiology and Biostatistics (NM) at Case Western Reserve University.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–55. doi: 10.1038/nrg2346. doi:nrg2346 [pii] 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association - DSM-5 Development. 299.00 Autistic Disorder. [Accessed February 01 2011];American Psychiatric Association. 2011 http://www.dsm5.org/ProposedRevisions/Pages/proposedrevision.aspx?rid=94.
- Bailey A, et al. Autism as a strongly genetic disorder: Evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- Benchek PH, Morris NJ. How meaningful are heritability estimates of liability? Hum Genet. 2013;132:1351–1360. doi: 10.1007/s00439-013-1334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder D, Pinborough-Zimmerman J, Miller J, McMahon W. Prenatal, perinatal, and neonatal factors associated with autism spectrum disorders. Pediatrics. 2009;123:1293–300. doi: 10.1542/peds.2008-0927. doi:123/5/1293 [pii] 10.1542/peds.2008-0927. [DOI] [PubMed] [Google Scholar]
- Bortolus R, Parazzini F, Chatenoud L, Benzi G, Bianchi MM, Marini A. The epidemiology of multiple births. Hum Reprod Update. 1999;5:179–87. doi: 10.1093/humupd/5.2.179. [DOI] [PubMed] [Google Scholar]
- Bruder CE, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–71. doi: 10.1016/j.ajhg.2007.12.011. doi:S0002-9297(08)00102-X [pii] 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Prevalence of Autism Spectrum Disorders — Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States, 2008. MMWR. 2012;61:1–19. [PubMed] [Google Scholar]
- Chandler S, et al. Validation of the social communication questionnaire in a population cohort of children with autism spectrum disorders. J Am Acad Child Adolesc Psychiatry. 2007;46:1324–32. doi: 10.1097/chi.0b013e31812f7d8d. doi:10.1097/chi.0b013e31812f7d8d S0890-8567(09)61851-7 [pii] [DOI] [PubMed] [Google Scholar]
- Chawarska K, Klin A, Paul R, Macari S, Volkmar F. A prospective study of toddlers with ASD: short-term diagnostic and cognitive outcomes. J Child Psychol Psychiatry. 2009;50:1235–45. doi: 10.1111/j.1469-7610.2009.02101.x. doi:JCPP2101 [pii] 10.1111/j.1469-7610.2009.02101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherney SS, DeFries JC, Fulker DW. Multiple regression analysis of twin data: A model-fitting approach. Behav Genet. 1992;22:489–497. doi: 10.1007/BF01066617. [DOI] [PubMed] [Google Scholar]
- Constantino JN. How continua converge in nature: cognition, social competence, and autistic syndromes. J Am Acad Child Adolesc Psychiatry. 2009;48:97–8. doi: 10.1097/CHI.0b013e318193069e. doi:10.1097/CHI.0b013e318193069e S0890-8567(09)60002-2 [pii] [DOI] [PubMed] [Google Scholar]
- Constantino JN, Gruber CP. Social Responsiveness Scale: Manual. Los Angeles, CA: Western Psychological Services; 2005. [Google Scholar]
- Constantino JN, Todd RD. Genetic structure of reciprocal social behavior. Am J Psychiatry. 2000;157:2043–2045. doi: 10.1176/appi.ajp.157.12.2043. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Todd RD. Autistic traits in the general population: A twin study. Arch Gen Psychiatry. 2003;60:524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- Constantino JN, Todd RD. Intergenerational transmission of subthreshold autistic traits in the general population. Biol Psychiatry. 2005;57:655–660. doi: 10.1016/j.biopsych.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Croen LA, Grether JK, Yoshida CK, Odouli R, Van de Water J. Maternal autoimmune diseases, asthma and allergies, and childhood autism spectrum disorders: a case-control study. Arch Pediatr Adolesc Med. 2005;159:151–7. doi: 10.1001/archpedi.159.2.151. doi:159/2/151 [pii] 10.1001/archpedi.159.2.151. [DOI] [PubMed] [Google Scholar]
- Davie AM. The ‘singles’ method for segregation analysis under incomplete ascertainment. Ann Hum Genet. 1979;42:507–512. doi: 10.1111/j.1469-1809.1979.tb00683.x. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Fulker DW. Multiple regression analysis of twin data. Behav Genet. 1985;15:467–473. doi: 10.1007/BF01066239. [DOI] [PubMed] [Google Scholar]
- DeFries JC, Fulker DW. Multiple regression analysis of twin data: Etiology of deviant scores versus individual differences. Acta Genet Med Gemellol (Roma) 1988;37:205–216. doi: 10.1017/s0001566000003810. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: A genetic study of 21 twin pairs. Journal of child psychology and psychiatry, and allied disciplines. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Frazier TW, et al. Autism spectrum disorders as a qualitatively distinct category from typical behavior in a large, clinically ascertained sample. Assessment. 2010;17:308–20. doi: 10.1177/1073191109356534. doi:1073191109356534 [pii] 10.1177/1073191109356534. [DOI] [PubMed] [Google Scholar]
- Frazier TW, et al. Validation of proposed DSM-5 criteria for autism spectrum disorder. J Am Acad Child Adolesc Psychiatry. 2012;51:28–40. doi: 10.1016/j.jaac.2011.09.021. e3 doi:S0890-8567(11)00890-2 [pii] 10.1016/j.jaac.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardener H, Spiegelman D, Buka SL. Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis. Pediatrics. 2011;128:344–55. doi: 10.1542/peds.2010-1036. doi:peds.2010-1036 [pii] 10.1542/peds.2010-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi: 10.1038/nature07953. doi:nature07953 [pii] 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory SG, et al. Genomic and epigenetic evidence for oxytocin receptor deficiency in autism. BMC Med. 2009;7:62. doi: 10.1186/1741-7015-7-62. doi:1741-7015-7-62 [pii] 10.1186/1741-7015-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether JK, Anderson MC, Croen LA, Smith D, Windham GC. Risk of autism and increasing maternal and paternal age in a large north American population. Am J Epidemiol. 2009;170:1118–26. doi: 10.1093/aje/kwp247. doi:kwp247 [pii] 10.1093/aje/kwp247. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–102. doi: 10.1001/archgenpsychiatry.2011.76. doi:archgenpsychiatry.2011.76 [pii] 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra RA, Bartels M, Verweij CJ, Boomsma DI. Heritability of autistic traits in the general population. Arch Pediatr Adolesc Med. 2007;161:372–7. doi: 10.1001/archpedi.161.4.372. doi:161/4/372 [pii] 10.1001/archpedi.161.4.372. [DOI] [PubMed] [Google Scholar]
- Holmes AS, Blaxill MF, Haley BE. Reduced levels of mercury in first baby haircuts of autistic children. Int J Toxicol. 2003;22:277–85. doi: 10.1080/10915810305120. doi:LP5KFYMAUK49Y7PM [pii] [DOI] [PubMed] [Google Scholar]
- Hu VW, Frank BC, Heine S, Lee NH, Quackenbush J. Gene expression profiling of lymphoblastoid cell lines from monozygotic twins discordant in severity of autism reveals differential regulation of neurologically relevant genes. BMC Genomics. 2006;7:118. doi: 10.1186/1471-2164-7-118. doi:1471-2164-7-118 [pii] 10.1186/1471-2164-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DG, Takahashi TN, Miles JH. Defining autism subgroups: a taxometric solution. J Autism Dev Disord. 2008;38:950–60. doi: 10.1007/s10803-007-0469-y. [DOI] [PubMed] [Google Scholar]
- Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 74:285–99. doi: 10.1016/j.neuron.2012.04.009. doi:S0896-6273(12)00340-6 [pii] 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip P, Wong V, Ho M, Lee J, Wong W. Mercury exposure in children with autistic spectrum disorder: case-control study. J Child Neurol. 2004;19:431–4. doi: 10.1177/088307380401900606. [DOI] [PubMed] [Google Scholar]
- Jiang YH, et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. Am J Hum Genet. doi: 10.1016/j.ajhg.2013.06.012. doi:S0002-9297(13)00281-4 [pii] 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd KK, Cavalli-Sforza LL. An analysis of the genetics of schizophrenia. Soc Biol. 1973;20:254–65. doi: 10.1080/19485565.1973.9988051. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Alarcon M, DeFries JC. Comorbidity of mathematics and reading deficits: evidence for a genetic etiology. Behav Genet. 1997;27:447–53. doi: 10.1023/a:1025622400239. [DOI] [PubMed] [Google Scholar]
- Lee H, et al. Accuracy of phenotyping of autistic children based on Internet implemented parent report. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1119–26. doi: 10.1002/ajmg.b.31103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–97. doi: 10.1016/j.neuron.2011.05.015. doi:S0896-6273(11)00396-5 [pii] 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Carlstrom E, Rastam M, Gillberg C, Anckarsater H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167:1357–63. doi: 10.1176/appi.ajp.2010.10020223. doi:appi.ajp.2010.10020223 [pii] 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- Logan JA, et al. Heritability across the distribution: an application of quantile regression. Behav Genet. 2012;42:256–67. doi: 10.1007/s10519-011-9497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Risi S, DiLavore PS, Shulman C, Thurm A, Pickles A. Autism from 2 to 9 years of age. Arch Gen Psychiatry. 2006;63:694–701. doi: 10.1001/archpsyc.63.6.694. doi:63/6/694 [pii] 10.1001/archpsyc.63.6.694. [DOI] [PubMed] [Google Scholar]
- Losh M, et al. Neuropsychological profile of autism and the broad autism phenotype. Arch Gen Psychiatry. 2009;66:518–26. doi: 10.1001/archgenpsychiatry.2009.34. doi:66/5/518 [pii] 10.1001/archgenpsychiatry.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom S, et al. Autism spectrum disorders and autistic like traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry. 2012;69:46–52. doi: 10.1001/archgenpsychiatry.2011.144. doi:69/1/46 [pii] 10.1001/archgenpsychiatry.2011.144. [DOI] [PubMed] [Google Scholar]
- Mandy WP, Charman T, Skuse DH. Testing the construct validity of proposed criteria for DSM-5 autism spectrum disorder. J Am Acad Child Adolesc Psychiatry. 2012;51:41–50. doi: 10.1016/j.jaac.2011.10.013. doi:S0890-8567(11)00951-8 [pii] 10.1016/j.jaac.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Martin JA, Hamilton BE, Osterman MJK. NCHS data brief. Vol. 80. Hyattsville, MD: National Center for Health Statistics; 2012. Three Decades of Twin Births in the United States, 1980-2009. [PubMed] [Google Scholar]
- McCanlies EC, et al. Parental Occupational Exposures and Autism Spectrum Disorder. J Autism Dev Disord. doi: 10.1007/s10803-012-1468-1. [DOI] [PubMed] [Google Scholar]
- Moss J, Magiati I, Charman T, Howlin P. Stability of the autism diagnostic interview-revised from pre-school to elementary school age in children with autism spectrum disorders. J Autism Dev Disord. 2008;38:1081–91. doi: 10.1007/s10803-007-0487-9. [DOI] [PubMed] [Google Scholar]
- Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. doi: 10.1038/nature11011. doi:nature11011 [pii] 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newschaffer CJ, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health. 2007;28:235–58. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- Newschaffer CJ, Fallin D, Lee NL. Heritable and nonheritable risk factors for autism spectrum disorders. Epidemiol Rev. 2002;24:137–153. doi: 10.1093/epirev/mxf010. [DOI] [PubMed] [Google Scholar]
- O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. doi:ng.835 [pii] 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. doi: 10.1038/nature10989. doi:nature10989 [pii] 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozonoff S, et al. Recurrence risk for autism spectrum disorders: a baby siblings research consortium study. Pediatrics. 2011;128:e488–95. doi: 10.1542/peds.2010-2825. doi:peds.2010-2825 [pii] 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Sham PC. A model-fitting implementation of the DeFries-Fulker model for selected twin data. Behav Genet. 2003;33:271–8. doi: 10.1023/a:1023494408079. [DOI] [PubMed] [Google Scholar]
- Reichenberg A, et al. Advancing paternal age and autism. Arch Gen Psychiatry. 2006;63:1026–32. doi: 10.1001/archpsyc.63.9.1026. doi:63/9/1026 [pii] 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- Rice C. Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ. 2009;58:1–20. doi:ss5809a1 [pii] [PubMed] [Google Scholar]
- Rietveld MJ, van Der Valk JC, Bongers IL, Stroet TM, Slagboom PE, Boomsma DI. Zygosity diagnosis in young twins by parental report. Twin Res. 2000;3:134–41. doi: 10.1375/136905200320565409. [DOI] [PubMed] [Google Scholar]
- Rijsdijk FV, Sham PC. Analytic approaches to twin data using structural equation models. Brief Bioinform. 2002;3:119–33. doi: 10.1093/bib/3.2.119. [DOI] [PubMed] [Google Scholar]
- Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142:74–7. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- Robinson EB, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%) Arch Gen Psychiatry. 2011;68:1113–21. doi: 10.1001/archgenpsychiatry.2011.119. doi:68/11/1113 [pii] 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EB, et al. A multivariate twin study of autistic traits in 12-year-olds: testing the fractionable autism triad hypothesis. Behav Genet. 2012;42:245–55. doi: 10.1007/s10519-011-9500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, et al. Genetic heterogeneity between the three components of the autism spectrum: A twin study. J Am Acad Child Adolesc Psychiatry. 2006a;45:691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Plomin R. The genetic relationship between individual differences in social and nonsocial behaviours characteristic of autism. Dev Sci. 2005;8:444–58. doi: 10.1111/j.1467-7687.2005.00433.x. doi:DESC433 [pii] 10.1111/j.1467-7687.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- Ronald A, Happe F, Price TS, Baron-Cohen S, Plomin R. Phenotypic and genetic overlap between autistic traits at the extremes of the general population. J Am Acad Child Adolesc Psychiatry. 2006b;45:1206–14. doi: 10.1097/01.chi.0000230165.54117.41. doi:10.1097/01.chi.0000230165.54117.41 S0890-8567(09)62375-3 [pii] [DOI] [PubMed] [Google Scholar]
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: A decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- Ronald A, Simonoff E, Kuntsi J, Asherton P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry. 2008;49:535–542. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–14. doi: 10.1001/archpediatrics.2009.98. doi:163/10/907 [pii] 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- Rutter M, Bailey A, Lord C. The Social Communication Questionnaire Manual. Los Angeles, CA: Western Psychological Services; 2003. [Google Scholar]
- Sanders SJ, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011a;70:863–85. doi: 10.1016/j.neuron.2011.05.002. doi:S0896-6273(11)00374-6 [pii] 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011b;70:863–85. doi: 10.1016/j.neuron.2011.05.002. doi:S0896-6273(11)00374-6 [pii] 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. doi:nature10945 [pii] 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandin S, Hultman CM, Kolevzon A, Gross R, MacCabe JH, Reichenberg A. Advancing maternal age is associated with increasing risk for autism: a review and meta-analysis. J Am Acad Child Adolesc Psychiatry. 2012;51:477–486. doi: 10.1016/j.jaac.2012.02.018. e1 doi:S0890-8567(12)00144-X [pii] 10.1016/j.jaac.2012.02.018. [DOI] [PubMed] [Google Scholar]
- Schaaf CP, et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorders. Hum Mol Genet. 2011;20:3366–75. doi: 10.1093/hmg/ddr243. doi:ddr243 [pii] 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, et al. Maternal periconceptional folic acid intake and risk of autism spectrum disorders and developmental delay in the CHARGE (CHildhood Autism Risks from Genetics and Environment) case-control study. Am J Clin Nutr. 96:80–9. doi: 10.3945/ajcn.110.004416. doi:ajcn.110.004416 [pii] 10.3945/ajcn.110.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton JF, Tancredi DJ, Hertz-Picciotto I. Independent and dependent contributions of advanced maternal and paternal ages to autism risk. Autism Res. 2010;3:30–9. doi: 10.1002/aur.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, Mandy WP, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry. 2005;187:568–72. doi: 10.1192/bjp.187.6.568. doi:187/6/568 [pii] 10.1192/bjp.187.6.568. [DOI] [PubMed] [Google Scholar]
- Spinath FM, Price TS, Dale PS, Plomin R. The genetic and environmental origins of language disability and ability. Child Dev. 2004;75:445–54. doi: 10.1111/j.1467-8624.2004.00685.x. doi:10.1111/j.1467-8624.2004.00685.x CDEV685 [pii] [DOI] [PubMed] [Google Scholar]
- Steffenburg S, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–16. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Stevenson J. Evidence for a genetic etiology in hyperactivity in children. Behav Genet. 1992;22:337–44. doi: 10.1007/BF01066665. [DOI] [PubMed] [Google Scholar]
- Stevenson J, Pennington BF, Gilger JW, DeFries JC, Gillis JJ. Hyperactivity and spelling disability: testing for shared genetic aetiology. J Child Psychol Psychiatry. 1993;34:1137–52. doi: 10.1111/j.1469-7610.1993.tb01779.x. [DOI] [PubMed] [Google Scholar]
- Stilp RL, Gernsbacher MA, Schweigert EK, Arneson CL, Goldsmith HH. Genetic variance for autism screening items in an unselected sample of toddler-age twins. J Am Acad Child Adolesc Psychiatry. 2010;49:267–76. doi: 10.1016/j.jaac.2009.11.012. doi:00004583-201003000-00010 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, et al. Sex differences in repetitive stereotyped behaviors in autism: implications for genetic liability. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:5–12. doi: 10.1002/ajmg.b.31238. [DOI] [PubMed] [Google Scholar]
- Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S. Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:844–9. doi: 10.1002/ajmg.b.30740. [DOI] [PubMed] [Google Scholar]
- Vaags AK, et al. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90:133–41. doi: 10.1016/j.ajhg.2011.11.025. doi:S0002-9297(11)00503-9 [pii] 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windham GC, Zhang L, Gunier R, Croen LA, Grether JK. Autism spectrum disorders in relation to distribution of hazardous air pollutants in the san francisco bay area. Environ Health Perspect. 2006;114:1438–44. doi: 10.1289/ehp.9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachor DA, Ben Itzchak E. Assisted reproductive technology and risk for autism spectrum disorder. Res Dev Disabil. 2011;32:2950–6. doi: 10.1016/j.ridd.2011.05.007. doi:S0891-4222(11)00166-1 [pii] 10.1016/j.ridd.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proceedings of the National Academy of Sciences. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


