Abstract
Damage of the zebrafish retina triggers a spontaneous regeneration response that is initiated by Müller Glia (MG) dedifferentiation and asymmetric cell division to produce multipotent progenitor cells. Subsequent expansion of the progenitor pool by proliferation is critical for retina regeneration. Pax6b expression in the progenitor cells is necessary for their proliferation, but exact regulation of its expression is unclear. Here, we show that miR-203 is downregulated during regeneration in proliferating progenitor cells. Elevated miR-203 levels inhibit progenitor cell expansion without affecting MG dedifferentiation or progenitor cell generation. Using GFP-reporter assays and gain and loss of function experiments in the retina, we show that miR-203 expression must be suppressed to allow pax6b expression and subsequent progenitor cell proliferation.
Keywords: miR-203, progenitor cell proliferation, pax6b, retina regeneration
Introduction
Retinal cell loss is the cause of many degenerative human diseases, including retinitis pigmentosa and age-related macular degeneration. In humans as well as other mammals, retinal cell loss is irreversible and ultimately leads to blindness. This is in stark contrast to the zebrafish retina, where damage by a variety of methods triggers a spontaneous regeneration response that restores not only lost retinal cell types, but also retina function (Fausett and Goldman, 2006; Vihtelic and Hyde, 2000; Yurco and Cameron, 2005). Intriguingly, Müller glia (MG), the cell type that initiates retina regeneration, are not specific to the zebrafish retina, but are common to all vertebrates (Lamba et al., 2008). However, mammalian MG normally respond to injury by undergoing reactive gliosis, which leads to scarring (Bringmann et al., 2009). Treatment of explants with transcription factors and select growth factors can induce mammalian MG to mount a regeneration response but the response is extremely limited (Karl et al., 2008; Ooto et al., 2004; Pollak et al., 2013). Understanding the mechanisms controlling retina regeneration in the zebrafish is a starting point to design strategies to trigger regeneration in the mammalian retina.
Recently, a number of genes and signaling pathways that control important steps in zebrafish retina regeneration have been identified. Ascl1a and Lin-28 activate MG dedifferentiation and cell cycle reentry (Ramachandran et al., 2010), while Pax6a and Pax6b are essential for proliferation of MG-derived progenitor cells (Thummel et al., 2010). Precise regulation of these genes is central to ensure efficient regeneration. miRNAs regulate gene expression by binding to recognition sites in the 3′ untranslated regions (UTRs) of target mRNAs triggering both mRNA decay and repression of translation (Kloosterman and Plasterk, 2006). Distinct subsets of miRNAs have been shown to regulate proliferation, maintenance of pluripotency, cell specification, and differentiation (Melton et al., 2010; Wang et al., 2008). miRNAs have also been implicated in regulating regeneration in a number of species (Sehm et al., 2009). In zebrafish, miRNAs regulate regeneration of a variety of tissues including the fin, heart, spinal cord and retina (Park et al., 2011; Ramachandran et al., 2010; Thatcher et al., 2008; Yin et al., 2012; Yin et al., 2008). In this study, RNA sequencing and analysis of specific cell types during zebrafish retina regeneration revealed that miR-203 expression must be downregulated during retina regeneration to enable proliferation of MG-derived progenitor cells. Downregulation of miR-203 allows upregulation of pax6b, which is required for increased progenitor cell proliferation and the formation of clusters of replicating progenitor cells associated with dedifferentiated MG. Our results demonstrate for the first time that miRNAs play a key role in the zebrafish retinal regeneration response after MG dedifferentiation and during the proliferation of MG-derived progenitor cells.
Materials and Methods
Fish maintenance
Zebrafish were maintained in 14h light and 10h dark cycles at 28.5°C. Albino fish were received from David Hyde (University of Notre Dame), tg:1016tuba1a:gfp were received from Daniel Goldman (University of Michigan) (Fausett and Goldman, 2006). Other fish lines used were tg:gfap:gfp (Bernardos and Raymond, 2006) and wild type AB. Embryos for microinjections were obtained from matings of AB fish. All experiments were performed with the approval of the Vanderbilt University Institutional Animal Care and Use Committee (Protocol # M/09/398).
Adult zebrafish light lesioning
Constant instense light lesioning was performed as previously described (Vihtelic and Hyde, 2000). Briefly, adult fish were dark adapted for 14 days, transferred to clear tanks placed between two fluorescent lights with light intensity at ~20,000 lux and the temperature maintained at 30–33°C. Zebrafish were subjected to light lesioning from 16h–3 days.
RNA isolation, RT-PCR, Taqman realtime PCR
Total RNA was isolated from control and light damaged zebrafish retinas using TRI Reagent®. For semi quantitative PCR, oligo-dT primers (Life Technologies) were used to synthesize cDNA using MMLV reverse transcriptase (Promega). PCR was performed using Phusion DNA polymerase (NEB) on a MyCycler thermal cycler (Biorad). GAPDH was used as a loading control. For quantitative real time PCR (qPCR), RNA was DNase treated (Rapid Out, Thermo Scientific), converted to cDNA using Maxima first strand cDNA synthesis kit (Thermo Scientific) and qPCR was performed using SYBR Green (Biorad). All qPCR primers spanned exon-exon junctions (IDT). miRNA realtime PCR was perfomed using Taqman probes as per the manufacturer’s instructions (Life Technologies). Relative RNA expression during regeneration was determined using the ΔΔCt method and normalized to 18s rRNA levels and U6 snRNA levels for mRNAs and miRNAs respectively. Real time PCR was performed on a Biorad CFX 96 Real time system. Primer sequences are listed in Supplemental Table 1.
Morpholino and miRNA injection and electroporation
Lissamine tagged morpholinos (MOs) (Gene Tools) were injected and electroporated into adult zebrafish eyes prior to light lesioning as described (Thummel et al., 2008b). The following 3′-Lissamine-tagged MOs were used:
| Gene Tools standard control MO: | 5′-CCTCTTACCTCAGTTACAATTTATA-3′ |
| pax6b MO: | 5′-CTGAGCCCTTCCGAGCAAAACAGTG-3′ |
| miR-203a MO: | 5′-CAAGTGGTCCTAAACATTTCAC-3′ |
Duplex mature miRNAs (Thermo scientific) were injected and electroporated into eyes either prior to start of light lesioning (Figure 2, 3, 5), or 51h after light lesioning (Figure 4) using the same procedure as the MOs but with reversed electrode polarity. For initial experiments, we used RNAs that contained a Dy-547 fluorescent tag at the 3′ end. Double stranded mature miRNAs were synthesized with 3′-UU overhangs for the following target sequences:
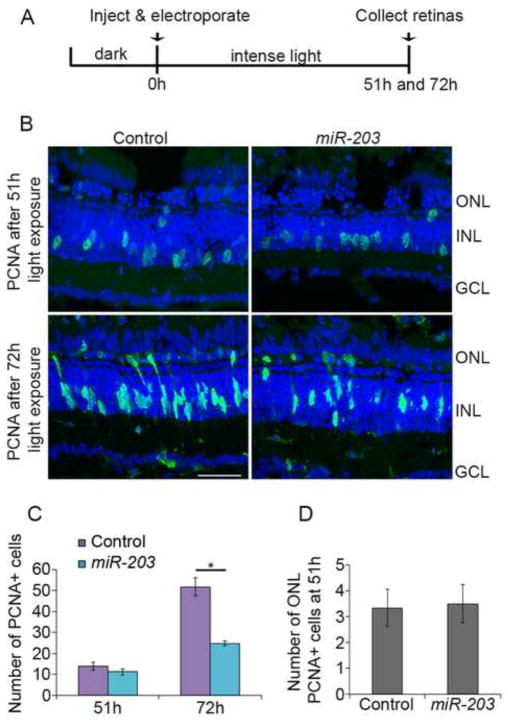
Figure 2. miR-203 overexpression affects proliferation during regeneration.
(A) Experimental scheme. (B) Control miRNA or miR-203 was injected and electroporated into the left eyes of albino zebrafish before intense light exposure. Retinas were collected after 51h and 72h of light exposure, sectioned and immunostained using an antibody against PCNA (green). Nuclei were counterstained with TOPRO (blue). (C) Quantification of INL PCNA+ cells. (D) Quantification of ONL PCNA+ cells at 51h. Data represent mean +/− s.e.m (n= 8 fish); *, p< 0.001 by Mann-Whitney U test. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
Figure 3. miR-203 gain-of-function does not affect MG dedifferentiation or proliferation.
Control miRNA or miR-203 was injected and electroporated into the left eyes of albino (A) or tg:1016tuba1a:gfp (D) zebrafish before intense light exposure. BrdU was injected intraperitoneally after 48h of light exposure and retinas were collected at 51h of light exposure, sectioned and immunostained with antibodies against BrdU and GS in (A) and BrdU, GFP and β-catenin in (D). In (A), BrdU+ nuclei (red) colabel with MG specific anti-glutamine synthetase (GS; blue) antibody (arrows). ONL rod precursors also incorporate BrdU (arrowheads). (B) Quantification of BrdU+/GS+ MG. (C) Quantification of total BrdU+ cells in INL and ONL. Data represent mean +/− s.e.m (n= 4 fish). No statistical significance was detected using Mann-Whitney U test. In (D), BrdU+ nuclei (red; arrows) colabel with β-catenin (blue) and GFP (green). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
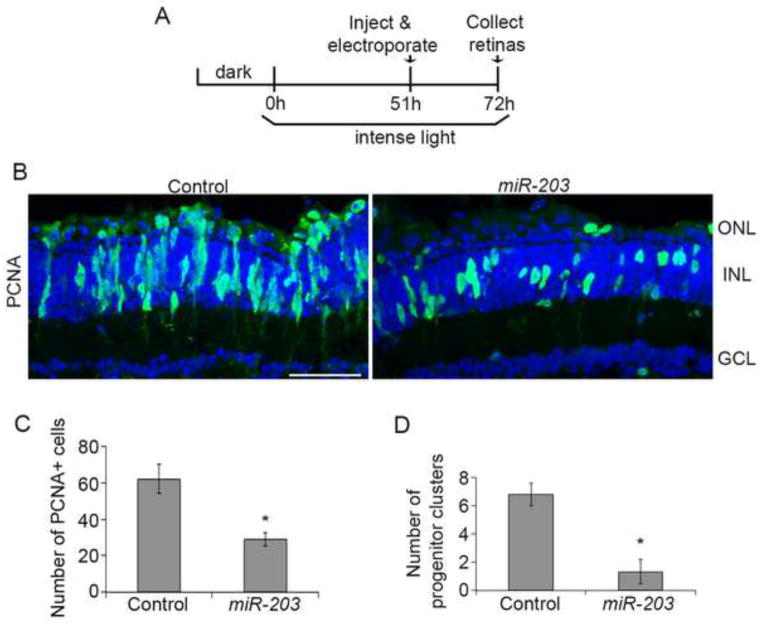
Figure 5. Excess miR-203 during progenitor cell proliferation phase impairs progenitor cluster formation.
(A) Experimental scheme. (B) Control miRNA or miR-203 was injected and electroporated into the left eyes of albino zebrafish after MG dedifferentiation and progenitor cell generation (51h of light). After 72h, retinas were collected, sectioned and immunostained with antibodies against PCNA (green). Nuclei were counterstained with TOPRO (blue). (C) Quantification of PCNA+ cells. (D) Quantification of progenitor clusters. Data represent mean +/− s.e.m (n= 3–5 fish); *, p< 0.03 by Mann-Whitney U test. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
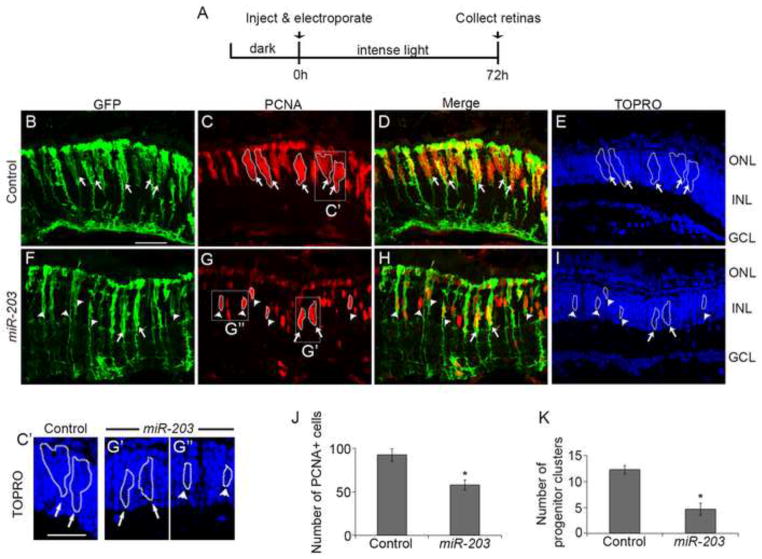
Figure 4. miR-203 gain-of-function reduces the number of progenitor clusters.
(A) Experimental scheme. (B–I) Control miRNA or miR-203 was injected and electroporated into the left eyes of tg:1016tuba1a:gfp fish prior to intense light exposure. Following 72h of light exposure, retinas were sectioned and stained with antibodies against GFP (green; panels B, F) and PCNA (red; panels C, G); nuclei are counterstained with TOPRO (blue; panels E, I). 1016tuba1a:gfp transgene expression was unchanged after miR-203 overexpression (B, F). In the control miRNA treated retinas, large clusters of PCNA+ cells associated along the processes of dedifferentiated MG (arrows in B–E). miR-203 overexpression reduced the number of dedifferentiated MG associated with large progenitor clusters (arrows in F–I) but increased the number of dedifferentiated MG associated with 1–3 PCNA+ cells (arrowheads in F–I). (C′ and G′) Higher magnification view of the progenitor clusters boxed in C and G respectively. (G″) Higher magnification view of boxed progenitors in G. (J) Quantification of PCNA+ cells. (K) Quantification of number of dedifferentiated MG associated with progenitor clusters. Data represent mean +/− s.e.m (n= 4 fish); *, p< 0.03 by Mann-Whitney U test. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar (panels B–I) 50um; (panels C′, G′ and G″) 30um.
miR-203: 5′-GUGAAAUGUUUAGGACCACUUG-3′
control: 5′-AAAAACAUGCAGAAAAUGCUG-3′
Electroporation was performed using the Gene Pulser Xcell™ Electroporation Systems (Biorad).
Immunohistochemistry, BrdU labeling, TUNEL assay and in situ hybridization
Adult zebrafish eyes were collected and fixed in 4% paraformaldehyde for 2–5h at room temperature. Following fixation, eyes were cryoprotected overnight in 30% sucrose/1X PBS at 4°C, before embedding in Shandon cryomatrix (Thermo Scientific) for sectioning. Embedded samples were kept at −80°C until sectioning. 10–12 micron sections were obtained using a cryostat (Leica), collected on charged Histobond slides (VWR), dried and stored at −80°C until used. For immunohistochemistry (IHC), slides were thawed for 30 min on a slide warmer, rehydrated in 1X PBS and blocked (3% Donkey serum, 0.1% TritonX-100 in 1X PBS) for 1–2h at room temperature before incubating with primary antibodies overnight at 4°C. Primary antibodies were mouse anti-PCNA monoclonal antibody (1:500, Sigma), mouse anti-glutamine synthetase monoclonal antibody-clone GS8 (1:200, Millipore), rat anti-BrdU monoclonal antibody (1:500, Abcam), rabbit anti-GFP polyclonal antiserum (1:1000, Torrey Pines Biolabs) and mouse anti-β-catenin antibody (1:500, BD Bioscience). After primary antibody incubation, sections were washed 3 times in 1X PBS/0.1% Tween-20 for 10 min each followed by 1–2h incubation in secondary antibody and nuclear stain TOPRO 3 (1:1000, Invitrogen) at room temperature. Secondary antibodies were donkey anti-mouse AF488 (1:200), donkey anti-mouse AF647 (1:200), donkey anti-rat Cy3 (1:100) and donkey anti-rabbit AF488 (1:200)(Jackson Immuno). Slides were washed in 1X PBS/0.1% Tween-20, 3 times for 10 mins, followed by a 5 min PBS wash before drying and coverslipping with Vectashield (Vector labs). Nuclear β-catenin was detected following citrate buffer antigen retrieval, as previously described (Ramachandran et al., 2011). For PCNA IHC, slides were boiled in 10mM sodium citrate buffer containing 0.05% Tween-20 (pH 6) for 20 mins, cooled at room temperature for 20 min and then quickly washed in 1X PBS prior to blocking. After IHC, slides were examined using an LSM 510 Meta inverted confocal microscope (Zeiss). For BrdU labeling, adult fish received a 25ul (20mM) IP injection of BrdU (Sigma) 3h prior to sacrifice. For BrdU IHC, sections were pretreated with 2N HCl for 20 min at 37°C and then quenched in 100mM sodium borate 2 times for 5 min each. For TUNEL assays, sections were processed as for IHC. TUNEL assays were performed using the TMR red in situ cell death detection kit (Roche). Digoxigenin labeled LNA probes (Exiqon) were used for miR-203 in situ hybridizations. A final probe concentration of 500nM and hybridization temperature of 42°C was used. Hybridization and wash conditions were previously described (Ramachandran et al., 2010). Slides were examined using a Leica DM6000B microscope.
Western blots
Protein lysates from 1dpf embryos were prepared as described previously (Flynt et al., 2007). Briefly, 1dpf embryos were dechorionated, deyolked and sonicated in lysis buffer supplemented with PMSF. For adult retinas, dorsal retinas were dissected from 7 fish per treatment group and sonicated in lysis buffer. Insoluble debris was removed by centrifugation and 20ug and 40ug of protein was separated on 12% polyacrylamide gels, for embryo lysates and adult retina lysates respectively. Proteins were transferred to PVDF membranes and probed with antibodies against GFP (1:1000, Torrey Pines), Pax6 (1:4000, Covance), Actin (1:100, Santa Cruz) and α-tubulin (1:1000, Abcam). For visualization, HRP-conjugated secondary antibodies (1:5000, GE Healthcare) were used followed by detection with ECL (Perkin Elmer). Imaging was performed on an LAS4000 Chemiluminescent CCD Imager (GE Healthcare) and blots were quantified using LAS4000 ImageQuant software (GE healthcare). GFP levels were normalized to α-tubulin levels and the GFP/tubulin ratio was determined for different injection conditions. Actin served as loading control for Pax6 Western blots.
Fluorescence activated cell sorting (FACS)
FACS was used to purify GFP+ cells from the retinas of undamaged tg:gfap:gfp fish and tg:1016tuba1a:gfp fish that were exposed to 72h of light damage following a procedure adapted from Qin et al. (Qin and Raymond, 2012). Briefly, retinas were pooled from 15 undamaged tg:gfap:gfp fish and 20, 72h light treated tg:1016tuba1a:gfp fish. Dissected retinas were treated with activated papain/dispase (Worthington) and incubated at 28°C for 30 min on a nutator for dissociation. Dissociated cells were pelleted by centrifugation, resuspended in DNaseI solution (Sigma) and gently tapped to complete tissue dissociation. Cells were triturated briefly, filtered through 35-micron filters and then sorted using BD FACSAria III (BD Biosciences). Approximately 27,000 GFP+ cells were obtained from 15 undamaged tg:gfap:gfp fish and 28,000 GFP+ cells were obtained from 20 light-damaged tg:1016tuba1a:gfp fish. As quality control for FACS, qPCR was used to confirm MG specific glutamine synthetase and progenitor cell specific ascl1a expression (Supplemental Figure 1). FACS was performed in the VUMC Flow Cytometry Shared Resource.
Molecular cloning and embryo microinjections
The pax6b 3′UTR was amplified from cDNA by PCR with the following primers:
pax6b-3′utr-fp: 5′-ACTAGTAAGGAACAACAGCCATTGTG-3′
pax6b-3′utr-rp: 5′ CTGTCTTGCAGATATTTCAATTTAACCTCGAG-3′
The 3′ UTR was cloned into the pCS2+ plasmid downstream of the GFP coding sequence. Capped RNA was transcribed from these reporter constructs using SP6 mMessage mMachine (Life Technologies). Zebrafish embryos at the single-cell stage were injected with 50pg of mRNA with or without 100pg of synthetic duplex miR-203a (Thermo Scientific). Titrations were performed to determine the lowest effective injection concentrations.
Cell counts and statistical analyses
For confocal microscopy, only retina sections that comprised optic nerves were used. All cell counts were done in the central-dorsal retina, at a linear distance of ~300 microns from the optic nerve. 3–8 retinas were used in every experiment. In Figures 4, 5 and 6, progenitor clusters were defined as groups of more than 3 closely adherent PCNA+ cells. In Figures 2–5 and 6O–P data are represented as mean +/− standard error of the mean (s.e.m) and significance was calculated using the non-parametric Mann-Whitney U test. Student-t tests were used to calculate significance for qPCR data and Western blots.
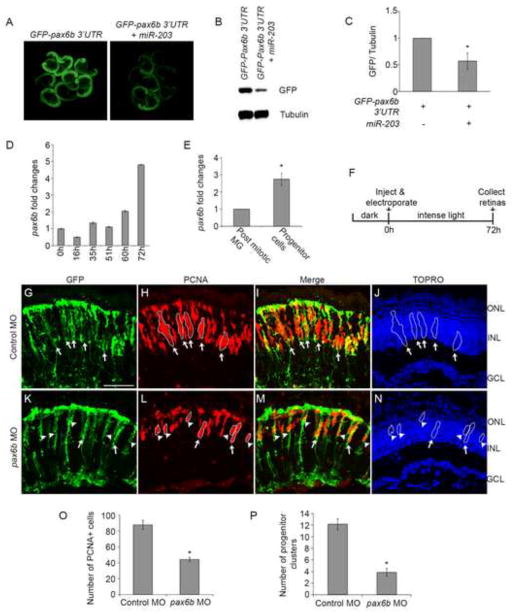
Figure 6. miR-203 regulates retina regeneration through pax6b.
(A–C) miR-203 targets the 3′ UTR of pax6b. The pax6b 3′UTR was fused to the GFP coding sequence. Single-cell zebrafish embryos were injected with in vitro transcribed mRNA from the GFP fusion construct in the presence or absence of miR-203. (A) Representative fluorescent images of injected embryos at 1dpf. (B) Western blot for GFP and α-tubulin control was performed on lysates prepared from the injected embryos in (A). (C) Quantification of GFP/tubulin ratios from multiple western blots as in (B). Data represent mean +/− s.d. (n=3 separate injection experiments); *, p< 0.008 by Student t-test. (D–E) pax6b mRNA is upregulated during retina regeneration. (D) qPCR was performed to quantitate pax6b levels from whole retina RNA. (E) qPCR for pax6b in FACS purified post mitotic MG and progenitor cells from 72h light treated fish. Data represent relative pax6b mRNA levels and show mean +/− s.e.m from 3 separate FACS experiments (n=3); *, p< 0.008 by Student t-test. For each FACS experiment post mitotic MG were purified from 15 undamaged fish and progenitor cells were purified from 20 light damaged fish. Samples were assayed in triplicate. 18s rRNA served as endogenous RNA control. (F) Experimental scheme for morpholino (MO) knockdown experiments in panels G–P. (G–N) Control MOs or pax6b MOs were injected and electroporated into the left eyes of tg:1016tuba1a:gfp fish prior to intense light exposure. Following 72h of light exposure, retinas were sectioned and stained with antibodies against GFP (green; panels G, K) and PCNA (red; panels H, L); nuclei were counterstained with TOPRO (blue; panels J, N). In the control MO treated retinas, large clusters of PCNA+ cells associated along the processes of dedifferentiated MG (arrows in G–J). pax6b MO treatment reduced the number of dedifferentiated MG associated with progenitor clusters (arrows in K–N) but increased the number of dedifferentiated MG associated with 1–3 PCNA+ cells (arrow heads in K–N). (O) Quantification of PCNA+ cells. (P) Quantification of number of dedifferentiated MG associated with progenitor clusters. Data represent mean +/− s.e.m (n= 6–7 fish); *, p< 0.002 by Mann-Whitney U test. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
Results
miR-203 is downregulated in proliferating progenitor cells during retina regeneration
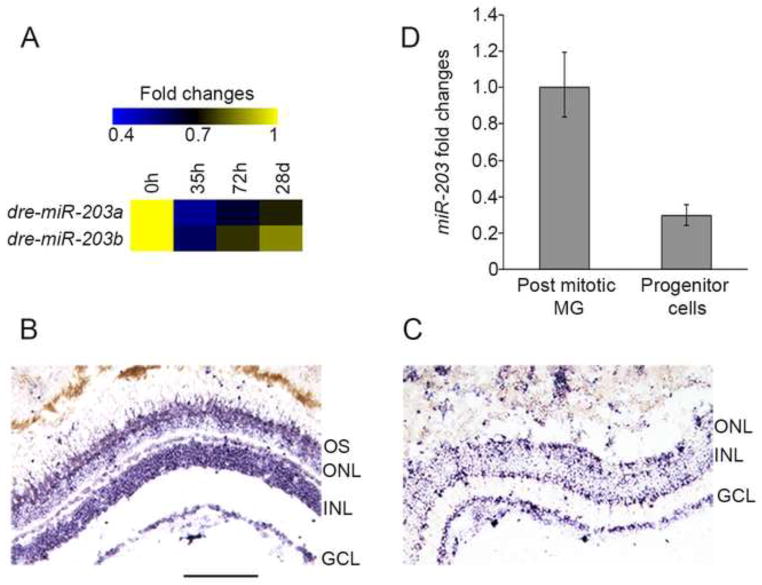
To identify differentially expressed miRNAs during regeneration of the light-damaged zebrafish retina, we performed a high-throughput sequencing screen. We identified 35 miRNAs that showed altered expression during regeneration (Harding, R., Rajaram, K., Bailey, T.J., Hyde, D.R., and Patton, J.G., manuscript in preparation). Among these, the miR-203 family was found to be downregulated (Figure 1A), a finding that intrigued us because we previously showed that miR-203 downregulation is necessary for zebrafish caudal fin regeneration (Thatcher et al., 2008). Similarly, miR-203 is downregulated in proliferating progenitor cells during mouse skin development and regeneration (Lena et al., 2008; Viticchie et al., 2012; Yi et al., 2008). To determine the role of miR-203 in retina regeneration, we examined its spatial expression pattern in undamaged and regenerating retinas using in situ hybridization with an LNA probe targeting mature miR-203 (Figure 1B, C). Sections from undamaged retinas showed miR-203 expression in the Ganglion Cell Layer (GCL), most of the Inner Nuclear Layer (INL) and the photoreceptor outer segments; with slightly lower expression in the Outer Nuclear Layer (ONL) (Figure 1B). In contrast, miR-203 expression was markedly reduced in light-damaged retinas that were actively regenerating (Figure 1C). The reduction was most striking in the INL, whereas the relative miR-203 levels in the GCL were overall similar or only slightly reduced between damaged and undamaged retinas.
Figure 1. miR-203 is downregulated in proliferating progenitor cells during retina regeneration.
(A) Heat map showing fold changes in deep sequencing reads for miR-203a and b during retina regeneration. Fold changes relative to 0h read number are shown. (B–C) LNA in situ hybridization for miR-203. Sections from undamaged retinas show strong miR-203 signals in the INL, GCL and photoreceptor outer segments (B). Staining is reduced in the INL of light damaged retinas (60h), but persists in the GCL (C). (D) qPCR for miR-203 in FACS purified post mitotic Müller glia (MG) and dedifferentiated MG-derived progenitor cells from 72h light treated fish. Data represent mean +/− s.e.m from 15 undamaged fish and progenitor cells were purified from 20 light damaged fish. OS, outer segments; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 100um.
Following light-induced injury, Müller glia respond by dedifferentiating and undergoing a single asymmetric division to produce progenitor cells that then continue to divide (Nagashima et al., 2013). As regeneration proceeds, clusters of proliferating progenitor cells occupy the INL along the processes of dedifferentiated MG (Fausett and Goldman, 2006; Nagashima et al., 2013; Thummel et al., 2008a). We sought to determine whether miR-203 levels are reduced in proliferating progenitor cells. To test this, we used fluorescence activated cell sorting (FACS) to isolate specific retinal cell types from two different transgenic lines: one expressing GFP in post-mitotic MG from undamaged retinas (tg:gfap:gfp) (Bernardos and Raymond, 2006), and the other expressing GFP in dedifferentiated MG and proliferating progenitor cells from actively regenerating retinas (tg:1016tuba1a:gfp) (Fausett and Goldman, 2006) (Supplemental Figure 1). Quantitative real-time PCR (qPCR) for miR-203 in these FACS-purified cell types revealed a ~70% reduction in miR-203 expression levels in proliferating progenitor cells relative to undamaged MG (Figure 1D). Together, these experiments indicate that miR-203 expression is downregulated in proliferating progenitor cells in the regenerating retina.
miR-203 overexpression affects proliferation during regeneration
Next, we examined if downregulation of miR-203 was necessary for retina regeneration. We performed gain-of-function studies (Supplemental Figure 2) by experimentally increasing the levels of miR-203 or a control miRNA prior to the onset of light damage. In our hands, 51h of light exposure in albino fish corresponds to the peak of dedifferentiated MG cell cycle reentry and the generation of progenitor cells, while 72h of light exposure corresponds to progenitor cell proliferation in the regenerating retina (unpublished). Thus, we overexpressed miR-203 or a control miRNA in dark-adapted albino fish retinas prior to the start of intense light damage (0h light exposure) and then assessed their effects on PCNA expression at 51h and 72h (Figure 2A). Compared to the control, miR-203 overexpression resulted in a significant decrease (p<0.001; n=8) in the number of PCNA+ proliferating progenitor cells at 72h, but not at 51h (Figure 2B,C). Additionally, ONL rod progenitor proliferation was unaffected by miR-203 gain-of-function (Figure 2D). These data suggested that miR-203 overexpression affects progenitor cell proliferation but not MG proliferation. To verify this, we injected zebrafish that were overexpressing either control miRNA or miR-203 with BrdU to specifically label proliferating cells at 51h. We labeled MG processes using an antibody against glutamine synthetase (GS). Immunohistochemistry revealed GS+ MG incorporating BrdU (Figure 3A, arrows). miR-203 gain-of-function did not affect MG proliferation (Figure 3B) or rod progenitor proliferation (Figure 3A, arrowheads; Figure 3C). Taken together, these data indicate miR-203 overexpression does not delay MG cell dedifferentiation or proliferation during retina regeneration, but specifically affects progenitor cell proliferation.
To test if suppression of miR-203 was sufficient to trigger retina regeneration, we injected and electroporated either control morpholinos (MOs) or MOs targeting mature miR-203 into undamaged 1016tuba1a:gfp transgenic fish eyes. This transgenic line specifically expresses GFP in dedifferentiated MG and progenitor cells. Compared to control MO treatment, loss of miR-203 did not cause significant changes in MG dedifferentiation or overall retinal proliferation (data not shown). Based on these results, we hypothesize that miR-203 downregulation is necessary for progenitor cell proliferation, but is not sufficient for initiation of MG dedifferentiation or for progenitor cell generation.
miR-203 inhibits progenitor cluster formation
To further test the hypothesis that progenitor cell proliferation and not progenitor cell generation is regulated by miR-203, we utilized the 1016tuba1a:gfp transgenic line that specifically expresses GFP in dedifferentiated MG and progenitor cells. Constant intense light exposure elicited a regeneration response in these fish that followed a timeline consistent with that observed in albino zebrafish (unpublished). Overexpression of miR-203 in these fish did not alter tuba1a:GFP transgene expression or BrdU incorporation at 51h (Figure 3D). We also detected β-catenin accumulation in GFP+/BrdU+ MG in both control and miR-203 overexpressing retinas (Figure 3D, arrows). β-catenin stabilization is required for MG proliferation and progenitor cell generation (Ramachandran et al., 2011). Thus MG proliferation and progenitor cell generation is unperturbed by miR-203 gain-of-function.
In contrast, miR-203 overexpression resulted in a significant reduction in the total number of PCNA+ progenitor cells at 72h (p<0.03; n=4) (Figure 4C, G, J). We also observed a striking reduction in the number of dedifferentiated MG associated with clusters of at least four PCNA+ progenitor cells (p< 0.03; n=4; Figure 4K; arrows in Figure 4B–I; nuclei in higher magnification Figure 4C′, G′). With increased miR-203, we found that most dedifferentiated MG were associated with only 1–3 PCNA+ progenitor cells (arrowheads in Figure 4F–I; nuclei in higher magnification Figure 4G″). These data are consistent with the hypothesis that miR-203 inhibits progenitor cell proliferation and the formation of large progenitor clusters. TUNEL staining demonstrated that these effects were not simply due to toxicity associated with miR-203 overexpression (Supplemental Figure 3).
Since MG dedifferentiation precedes progenitor cell proliferation during regeneration, we altered the timing of miR-203 overexpression to more precisely define its role. Thus, we overexpressed miR-203 at 51h, a time point after most MG have dedifferentiated and asymmetrically divided to generate progenitor cells (Figure 5A). Under these conditions, miR-203 overexpression resulted in a significant reduction (p<0.03; n=3–5 retinas; Figure 5C) in the number of PCNA+ progenitor cells compared to control miRNA injections (Figure 5B). Large clusters (>3) of PCNA+ progenitor cells with elongated cell bodies were almost undetectable in miR-203 overexpressing retinas (p<0.03; n=3–5; Figure 5D; Figure 5B). Taken together, these results support the hypothesis that miR-203 inhibits progenitor cell proliferation, and that downregulation of miR-203 is necessary to generate large clusters of progenitor cells during retina regeneration.
miR-203 regulates retina regeneration through pax6b
To determine what mRNA targets are regulated by miR-203 during progenitor cell proliferation, we used target prediction algorithms including Targetscan (Lewis et al., 2005). Several potential miR-203 targets were identified and tested for direct targeting (see Discussion; Supplemental Table 2). Among these, pax6b emerged as a likely target (Supplemental Figure 4B). Previous studies showed that Pax6b is expressed in proliferating progenitor cells during retina regeneration and is required for the formation of large clusters of progenitor cells (Thummel et al., 2008a). To determine if miR-203 can directly regulate pax6b, we performed GFP reporter assays in zebrafish embryos (Flynt et al., 2007) (Figure 6A–C, Supplemental Figure 4B). The 3′UTR of pax6b was cloned downstream of the GFP coding sequence and in vitro transcribed mRNAs from these reporter constructs were injected into single-cell stage zebrafish embryos in the presence or absence of exogenous miR-203. At 1dpf, co-injection with miR-203 reduced GFP levels by ~40% relative to the control injections (Figure 6B,C) (p<0.008, n=3). Consistent with previous studies (Thummel et al., 2010; Thummel et al., 2008a) and consistent with our finding that miR-203 levels decrease during regeneration, we detected upregulation of pax6b transcripts in RNA isolated from whole retinas during regeneration (Figure 6D and Supplemental Figure 5). More precisely, we determined the levels of pax6b in FACS-purified cells and observed significant enrichment of pax6b mRNA in proliferating progenitor cells compared to post-mitotic MG from undamaged retinas (p < 0.008; n=3) (Figure 6E). These data are consistent with the hypothesis that miR-203 regulates pax6b.
If miR-203 regulates pax6b, then overexpression of miR-203 should mimic morpholino knockdown of pax6b. Previously, Thummel et al. (2010) showed that loss of pax6b reduced the number of PCNA+ and BrdU+ cells in albino fish subject to intense light damage (Thummel et al., 2010). To test whether overexpression of miR-203 only affects progenitor cell proliferation by regulating pax6b, we injected and electroporated either standard control MOs or pax6b MOs into the eyes of 1016tuba1a:gfp transgenic zebrafish prior to the start of light lesioning (Figure 6F). Under these conditions, MG dedifferentiation was unaffected (Figure 6G, K), and consistent with previous results, there were fewer PCNA+ progenitor cells in the pax6b MO-treated retinas at 72h compared to controls (p<0.002; n=6–7; Figure 6H, L, O). Interestingly, there was a significant decrease in the number of progenitor clusters (arrows in Figure 6I, M) associated along the processes of dedifferentiated MG in pax6b MO-injected retinas (p<0.002; n=6–7; Figure 6P). Additionally, knockdown of pax6b yielded clusters with only 1–3 progenitor cells per dedifferentiated MG (arrowheads in Figure 6M) relative to the larger clusters observed in standard control morphants. This indicates that loss of Pax6b significantly reduces progenitor cell proliferation and phenocopies the miR-203 gain-of-expression result.
The reporter experiments and the Pax6b loss-of-function experiments indicate the miR-203 regulates pax6b. To test whether overexpression of miR-203 would decrease Pax6b levels in vivo, we analyzed Pax6b protein levels in the dorsal retina after miR-203 or control miRNA gain-of-function. Consistent with the hypothesis that miR-203 regulates Pax6b, we observed a ~30% reduction in Pax6b protein levels (Supplemental Figure 6). We attempted to directly rescue the miR-203 gain-of-function phenotype by injection and electroporation of pax6b mRNA but unfortunately we could not achieve mRNA delivery.
Discussion
Here, we identify miR-203 as a novel regulator of zebrafish retina regeneration. We demonstrate that miR-203 represses pax6b, which is necessary for progenitor cell expansion. During retina regeneration, miR-203 levels must be reduced to allow progenitor cell amplification and cluster formation via pax6b expression. Elevated miR-203 or loss of pax6b expression during retina regeneration inhibits progenitor cell proliferation and significantly impairs progenitor cluster formation. This reveals a novel regulatory mechanism that controls progenitor cell proliferation, a key step in retina regeneration.
miR-203 inhibits proliferation
miR-203 functions as negative regulator of proliferation and stem cell properties in a number of cancers (Viticchie et al., 2011; Wang et al., 2013; Wang et al., 2012; Yu et al., 2013), as well as in mouse skin development and regeneration (Jackson et al., 2013; Lena et al., 2008; Viticchie et al., 2012; Yi et al., 2008). miR-203 is also important for zebrafish regeneration. We previously showed that downregulation of miR-203 is necessary to initiate blastema formation following caudal fin amputation (Thatcher et al., 2008). During retina regeneration, in situ localization showed that miR-203 levels are significantly reduced in the INL upon light damage and qPCR revealed a nearly 70% reduction in miR-203 levels in progenitor cells compared to undamaged post-mitotic MG (Figure 1). This is highly reminiscent of the role of miR-203 in mouse skin development, where miR-203 expression is restricted to the differentiated and basal layers of newly stratified skin epithelium, but conspicuously absent in the proliferating progenitor compartment (Yi et al., 2008). Overexpression of miR-203 induced skin progenitors to exit the cell cycle, which severely reduced the pool of proliferating progenitors (Yi et al., 2008). During zebrafish retina regeneration, miR-203 downregulation in progenitor cells is also necessary for their proliferation (Figures 2, 4, 5). We restricted our analysis of the effects of miR-203 to the first 72h post injury. It will be interesting to determine the fate of the proliferating progenitor cells in retinas overexpressing miR-203, as well as the effects of long-term suppression of miR-203 on regeneration.
miR-203 inhibits progenitor expansion by targeting pax6b
Intense light treatment causes dying photoreceptors to express TNFα, which induces MG dedifferentiation (Nelson et al., 2013). Dedifferentiated MG express ascl1a, activate Wnt signaling, and undergo asymmetric cell division to produce progenitor cells (Nagashima et al., 2013; Ramachandran et al., 2011). These progenitors undergo multiple rounds of cell division and form clusters, which associate along the processes of the dedifferentiated MG and span the length of the INL (Thummel et al., 2010). Ectopic expression of miR-203 before starting intense light exposure in the transgenic 1016:tuba1a:gfp line did not affect overall MG dedifferentiation or the first cell divisions needed to generate progenitor cells (Figure 3 and Supplemental Figure 3). However, relative to control retinas, we detected a significant reduction in the number of dedifferentiated MG associated with progenitor clusters that had >3 associated progenitor cells (Figure 4). Instead, most of the dedifferentiated MG were associated with only 1–3 progenitor cells. This reduction was apparent when miR-203 was overexpressed specifically during the progenitor expansion phase in the regenerating retina (Figure 5).
Progenitor cell proliferation during retina regeneration is under the control of pax6 genes. While pax6b controls the first cell division of the MG-derived progenitor cells to generate 4 cell clusters, pax6a is required for further amplification of these 4 cell clusters (Thummel et al., 2010). It was previously reported that MO-mediated loss of pax6b prevents cluster formation by inhibiting the first cell division of the MG-derived progenitor cells. This is similar to the inhibition of progenitor cell proliferation and cluster formation phenotype we observed with miR-203 gain-of-function (Figures 4, 5), and consistent with miR-203 targeting of pax6b using reporter and qPCR assays (Figure 6). Similar to miR-203 gain-of-function, we detected a reduction in progenitor cluster number upon MO-mediated reduction of pax6b (Figure 6). Moreover, overexpression of miR-203 during the progenitor cell proliferation phase resulted in a ~30% reduction in Pax6b protein levels in dorsal retinas (Supplemental Figure 6). A more elegant approach would be to rescue the miR-203 overexpression phenotype by co-injection of pax6b mRNA. Unfortunately the ability to achieve delivery and expression of mRNAs by injection and electroporation into retinas has not been reported and proved technically challenging in our hands.
Regulation of Pax6b
Although the role of Pax6b in progenitor cell expansion is well established, the exact mechanism that regulates its expression is not clear. Knockdown of HB-EGF and ascl1a reduced the total amount of pax6b mRNA in the retina (Ramachandran et al., 2011; Wan et al., 2012) and stabilization of β-catenin upregulated pax6b levels (Ramachandran et al., 2011). Since pax6b is upregulated in proliferating progenitor cells, regulatory mechanisms must exist to precisely control its expression in these MG-derived progenitors. An intriguing hypothesis is that Wnt activation in asymmetrically derived progenitor cells suppresses miR-203 leading to upregulation of pax6b and subsequent progenitor cell expansion.
Regulation of miR-203
While miR-203 must be downregulated to allow progenitor cell proliferation, the mechanism of its downregulation remains unknown. In human keratinocytes, miR-203 expression is activated by PKC and inhibited by EGF treatment (Sonkoly et al., 2010). We previously showed that during zebrafish development, miR-203 expression is activated by suppressing hedgehog signaling (Thatcher et al., 2007). Studies in cancer also suggest epigenetic control of miR-203 expression via promoter DNA methylation (Bueno et al., 2008; Diao et al., 2013; Taube et al., 2013). In zebrafish, both miR-203 genes (miR-203a and miR-203b; Supplemental Figure 4A) are monocistronic, contain their own promoters and reside on separate chromosomes, suggesting possible transcriptional and epigenetic control of miR-203 gene expression. A recent study detected the existence of dynamic DNA methylation in MG and progenitor cells during zebrafish retina regeneration (Powell et al., 2013). Inhibition of DNA methylation during progenitor cell expansion inhibited proliferation, suggesting that genes that inhibit progenitor cell proliferation might be epigenetically suppressed. Moreover, HB-EGF induction in the dedifferentiated MG is necessary for progenitor cell production and retina regeneration (Wan et al., 2012). It is tempting to speculate that miR-203 expression in progenitor cells could be negatively regulated via both HB-EGF and promoter methylation, thus allowing progenitor cell proliferation and cluster formation.
Targets of miR-203
Several genes that are critical for retina regeneration are predicted to be miR-203 targets, including HB-EGF, Lin-28, and Insm1a (Supplemental Table 2). We found that pcna and lef1, which are both important for regeneration, are bona fide miR-203 targets (unpublished data) (Thatcher et al., 2008). Interestingly, miR-203 gain-of-function prior to retina damage does not inhibit PCNA expression early (51h of light), but does reduce PCNA expression at later stages (72h of light). This suggests that PCNA may not be a primary miR-203 target during retina regeneration, but rather is a consequence of miR-203 repressing DNA replication through its inhibition of the progenitor cell cycle. During zebrafish caudal fin regeneration, targeting of lef1 by miR-203 was central to both initiation and termination of regeneration (Thatcher et al., 2008). However, we have not found that targeting of lef1 by miR-203 affects retina regeneration (data not shown). In mouse skin development, the main target for miR-203 is ΔNp63, a member of the p53 family and a regulator of proliferation in epithelial cells and various cancers (Dotto, 2012; Lee and Kimelman, 2002; Parsa et al., 1999; Rivetti di Val Cervo et al., 2012; Senoo et al., 2007; Truong et al., 2006; Yang et al., 1999). In zebrafish, p63 expression is upregulated during optic nerve regeneration (Saul et al., 2010). However, we detected very low levels of p63 expression in FACS-purified post mitotic MG and progenitor cells from undamaged and regenerating retinas, respectively, suggesting that p63 is not an important target for miR-203 in the retina.
Limitations and implications of miRNA gain-of-function experiments
A caveat of our miR-203 overexpression studies is that all retinal cells, not just MG, take up the miRNA (Supplemental Figure 2). Hence, it is formally possible that inhibition of progenitor cell proliferation is a result of gain of miR-203 in cell types other than the MG-derived progenitor cells, that the effects we observe may not be cell autonomous. Since miR-203 can regulate many targets and Pax6 proteins are also expressed in amacrine and bipolar cells, it is possible that miR-203 inhibition of Pax6b or additional targets in these retinal neurons feeds back and inhibits progenitor cell proliferation by additional secondary mechanisms. It is therefore possible that the loss of progenitor cell proliferation due to excess miR-203 could be the result of the function of miR-203 in progenitor cells, retinal neurons or both.
miRNAs and regeneration
Early zebrafish retina regeneration can be divided into distinct steps requiring precise gene expression changes as MG dedifferentiate, undergo asymmetric division, and generate progenitor cells (Fausett and Goldman, 2006; Nagashima et al., 2013; Thummel et al., 2008a). We show for the first time that miR-203 must be downregulated to derepress pax6b and allow progenitor cell proliferation. Coupled with previous work showing that let-7 plays a key role regulating MG dedifferentiation (Ramachandran et al., 2010), it seems likely that additional miRNAs will be identified that regulate not only early steps during regeneration, but also migration and re-differentiation as lost cell types are replaced. miR-203 is an especially interesting case as it has now been demonstrated to play a key regulatory role in progenitor proliferation during development (Jackson et al., 2013; Lena et al., 2008; Yi et al., 2008), regeneration (Thatcher et al., 2008), and possibly adult neurogenesis.
Supplementary Material
FACS isolated post-mitotic MG and proliferating progenitor cells were analyzed by qPCR. Relative mRNA levels of MG-specific glutamine synthetase (GS) and progenitor cell-specific Ascl1a are shown. Data represent mean +/− s.e.m. (n=3 independent FACS).
Albino fish retinas were injected and electroporated with Dy-547 tagged control miRNA (A–C) or miR-203 (D–F) prior to the start of light exposure (0h). After 72h, retinas were assessed for PCNA expression (green) and Dy-547 fluorescent tag localization (red). Nuclei were counterstained with TOPRO (blue). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
TUNEL staining was performed on 72h light treated retina sections from control miRNA (A) or miR-203 (B) gain-of-function. Some TUNEL+ nuclei (red) were detected in the ONL. DNaseI treated retinas were used as positive controls for the TUNEL assay and showed TUNEL+ nuclei in all retinal layers (C). No cell death was detected in the negative control retina sections, which were subjected to the TUNEL protocol in the absence of terminal transferase (D). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
(A) Mature miRNA sequences for miR-203a and miR-203b. The miRNA ‘seed’ is highlighted in red. The single nucleotide that differs between the two family members is highlighted in magenta. (B) The pax6b 3′ UTR was cloned downstream of GFP. Pairing between miR-203a and its miRNA recognition element (MRE) in the pax6b 3′ UTR is shown.
RT-PCR was performed for pax6b, ascl1a and gapdh from whole retinas during retina regeneration. pax6b is upregulated during regeneration. ascl1a served as a positive control for the progression of retina regeneration. gapdh served as loading control.
Control miRNA or miR-203 was injected and electroporated into the left eyes of albino zebrafish after MG dedifferentiation and progenitor cell generation (51h of light). After 72h, dorsal retinas were collected from control miRNA and miR-203 injected fish and protein lysates were prepared for Western blots. Pax6b protein levels were probed and Actin served as loading control.
Highlights.
miR-203 expression is downregulated during retina regeneration
Downregulation of miR-203 is necessary for progenitor cell proliferation
Excess miR-203 impairs formation of progenitor clusters
miR-203 regulates proliferation by suppressing Pax6b expression
Acknowledgments
We thank Qiang Guan for zebrafish maintenance at the Vanderbilt University Stevenson Center Fish Facility. This study was supported in part by grants from the National Eye Institute–National Institutes of Health to J.G.P (R21 EY 019759) and D.R.H. (R01-EY018417) and the Center for Zebrafish Research, University of Notre Dame. K.R. was supported in part by the Vanderbilt International Scholar Program and by the Gisela Mosig fund in the Department of Biological Sciences.
Footnotes
Competing Interests
The authors declare no financial or non-financial competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernardos RL, Raymond PA. GFAP transgenic zebrafish. Gene Expr Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Iandiev I, Pannicke T, Wurm A, Hollborn M, Wiedemann P, Osborne NN, Reichenbach A. Cellular signaling and factors involved in Muller cell gliosis: neuroprotective and detrimental effects. Progress in Retinal and Eye Research. 2009;28:423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Diao Y, Guo X, Jiang L, Wang G, Zhang C, Wan J, Jin Y, Wu Z. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in Rhabdomyosarcoma. The Journal of Biological Chemistry. 2013;288:18923–18938. doi: 10.1074/jbc.M113.494716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP. p63 and FGFR: when development meets proliferation. EMBO Molecular Medicine. 2012;4:165–167. doi: 10.1002/emmm.201200202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Muller glia in regeneration of the injured zebrafish retina. Journal of Neuroscience. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SJ, Zhang Z, Feng D, Flagg M, O’Loughlin E, Wang D, Stokes N, Fuchs E, Yi R. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140:1882–1891. doi: 10.1242/dev.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl MO, Hayes S, Nelson BR, Tan K, Buckingham B, Reh TA. Stimulation of neural regeneration in the mouse retina. Proc Natl Acad Sci U S A. 2008;105:19508–19513. doi: 10.1073/pnas.0807453105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Lamba D, Karl M, Reh T. Neural Regeneration and Cell Replacement: A view from the eye. Cell Stem Cell. 2008;2:538–549. doi: 10.1016/j.stem.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kimelman D. A dominant-negative form of p63 is required for epidermal proliferation in zebrafish. Dev Cell. 2002;2:607–616. doi: 10.1016/s1534-5807(02)00166-1. [DOI] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, Candi E. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima M, Barthel LK, Raymond PA. A self-renewing division of zebrafish Muller glial cells generates neuronal progenitors that require N-cadherin to regenerate retinal neurons. Development. 2013;140:4510–4521. doi: 10.1242/dev.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci U S A. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Kogure T, Nuovo GJ, Jiang J, He L, Kim JH, Phelps MA, Papenfuss TL, Croce CM, Patel T, et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. The Journal of Investigative Dermatology. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- Pollak J, Wilken MS, Ueki Y, Cox KE, Sullivan JM, Taylor RJ, Levine EM, Reh TA. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development. 2013;140:2619–2631. doi: 10.1242/dev.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell C, Grant AR, Cornblath E, Goldman D. Analysis of DNA methylation reveals a partial reprogramming of the Muller glia genome during retina regeneration. Proc Natl Acad Sci U S A. 2013;110:19814–19819. doi: 10.1073/pnas.1312009110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Raymond PA. Microarray-based gene profiling analysis of Muller glia-derived retinal stem cells in light-damaged retinas from adult zebrafish. Methods Mol Biol. 2012;884:255–261. doi: 10.1007/978-1-61779-848-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Zhao XF, Goldman D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc Natl Acad Sci U S A. 2011;108:15858–15863. doi: 10.1073/pnas.1107220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, Saintigny G, Dellambra E, Odorisio T, Mahe C, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul KE, Koke JR, Garcia DM. Activating transcription factor 3 (ATF3) expression in the neural retina and optic nerve of zebrafish during optic nerve regeneration. Comparative biochemistry and physiology Part A, Molecular & Integrative Physiology. 2010;155:172–182. doi: 10.1016/j.cbpa.2009.10.042. [DOI] [PubMed] [Google Scholar]
- Sehm T, Sachse C, Frenzel C, Echeverri K. miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Dev Biol. 2009;334:468–480. doi: 10.1016/j.ydbio.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Wei T, Pavez Lorie E, Suzuki H, Kato M, Torma H, Stahle M, Pivarcsi A. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. The Journal of Investigative Dermatology. 2010;130:124–134. doi: 10.1038/jid.2009.294. [DOI] [PubMed] [Google Scholar]
- Taube JH, Malouf GG, Lu E, Sphyris N, Vijay V, Ramachandran PP, Ueno KR, Gaur S, Nicoloso MS, Rossi S, et al. Epigenetic silencing of microRNA-203 is required for EMT and cancer stem cell properties. Scientific Reports. 2013;3:2687. doi: 10.1038/srep02687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher EJ, Flynt AS, Li N, Patton JR, Patton JG. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn. 2007;236:2172–2180. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- Thatcher EJ, Paydar I, Anderson KK, Patton JG. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci USA. 2008;105:18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Enright JM, Kassen SC, Montgomery JE, Bailey TJ, Hyde DR. Pax6a and Pax6b are required at different points in neuronal progenitor cell proliferation during zebrafish photoreceptor regeneration. Exp Eye Res. 2010;90:572–582. doi: 10.1016/j.exer.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Enright JM, Nelson CM, Montgomery JE, Hyde DR. Characterization of Muller glia and neuronal progenitors during adult zebrafish retinal regeneration. Exp Eye Res. 2008a;87:433–444. doi: 10.1016/j.exer.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel R, Kassen SC, Montgomery JE, Enright JM, Hyde DR. Inhibition of Muller glial cell division blocks regeneration of the light-damaged zebrafish retina. Developmental Neurobiology. 2008b;68:392–408. doi: 10.1002/dneu.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Viticchie G, Lena AM, Cianfarani F, Odorisio T, Annicchiarico-Petruzzelli M, Melino G, Candi E. MicroRNA-203 contributes to skin re-epithelialization. Cell Death & Disease. 2012;3:e435. doi: 10.1038/cddis.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viticchie G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, Bernardini S, Mauriello A, Miano R, Spagnoli LG, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- Wan J, Ramachandran R, Goldman D. HB-EGF is necessary and sufficient for Muller glia dedifferentiation and retina regeneration. Dev Cell. 2012;22:334–347. doi: 10.1016/j.devcel.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, Wang N, Zhang S, Zen K, Zhang C, et al. miR-203 Inhibits Cell Proliferation and Migration of Lung Cancer Cells by Targeting PKCalpha. PloS one. 2013;8:e73985. doi: 10.1371/journal.pone.0073985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zheng X, Shen C, Shi Y. MicroRNA-203 suppresses cell proliferation and migration by targeting BIRC5 and LASP1 in human triple-negative breast cancer cells. Journal of Experimental & Clinical Cancer Research. 2012;31:58. doi: 10.1186/1756-9966-31-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol. 2012;365:319–327. doi: 10.1016/j.ydbio.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li H, Jiang X, Guo L, Jiang W, Lu SH. MiR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells and Development. 2013 doi: 10.1089/scd.2013.0308. (available online) [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA. Responses of Muller glia to retinal injury in adult zebrafish. Vision Research. 2005;45:991–1002. doi: 10.1016/j.visres.2004.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FACS isolated post-mitotic MG and proliferating progenitor cells were analyzed by qPCR. Relative mRNA levels of MG-specific glutamine synthetase (GS) and progenitor cell-specific Ascl1a are shown. Data represent mean +/− s.e.m. (n=3 independent FACS).
Albino fish retinas were injected and electroporated with Dy-547 tagged control miRNA (A–C) or miR-203 (D–F) prior to the start of light exposure (0h). After 72h, retinas were assessed for PCNA expression (green) and Dy-547 fluorescent tag localization (red). Nuclei were counterstained with TOPRO (blue). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
TUNEL staining was performed on 72h light treated retina sections from control miRNA (A) or miR-203 (B) gain-of-function. Some TUNEL+ nuclei (red) were detected in the ONL. DNaseI treated retinas were used as positive controls for the TUNEL assay and showed TUNEL+ nuclei in all retinal layers (C). No cell death was detected in the negative control retina sections, which were subjected to the TUNEL protocol in the absence of terminal transferase (D). ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar 50um.
(A) Mature miRNA sequences for miR-203a and miR-203b. The miRNA ‘seed’ is highlighted in red. The single nucleotide that differs between the two family members is highlighted in magenta. (B) The pax6b 3′ UTR was cloned downstream of GFP. Pairing between miR-203a and its miRNA recognition element (MRE) in the pax6b 3′ UTR is shown.
RT-PCR was performed for pax6b, ascl1a and gapdh from whole retinas during retina regeneration. pax6b is upregulated during regeneration. ascl1a served as a positive control for the progression of retina regeneration. gapdh served as loading control.
Control miRNA or miR-203 was injected and electroporated into the left eyes of albino zebrafish after MG dedifferentiation and progenitor cell generation (51h of light). After 72h, dorsal retinas were collected from control miRNA and miR-203 injected fish and protein lysates were prepared for Western blots. Pax6b protein levels were probed and Actin served as loading control.