Abstract
The vertebrate T-box transcription factor gene Tbx18 performs a vital role in development of multiple organ systems. Tbx18 insufficiency manifests as recessive phenotypes in the upper urinary system, cardiac venous pole, inner ear, and axial skeleton; homozygous null mutant animals die perinatally. Here, we report a new regulatory mutation of Tbx18, a reciprocal translocation breaking 78 kbp downstream of the gene. 12Gso homozygotes present urinary and vertebral defects very similar to those associated with Tbx18-null mutations, but 12Gso is clearly not a global null allele since homozygotes survive into adulthood. We show that 12Gso down-regulates Tbx18 expression in a manner that is both spatially- and temporally-specific; combined with other data, the mutation points particularly to the presence of an essential urogenital enhancer located near the translocation breakpoint site. In support of this hypothesis, we identify a distal enhancer element, ECR1, which is active in developing urogenital and other tissues; we propose that disruption of this element leads to premature loss of Tbx18 function in 12Gso mutant mice. These data reveal a long-range regulatory architecture extending far downstream ofTbx18, identify a novel and likely essential urogenital enhancer, and introduce a new tool for dissecting postnatal phenotypes associated with dysregulation of Tbx18.
Keywords: T-Box transcription factor, urogenital development, chromosome translocation, regulatory mutation
Introduction
Tbx18 is a member of the ancient T-box gene family, encoding a transcription factor with critical roles in the development of the ureter, heart, inner ear, and somites (Airik et al., 2006; Bussen et al., 2004; Christoffels et al., 2006; Trowe et al., 2008). Tbx18-null mutant animals display recessive malformations of the axial skeleton, reflecting the gene’s important role in maintenance of anterior-posterior identity in the developing somites (Bussen et al., 2004; Farin et al., 2008). Newborn Tbx18−/− mice and late-stage embryos also display hydronephrosis and hydroureter, resulting from incomplete and irregular formation of the smooth muscle layer of the ureter required for proper functioning (Airik et al., 2006; 2010; Nie et al., 2010). The homozygous mutant animals also display complex abnormalities of the heart (Christoffels et al., 2006; Wiese et al., 2009). These animals die soon after birth with signs of respiratory distress implicating their severely malformed thoracic skeletons (Bussen et al., 2004; Christoffels et al., 2006). This suite of severe developmental phenotypes reflects the importance of Tbx18 expression during multiple developmental time points, and in a diverse range of tissues.
Tbx18 expression is first detected in presomitic mesoderm at embryonic day 7.75 (E7.75) of mouse development, and later appears in the urogenital ridge, the proepicardium of the heart, limb buds, dermis, otic mesenchyme, and other tissues (Bin Zeng et al., 2011; Bohnenpoll et al., 2013; Grisanti et al., 2012; Kraus et al., 2001; Trowe et al., 2008). This dynamic pattern of tissue-specific expression suggests the existence of complex gene regulatory mechanisms involving multiple tissue- and stage-specific enhancers. Supporting this idea, Tbx18 is embedded in a “gene desert” region, which is highly conserved from mouse to human, with nearest upstream and downstream neighbors residing more than 500 kbp away. Genomic neighborhoods like this one are found primarily around genes which, like Tbx18, encode developmental regulators with dynamic patterns of spatial and temporal expression (Ovcharenko et al., 2005). These extended gene deserts house multiple enhancers and silencing elements that interact through long-range chromatin loops; mutations that disrupt the structure of such domains can alter or ablate gene expression, even when they occur long distances away from the gene. Mutations like these, primarily in the form of reciprocal translocations, have revealed the existence of distant regulatory elements that would have been difficult to identify by other means. For example, translocations and deletions located in a region extending more than 1 Mbp upstream of Sox9 have been linked to campomelic dysplasia; the more distant translocations yield phenotypes that recapitulate only subsets of the disease phenotypes indicating tissue-specific disruption of the gene (Gordon et al., 2009). Translocations associated with Pax6, Shh and other important developmental regulators have also been crucial to the discovery of regulatory elements for those genes (Kleinjan and Coutinho, 2009; Kleinjan and van Heyningen, 1998; Lettice, 2003).
The structure of the Tbx18 gene desert locus suggests that this gene could also be regulated by a system of long-range enhancers. In one recent study, a bacterial artificial chromosome (BAC) reporter construct was shown to recapitulate most but not all aspects of the Tbx18 expression pattern; these data indicated that most of the important regulatory sequences would be found within an approximately 209 kbp region surrounding the gene (Wang et al., 2009). However, beyond this study, very little is known about the organization and function of the regulatory elements directing the complex developmental expression patterns of Tbx18 or other members of the extended T-box transcription factor family.
Here, we report the molecular and phenotypic characterization of a mouse mutation, 12Gso, a T(4;9) reciprocal translocation corresponding to a clean breakage-and-reunion event located 78 kbp downstream of Tbx18. 12Gso homozygotes display ureter and axial skeleton abnormalities very similar to those seen in Tbx18−/− mice, and complementation tests confirm that these recessive phenotypes are due to Tbx18 loss-of-function. The translocation breaks within a cluster of highly conserved sequences roughly 4.5 kbp in length, and a transgenic reporter assay confirms that sequences surrounding the breakpoint site contain at least one enhancer that is active in the urogenital mesenchyme. The 12Gso ureter phenotype and Tbx18 gene expression pattern strongly suggests that this downstream enhancer region is required in cis to activate the Tbx18 promoter during critical stages of urogenital development. Our data also indicate that additional enhancer(s) essential to somite development will be found further downstream of the gene. These data provide new insight to the Tbx18 regulatory structure, identify a novel developmental enhancer region that directs expression of a reporter in the urogenital mesenchyme, and define 12Gso as a novel regulatory mutation of Tbx18.
Results
Basic characterization of 12Gso mutants
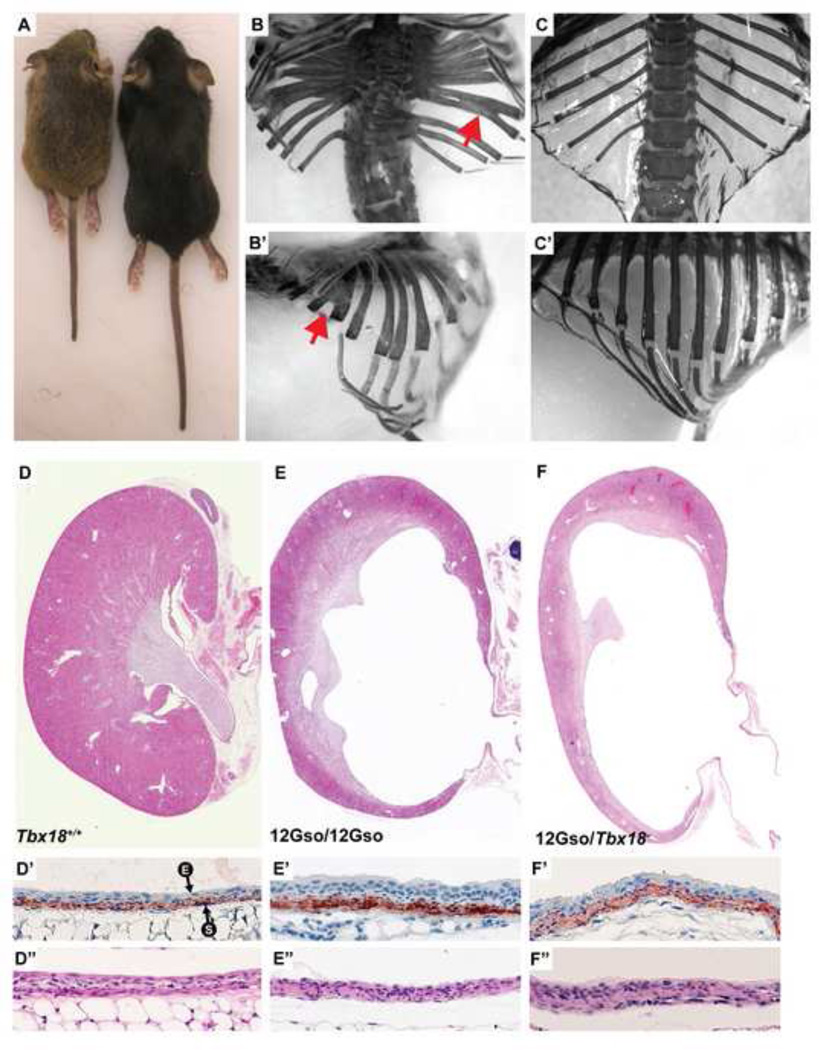
The 12Gso mutation arose in the offspring of male mice treated with bis-acrylamide in late meiotic stages, a treatment known to generate translocations at high frequencies (reviewed in (Culiat et al., 1997). The founder animal and its offspring were identified as translocation carriers by breeding tests, which revealed semi-sterility; this identification was confirmed by analysis of Giemsa-stained metaphase chromosomes, which revealed that the animals carried a reciprocal translocation affecting chromosome 4 (chr4) band B2 and chr9, band E2 (Chittenden, 2002). Heterozygous 12Gso animals are fertile and long-lived with no apparent phenotype other than translocation-related semi-sterility. However, homozygous 12Gso animals were found to display abnormalities of the axial skeleton including vertebral fusion, leading to a significant shortening of the body and often a kinked tail, as well as bifid ribs (Fig. 1D,I,N). Upon closer inspection, we observed kidney pathologies including increased kidney size, hydroureter and early stages of hydronephrosis at birth (Fig. 1S,X). Hydronephrosis and hydroureter was fully penetrant in both kidneys in all animals of both genders, although the severity at birth varied by individual. In adults the hydronephrosis was more severe, distending the entirety of the kidney and proximal ureter. While some 12Gso homozygotes survive well past weaning (Fig. 2), many animals die as juveniles and some within a few days of birth. These phenotypes indicated that the mutation disrupted the function of a gene that is critical in the development of the somites and urinary system, and possibly other tissues.
Fig. 1.
The 12Gso chromosome translocation is an allele of Tbx18 and produces recessive phenotypes that are similar to the Tbx18-null mice. (A–E) H&E stained sections of newborn thoracic vertebrae. Fused and malformed vertebrae are indicated with black arrow. (F–J) Alizarin red and alcian blue stained axial skeletons from newborn mice. Expanded proximal ribs are indicated with red arrows. (K–O) Lateral views of newborn thoracic skeletons. Fused pedicles of adjacent vertebrae are indicated with red arrows. (P–T) Hematoxylin and Eosin stained sections of newborn kidneys and ureters. 12Gso/12Gso and 12Gso/Tbx18− newborns display patent hydroureter (black arrows). (U–Y) Newborn urogenital systems in whole mount. The dilated ureters are outlined in black for clarity.
Fig. 2.
12Gso mutants can survive into adulthood. (A) Three week old 12Gso/Tbx18− female (left), and wild type female littermate (right) demonstrating the shortened body length of the mutants. (B-B’) Thoracic skeletons of adult 12Gso/Tbx18− animal and (C-C’) wild type littermate. Red arrows indicate bifid ribs. (D–F) H&E stained kidneys from wild type, 12Gso/12Gso, and 12Gso/Tbx18− respectively. (D’–F’) Anti-SMA (red) stained proximal ureters. “E” and “S” with arrows indicate urinary epithelium and smooth muscle, respectively. (D’’–F’’) H&E stained proximal ureters.
I would indicate the genotypes in the figure. It is much easier for the reader.
Mapping and sequencing the mutation
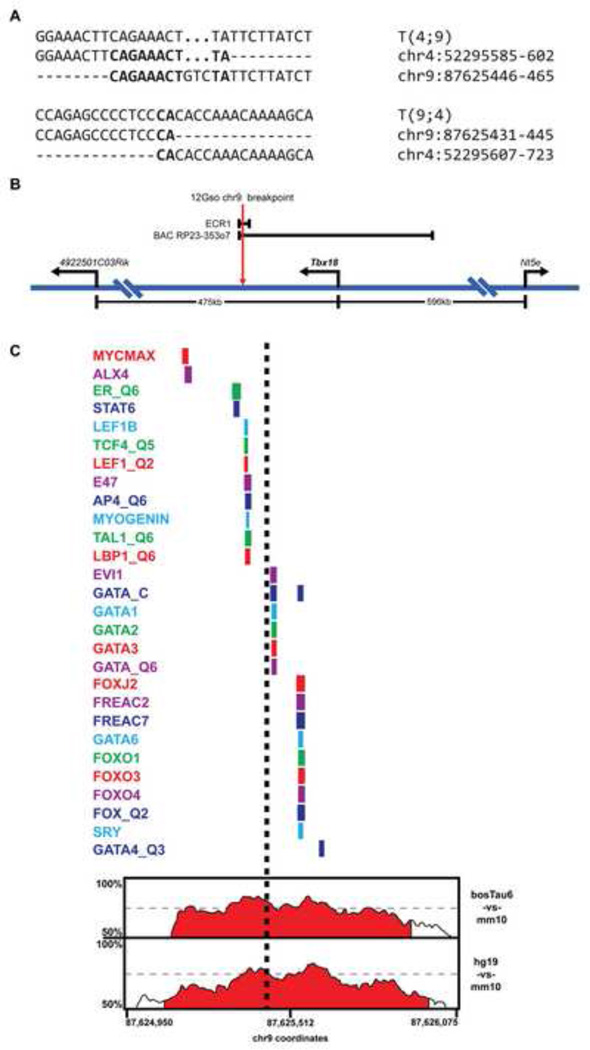
The 12Gso chr4 breakpoint had previously been mapped to a region spanned by a single bacterial artificial chromosome (BAC) clone, RP23-353g1, with fluorescent in situ hybridization (FISH). The translocation breakpoint was shown to disrupt the Abca1 gene, which spans most of the BAC region, and was further localized to a 476 bp region corresponding to intron 34 of the gene (Chittenden, 2002). We designed primers within the 3’ edge of exon 34 and the 5’ edge of exon 35 region to isolate T(4;9) and T(9;4) junction fragments from 12Gso mutant DNA by “genome walking” (see (Chittenden, 2002) for details). We sequenced the junction fragments and then designed primers flanking the chr9 breakpoint region to sequence DNA surrounding the 12Gso mutation sites on both chromosomes and compared them to wild type DNA. The results indicated that the mutation occurred as a clean breakage-and-reunion event (Fig. 3A) very similar to those documented in the analysis of other germline translocations (Elso et al., 2008; 2013; Elso, 2004).
Fig. 3.
The 12Gso breakpoint. (A) Sequence of the 12Gso breakpoints on Mmu4;9 and Mmu9;4. Bold text indicates sequence homology between the two chromosomes. (B) Map of the Mmu9 Tbx18 locus (not drawn to scale). The 12Gso breakpoint is located 78 kbp downstream of the 3’ end of Tbx18. The enhancer ECR1 spans the 12Gso breakpoint and contains a highly conserved cluster of elements. The BAC RP23-353o7, which spans 210 kbp of the Tbx18 locus was reported to contain a somite and ureter enhancer, and extends just past the 12Gso Mmu9 breakpoint. (C) A map of mammalian conservation and conserved transcription factor binding motifs surrounding the 12Gso breakpoint within ECR1. At the bottom of the figure is a percent identity plot (PIP) from the ECR browser (Ovcharenko et al., 2004) showing sequence similarity across a ~1 kbp region surrounding the 12Gso breakpoint within the ECR1 sequence. Percent identity (between 50–100% in 100 bp windows) is traced over the region to identify regions with significant sequence homology between the mouse sequence and regions in the human (hg19), bovine (bostTau6), and opossum (monDom5) genomes. The conserved regions are filled with red by default in the ECR browser if they include sequence identity above 70% in 100 bp windows. Above the PIP plot is a map of the locations of transcription factor binding motifs, identities of which are listed at left, that are conserved in all four species. The approximate location of the 12Gso breakpoint is shown as a black dashed line.
The chr9 breakpoint is within a large “gene desert” region, located approximately 78 kbp downstream of Tbx18 (105 kbp from the annotated Tbx18 promoter; Fig. 3B). Because of the striking similarities between the 12Gso phenotype and the skeletal and kidney abnormalities observed in Tbx18-null mutant mice (Airik et al., 2006; Bussen et al., 2004), we hypothesized that the 12Gso mutation disrupts Tbx18 expression in those tissues.
Complementation tests confirm that 12Gso is an allele of Tbx18
To test the relationship between 12Gso, Abca1 and Tbx18, we used targeted null mutations that are available for both genes (Bussen et al., 2004; Christiansen-Weber et al., 2000). Abca1 encodes a lipid transporter gene that plays an important role in cholesterol metabolism in both humans and mice, but Abca1−/− mice are long-lived and relatively healthy, with no recorded defects of the axial skeleton or ureter (Christiansen-Weber et al., 2000). We crossed Abca1−/+ with 12Gso/+ animals to generate several litters containing compound 12Gso/Abca1− mice. All of the compound animals (n=10) were born with normal skeletons and no apparent urinary defects when sacrificed at 0 days post-natal (Fig. 1C,H,M,R,W).
On the other hand, crosses between Tbx18−/+ and 12Gso/+ mice generated compound mutants (12Gso/Tbx18−) with skeletal defects, including fused vertebrae, bifid ribs and hydroureter with concomitant hydronephrosis that are typical of the recessive defects associated with each mutation (Fig. 1E,J,O,T,Y). We genotyped all pups born from these crosses, and all compound mutants (19 of 60 animals genotyped) but none of the carriers or wild type littermates (41/60) displayed the somite and kidney defects. These data confirm that 12Gso is a loss-of-function allele of Tbx18, affecting gene function in both somite and urinary development.
On the genetic background we used for this study (see Methods), the axial skeleton phenotype associated with Tbx18−/− was less severe than has previously been reported (Fig. 1S)(Bussen et al., 2004). On this background, we observed that 12Gso homozygous mutants and Tbx18−/− skeletons were very similar, and equally malformed (Fig. 1)(Fig. 1S).
In contrast to Tbx18-null mutants (Bussen et al., 2004), 12Gso animals can survive well into adulthood, and some 12Gso/Tbx18− compound mutants also share this relative longevity (Fig. 2). This fact suggests that Tbx18 expression is only partially abrogated by the 12Gso translocation, suggesting that the mutation is either a global hypomorph (with lower levels of expression in all tissues) or that the mutation affects Tbx18 expression only in specific tissues or at certain developmental time points or some combination of both possibilities.
Tbx18 expression is significantly down-regulated in 12Gso mice
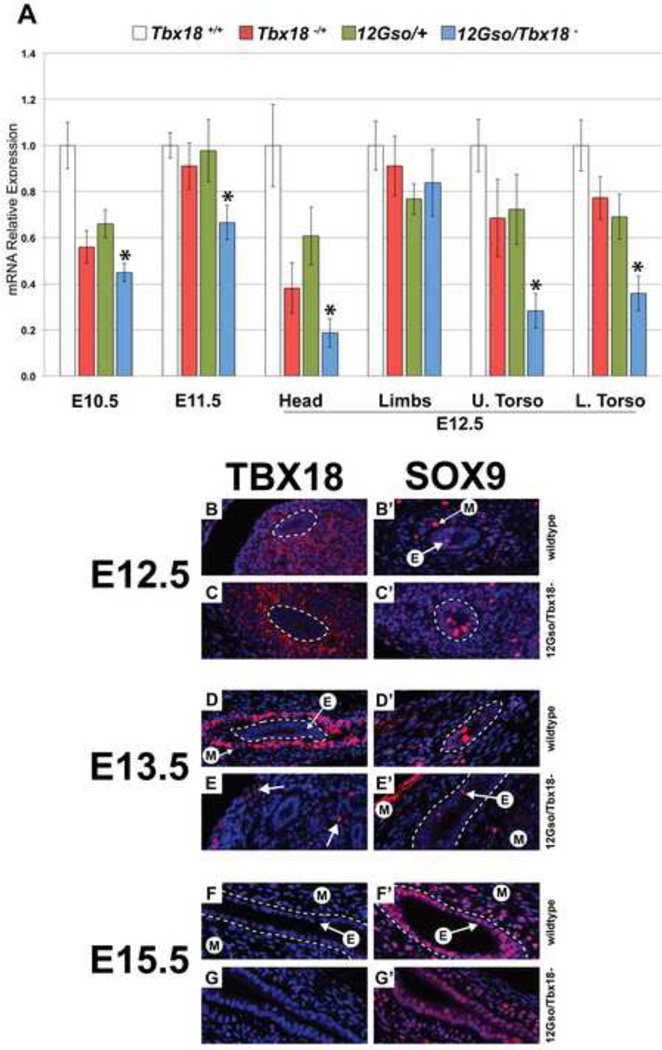
To examine Tbx18 expression, we collected 12Gso and Tbx18 heterozygotes, 12Gso/Tbx18− compound heterozygotes, and wild type littermates at different embryonic stages. First, we isolated RNA from whole embryos collected at embryonic day 10.5 (E10.5) and E11.5 and performed quantitative RT-PCR (qRT-PCR) to measure and compare overall levels of Tbx18 expression. Tbx18 mRNA levels are reduced by 12Gso at these stages but not entirely lost (Fig. 4A). 12Gso and Tbx18 heterozygotes showed a roughly 40% reduction in Tbx18 mRNA quantities at E10.5, but no significant change at E11.5. 12Gso/Tbx18− compound heterozygotes were significantly down-regulated compared to wild type mice and heterozygotes at these stages. At E12.5, we dissected embryo heads, upper torsos, lower torsos, and limbs. Tbx18 was reliably detected in the wild type embryos of each stage as well as in all of the dissected tissue samples we tested. Compared to wild type littermates, we found Tbx18 to be significantly down-regulated in heads, upper torsos, and lower torsos, but not changed in the limbs of the compound heterozygotes where Tbx18 expression is particularly high at this developmental stage (Kraus et al., 2001)(Fig. 4A).
Fig. 4.
Expression patterns of Tbx18 in 12Gso mutant mouse embryos. (A) Quantitative reverse transcription PCR in whole E10.5, E11.5 and E12.5 heads, limbs, upper torsos, and lower torsos. Tbx18 mRNA levels are significantly reduced in E10.5, E11.5, and E12.5 heads, and torsos, but not in E12.5 limbs. Asterisks indicate tissues in which Tbx18 is significantly reduced in compound heterozygotes compared to wild type embryos. For all genotypes, stages, and tissues, n ≥ 4, error bars are Standard Error of the Mean. (B–G) TBX18 IHC on wild type and 12Gso/Tbx18− embryos at E12.5, E13.5, and E15.5; (B’–G’) SOX9 IHC on adjacent sections. (B–C) At E12.5 TBX18 is detected strongly in the forming ureter mesenchyme in both genotypes. TBX18 is not detected in the ureter epithelium. White dashed lines encircle the ureter epithelium in all panels for clarity. (D–E) By E13.5, the protein is still detected but only in cells located distally from the ureter epithelium of mutants (arrows in E) while being strongly expressed in condensed mesenchyme adjacent to the epithelium in wild type animals. At this stage, SOX9 is present in mesenchyme (“M” with arrow) and epithelium (“E” with arrow). (F–G) TBX18 is not detected in ureter mesenchyme by E15.5 in mutants and in wild type embryos but SOX9 expression is still present in both genotypes (F’–G’).
To examine protein expression at the cellular level, we generated a polyclonal antibody to a unique epitope of the protein (see Methods) and carried out immunohistochemistry (IHC) in sectioned E12.5, E13.5, and E15.5 mutant and wild type embryos (Fig. 4B–G). Because 12Gso mutants display a clear ureter phenotype, we focused primarily on expression in that tissue. In addition, we tested expression of the SOX9 protein at these stages (Fig. 4B’–G’). Sox9 has been shown to act downstream of Tbx18 in the ureteric mesenchyme; the loss of TBX18 in the condensing mesenchyme results in the loss of Sox9 gene expression and animals inheriting a Tbx18+/Cre,Sox9flox/flox genotype present extreme hydroureter (Airik et al., 2010). SOX9 thus provides an important and relevant marker for this tissue during this period of embryonic development.
We detected significant levels TBX18 protein in the mesenchyme surrounding the ureter epithelium in both compound mutant and wild type embryos at E12.5 (Fig. 4B,C; dashed lines encircle ureter epithelium in all panels). However, clear differences were detected between mutant and wild type littermates by E13.5.
At this stage in wild type animals, TBX18 protein was detected in the nuclei of cells concentrated in the inner ring of condensing mesenchyme adjacent to the epithelium (Fig. 4D). In striking contrast, TBX18-expressing cells were significantly reduced in number, and primarily limited to the distal perimeter of mutant ureters (arrows in Fig. 4E); with a few isolated cells located in the mesenchyme proximal to the ureter epithelium. By E15.5 TBX18 protein was not detected in the developing ureter of either genotype, consistent with the reported down-regulation of the gene in this location (Airik et al., 2006)(Fig. 4F,G). At the earlier stages SOX9 was detected in scattered cells in the ureter epithelium and the mesenchyme consistent with previous reports (Airik et al., 2010)(Fig. 4B’ with arrows). We detected no obvious differences in SOX9 expression in the 13.5 mutant ureters despite the premature loss of TBX18 (Fig. 4B’–E’). Indeed, SOX9 expression was still robust in both epithelial and mesenchymal compartments at E15.5 in both wild type and mutant mice (Fig. 4F’,G’). At these stages and in this tissue at least, 12Gso thus appears to have little if any effect on expression of the Sox9 gene.
Taken together, the qRT-PCR and IHC expression data indicate that the 12Gso mutation has both spatial (tissue-specific) and temporal (developmental stage-specific) effects on Tbx18 expression.
Generating a Tbx18 enhancer reporter construct
Mapping, complementation and expression results suggested that the 12Gso mutation separates Tbx18 from critical downstream enhancers that are required for appropriate function in somites and ureteric mesenchyme, and perhaps other tissues. Theoretically at least, these putative enhancers could be located anywhere within the 475 kbp between Tbx18 and the nearest neighboring gene, 4922501C03Rik, or possibly even further downstream of the gene. However, recent data made it likely that we would find a critical enhancer element much closer to the breakpoint site. In particular, a 209 kbp BAC reporter containing Tbx18 coding sequences has been shown to recapitulate much of the normal pattern of embryonic and neonatal expression displayed by the native Tbx18 gene (Wang et al., 2009). This BAC clone, RP23-353o7, contains and extends just past the 12Gso breakpoint sequence (Fig. 3B).
Since the 12Gso mutation is associated with clear somite and ureter phenotypes, we hypothesized that enhancer(s) active in one or both of these tissues would be found in the region directly surrounding the 12Gso breakpoint site. However, given that the RP23-353o7 reporter faithfully recapitulates Tbx18 urogenital tissue but not in anterior somites (Wang et al., 2009), this region seemed particularly likely to harbor elements active in urogenital tissues. Therefore, we focused on a 4.5 kbp region spanning the mutation site, and extending to the end of the region spanned by RP23-353o7 (chr9:87624100-87628673 in the Mm10 genome sequence build); we will hereafter refer to this sequence as ECR1. ECR1 includes several subregions that are highly conserved in mammals; the 12Gso breakpoint interrupts one of these conserved genomic regions (Fig. 3C). The breakpoint falls between two clusters of transcription factor binding sites (TFBS) that are conserved in the mouse, human, and bovine genomes; 12Gso thus separates these two closely apposed TFBS clusters. On the distal side of the breakpoint lies motifs for ALX4, LEF1, and Myogenin, E2A and LBP1 – (the latter three proteins recognizing the same E-box like conserved motif (CAGCTG)). About 140 bp away, but on the Tbx18-proximal side of the 12Gso mutation, lies a conserved EVI1 motif and sites for GATA protein binding adjacent to or overlapping the shared binding sites for FOXO proteins (FOXO1,2, and 4), SRY (all including TAAACA) (Fig. 3C). The homologous human region (chr6:85350507-85351466 in the hg19 genome build) contains a peak of DNAse hypersensitivity but has not been identified as a binding site for any of the transcription factors thus far tested with ChIP in human or mouse cells by the ENCODE consortium (not shown).
We PCR amplified and cloned the ECR1 sequence into an Hsp68 promoter-LacZ reporter plasmid (Kothary et al., 1988; Nobrega et al., 2003) and injected the construct into mouse embryos to generate transgenic lines. We collected embryos from multiple transgenic lines, testing β-galactosidase (β-gal) staining in whole mount preparations. Very similar patterns of expression were detected in five independent lines; we used the most intensely active transgenic line ECR1-2870 for in-depth analysis of developmental expression.
ECR1 drives reporter expression in urogenital and other tissues
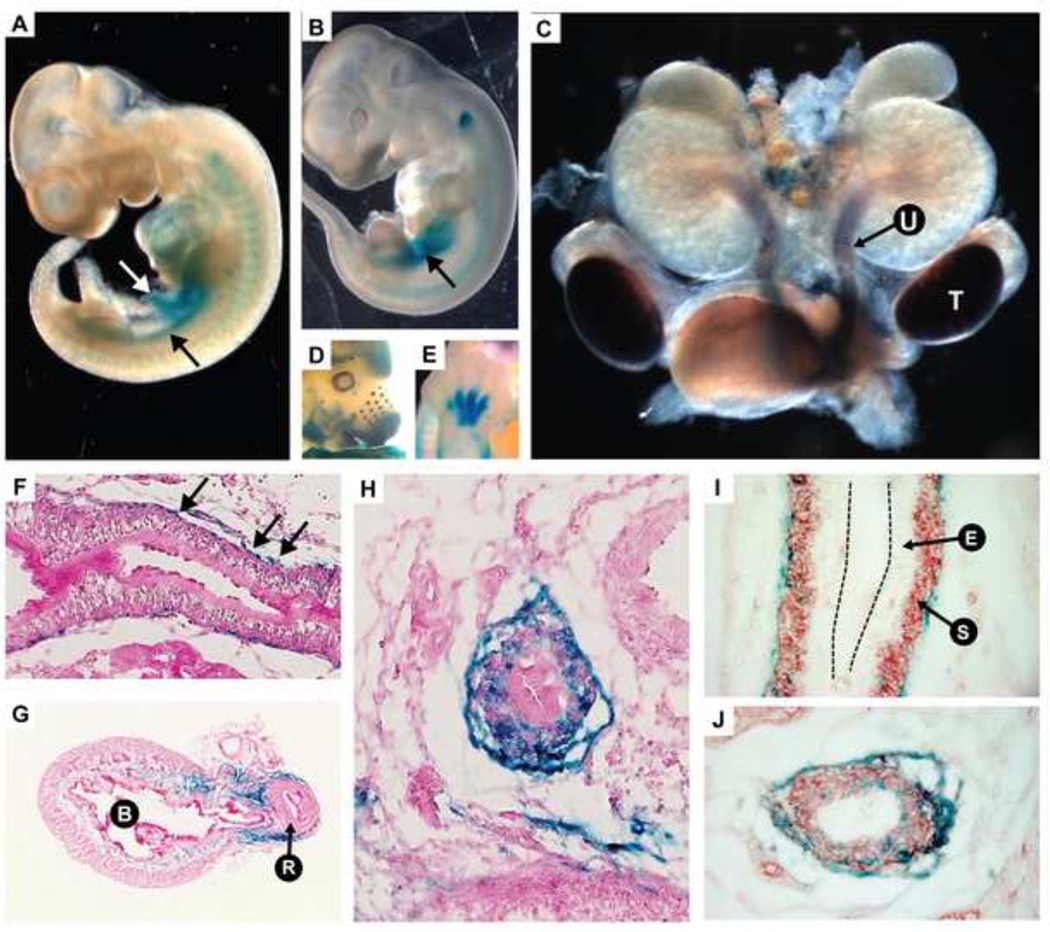
We collected transgenic embryos carrying the ECR1 reporter at multiple stages of development and utilized the LacZ transgene to detect tissue specificity of the ECR by staining embryos or isolated tissues as whole-mount preparations or generating frozen sections for β-gal staining (see Methods). ECR1-transgenic embryos consistently showed strong β-gal activity in urogenital tissues. Spatial expression was consistent with known domains of native Tbx18 expression, although in many cases the timing of expression during development was not entirely consistent with the native expression of Tbx18. For example, ECR1 staining in the ureter persists through birth (not shown) while the Tbx18 mRNA is depleted at this stage (Fig. 4F–H and (Airik et al., 2006)).
Native Tbx18 is expressed very early in development within the urogenital ridge (Bohnenpoll et al., 2013; Kraus et al., 2001). In ECR1-2870, we observed β-gal staining beginning at E10.5 in the urogenital ridge, in a manner very similar to native Tbx18 expression (Fig. 5A). At E11.5 staining had expanded in the region of the urogenital sinus (Fig. 5B). By E14.5, staining had localized to the perimeter of the ureter in the region of the forming smooth muscle layer (Fig. 5C). Staining persists in the ureter through E15.5 but is then reduced to small clusters of cells coating the ureter perimeter (Fig. 5F–J). Using a smooth muscle actin (SMA) antibody, we stained late gestation ureter sections that had previously been stained by β-gal. Most of the β-gal stained cells were in the periureteric mesenchyme, but many co-stained with SMA within the smooth muscle layer (Fig. 5I,J).
Fig. 5.
The ECR1 enhancer which spans the 12Gso breakpoint contains a urogenital-active enhancer element. (A) At embryonic day E10.5, the enhancer directs LacZ expression in the urogenital ridge (black arrow) and the vitelline vein (white arrow). (B) Expression at E11.5 is primarily localized to the urogenital mesenchyme (black arrow). (C) E14.5 dissected urogenital system shows strong staining in the ureter mesenchyme “U” and testes “T”. (D–E) LacZ is also expressed in the developing whisker buds, maxilla and mandible, eyecup, forebrain, and limbs as previously reported for the Tbx18 mRNA (Airik et al., 2006; Kraus et al., 2001). (F–H) Sections of LacZ stained E16.5 ureters and bladder showing expression in the ureter mesenchyme. (F) Black arrows indicating β-galactosidase stained cells along the length of the ureter. (G) Bladder indicated “B” and urethra is indicated by “R”. Blue staining is observed in a portion of the bladder stroma. (I) The urinary epithelium is labeled “E” and the differentiated smooth muscle layer is labeled “S”. The black dotted lines indicate the apical surface the urinary epithelium. (I–J) E16.5 ureters co-stained with LacZ (blue) and anti-SMA (red). (J) In the ureter section closest to the bladder, LacZ positive cells are dispersed through the mesenchyme.
By E13.5 we also detected ECR1-driven LacZ activity in several other tissues known to express Tbx18. For example, strong staining was detected in the interdigital mesenchyme and portions of the limb (Fig. 5E). Expression was also detected in the vibrissae (Fig. 5D), and the testes (Fig. 5C), which are known sites of late Tbx18 expression (Bohnenpoll et al., 2013; Kraus et al., 2001; Wang et al., 2009). We also observed staining in the bladder, which has been recently reported as containing Tbx18-positive derived cells (Bohnenpoll et al., 2013) and in developing ovaries (not shown). In contrast to ureter, clear phenotypes have not been reported in any of these tissues in Tbx18 null mutants (Bohnenpoll et al., 2013), and we did not identify any obvious abnormalities in 12Gso mice (not shown).
Discussion
Data presented here identify the 12Gso translocation as a regulatory mutation of Tbx18, affecting the developmental expression both spatially and temporally from a distance of approximately 78 kbp downstream of the T-box gene. As demonstrated by quantitative RT-PCR and IHC, 12Gso does not completely ablate Tbx18 expression but does alter levels of Tbx18 mRNA and protein with differential tissue- and at least in ureter, developmental time-specific effects (Fig. 4). Concordantly, phenotypes associated with 12Gso also vary in their severity relative to the Tbx18-null allele. For example, patent hydronephrosis is detected later in 12Gso mice than in Tbx18−/− animals, and 12Gso and compound-mutant animals can be long-lived (Fig. 2). However, all 12Gso homozygotes eventually develop hydronephrosis, and all display severe skeletal malformations that are nearly indistinguishable from Tbx18−/− mice (Fig. 1 and Fig. 1S). These data pointed to the importance of one or more critical Tbx18 enhancers required for proper urogenital and somite expression and located downstream of the gene, near or beyond the 12Gso breakpoint site.
A recent study using a Tbx18 BAC reporter provided strong evidence that most of the enhancers required to drive Tbx18’s pattern of developmental expression should be contained within a 209 kbp region extending upstream and downstream of the gene (Wang et al., 2009). Indeed, expression in urogenital tissues, heart, limbs, vibrissae, and other tissues were largely recapitulated by the BAC reporter. The one exception to faithful expression was noted in somites: BAC reporter activity was mostly limited to tail somites and was not seen in the regions anterior to the tail (Wang et al., 2009) whereas native Tbx18 expression is also expressed strongly in the anterior somites (Bussen et al., 2004; Kraus et al., 2001). These data suggested that whereas somite enhancers might lie outside the region spanned by the BAC, an enhancer critical to ureter expression should be located in the narrow interval defined by the proximal 12Gso breakpoint and by the end of the region spanned by RP23-353o7 on the distal side.
This ~4 kbp region, which we call ECR1, includes patches of DNA sequence that are highly conserved in mammals; 12Gso disrupts one of these conserved patches including binding sites for TF proteins with known roles in urogenital and smooth muscle development, including GATA proteins, GATA2, 3, 4, and 6 and LBP1 (also called TFCP2) (Fig. 3) (Kang et al., 2004; Trowe et al., 2012; Wada, 2000; Zhou et al., 1998). Using a transgenic reporter, we have confirmed the presence of at least one enhancer in the ECR1 region directing reporter gene expression in urogenital tissues. ECR1 also drives reporter expression in a portion of the limbs, the vitelline veins, and vibrissae in a manner that matches previously published Tbx18 expression domains (Fig. 5) (Kraus et al., 2001; van Wijk et al., 2009).
The expression pattern driven by ECR1 also differs in subtle ways from native Tbx18 expression; we posit that these discrepancies are due to the isolation of ECR1 from its natural genomic context in the transgenic mice, and the importance of interactions between ECR1 and other regulatory elements in the intact context of this extended locus. Indeed, it is likely that ECR1 includes just a subset of the active enhancers controlling Tbx18 expression in the urogenital tract and in other tissues throughout development. Interactions between these elements are likely to confer nuanced specificity to Tbx18 expression that cannot be recapitulated with an isolated element in transgenic mice. For example, both ECR1 and BAC RP23-353o7 drive continued expression of the reporter gene throughout gestation, and thus beyond the time when native Tbx18 mRNA and protein is detected (Fig. 4B–G and Airik et al., 2006). One explanation could be that regulatory elements contained in both ECR1 and RP23-353o7 are required to activate or maintain Tbx18 in the ureter mesenchyme, but are eventually repressed by elements located outside the RP23-353o7 region.
However, the notion that ECR1 might function to maintain Tbx18 expression during later prenatal stages is especially intriguing, given the expression pattern we observed during ureter development in 12Gso/Tbx18− compound mutant mice. Specifically, our data suggest that TBX18 protein is expressed normally until approximately E12.5, but disappears from developing ureter tissues in mutants around E13.5 (Fig. 4). Although we cannot rule out a role for more distant enhancers or silencing elements, we hypothesize that ECR1 is required in cis for full expression and function of Tbx18 during ureter development. Specifically we propose that ECR1 functions to maintain Tbx18 expression beyond E12.5 in this tissue, and that the separation of this element from the Tbx18 gene underlies the premature loss of Tbx18 expression in 12Gso mutant ureters.
How ECR1 might function in this capacity remains uncertain. However, recent work has shown that Tbx18 expression is maintained after E12.5 in the ureteric mesenchyme through the action of epithelial Wnt7b and Wnt9b (Bohnenpoll et al., 2013; Trowe et al., 2012). These reports raise the intriguing possibility that WNT signaling might be mediated directly through interactions with ECR1. Supporting this notion, the WNT-activated transcription factor, LEF1, is one of the TFs with a conserved motif in ECR1; furthermore, this motif is positioned just distal of the 12Gso breakpoint site (Fig. 3C). It is thus enticing to speculate that 12Gso directly disrupts essential and conserved WNT-mediated signaling events by severing the connection between ECR1 and the Tbx18 gene. This hypothesis can be tested directly by chromatin immunoprecipitation and other experiments in future studies.
The expression pattern we documented and the hypothesis that ECR1 is required to maintain, but not to initiate Tbx18 expression during development may also help explain the milder ureter phenotype observed in 12Gso compared to Tbx18−/− mice. For example, unlike Tbx18−/− mice, 12Gso/12Gso newborns exhibit only mild hydronephrosis although with patent hydroureter (Fig. 1). The hydronephrosis phenotype is more distinct in the compound mutant than in 12Gso homozygotes, but still not as severe as that seen in Tbx18−/− embryos. The expression of Tbx18 in the correct cells initially, but restriction to a subset of cells in the ureteric mesenchyme by E13.5 in 12Gso mutants, suggests a cell-type specific effect that could explain the milder 12Gso phenotype (Fig. 4). Specifically, the 12Gso mutation attenuates or abrogates expression of Tbx18 in the mesenchymal cells proximal to the ureter epithelium but not distally, in a time-dependent manner. Expression in these cells is clearly not sufficient to direct normal smooth muscle development, since adult 12Gso/12Gso and 12Gso/Tbx18− compound mutant animals show disorganized smooth muscle and urothelium in the ureter proximal to the kidney (Fig. 2). Interestingly, the small amount of TBX18 that does exist in mutants appears to be capable of activating Sox9 downstream in the mesenchyme and epithelium (Fig. 4B–G’). We found SOX9 protein to be abundant in both cell layers of 12Gso/Tbx18− compound mutant ureters at E15.5; this is not the case in Tbx18−/− animals, where Sox9 is absent (Airik et al., 2010). As previously remarked, the expression patterns of Tbx18 and Sox9 in the developing ureter are similar in location and timing, and both are essential to proper patterning. Data presented here suggest that the temporary activation of TBX18 expression around E12.5 may be sufficient to permit continued expression of SOX9 in the E15.5 12Gso/Tbx18− mutant ureter, likely contributing to the lesser severity of the ureter phenotype in 12Gso mice.
ECR1 therefore likely plays an important role in the 12Gso ureter phenotype, but since ECR1 is not expressed in somites, the disruption of this element cannot explain the skeletal abnormalities that are very clearly displayed by 12Gso mutants and 12Gso/Tbx18− compound mutant mice. This fact, and previous reports that the BAC RP11-353o7 reporter transgene also does not completely recapitulate Tbx18 somite expression (Wang et al., 2009), leads us to surmise that enhancer(s) critical to anterior somite expression will be found beyond the boundaries of the BAC reporter and of ECR1, and thus further downstream of the Tbx18 gene.
Additional Tbx18 enhancer elements may also be found in this distal downstream region. For example, qRT-PCR experiments indicate reduced quantities of Tbx18 in the whole torso by E12.5, although IHC shows that TBX18 protein is present at significant levels in the ureteric mesenchyme at this point (Fig. 4). Thus, Tbx18 transcripts may be depleted in other tissues in the lower torso. This difference, and others, could possibly contribute to the relative longevity of 12Gso compared to Tbx18-null animals. In other words, the translocation may allow Tbx18 to be expressed in certain cell types that are essential to postnatal survival. Specifically, while virtually all Tbx18-null mutants die perinatally (Bussen et al., 2004), 12Gso homozygotes and 12Gso/Tbx18− animals survive at least to neonatal stages and some live well into adulthood. As we have noted, the ureter phenotype of 12Gso mutants appears later, and is less severe than that seen Tbx18−/− mice, and this difference could possibly enable postnatal survival. However, it is likely that expression in other cell types plays an as yet undiscovered role. Further studies will be required to unravel the specific causes of Tbx18-linked perinatal lethality and the biological differences that explain the relative health and longevity of 12Gso mutant mice.
Materials and methods
Breakpoint mapping
Based on previously published data (Chittenden, 2002) we designed primers within exons 34 and 35 of the Abca1 gene for genome walking (Genome Walker Kit, Clontech Laboratories, Mountain View CA USA; primers abca1-34 GCAGGGCTGGATACGAAAAACAATGTCAAG and abca1-35 ATAGCATGCCAGCCCTTGTTATTGAACCAC). DNA fragments retrieved from these experiments were sequenced and chr9 junction sequences were identified by BLAST. We designed primers from the chr9 flanking region (CTGAAGGTGCTTTCTGCCTC) and used this primer with the abca1-34 primer to genotype mutant animals and embryos.
Histology and immunohistochemistry
The TBX18-2 polyclonal antibody was generated under contract with Abgent, Inc (San Diego, CA) by attaching the peptide sequence (qqqlqkkrrklateea) to KLH and immunizing rabbits. Adult tissues were preserved in 4% paraformaldehyde in PBS. Post-fixed tissues were dehydrated, embedded in paraffin and sectioned for staining. Thin-sectioned tissues were stained with Hematoxylin and Eosin as previously described. Embryos were fixed in 4% PFA overnight at 4°C. They were then dehydrated and embedded in paraffin for sectioning. Four micron sections were deparaffinized, boiled for 30 minutes in citrate target retrieval solution pH 6.0, then treated with 3% H2O2 for thirty minutes. Slides were blocked with goat blocking serum for 1 hour, and then the TBX18-2 (1:800), Sox9 (1:400)(Abcam ab3697) and alpha-Smooth Muscle Actin (1:1000)(Abcam ab5694) antibodies were incubated overnight at 4°C. Sections were washed and a goat-anti-rabbit-HRP (Jackson Immunoresearch) secondary antibody was applied at 1:400 for one hour at room temperature. Sections were then stained with Perkin-Elmer TSA-Rhodamine for three minutes, washed and counterstained with Hoechst 33342.
Blue/red skeletal preparations
Animals were sacrificed with CO2 to prevent damage to the skeletal structure. Dead animals were skinned and eviscerated. The bodies were stored in 95% ethanol overnight with rocking for fixing. Post-fixed carcasses were stored in 100% acetone for 24 hours to dissolve adipose stores. For 1–5 days, the body was washed in 1% KOH with frequent changes, until most of the skeletal muscle was transparent. The solution was changed to blue stain (0.05% alcian blue, 80% ethanol, 20% glacial acetic acid) for 24–48 hours depending on the age of the mouse. The skeletons were then cleared in 1% KOH overnight. Then the solution was changed to red stain (0.03% alizarin red, 1%KOH) for 30 minutes. After red staining, they were washed in water and cleared in 1% KOH.
ECR1 construct and transgenic mice
The ECR1 transgenic construct was created using the Hsp68-LacZ vector (a generous gift from Len Pennacchio). The ECR was amplified out of BAC RP23-461h11 with Invitrogen Pfx high fidelity polymerase, and the isolated 4.5 kbp PCR product was cloned into pGemT-EZ. The insert was cut out of pGemT-EZ with SmaI and XhoI, and ligated into (ECR1)-Hsp68-LacZ. Digested product was injected into C57BL/6 X C3HF-F1 embryos and implanted in Swiss-Webster surrogates. Transgenic mice were genotyped by PCR for the LacZ gene. β-Gal staining was performed essentially as outlined in (Loughna and Henderson, 2007). Incubations were performed at room temperature on a rocking plate. Embryos were tested for the presence of the transgenic construct by PCR for the LacZ transgene with primers TTTCCATGTTGCCACTCGC and AACGGCTTGCCGTTCAGCA to produce a 350bp product.
Tbx18 qRT-PCR
RNA was collected by homogenizing embryos in Invitrogen Trizol reagent. RNA was DNase treated with Qiagen on-column protocol on RNeasy columns. Invitrogen Superscript III reverse transcriptase was used to generate cDNA. Tbx18 mRNA levels were detected by qRT-PCR with Qiagen Tbx18 QPCR primers Cat#QT00133140. Mouse Gapdh QPCR primers (QT01658692) were used for normalization. An Applied Biosciences 7900HT thermocycler was used for amplification. The delta-delta-Ct method was employed to establish quantity differences. Embryos were dissected into four parts, full limbs were separated from the torso, heads were removed at the base of the neck, and torsos were bisected above the liver to produce upper and lower torso halves.
Animal Breeding and Welfare
Because 12Gso is a reciprocal translocation, the carrier animals are naturally semisterile; breeding rates are particularly low when these mutations are maintained on an inbred background (Elso et al., 2008). For that reason, we maintained 12Gso animals, and all of the other genotypes used in this study, on an F1 hybrid background (C3H/HeJ × C57BL/6J). Specifically, we maintained the mutant line by breeding heterozygotes to B6C3-F1 animals at each generation.
Animal work described in this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Illinois (Animal Assurance Number: A3118-01; approved IACUC protocol number 11030)
Supplementary Material
Highlights.
12Gso, a T(T4;9) reciprocal translocation, is disrupts developmental regulation of Tbx18
The 12Gso mutation disrupts Tbx18 somite and urogenital functions
12Gso breaks 78 kb downstream of Tbx18, disrupting an extended regulatory structure
We identify an enhancer that, we propose, is essential to Tbx18 urogenital function
Acknowledgments
We would like to thank Len Pennacchio for generously contributing the Hsp68-LacZ reporter vector. We also thank Laura Chittenden for helpful discussions and early work on breakpoint sequencing, Heather Thompson for expert technical assistance, and Hillary Thurkow and Dina Leiding for their persistent dedication to our mice. Thank you to Derek Caetano-Anolles for critical reading and comments. This work was supported primarily by a grant from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (grant number RO1 DK095685; awarded to L.S.). The work in the laboratory of A.K. is supported by a grant from the Deutsche Forschungsgemeinschaft (German Research Foundation) (DFG Ki728/7-1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Airik R, Bussen M, Singh M, Petry M, Kispert A. Tbx18 regulates the development of the ureteral mesenchyme. Journal of Clinical Investigation. 2006;116:663–674. doi: 10.1172/JCI26027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airik R, Trowe M-O, Foik A, Farin HF, Petry M, Schuster-Gossler K, Schweizer M, Scherer G, Kist R, Kispert A. Hydroureternephrosis due to loss of Sox9-regulated smooth muscle cell differentiation of the ureteric mesenchyme. Human Molecular Genetics. 2010;19:4918–4929. doi: 10.1093/hmg/ddq426. [DOI] [PubMed] [Google Scholar]
- Bin Zeng, Ren X-F, Cao F, Zhou X-Y, Zhang J. Developmental patterns and characteristics of epicardial cell markers Tbx18 and Wt1 in murine embryonic heart. Journal of Biomedical Science. 2011;18:67. doi: 10.1186/1423-0127-18-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnenpoll T, Bettenhausen E, Weiss A-C, Foik AB, Trowe M-O, Blank P, Airik R, Kispert A. Tbx18 expression demarcates multipotent precursor populations in the developing urogenital system but is exclusively required within the ureteric mesenchymal lineage to suppress a renal stromal fate. Developmental Biology. 2013:1–12. doi: 10.1016/j.ydbio.2013.04.036. [DOI] [PubMed] [Google Scholar]
- Bussen M, Petry M, Schuster-Gossler K, Leitges M, Gossler A, Kispert A. The T-box transcription factor Tbx18 maintains the separation of anterior and posterior somite compartments. Genes & Development. 2004;18:1209. doi: 10.1101/gad.300104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden LR. PhD Dissertation. University of Tennessee; 2002. Molecular Characterization of the T(4;9)12Gso Mutation and Analysis of the Associated Fitness, Skeletal, and Lymphoproliferative Phenotypes; pp. 1–196. [Google Scholar]
- Christiansen-Weber TA, Voland JR, Wu Y, Ngo K, Roland BL, Nguyen S, Peterson PA, Fung-Leung WP. Functional loss of ABCA1 in mice causes severe placental malformation, aberrant lipid distribution, and kidney glomerulonephritis as well as high-density lipoprotein cholesterol deficiency. Am J Pathol. 2000;157:1017–1029. doi: 10.1016/S0002-9440(10)64614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V, Mommersteeg M, Trowe M, Prall O, de Gier-De Vries C, Soufan A, Bussen M, Schuster-Gossler K, Harvey R, Moorman A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circulation Research. 2006;98:1555. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- Culiat CT, Carver EA, Walkowicz M, Rinchik EM, Cacheiro NL, Russell LB, Generoso WM, Stubbs L. Induced mouse chromosomal rearrangements as tools for identifying critical developmental genes and pathways. Reprod Toxicol. 1997;11:345–351. doi: 10.1016/s0890-6238(96)00147-5. [DOI] [PubMed] [Google Scholar]
- Elso C, Lu X, Morrison S, Tarver A, Thompson H, Thurkow H, Yamada NA, Stubbs L. Germline translocations in mice: unique tools for analyzing gene function and long-distance regulatory mechanisms. J Natl Cancer Inst Monographs. 2008;2008:91–95. doi: 10.1093/jncimonographs/lgn008. [DOI] [PubMed] [Google Scholar]
- Elso C, Lu X, Weisner PA, Thompson HL, Skinner A, Carver E, Stubbs L. A reciprocal translocation dissects roles of Pax6alternative promoters and upstream regulatory elements in the development of pancreas, brain, and eye. genesis n/a–n/a. 2013 doi: 10.1002/dvg.22409. [DOI] [PubMed] [Google Scholar]
- Elso CM. Heightened susceptibility to chronic gastritis, hyperplasia and metaplasia in Kcnq1 mutant mice. Human Molecular Genetics. 2004;13:2813–2821. doi: 10.1093/hmg/ddh307. [DOI] [PubMed] [Google Scholar]
- Farin H, Mansouri A, Petry M, Kispert A. T-box protein Tbx18 interacts with the paired box protein Pax3 in the development of the paraxial mesoderm. Journal of Biological Chemistry. 2008;283:25372. doi: 10.1074/jbc.M802723200. [DOI] [PubMed] [Google Scholar]
- Gordon CT, Tan TY, Benko S, Fitzpatrick D, Lyonnet S, Farlie PG. Long-range regulation at the SOX9 locus in development and disease. J. Med. Genet. 2009;46:649–656. doi: 10.1136/jmg.2009.068361. [DOI] [PubMed] [Google Scholar]
- Grisanti L, Clavel C, Cai X, Rezza A, Tsai S-Y, Sennett R, Mumau M, Cai C-L, Rendl M. Tbx18 Targets Dermal Condensates for Labeling, Isolation, and Gene Ablation during Embryonic Hair Follicle Formation. 2012:1–10. doi: 10.1038/jid.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HC, Chae JH, Kim BS, Han SY, Kim S-H, Auh C-K, Yang S-I, Kim CG. Transcription factor CP2 is involved in activating mBMP4 in mouse mesenchymal stem cells. Mol. Cells. 2004;17:454–461. [PubMed] [Google Scholar]
- Kleinjan D, Coutinho P. Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Briefings in Functional Genomics & Proteomics. 2009;8:317. doi: 10.1093/bfgp/elp022. [DOI] [PubMed] [Google Scholar]
- Kleinjan D, van Heyningen V. Position effect in human genetic disease. Human Molecular Genetics. 1998;7:1611. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Brown A, Campbell R, Peterson A, Rossant J. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. nature. 1988;335:435–437. doi: 10.1038/335435a0. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- Lettice LA. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Human Molecular Genetics. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- Loughna S, Henderson D. Methodologies for staining and visualisation of betagalactosidase in mouse embryos and tissues. Methods Mol Biol. 2007;411:1–11. doi: 10.1007/978-1-59745-549-7_1. [DOI] [PubMed] [Google Scholar]
- Nie X, Sun J, Gordon R, Cai C, Xu P. SIX1 acts synergistically with TBX18 in mediating ureteral smooth muscle formation. Development. 2010;137:755. doi: 10.1242/dev.045757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots G, Nobrega M, Hardison R, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Research. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Research. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowe M, Maier H, Schweizer M, Kispert A. Deafness in mice lacking the T-box transcription factor Tbx18 in otic fibrocytes. Development. 2008;135:1725. doi: 10.1242/dev.014043. [DOI] [PubMed] [Google Scholar]
- Trowe MO, Airik R, Weiss AC, Farin HF, Foik AB, Bettenhausen E, Schuster-Gossler K, Taketo MM, Kispert A. Canonical Wnt signaling regulates smooth muscle precursor development in the mouse ureter. Development. 2012;139:3099–3108. doi: 10.1242/dev.077388. [DOI] [PubMed] [Google Scholar]
- van Wijk B, van den Berg G, Abu-Issa R, Barnett P, van der Velden S, Schmidt M, Ruijter JM, Kirby ML, Moorman AFM, van den Hoff MJB. Epicardium and Myocardium Separate From a Common Precursor Pool by Crosstalk Between Bone Morphogenetic Protein- and Fibroblast Growth Factor-Signaling Pathways. Circulation Research. 2009;105:431–441. doi: 10.1161/CIRCRESAHA.109.203083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H. A p300 Protein as a Coactivator of GATA-6 in the Transcription of the Smooth Muscle-Myosin Heavy Chain Gene. Journal of Biological Chemistry. 2000;275:25330–25335. doi: 10.1074/jbc.M000828200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tripathi P, Guo Q, Coussens M, Ma L, Chen F. Cre/lox recombination in the lower urinary tract. genesis. 2009;47:409–413. doi: 10.1002/dvg.20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Grieskamp T, Airik R, Mommersteeg M, Gardiwal A, de Gier-De Vries C, Schuster-Gossler K, Moorman A, Kispert A, Christoffels V. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circulation Research. 2009;104:388. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lim K, Onodera K, Takahashi S, Ohta J, Minegishi N, Tsai F, Orkin S, Yamamoto M, Engel J. Rescue of the embryonic lethal hematopoietic defect reveals a critical role for GATA-2 in urogenital development. The EMBO Journal. 1998;17:6689–6700. doi: 10.1093/emboj/17.22.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.