Abstract
The effective delivery of DNA locally would increase the applicability of gene therapy in tissue regeneration, where diseased tissue is to be repaired in situ. One promising approach is to use hydrogel scaffolds to encapsulate and deliver plasmid DNA in the form of nanoparticles to the diseased tissue, so that cells infiltrating the scaffold are transfected to induce regeneration. This study focuses on the design of a DNA nanoparticle loaded hydrogel scaffold. In particular, this study focuses on understanding how cell-matrix interactions affect gene transfer to adult stem cells cultured inside matrix metalloproteinase degradable hyaluronic acid hydrogel scaffolds. Hyaluronic acid was crosslinked to form a hydrogel material using a matrix metalloproteinase degradable peptide and Michael addition chemistry. Gene transfer inside these hydrogel materials was assessed as a function of polyplex nitrogen to phosphate ratio (N/P = 5 to 12), matrix stiffness (100 to 1700 Pa), RGD concentration (10 to 400 µM) and RGD presentation (.2 to 4.7 RGDs per HA molecule). All variables were found to affect gene transfer to mouse mensenchymal stem cells culture inside the DNA loaded hydrogels. As expected higher N/P ratios lead to higher gene transfer efficiency but also higher toxicity, softer hydrogels resulted in higher transgene expression than stiffer hydrogels, an intermediate RGD concentration and RGD clustering resulted in higher transgene expression. We believe that the knowledge gained through this in vitro model can be utilized to design better scaffold-mediated gene delivery for local gene therapy.
Keywords: Hydrogel scaffolds, non-viral gene delivery, poly(ethylene imine), hyaluronic acid, mesenchymal stem cell
Introduction
The natural extracellular matrix (ECM) is a dynamic network consisting of protein fibers and polysaccharides that support cell fate and provide biophysical and biochemical cues to cells through cell surface receptors, such as integrins1, 2. Naturally cells degrade the ECM through proteases during their migration and tissue remodeling. Synthetic ECM hydrogels that aim to provide an environment in which to direct stem cell fate should recapitulate key features of the natural ECM such as integrin mediated cell binding and growth factor mediated signaling, while also allowing for cell migration and proliferation. Hyaluronic acid (HA), an anionic, non-sulfated glycoaminoglycan and major component of the ECM, is widely distributed in connective, epithelial and neural tissue3. HA has recently gained popularity as a biomaterial for tissue engineering and tissue regeneration due to its high biocompatibility and low immunogenicity4–7. Several studies have demonstrated that HA-based hydrogels are good candidates for culturing stem cells8–12. HA specifically interacts with cell surface receptors, such as CD44, RHAMM (receptor for HA mediated motility) and ICAM-1 (intercellular adhesion molecule 1), and contributes to tissue hydrodynamics, cell proliferation and migration13, 14. Semi-synthetic hyaluronic acid (HA) hydrogels which are also degradable by matrix metalloproteinases (MMPs) have previously been developed for culturing mouse mesenchymal stem cells in 3-dimensions15. While peptides and growth factors can be easily incorporated within these hydrogels, rapid degradation by proteases generally limits their effectiveness in long-term cell culture. Non-viral gene delivery is an ideal alternative for the incorporation of bioactive signals within tissue-engineering constructs.
Local gene delivery using hydrogel scaffolds has been studied for over a decade primarily through the encapsulation of naked DNA during hydrogel formation. Naked DNA has been successfully incorporated inside collagen16, pluronic-hyaluronic acid17, PEG-poly(lactic acid)-PEG18, alginate19, oligo(polyethylene glycol) fumarate20 and engineered silk elastin21. Although naked DNA has shown gene expression and ability to guide regeneration in vivo16, 22, limitations with low gene transfer efficiency and rapid diffusion of the DNA from the hydrogel scaffold has motivated the use of DNA nanoparticles instead of naked DNA. DNA condensed either with cationic peptides, lipids, or polymers has been introduced into fibrin hydrogels23–27, enzymatically degradable PEG hydrogels23 and PEG-hyaluronic acid hydrogels24.
Here we aim to study gene transfer to stem cells as a function of cell-matrix interactions since the effects of cell-matrix interactions on non-viral gene transfer are not well-established and required for further progress to be made with hydrogel-mediated transfection. For cells seeded in two dimensions matrix stiffness has been shown to influence transgene expression, with stiffer surfaces resulting in enhanced transgene expression28. Further, for cells seeded in two dimensions RGD presentations were also shown to influence transgene expression, with RGD clustering resulting in enhanced gene transfer compared to homogeneously distributed peptides29. Studies for gene transfer in three dimensions have shown that the migration rates as well as the matrix degradation are important parameters that influence transgene expression30, 31.
We previously reported on a matrix metalloproteinase degradable hyaluronic acid hydrogel scaffold for stem cell culture15. Here we use this hydrogel scaffold to investigate how cell-matrix interactions modulate gene transfer. Matrices with different mechanical properties and RGD presentations are used to determine how these parameters affect gene transfer to cells seeded in three dimensions (inside the hydrogel scaffold). For these studies poly(ethylene imine) (PEI) was used as a transfection reagent. PEI is a cationic polymer that has been widely utilized for non-viral gene delivery. PEI is able to condense DNA through electrostatic interactions between the positively charged amines on the PEI and the negatively charged phosphates on the DNA, forming nanoparticles (polyplexes) in the range of 50 to 200 nm32. PEI has been successfully used in vivo delivering DNA or siRNA to the brain33, 34, lungs35–38, abdomen39, and tumors40–42. We believe that the use of gene based bioactive signals to guide stem cell differentiation or cell trans-differentiation in 3D hydrogel scaffolds will be a powerful approach for tissue engineering and tissue regeneration applications.
Materials and Methods
Materials
Peptides Ac-GCRDGPQGIWGQDRCG-NH2 (MMPxl) and Ac-GCGWGRGDSPG-NH2 (RGD) were obtained from (Genescript, Piscataway, NJ). Sodium hyaluronan was a gift from Genzyme Corp. (Boston, MA). Gaussia luciferase expression vector (pGluc, New England BioLabs, Ipswich, MA) was expanded using an endotoxin free Giga Prep kit from Qiagen following the manufacturer’s instructions. Linear PEI (25 kg/mol) was purchased from Polysciences (Warrington, PA). All other products were purchased from Fisher Scientific unless noted otherwise.
Cell Culture
Mouse bone marrow cloned mesenchymal stem cells (mMSCs, D1, CRL12424) were purchased from ATCC (Manassas, VA) and cultured in DMEM (Invitrogen) supplemented with 10% bovine growth serum (BGS, Hyclone, Logan, UT) and 1% penicillin/streptomycin (Invitrogen, Grand Island, NY) at 37°C and 5% CO2. The cells were split using trypsin following standard protocols.
Modification of hyaluronic acid
Acrylated hyaluronic acid (HA-AC) was prepared using a two-step synthesis as previously described15. Briefly, hyaluronic acid (60,000 Da, Genzyme Corporation, Cambridge, MA) (2.0 g, 5.28 mmole, 60 kDa) was reacted with 18.0 g (105.5 mmole) adipic dihydrazide (ADH) at pH 4.75 in the presence of 4.0 g (20 mmole) 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) overnight and purified through dialysis (8000 MWCO) in DI water for 2 days. The purified intermediate (HA-ADH) was lyophilized and stored at −20 °C until used. 40.46% of the carboxyl groups were modified with ADH based on the trinitrobenzene sulfonic acid (TNBSA, Pierce, Rockford, Illinois) assay. HA-ADH (1.9 g) was reacted with N-Acryloxysuccinimide (NHS-AC) (1.33 g, 4.4 mmole) in HEPES buffer (pH 7.2) overnight and purified through dialysis in DI water for 2 days before lyophilization. The degree of acrylation of 10% was determined using 1H-NMR (D2O) by taking the ratio of multiplet peak at δ = 6.2 corresponding to the cis and trans acrylate Hs to the singlet peak of the acetyl methyl protons in HA (δ = 1.6).
DNA loaded HA hydrogel synthesis and characterization
HA hydrogels were formed by Michael-type addition of bis-cysteine containing MMPxl peptides onto HA-AC functionalized with cell adhesion peptides (RGD). A lyophilized aliquot of RGD peptides (0.1 mg) was dissolved in 15 µL of .3M TEOA buffer (pH=8.7), mixed with HA-AC and allowed to react for 20 minutes at room temperature. The HA-RGD solution was kept in ice until used. DNA/PEI polyplexes were formed by mixing 5µg plasmid DNA with 4.57µg PEI in nuclease-free water, vortexing for 15 s and incubating for 10 min at room temperature. Polyplexes were cooled in ice prior to gelation. Immediately before adding the polyplex solution to the hydrogel precursors ice cooled 3M TEOA (pH=8.2) was added to bring the final concentration of buffer in the polyplexes to .3M TEOA. A lyophilized aliquot of the crosslinker (0.91 mg MMPxl) was then diluted in 18.2 µL of .3M TEOA buffer (pH=8.2) immediately before mixing with DNA/PEI polyplexes, HA-RGD (final concentration of 100 µM RGD) and the cell solution (500,000 cells per 100 µl final gel volume). The final gel solution had a pH=8.1 and all precursors were kept on ice prior to mixing. Gelation was achieved by placing a drop of the precursor solution between sigmacoted glass slides for 30 min at 37 °C. The final gel was placed inside 96-well plates for culture. Thorough mixing was used to ensure the polyplexes were uniformly distributed throughout the hydrogel.
Hydrogels with variable stiff nesses were prepared by changing the ratio of thiols to acrylates (r ratio) or the concentration of HA used (see Table 1 for conditions and resulting storage moduli). To generate hydrogels with different RGD presentations different portions of HA-AC were first mixed with the constant amount of RGD peptide. The HA-RGD was then mixed with unmodified HA-AC. Thus homogenous RGD represents the condition were 100% of the HA-AC was mixed with the RGD peptide. For the RGD presentation studies a constant concentration of 100 µM RGD was used.
Table 1. Hydrogel mechanical properties.
Hydrogel formation conditions and overall storage modulus
| Hydrogel ID |
HA% | r ratio | G’ (+DNA) |
|---|---|---|---|
| 1 | 3 | 0.7 | 100.0 ± 2.11 |
| 2 | 3 | 1.05 | 260.0 ± 1.30 |
| 3 | 3.5 | 1.05 | 839 ± 12.30 |
| 4 | 4 | 1.05 | 1360 ± 11.73 |
| 5 | 5 | 1.05 | 1730 ± 45.62 |
r ratio represents the moles of –SH over the moles of AC
To visualize the distribution of the polyplexes inside the HA hydrogels, gels were formed using the same protocol as described above but in the absence of cells. The hydrogels were stained with ethidium bromide post hydrogel formation and imaged using a fluorescence inverted microscope. The images were taken using the fluorescent (Observer Z1 Zeiss) microscope with 10× magnification.
Characterization of HA hydrogel mechanical properties
The storage and loss modulus were measured with a plate-to-plate rheometer (Physica MCR, Anton Paar, Ashland, VA) using a 8 mm plate under a constant strain of 0.03 and frequency ranging from 0.1 to 10 rad/s. Hydrogels were made as detailed above and cut to a size of 8.0 mm in diameter to fit the plate. A humid hood was used to prevent the hydrogel from drying and the temperature was kept at (37°C).
Radiolabeling DNA
Plasmid DNA was radiolabeled with 3H-dCTP (100 µCi, MP Biomedicals, Santa Ana, CA) using a Nick translation kit (Roche, Indianapolis, IN) as per the manufacturer’s protocol. Briefly, an equimolar mixture of dATP, dGTP, dTTP, and 3H-dCTP was prepared and added to the DNA (1 µg) solution. Once the enzyme solution was added to the mixture, the final solution (200 µl) was gently mixed by pipetting and incubated for 2 h at 15 °C. The reaction was stopped by addition of 10 µl 0.2 M EDTA (pH=8.0) and heating to 65 °C for 10 min. The DNA was purified using the mini Prep kit from Qiagen following the manufacturer’s instructions. The final DNA concentration was 0.04 µg/µl.
DNA/PEI polyplex release kinetics and activity
In order to determine the extent of release of the encapsulated polyplexes and their activity post encapsulation, gels were formed using the protocols indicated above with 1% radiolabeled DNA. To test the release kinetics, the gels were swelled in PBS for 2 h and the swelling solution was collected. The gels were then placed in 150 µL of release solution (PBS, 10 U/ml HAase and 0.50 U/ml Col I). At the indicated time points, 150 µL of the solution was removed and an additional 150 µL of fresh release medium was added. After 192 h, the Col I concentration was increased to 1 U/ml. Following the final release medium collection, the gels were incubated with 0.25% trypsin/EDTA to result in complete release of the DNA from the gel upon degradation. DNA concentrations were measured using a scintillation counter at the UCLA Chemistry core facility. The readout was analyzed using a standard curve.
To determine the activity of the encapsulated DNA/PEI polyplexes, a HA gel was prepared and swelled as indicated above using DNA encoding for Gaussia luciferase (pGluc). After swelling in PBS, the gel was degraded through incubation with 100 µL 0.25% trypsin at 37 °C for 10 min. To determine how much release solution should be added to add 0.5µg of DNA to mMSCs cultured on a 48-well plate, the DNA concentration in the sample was measured using HOECHST dye (H33258). In a typical measurement, 100 µl heparin (10 mg/mL in PBS) was incubated with 10 µL of the degraded sample for 10 min at room temperature to displace the DNA from DNA/PEI complex.10 µL of the above solution was then mixed with 100 µL H33258 assay solution (0.1 mg/mL H33258 in TNE buffer (0.2 M NaCl, 10 mM Tris, 1.3 mM EDTA) and the fluorescence light intensity was read using a fluorometer equipped with a UV filter (Turner Biosystems, Sunnyvale, CA). The readout was analyzed using a standard curve measured using complexed DNA. The collected polyplexes from the degraded hydrogel sample was then used for a bolus transfection (0.5µg DNA for a 48 well-plate) and compared to freshly made polyplexes. The cell media was collected after 48 h and transgene expression was measured using the Gaussia Luciferase Assay Kit (New England BioLabs. Ipswich, MA) as described below (Gene transfer section).
Cell viability, spreading and proliferation
Cell viability in these hydrogels was studied using LIVE/DEAD viability/cytotoxicity kit (Molecular Probes, Eugene, OR). Briefly, 2 µl of ethidium homodimer-1 and 0.5 µL of calcein AM from the kit were diluted with 1 mL DMEM. Each gel was stained with 150 µL of this staining solution for 30 min at 37 °C in the dark.
To better analyze cell spreading, separate gels were fixed for 30 min at RT using 4 % paraformaldehyde, rinsed with PBS, treated with 0.1 % triton-X for 10 min and stained for 90 min in the dark with DAPI for cell nuclei (500× dilution from 5 mg/ml stock) and rhodamine-phalloidin (5 µl per 200 µl final stain solution) in 1 % bovine serum albumin solution. The samples were then washed with 0.05 % tween-20. For both cell viability and cell spreading, an inverted Observer Z1 Zeiss fluorescence microscope was used to visualize samples. To better visualize the distribution throughout the hydrogel, multiple z-stacks 12–13 µm thick were taken for each image, deconvoluted to minimize background, and presented as maximum intensity projections.
MTT assay (CellTiter 96R AQueous One Solution Cell Proliferation Assay, Promega, Madison, WI) was used to quantify the cell proliferation rate. 20 µL MTT reagent with 100 µL DMEM was added to each well (96-well plate) and incubated at 37 °C for 2 hrs. The cells were lysed after 2 h with addition of 10 % sodium dodecyl sulfate. The solutions were transferred to a new plate and absorbance was measured at 490 nm using a standard plate reader. Three gels for each condition were analyzed at each time point. Readings were normalized to day 2 readings for gels containing no DNA.
Gene transfer
pGluc/PEI nanoparticle loaded hydrogels with mMSCs were made as described above. Each day the media was collected and frozen immediately at –20°C and fresh media was added to each gel. To quantify secreted Gaussia luciferase levels in the media, the samples were thawed on ice and assayed using a BioLuxTM Gaussia Luciferase Assay Kit (New England Biolabs, Ipswich, MA) as per the manufacturer’s protocol. Briefly, 20 µl sample was mixed with 50 µl 1× substrate solution, pipetted for 2–3 sec, and read for luminescence with a 5 sec integration. Background was determined with media from gels that did not contain any DNA and values were expressed as relative light units (RLU).
Results
Hydrogel preparation, DNA loading and characterization
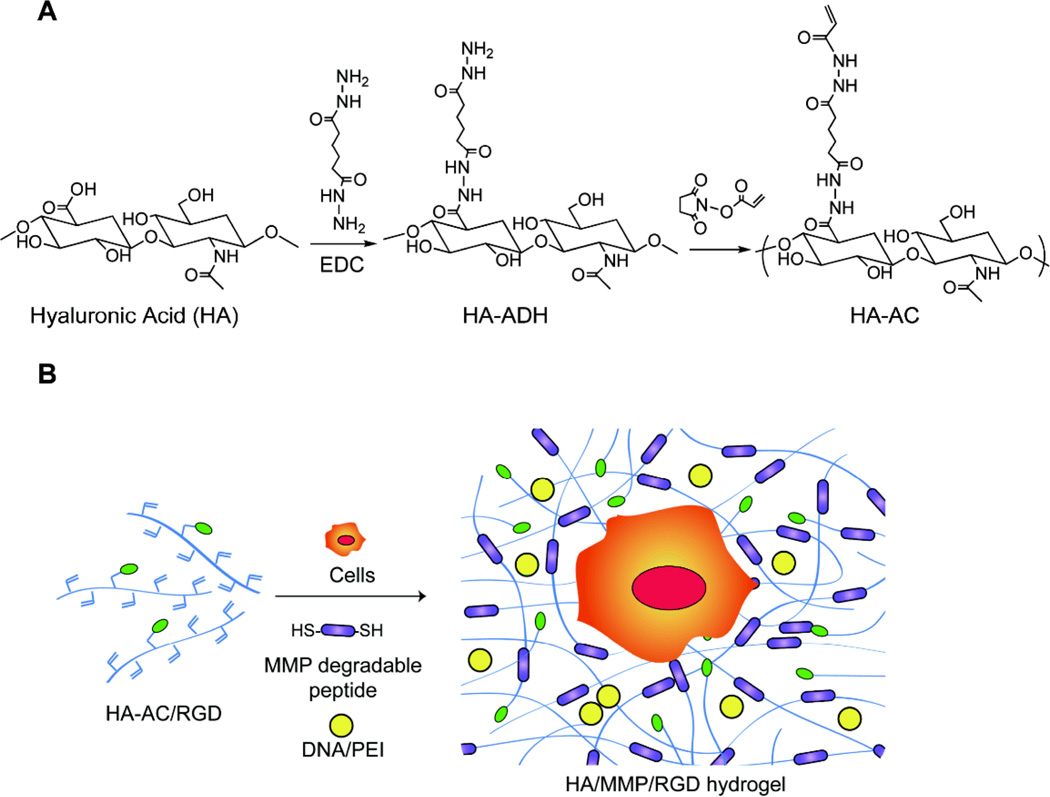
Acrylates were conjugated onto the HA backbone through a two-step process (Scheme 1A). HA was first modified with adipic acid dihydrazide (ADH) using carboxylic acid/amine chemistry and the resulting hydrazide groups were then modified with NHS-acrylate (NHS-AC) to get acrylamide functionalities. An amine content assay (TNBSA assay) showed that 40.46% of the carboxylic acids were modified with ADHs. After reacting the HA-ADH with NHS-AC at pH 7.2 overnight, 1H-NMR showed that 10% of the hydrazide groups were modified with acrylate groups on the final products, resulting in approximately 15 acrylates per HA chain.
Scheme 1.
chematic of HA modification and hydrogel formation. (A) HA-acrylate synthesis is a two-step process first reacting HA with ADH and then using the pendant hydrazide to react with NHS-acrylate. (B) Schematic of DNA-loaded hydrogel formation. Liquid HA-AC is first modified with RGD peptides using Michael type addition. HA-RGD is then crosslinked using an MMP degradable peptide in the presence of DNA/PEI polyplexes.
RGD adhesion peptides were incorporated through Michael-type addition of the cysteine side chain in the peptide to the acrylate groups on the HA backbone. The crosslinker was then added to form the hydrogels (Scheme 1B). Polyplexes were incorporated into the hydrogel during hydrogel gelation. Polyplexes were formed in a final volume of 20 µL by mixing equal volumes of PEI and DNA, vortexing for 15 seconds and incubating for 15 minutes. The full volume of the polyplexes was incorporated into 100 µL of hydrogel precursor solution to achieve a final DNA concentration of 0.05 µg DNA/µL hydrogel. Cells were also incorporated during gelation.
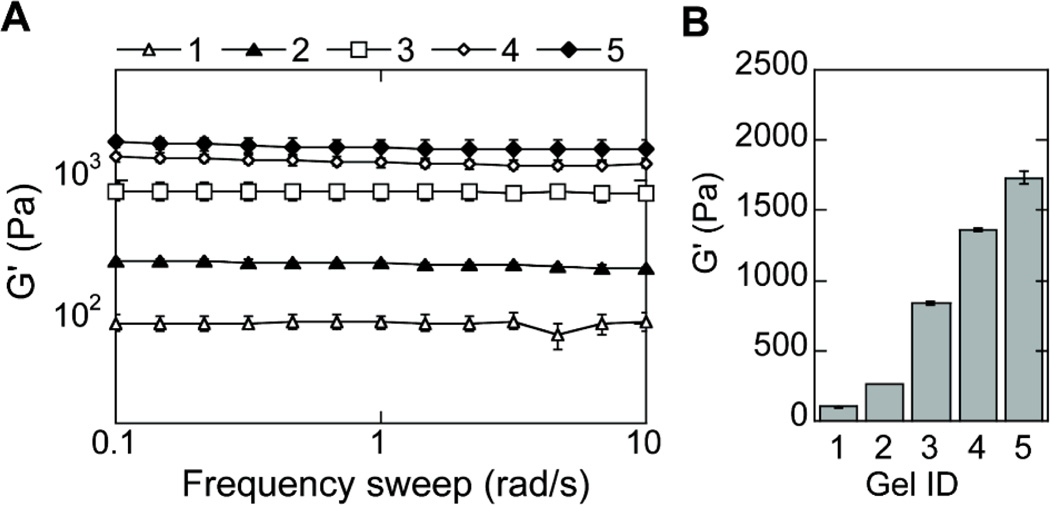
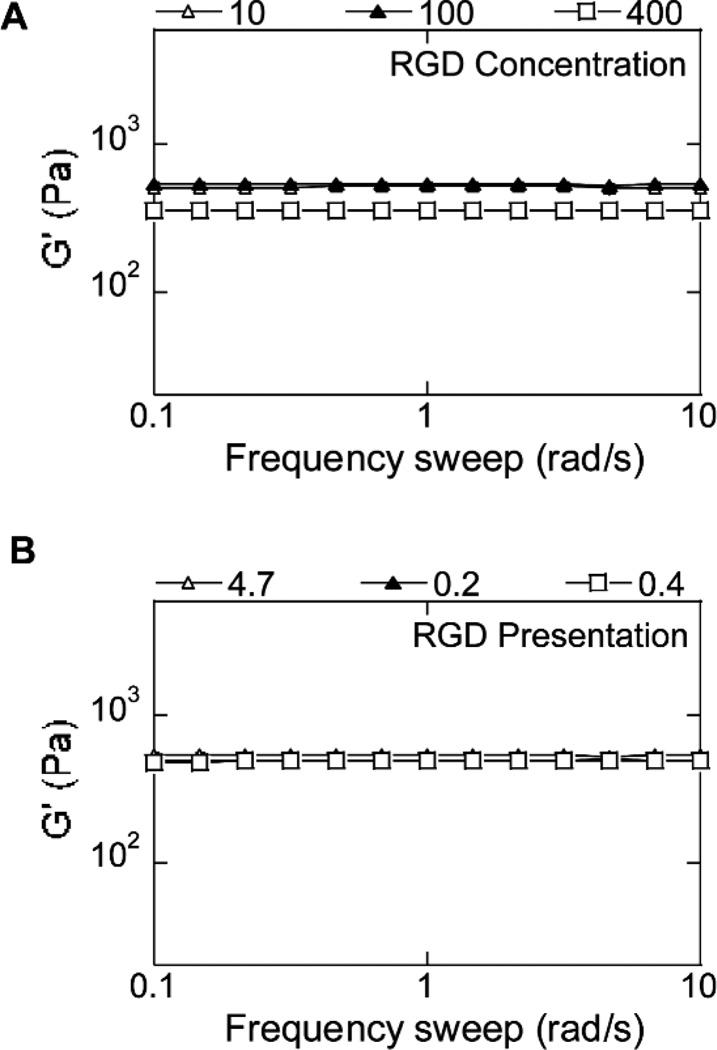
The storage (G') and loss moduli (G") of hydrogels with DNA polyplexes were measured at 37°C using plate-to-plate rheology with an 8 mm geometry. An evaporation blocker was utilized to avoid drying of the hydrogel sample. Results showed that the G' and G" did not cross at any measured frequency (0.1 to 10 Hz) and were frequency independent (Fig. 1A), both of which are consistent with typical hydrogel characteristics. The loss tangent values (ratio of G" to G') were lower than 0.06 for all hydrogels tested, indicating that the hydrogels were highly elastic. The mechanical properties of the hydrogels were controlled by varying the percent of the HA solution or the r ratio, resulting in a final range of mechanical properties from 100 to 1700Pa (Table 1, Fig. 1B).
Figure 1.
Hydrogel mechanical properties. The mechanical properties of the hydrogels were determined using plate-to-plate rheometry Storage (A) and average (B) modulus over a frequency range of 0.1 to 10 rad/s at a constant strain of .03 are shown for increasingly stiff hydrogels (Gel ID 1 < 2 < 3 < 4 < 5).
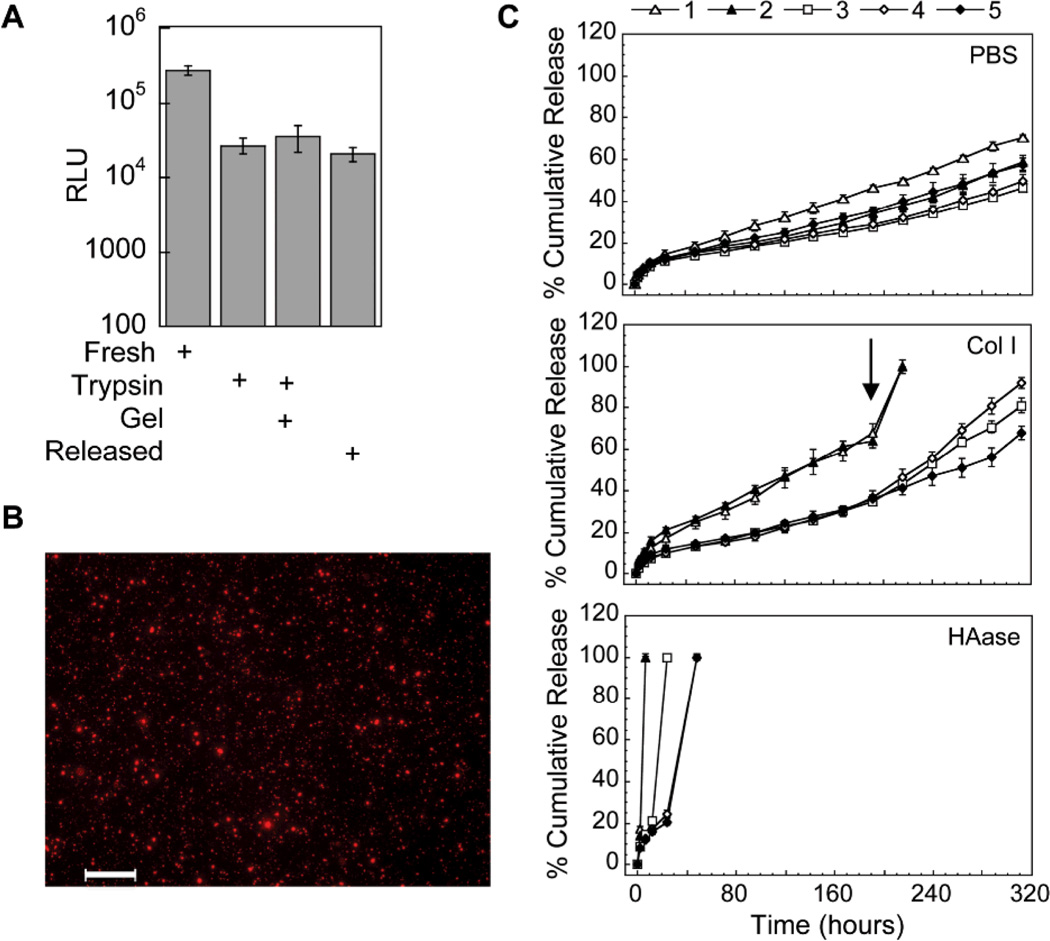
The activity of the entrapped polyplexes was estimated by synthesizing a polyplex-loaded hydrogel, degrading the hydrogel with trypsin and performing a bolus transfection with the released polyplexes (Fig. 2A). The observed transfection was compared to that of fresh polyplexes, fresh polyplexes with trypsin added, and fresh polyplexes with a degraded hydrogel added. The amount of transgene expression decreased with the addition of trypsin but was the same for that of polyplexes exposed to hydrogel degradation products and released polyplexes indicating that the polyplexes were active inside the hydrogel. The distribution of the polyplexes inside the hydrogel scaffold was determined through the staining of the hydrogel with ethidium bromide post hydrogel formation. Polyplexes were observed mostly as unaggregated particles and uniformly distributed throughout the gel (Fig. 2B).
Figure 2.
Polyplex activity, distribution inside hydrogel scaffolds and release. (A) Activity of the entrapped polyplexes was determined through the release of the polyplexes post hydrogel formation using trypsin and a subsequent bolus transfection with the released polyplexes. The gene transfer of the released polyplexes was compared to fresh polyplexes with trypsin added and fresh polyplexes with gel degradation products added. (B) DNA/PEI polyplexes were stained with ethidium bromide post hydrogel formation and imaged with a fluorescence microscope equipped with z-stack capability. Scale bar = 100µm. (C) DNA release was determined using radiolabeled DNA. DNA/PEI loaded hydrogels were incubated in different release solutions and at predetermined time points samples were gathered and analyzed for radioactivity using a scintillation counter. At the final day of the release assay the hydrogel was fully degraded with trypsin and the final activity measured. Data is plotted as the % cumulative release.
To determine the release kinetics of the entrapped polyplexes, radiolabeled DNA was used. Release studies indicated that the hydrogel mechanical properties modulated the release rate of the polyplexes (Fig. 2C). In general, stiffer materials resulted in lower release rates in PBS, collagenase I and hyaluronidase. DNA release in collagenase I and hyaluronidase were directly modulated by hydrogel stiffness, with hydrogels with 100 and 260 Pa resulting in faster release than hydrogels with higher storage modulus. At 192 hours a higher collagenase I concentration was added to induce faster release. However, even with this higher concentration stiff hydrogels did not result in complete hydrogel degradation and DNA release by 312 hours, with 81%, 92% or 68%, released for 839, 1360 and 1730 Pa gels, respectively. DNA release in PBS was also seen with the trend 70%, 59%, 46.4%, 50%, and 58%, for 100, 260, 839, 1360, 1730 Pa hydrogels respectively which was not as heavily dependent of the hydrogel stiffness.
Cell viability, proliferation and gene transfer as a function of N/P ratio
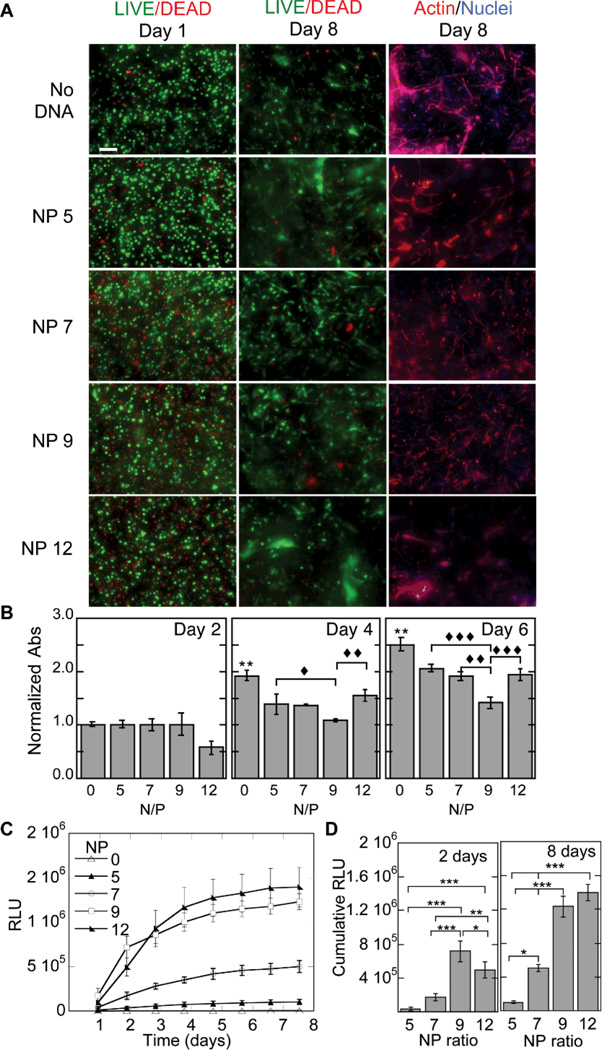
The toxicity of the DNA/PEI polyplexes was assessed using both the LIVE/DEAD and MTT assays as a function of nitrogen to phosphate (N/P) ratio (Fig. 3A,B). Although all conditions contained some dead (red) cells, we observed minimal toxicity for cells cultured in hydrogels that did not contain polyplexes and for cells cultured inside hydrogels with polyplexes made with N/P 5 and N/P 7 ratios. However, by 8 days the images showed lower numbers of cells for the N/P 9 and N/P of 12 conditions. Cell spreading as a function of N/P ratio was also investigated (Fig. 3A). Cell spreading was observed for all the conditions tested; however, N/P of 5, 7 and 9 had the most abundant cell spreading at day 8. The MTT assay was performed at days 2, 4 and 6 of transfection, supporting the results from the LIVE/DEAD and spreading images. It showed that the no DNA, N/P 5 and N/P 7 conditions contained more cells than the N/P 9 and 12 conditions. At day 2 there were a statistically lower number of cells in the N/P 12 condition (p < 0.05); however, all other conditions contained the same number of cells. By day 4 and 6 the no DNA condition contained statistically more cells than any other condition, while N/P 5 and 7 have statistically the same cell number with respect to each other but have statistically more cells than the N/P 9 condition (p < at least 0.05). Surprisingly the N/P 12 condition recovered in cell number quicker than the N/P 9 condition suggesting that the polyplexes were released faster from this hydrogel and allowed the cells to grow again.
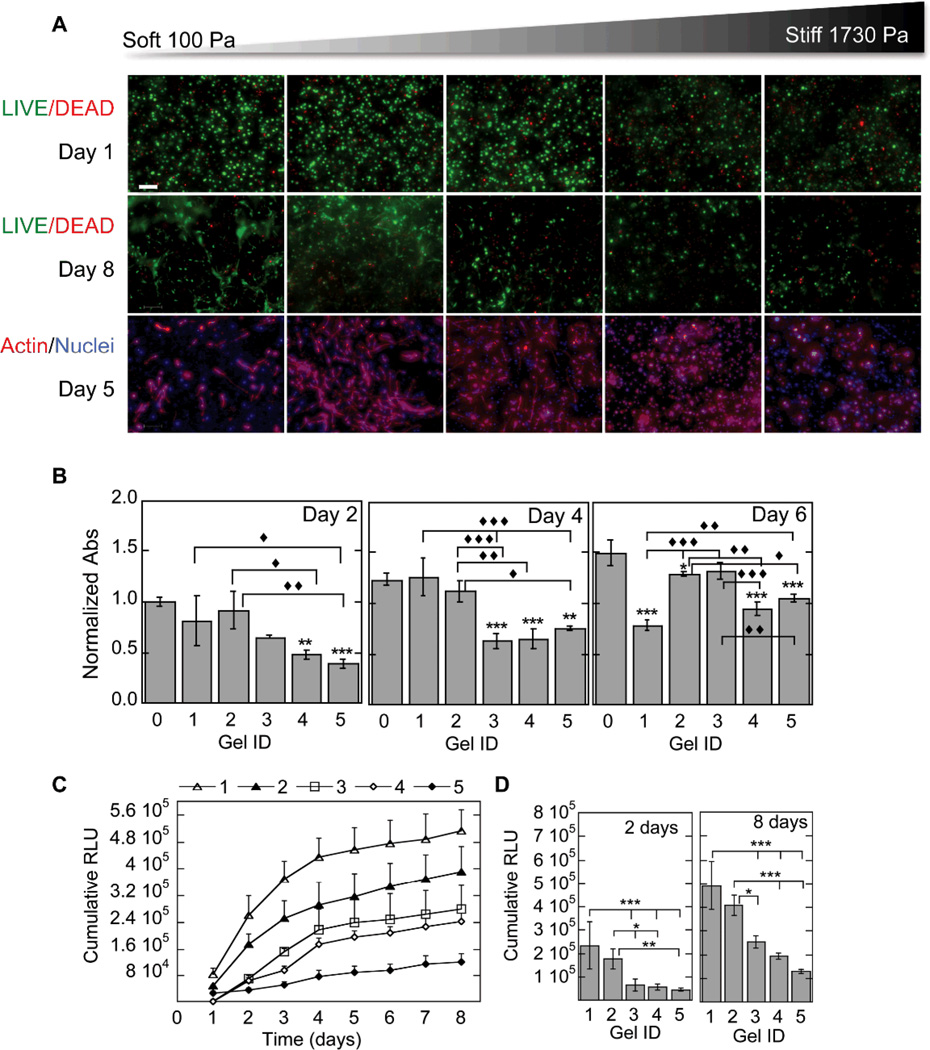
Figure 3.
Gene transfer as a function of N/P ratio. The effect of N/P ratio on transgene expression was studied for cells cultured inside MMP degradable HA hydrogels. For these studies a 3% hydrogel with an r ratio of 0.3 was used. The cell viability, ability of the cells to spread and the metabolic activity of the cells were studied using the LIVE/DEAD assay, phalloidin staining (A) and MTT assay (B). Gene expression was determined over time using a reporter plasmid, gaussia luciferase, which is secreted by the cell when expressed (C). The cumulative expression at days 2 and 8 is plotted for ease of comparison (D). Statistical significance was determined using multiple comparisons and either the Dunnett or the Tukey multiple comparison’s tests. The symbol ** indicates statistical significance at the level of 0.01 between the indicated condition and the corresponding no DNA control in (B) or between the indicated conditions in (D). The symbols ♦, ♦♦, ♦♦♦ indicate statistical significance at the level of 0.05, 0.01 and 0.001 between the indicated conditions in (B). N/P = 0 represents the condition with no DNA polyplexes added to the hydrogel. Scale bar = 100µm.
Gene transfer was also assessed as a function of N/P ratio while keeping the amount of DNA constant at 5µg per 100µL of hydrogel. In general, transgene expression increased with increasing N/P (Fig. 3C), with N/P of 12 and 9 achieving significantly more transgene expression than N/P of 5 and 7 (Fig. 3D). At day 2, N/P of 9 and 12 transgene expression was statistically significantly more than N/P 5 and 7 (p < at least 0.01) and N/P 9 was significantly more than N/P of 12 (p < 0.05). Similarly at day 8, transgene expression for N/P of 9 and 12 was statistically significantly more than N/P 5 and 7 (p < 0.001); however, there was also a significant difference between N/P 7 and N/P 5 (p < 0.05) with no statistical difference between N/P 9 and 12. To balance toxicity with gene transfer in our subsequent experiments N/P of 7 was used.
Hydrogel mechanical properties influence cell proliferation, spreading and transgene expression
Hydrogels with different mechanical properties were synthesized and analyzed for their ability to modulate non-viral gene transfer. The viability of the cells at days 1 and 8 was assessed using the LIVE/DEAD stain and at 2, 4, and 6 days using the MTT assay (Fig. 4A,B). The LIVE/DEAD stain showed minimal cell death at day 1 indicating that the hydrogel synthesis conditions did not affect cell viability. However, by day 8 significantly fewer cells were observed for the highest modulus hydrogels (gel ID 4 and 5). Cell spreading at day 5 was affected by hydrogel stiffness with 100 to 839Pa hydrogels resulting in substantial cell spreading, while hydrogel stiffness of 1350 and 1730 Pa resulted in reduced cell spreading (Fig. 4A). The MTT assay agreed with the live/dead data (Fig. 4B), showing significantly more metabolic activity at days 2 and 4 for the 100 Pa and 260Pa hydrogels compared to 1360 and 1730Pa hydrogels (p at least < 0.05). Further, at days 2 and 4 there was no statistically significant difference between the no DNA condition (gel ID 0) and the softest hydrogels (gel ID 1,2), while there was a statistical difference between no DNA and the stiffest samples (gel ID 4,5). By day 6 the softest hydrogel was significantly degraded, which resulted in the loss of cells and, therefore, showed a lower cell number. The 260Pa (gel ID 2) hydrogel remained higher in cell number than the two stiffest samples (gel ID 4, 5).
Figure 4.
Gene transfer as a function of hydrogel stiffness. The effect of hydrogel stiffness on the ability of cells seeded inside the hydrogel to become transfected was studied for hydrogels with storage modulus ranging from 100 Pa to 1730 Pa. The cell viability, ability of the cells to spread and the metabolic activity of the cells was studied using the LIVE/DEAD assay, phalloidin staining (A) and MTT assay (B). None of the cell stiffness resulted in lower cellular viability. However, cell spreading was inhibited for stiffer hydrogels. Gene expression was determined over time using a reporter plasmid, gaussia luciferase, which is secreted by the cell when expressed (C). The cumulative expression at days 2 and 8 is plotted for ease of comparison (D). Matrix stiffness influenced transgene expression. The numbers 1–5 represent different hydrogel stiffness. 1 = 100 Pa, 2 = 260 Pa, 3 = 839 Pa, 4 = 1360 Pa, 5 = 1730 Pa. Statistical significance was determined using multiple comparisons and either the Dunnett or the Tukey multiple comparison’s tests. The symbols ** and *** indicate statistical significance at the level of 0.05, 0.01 and 0.001 between the indicated condition and the corresponding no DNA control in (B) or between the indicated conditions in (D). The symbols ♦, ♦♦, ♦♦♦ indicate statistical significance at the level of 0.05, 0.01 and 0.001 between the indicated conditions in (B). Gel ID 0 represents the condition with no DNA polyplexes added to the hydrogel. Scale bar = 100µm.
Transgene expression was a direct function of the hydrogel stiffness with softer hydrogels resulting in enhanced gene transfer compared to stiffer hydrogels (Fig. 4C). Transgene expression at days 2 and 8 for 200 and 260 Pa hydrogels was also statistically significantly higher compared to the three stiffest hydrogels, with p < 0.001 and p < at least 0.05 for 200 and 260 Pa comparisons, respectively (Fig. 4D).
Effect of RGD concentration and presentation on gene transfer
In addition to the mechanical properties we also investigated how the concentration and presentation of RGD peptides on the hydrogel would affect the process of non-viral gene transfer. RGD concentrations of 10, 100 and 400 µM for the conditions containing DNA and 100µM for the condition with no DNA were tested. To look at the effect of RGD presentation the concentration of RGD was kept constant at 100µM but the fraction of the total HA it was reacted with was modulated to result in different numbers of RGD molecules per HA chain. For homogeneously dispersed RGD 100% of the HA was modified with RGD, however, for increasing degrees of RGD clustering lower and lower percentages of HA were reacted with the RGD. Thus, the trend of 0.2 < 0.4 < 4.7 for number of RGD per HA molecule represents an increase in clustering of RGD, with 4.7 RGD/HA representing the most clustered RGD condition. To ensure comparisons in transfection were due to the RGD concentration and presentation and not hydrogel stiffness, mechanical properties were tested. Regardless of RGD clustering and at low RGD concentrations (10 and 100µM) the mechanical properties were independent of RGD presentation and concentration (Fig. 5A,B). However, 400 µM RGD hydrogels were slightly softer (360 Pa for 400 µM compared to 516 and 541 Pa for 10 and 100 µM, respectively). This slight difference in modulus could not be avoided in order to be able to test a complete range of RGD concentrations.
Figure 5.
Hydrogel mechanical properties for hydrogels with different RGD concentrations and presentations. The mechanical properties of the hydrogels were determined using plate-to-plate rheometry Storage (A, B)modulus over a frequency range of 0.1 to 10 rad/s at a constant strain of 0.03 are shown for hydrogels with various RGD concentrations and presentations, respectively. RGD presentation is displayed as number of RGD/HA molecule with 4.7 RGD/HA being the most clustered condition and .2 RGD/HA being the least clustered/homogeneously distributed condition.
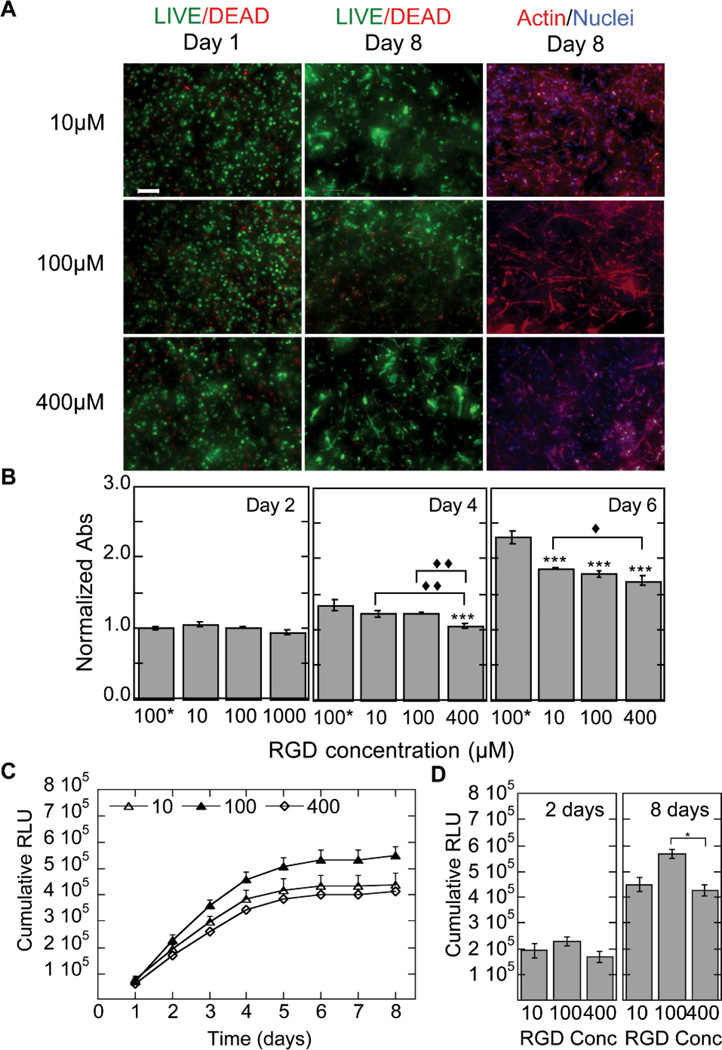
The viability of the cells at days 1 and 8 was assessed using the LIVE/DEAD stain and at 2, 4, and 6 days using the MTT assay (Fig. 6A,B). The LIVE/DEAD stain showed minimal cell death at day 1 indicating that the RGD conditions did not affect cell viability. By day 8 spreading was observed in all conditions (Fig. 6A). The MTT assay for various RGD concentrations showed that the number of cells in the hydrogel was the same as the no DNA condition for low concentrations of RGD peptide at days 2 and 4, while at day 4 the highest concentration of RGD tested showed a lower metabolic activity than the no DNA condition (Fig. 6B). At day 4 the 10 and 100µM RGD concentrations were statistically higher in metabolic activity than the 400µM condition. By day 6 all conditions showed lower metabolic activities than the sample with no DNA (p < 0.001), although the 10µM condition continued to show a higher metabolic activity than the 400µM condition (p < 0.05).
Figure 6.
Gene transfer as a function of RGD concentration. The effect of RGD concentration on the ability of cells seeded inside the hydrogel to become transfected was studied for hydrogels with RGD ranging from 10 µM to 400 µM. The cell viability, ability of the cells to spread and the metabolic activity of the cells was studied using the LIVE/DEAD assay, phalloidin staining (A) and MTT assay (B). Gene expression was determined over time using a reporter plasmid, gaussia luciferase, which is secreted by the cell when expressed (C). The cumulative expression at days 2 and 8 is plotted for ease of comparison (D). Different RGD concentration influenced transgene expression. Statistical significance was determined using multiple comparisons and either the Dunnett or the Tukey multiple comparison’s tests. The symbol *** indicates statistical significance at the level of 0.001 between the indicated condition and the no DNA control in (B) or between the indicated conditions in (D). The symbols ♦, ♦♦, ♦♦♦ indicate statistical significance at the level of 0.05, 0.01 and 0.001 between the indicated conditions in (B). 100* represents the condition with no DNA polyplexes added to the hydrogel. Scale bar = 100µm.
RGD concentration affected transgene expression with the 100µM RGD concentration showing higher levels of transgene expression followed by 10, which was followed by 400µM RGD (Fig. 6C). At day 8 the 100µM RGD concentration was statistically higher than the 400µM condition (p < 0.05, Fig. 6D).
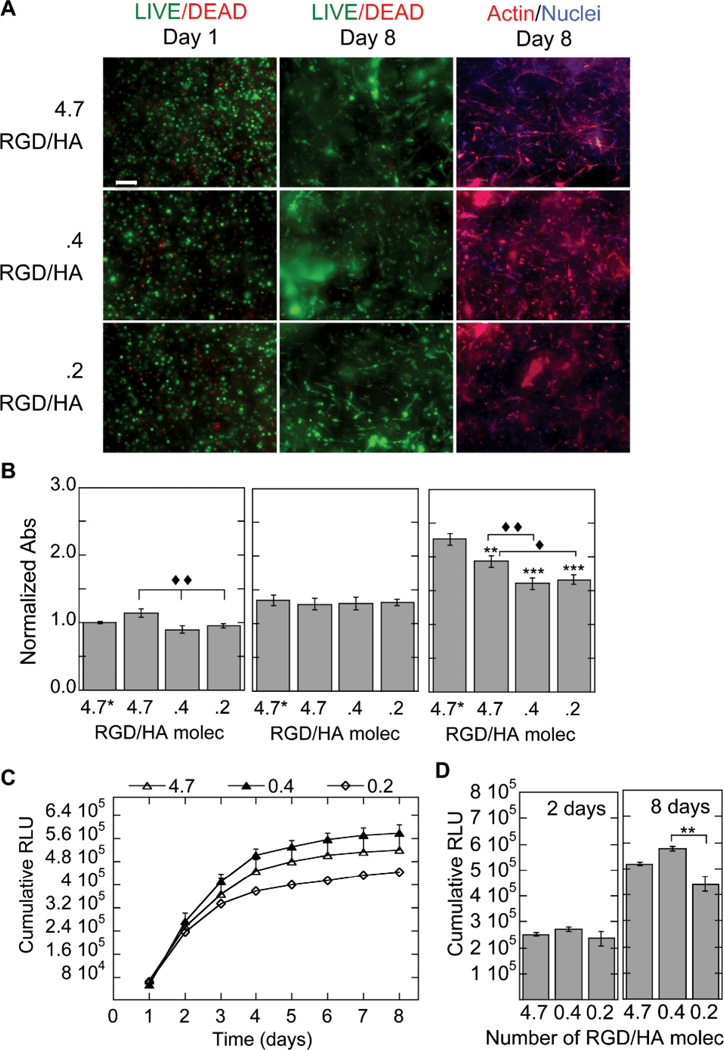
RGD presentation was also assessed as a possible factor to influence cell viability, proliferation, and transfection efficiency. The viability of the cells at days 1 and 8 was assessed using the LIVE/DEAD stain and at 2, 4, and 6 days using the MTT assay (Fig. 7A,B). The LIVE/DEAD stain showed minimal cell death at day 1 indicating that RGD clustering conditions did not affect cell viability. By day 8 spreading was also observed in all conditions (Fig. 7A). The MTT assay for RGD presentation showed that the degree of RGD clustering influenced the metabolic activity with the highest clustered condition (4.7 RGD/HA molecule) resulting in higher MTT readings than the lower clustering conditions (.4 and .2 RGD/HA molecule, Fig. 7B). At day 2 there is a significantly higher MTT reading than for the 4.7 RGDs per HA molecule than the 0.4 and 0.2 RGD clustering conditions (p < 0.01). By day 6 all the conditions were again lower than the no DNA condition (p at least < 0.01) and the highest clustered condition continued to have higher MTT values than the lower clustering conditions.
Figure 7.
Gene transfer as a function of RGD presentation. The effect of RGD presentation on the ability of cells seeded inside the hydrogel to become transfected was studied for hydrogels with 100 µM RGD displayed either homogeneously (100% HA-RGD, .2 RGD/HA molecule) or as RGD clusters (52% to 4.3% HA-RGD, .4 and 4.7 HA/RGD molecule, respectively). RGD clustering is achieved by reacting different amounts of HA-AC with the same amount of RGD and then mixing the resulting HA-RGD with unmodified HA. The cell viability, ability to of the cells to spread and the metabolic activity of the cells were studied using the LIVE/DEAD assay, phalloidin staining (A) and MTT assay (B). Gene expression was determined over time using a reporter plasmid, gaussia luciferase, which is secreted by the cell when expressed (C). The cumulative expression at days 2 and 8 is plotted for ease of comparison (D). RGD presentation influenced transgene expression. Statistical significance was determined using multiple comparisons and either the Dunnett or the Tukey multiple comparison’s tests. The symbols ** and *** indicates statistical significance at the level of 0.01 and 0.001 between the indicated condition and the corresponding no DNA control in (B) and between the indicated conditions in (D). The symbols ♦, ♦♦ indicate statistical significance at the level of 0.05 and 0.01 between the indicated conditions in (B). 4.7* represents the condition with no DNA polyplexes added to the hydrogel. Scale bar = 100µm.
Transgene expression was also affected by the RGD presentation in the scaffold, with an intermediate RGD clustering resulting in optimal transgene expression (Fig. 7C). The trend observed was 0.4 > 4.7 > 0.2 RGD per HA molecule, with .4 RGD/HA resulting in the greatest transgene expression. The 0.4 clustering condition was statistically higher than the 0.2 (homogeneous RGD) clustering condition (p < 0.01, Fig. 7D).
Discussion
Cell-matrix interactions are increasingly being utilized to guide stem cell differentiation43–45 and have been previously implicated with DNA/PEI non-viral gene transfer28, 29 for cells plated on top of hydrogel materials (in two dimensions). We are interested in introducing gene based bioactive signals to aid in the differentiation of either transplanted or endogenous stem cells from within 3-dimensional hydrogel scaffolds. Here HA-RGD was crosslinked with a MMP labile peptide using Michael type addition to form hydrogels that were degradable through a combination of hyaluronidases and MMPs. In addition, DNA/PEI polyplexes and mouse mesenchymal stem cells were encapsulated during gelation. The role of cell-matrix interactions on DNA/PEI non-viral gene transfer for cells seeded within hydrogel scaffolds was investigated here (in three dimensions). Using a range of mechanical properties from 100 ± 2.11 to 1730 ± 45.62 Pa, we observed that gene transfer was strongly influenced by mechanical properties, with softer hydrogels resulting in optimal transgene expression. We further studied if the concentration and presentation of RGD cell adhesion peptides affected gene transfer in three dimensions. An intermediate RGD clustering resulted in the most effective transgene expression.
The activity of the entrapped polyplexes was as high as fresh polyplexes mixed with trypsin and hydrogel degradation products, indicating that the encapsulation process does not inactivate the polyplexes (Fig. 2A). However, compared to fresh polyplexes without trypsin and hydrogel degradation products the activity was lower. This indicates that the presence of trypsin in the polyplex solution negatively affects transfection. The distribution of the polyplexes inside the hydrogel was studied through the labeling of the DNA post hydrogel formation. The presence of DNA clusters indicates that the polyplexes are present in the hydrogel (Fig. 2A). Naked DNA would show a uniform fluorescence inside the hydrogel (data not shown). Although some larger aggregates are observed the majority of particles were found to not be aggregated. This result is consistent with our previous studies showing that concentrations of DNA below 0.1µg DNA/µL hydrogel do not result in severe polyplex aggregation23, 31. As expected the polyplex physical properties affected the rate of non-viral gene transfer for cells seeded inside hydrogel scaffolds with increasing N/P ratio resulting in higher transgene expression, but also higher toxicity (Fig. 3). This is the same trend observed for polyplexes grown in two-dimensional surfaces46, 47. Thus, although we observed two fold higher transgene expression for N/P of 9 and 12, an N/P of 7 was used in the remaining studies to ensure minimal toxicity was observed.
For cells cultured on two-dimensional surfaces cell area28, 29 has been previously been linked to non-viral gene transfer, with increased cell area also increasing transgene expression. For cells cultured in three-dimensions the migration rate30, 31 of cells through the material has previously been implicated with effective non-viral gene transfer. Based on these findings we postulated that hydrogel scaffolds that allowed for extensive cell spreading and migration would result in enhanced gene transfer over hydrogels that did not allow for substantial cell spreading or migration. We previously published on culture of MSCs inside HA/MMP hydrogels. In that study it was observed that soft hydrogel materials allowed for extensive spreading and migration, while stiff materials did not15. Here we find that soft hydrogel materials result in more effective transfection of encapsulated cells compared to those within stiff hydrogels (Fig. 4), suggesting that cell spreading and migration also play a role in gene transfer to cells seeded inside hydrogels. It should be noted that for cells seeded on top of the hydrogel material (in two dimensions), stiff substrates have been shown to promote more efficient gene transfer than cells cultured on soft substrates28.
The lack of efficient gene transfer on stiff hydrogels is attributed to decreased migration rates, spreading, and actin polymerization, all of which have been implicated with enhanced transfection28–31, 48. Further, to achieve stiff hydrogels higher HA percentages were used to form the hydrogels, which decreased the pore size. Higher stiffness did slow down cellular metabolic activity and cell spreading; thus, nutrient diffusion and slowed matrix degradation could be a reason for lower gene transfer specially at early time points.
One possibility to enhance transfection further in our soft hydrogels is to prevent polyplex release from our hydrogel scaffolds. Extensive release was observed even when release was performed in PBS. Thus, if more DNA were retained throughout the 8-day incubation period (and beyond) more transgene expression might be observed.
We next wanted to study the role of the concentration and presentation of RGD peptide on non-viral gene transfer in three-dimensions. As mentioned RGD presentation was found to be important for gene transfer in 2 dimensions29. Because each molecule of HA in our system has on average 64 acrylate groups and < 7% of those groups are used for conjugating RGD many sites are still available to crosslink without compromising mechanical properties. As expected the mechanical properties of these hydrogels remained approximately constant (Fig. 5). An intermediate RGD concentration of 100µM was found to achieve the most efficient transgene expression compared to 10µM and 400µM, suggesting that there is an optimal degree of spreading and migration rate associated with transgene expression inside hydrogel scaffolds (Fig. 6C,D). RGD concentration was not found to affect cell viability as determined by the LIVE/DEAD assay, however, the MTT metabolic activity assay showed lower metabolic activity for 400µM RGD compared to lower RGD concentrations. This could explain why 400µM RGD resulted in the lowest transgene expression of the three tested RGD concentrations.
To cluster RGD we used a similar approach used by the Mooney group to cluster RGD in alginate hydrogels29, 45. In this approach, a fraction of the total polymer is modified with RGD and then mixed with unmodified polymer. By changing the ratio of modified to unmodified polymer, while keeping the overall concentration constant, the RGD presentation is modulated. Using 100% modified HA would result in homogeneously displayed RGD and 0.2 RGDs per HA molecule, while using 4.3% modified HA and 95.7% unmodified HA would result in our most RGD clustered condition with 4.7 RGDs per HA molecule. RGD presentation was found to significantly influenced transgene expression with an intermediate clustering achieving the mot efficient transgene expression (Fig. 7C,D). Metabolic activity was the highest for the most clustered condition, however, this condition did not achieve the most efficient gene transfer. Another complementary explanation for the effect of RGD presentation ion transgene expression is integrin clustering. Clustering of integrins has been implicated with stem cell differentiation49, enhanced bone formation in vivo50, enhanced nanoparticle targeting51 and enhanced transgene expression29, 52, suggesting that the clustering of integrins may be playing a role in making the cells more susceptible to transfection.
In conclusion, we explored the role of matrix stiffness and RGD presentation on gene transfer to cells seeded inside hydrogel scaffolds. Both matrix stiffness and RGD presentation significantly influenced transgene expression with an intermediate stiffness and RGD clustering resulting in maximal transgene expression. We believe that the knowledge gained through this in vitro model can be utilized to design better scaffold-mediated gene delivery for local gene therapy for in vivo applications.
Acknowledgements
The authors would like to thank Jonathan Lam, Charlie Steingard, Kyoung-Joo Jenny Park, Sarah Kaushal for their continuous assistance with this project. The authors acknowledge the NIH (1R21EB007730-01), NSF (CAREER 0747539), and CRCC for funding this work. T. Tokatlian also acknowledges the NIH Biotechnology Training Grant (T32GM067555) for funding.
References
- 1.Kleinman HK, Philp D, Hoffman MP. Role of the extracellular matrix in morphogenesis. Curr Opin Biotechnol. 2003;14(5):526–532. doi: 10.1016/j.copbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 2.Bottaro DP, Liebmann-Vinson A, Heidaran MA. Molecular signaling in bioengineered tissue microenvironments. Ann N Y Acad Sci. 2002;961:143–153. doi: 10.1111/j.1749-6632.2002.tb03068.x. [DOI] [PubMed] [Google Scholar]
- 3.Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242(1):27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- 4.Leach JB, Bivens KA, Patrick CW, Schmidt CE. Photocrosslinked hyaluronic acid hydrogels: Natural, biodegradable tissue engineering scaffolds. Biotechnology and Bioengineering. 2003;82(5):578–589. doi: 10.1002/bit.10605. [DOI] [PubMed] [Google Scholar]
- 5.Park YD, Tirelli N, Hubbell JA. Photopolymerized hyaluronic acid-based hydrogels and interpenetrating networks. Biomaterials. 2003;24(6):893–900. doi: 10.1016/s0142-9612(02)00420-9. [DOI] [PubMed] [Google Scholar]
- 6.Shu XZ, Liu YC, Palumbo FS, Lu Y, Prestwich GD. In situ crosslinkable hyaluronan hydrogels for tissue engineering. Biomaterials. 2004;25(7–8):1339–1348. doi: 10.1016/j.biomaterials.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Yeo Y, Highley CB, Bellas E, Ito T, Marini R, Langer R, Kohane DS. In situ cross-linkable hyaluronic acid hydrogels prevent post-operative abdominal adhesions in a rabbit model. Biomaterials. 2006;27(27):4698–4705. doi: 10.1016/j.biomaterials.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Chung C, Burdick JA. Influence of Three-Dimensional Hyaluronic Acid Microenvironments on Mesenchymal Stem Cell Chondrogenesis. Tissue Engineering Part A. 2009;15(2):243–254. doi: 10.1089/ten.tea.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerecht S, Burdick JA, Ferreira LS, Townsend SA, Langer R, Vunjak-Novakovic G. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(27):11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Park Y, Tae G, Lee KB, Hwang CM, Hwang SJ, Kim IS, Noh I, Sun K. Characterization of low-molecular-weight hyaluronic acid-based hydrogel and differential stem cell responses in the hydrogel microenvironments. Journal of Biomedical Materials Research Part A. 2009;88A(4):967–975. doi: 10.1002/jbm.a.31947. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Park Y, Tae G, Lee KB, Hwang SJ, Kim IS, Noh I, Sun K. Synthesis and characterization of matrix metalloprotease sensitive-low molecular weight hyaluronic acid based hydrogels. Journal of Materials Science-Materials in Medicine. 2008;19(11):3311–3318. doi: 10.1007/s10856-008-3469-3. [DOI] [PubMed] [Google Scholar]
- 12.Pan LJ, Ren YJ, Cui FZ, Xu QY. Viability and Differentiation of Neural Precursors on Hyaluronic Acid Hydrogel Scaffold. Journal of Neuroscience Research. 2009;87(14):3207–3220. doi: 10.1002/jnr.22142. [DOI] [PubMed] [Google Scholar]
- 13.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61(7):1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 14.Sherman L, Sleeman J, Herrlich P, Ponta H. Hyaluronate Receptors - Key Players in Growth, Differentiation, Migration and Tumor Progression. Current Opinion in Cell Biology. 1994;6(5):726–733. doi: 10.1016/0955-0674(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 15.Lei Y, Gojgini S, Lam J, Segura T. The spreading, migration and proliferation of mouse mesenchymal stem cells cultured inside hyaluronic acid hydrogels. Biomaterials. 2011 doi: 10.1016/j.biomaterials.2010.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonadio J, Smiley E, Patil P, Goldstein S. Localized, direct plasmid gene delivery in vivo: prolonged therapy results in reproducible tissue regeneration. Nat Med. 1999;5(7):753–759. doi: 10.1038/10473. [DOI] [PubMed] [Google Scholar]
- 17.Chun KW, Lee JB, Kim SH, Park TG. Controlled release of plasmid DNA from photo-cross-linked pluronic hydrogels. Biomaterials. 2005;26(16):3319–3326. doi: 10.1016/j.biomaterials.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 18.Quick DJ, Anseth KS. DNA delivery from photocrosslinked PEG hydrogels: encapsulation efficiency, release profiles, and DNA quality. Journal of Controlled Release. 2004;96(2):341–351. doi: 10.1016/j.jconrel.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Kong HJ, Kim ES, Huang YC, Mooney DJ. Design of biodegradable hydrogel for the local and sustained delivery of angiogenic plasmid DNA. Pharmaceutical Research. 2008;25(5):1230–1238. doi: 10.1007/s11095-007-9526-7. [DOI] [PubMed] [Google Scholar]
- 20.Kasper FK, Jerkins E, Tanahashi K, Barry MA, Tabata Y, Mikos AG. Characterization of DNA release from composites of oligo(poly(ethylene glycol) fumarate) and cationized gelatin microspheres in vitro. Journal of Biomedical Materials Research Part A. 2006;78A(4):823–835. doi: 10.1002/jbm.a.30736. [DOI] [PubMed] [Google Scholar]
- 21.Megeed Z, Haider M, Li D, O'Malley BW, Jr, Cappello J, Ghandehari H. In vitro and in vivo evaluation of recombinant silk-elastinlike hydrogels for cancer gene therapy. J Control Release. 2004;94(2–3):433–445. doi: 10.1016/j.jconrel.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 22.Jang JH, Rives CB, Shea LD. Plasmid delivery in vivo from porous tissue-engineering scaffolds: transgene expression and cellular transfection. Mol Ther. 2005;12(3):475–483. doi: 10.1016/j.ymthe.2005.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei P, Padmashali RM, Andreadis ST. Cell-controlled and spatially arrayed gene delivery from fibrin hydrogels. Biomaterials. 2009;30(22):3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieland JA, Houchin-Ray TL, Shea LD. Non-viral vector delivery from PEG-hyaluronic acid hydrogels. Journal of Controlled Release. 2007;120(3):233–241. doi: 10.1016/j.jconrel.2007.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saul JM, Linnes MP, Ratner BD, Giachelli CM, Pun SH. Delivery of non-viral gene carriers from sphere-templated fibrin scaffolds for sustained transgene expression. Biomaterials. 2007;28(31):4705–4716. doi: 10.1016/j.biomaterials.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 26.Trentin D, Hall H, Wechsler S, Hubbell JA. Peptide-matrix-mediated gene transfer of an oxygen-insensitive hypoxia-inducible factor-1 alpha variant for local induction of angiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2506–2511. doi: 10.1073/pnas.0505964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trentin D, Hubbell J, Hall H. Non-viral gene delivery for local and controlled DNA release. Journal of Controlled Release. 2005;102(1):263–275. doi: 10.1016/j.jconrel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 28.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4(6):460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 29.Kong HJ, Hsiong S, Mooney DJ. Nanoscale cell adhesion ligand presentation regulates nonviral gene delivery and expression. Nano Lett. 2007;7(1):161–166. doi: 10.1021/nl062485g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepard JA, Huang A, Shikanov A, Shea LD. Balancing cell migration with matrix degradation enhances gene delivery to cells cultured three-dimensionally within hydrogels. J Control Release. 2010;146(1):128–135. doi: 10.1016/j.jconrel.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lei YG, Segura T. DNA delivery from matrix metal lop roteinase degradable poly(ethylene glycol) hydrogels to mouse cloned mesenchymal stem cells. Biomaterials. 2009;30(2):254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lungwitz U, Breunig M, Blunk T, Gopferich A. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60(2):247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Abdallah B, Hassan A, Benoist C, Goula D, Behr JP, Demeneix BA. A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: Polyethylenimine. Human Gene Therapy. 1996;7(16):1947–1954. doi: 10.1089/hum.1996.7.16-1947. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Ma N, Gao SJ, Yu H, Leong KW. Transgene expression in the brain stem effected by intramuscular injection of polyethylenimine/DNA complexes. Molecular Therapy. 2001;3(5):658–664. doi: 10.1006/mthe.2001.0324. [DOI] [PubMed] [Google Scholar]
- 35.Wiseman JW, Goddard CA, McLelland D, Colledge WH. A comparison of linear and branched polyethylenimine (PEI) with DCChol/DOPE liposomes for gene delivery to epithelial cells in vitro and in vivo. Gene Therapy. 2003;10(19):1654–1662. doi: 10.1038/sj.gt.3302050. [DOI] [PubMed] [Google Scholar]
- 36.Kichler A, Chillon M, Leborgne C, Danos O, Frisch B. Intranasal gene delivery with a polyethylenimine-PEG conjugate. Journal of Controlled Release. 2002;81(3):379–388. doi: 10.1016/s0168-3659(02)00080-9. [DOI] [PubMed] [Google Scholar]
- 37.Rudolph C, Schillinger U, Plank C, Gessner A, Nicklaus P, Muller RH, Rosenecker J. Nonviral gene delivery to the lung with copolymer-protected and transferrin-modified polyethylenimine. Biochimica Et Biophysica Acta-General Subjects. 2002;1573(1):75–83. doi: 10.1016/s0304-4165(02)00334-3. [DOI] [PubMed] [Google Scholar]
- 38.Gautam A, Densmore CL, Golunski E, Xu B, Waldrep JC. Transgene expression in mouse airway epithelium by aerosol gene therapy with PEI-DNA complexes. Molecular Therapy. 2001;3(4):551–556. doi: 10.1006/mthe.2001.0300. [DOI] [PubMed] [Google Scholar]
- 39.Segura T, Schmokel H, Hubbell JA. RNA interference targeting hypoxia inducible factor 1alpha reduces post-operative adhesions in rats. J Surg Res. 2007;141(2):162–170. doi: 10.1016/j.jss.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 40.Iwai M, Harada Y, Tanaka S, Muramatsu A, Mori T, Kashima K, Imanishi J, Mazda O. Polyethylenimine-mediated suicide gene transfer induces a therapeutic effect for hepatocellular carcinoma in vivo by using an Epstein-Barr virus-based plasmid vector. Biochemical and Biophysical Research Communications. 2002;291(1):48–54. doi: 10.1006/bbrc.2002.6383. [DOI] [PubMed] [Google Scholar]
- 41.Aoki K, Furuhata S, Hatanaka K, Maeda M, Remy JS, Behr JP, Terada M, Yoshida T. Polyethylenimine-mediated gene transfer into pancreatic tumor dissemination in the murine peritoneal cavity. Gene Therapy. 2001;8(7):508–514. doi: 10.1038/sj.gt.3301435. [DOI] [PubMed] [Google Scholar]
- 42.Coll JL, Chollet P, Brambilla E, Desplanques D, Behr JP, Favrot M. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10(10):1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 43.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95(9):4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 45.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9(6):518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Lei Y, Dhaliwal A, Ng QK, Du J, Yan M, Lu Y, Segura T. Protein-polymer nanoparticles for nonviral gene delivery. Biomacromolecules. 2011;12(4):1006–1014. doi: 10.1021/bm101354a. [DOI] [PubMed] [Google Scholar]
- 47.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. Journal of Controlled Release. 1999;60(2–3):149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]
- 48.Dhaliwal A, Maldonado M, Han Z, Segura T. Differential uptake of DNA-poly(ethylenimine) polyplexes in cells cultured on collagen and fibronectin surfaces. Acta Biomater. 2010;6(9):3436–3447. doi: 10.1016/j.actbio.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsiong SX, Huebsch N, Fischbach C, Kong HJ, Mooney DJ. Integrin-adhesion ligand bond formation of preosteoblasts and stem cells in three-dimensional RGD presenting matrices. Biomacromolecules. 2008;9(7):1843–1851. doi: 10.1021/bm8000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrie TA, Raynor JE, Dumbauld DW, Lee TT, Jagtap S, Templeman KL, Collard DM, Garcia AJ. Multivalent integrin-specific ligands enhance tissue healing and biomaterial integration. Sci Transl Med. 2010;2(45):45ra60. doi: 10.1126/scitranslmed.3001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng QK, Su H, Armijo AL, Czernin J, Radu CG, Segura T. Clustered arg-gly-asp peptides enhances tumor targeting of nonviral vectors. ChemMedChem. 2011;6(4):623–627. doi: 10.1002/cmdc.201000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng QK, Sutton MK, Soonsawad P, Xing L, Cheng H, Segura T. Engineering clustered ligand binding into nonviral vectors: alphavbeta3 targeting as an example. Mol Ther. 2009;17(5):828–836. doi: 10.1038/mt.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]