Abstract
Mitochondria play numerous, essential roles in the life of eukaryotes. Disruption of mitochondrial function in humans is often pathological or even lethal. Surprisingly, in some organisms mitochondrial dysfunction can result in life extension. This paradox has been studied most extensively in the long-lived Mit mutants of the nematode Caenorhabditis elegans. In this review, we explore the major responses that are activated following mitochondrial dysfunction in these animals and how these responses potentially act to extend their life. We focus our attention on five broad areas of current research – reactive oxygen species signaling, the mitochondrial unfolded protein response, autophagy, metabolic adaptation, and the roles played by various transcription factors. Lastly, we also examine why disruption of complexes I and II differ in their ability to induce the Mit phenotype and extend lifespan.
Keywords: C. elegans, Mitochondria, Mit Mutant, Lifespan, Aging, Metabolism
1. Introduction
In four billion years, complex life appears to have arisen only once, and from an unlikely endosymbiosis between two primordial prokaryotes. By endowing their host with vastly expanded respiratory capacity, the endosymbionts enabled their host to break through the bioenergetic constraints on genome size and to evolve a vast repertoire of genes (Lane and Martin 2010). Ultimately, this new chimeric cell type gave rise to all eukaryotic life and the ancient endosymbionts became today’s mitochondria. For many eukaryotes, including humans, oxidative phosphorylation by the mitochondria is the primary mode of generating ATP and complete disruption of this process is lethal. Thus, it was a surprise to find that crippling the very organelle that enables our own existence can, in some species, prolong life. Exemplifying this are the Mit mutants of the nematode Caenorhabditis elegans.
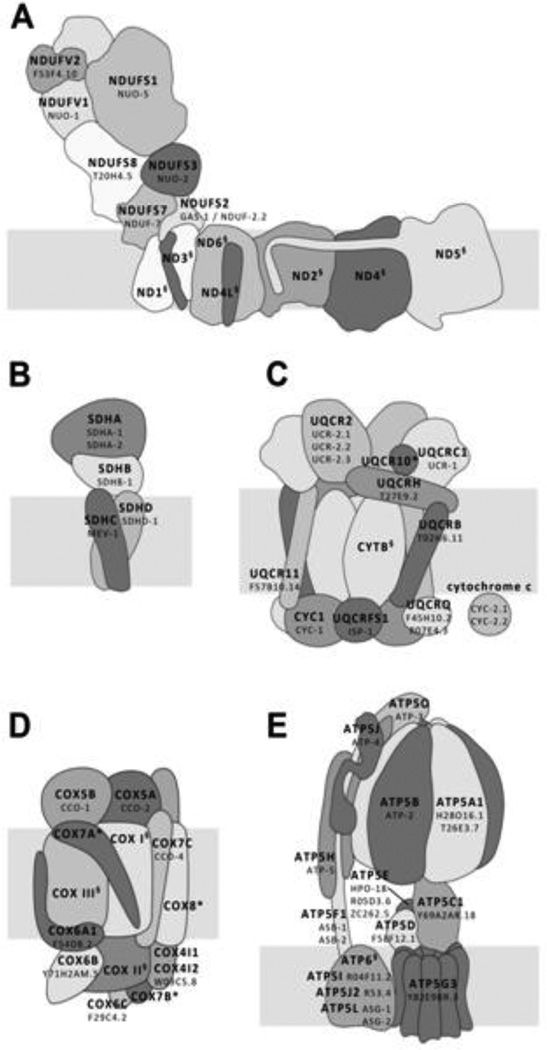
The first Mit mutant discovered was clk-1(e2519) (Wong and others 1995), so named because this mutant displayed disrupted timing of several developmental and behavioral processes, leading to the mistaken hypothesis that the clk-1 gene specified some sort of biological clock. In fact, clk-1 encodes the C. elegans ortholog of demethoxyubiquinone hydroxylase. This protein is required for the synthesis of ubiquinone, the molecule which transports electrons from both complexes I and II, as well as other metabolic enzymes (see section 4.3), to complex III of the mitochondrial electron transport chain (ETC) (Miyadera and others 2001). Since this initial discovery, it has been found that disruption of almost any subunit of the ETC, or of enzymes needed for assembly of the ETC and its co-factors, can extend lifespan in C. elegans. RNAi knockdown of genes encoding subunits of complexes I, III, or IV increase lifespan, on average by 50% (Zuryn and others 2010). Lifespan can also be extended by low doses of mitochondrial toxins such as antimycin A (Dillin and others 2002), ethidium bromide (Tsang and Lemire 2002), rotenone (Dillin and others 2002; Schmeisser and others 2013a), and arsenite (Schmeisser and others 2013b). Given the apparent non-specificity of the nature of mitochondrial ETC disruption in extending life, it is surprising that lifespan is never increased in response to knockdown of any of the subunits of complex II (Ichimiya and others 2002; Kuang and Ebert 2012; Rea and others 2007). Possible reasons for this are discussed in section 4. Nuclear DNA-encoded mitochondrial proteins for which the effect of mutational or RNAi knockdown on lifespan have been reported are listed in Table 1. The positions of the ETC subunits within the respiratory complexes are shown schematically in Figure 1. Unless specified explicitly, in this review, we refer to all long-lived animals with either mutational- or RNAi-induced knockdown of mitochondrial components as Mit “mutants”.
Table 1.
| C. elegans gene | Human Ortholog |
Life Extension * |
Method of Knockdown |
Reference ** |
|---|---|---|---|---|
| COMPLEX I | ||||
| K09A9.5 (gas-1) | NDUFS2 | decrease | genetic | [1, 2, 3‡] |
| ZK973.10 (lpd-5) | NDUFS4 | |||
| T26A5.3 (nduf-2.2) | NDUFS2 | √ | RNAi | [4] |
| Y54E10BL.5 (nduf-5) | NDUFS5 | |||
| F22D6.4 (nduf-6) | NDUFS6 | |||
| W10D5.2 (nduf-7) | NDUFS7 | |||
| C09H10.3 (nuo-1) | NDUFV1 | √ | genetic & RNAi | [5, 6‡, 7‡, 8] |
| T10E9.7 (nuo-2) | NDUFS3 | √ | RNAi | [9, 10, 11] |
| Y57G11C.12 (nuo-3) | NDUFA6 | √ | RNAi | [4, 7‡, 10] |
| K04G7.4 (nuo-4) | NDUFA10 | √ | RNAi | [4, 10, 12‡] |
| Y45G12B.1 (nuo-5) | NDUFS1 | √ | RNAi | [10] |
| W01A8.4 (nuo-6) | NDUFB4 | √ | genetic & RNAi | [13] |
| C16A3.5 | NDUFB9 | |||
| C18E9.4 | NDUFB3 | √ | RNAi | [7‡] |
| C25H3.9 | NDUFB5 | √ | RNAi | [7‡] |
| C33A12.1 | NDUFA5 | √ | RNAi | [4, 7‡] |
| C34B2.8 | NDUFA13 | |||
| D2030.4 | NDUFB7 | √ | RNAi | [12‡, 14‡] |
| F31D4.9 | NDUFA1 | |||
| F37C12.3 | NDUFAB1 | |||
| F42G8.10 | NDUFB11 | |||
| F44G4.2 | NDUFB2 | |||
| F45H10.3 | NDUFA7 | |||
| F53F4.10 | NDUFV2 | |||
| F59C6.5 | NDUFB10 | √ | RNAi | [5‡] |
| T20H4.5 | NDUFS8 | √ | RNAi | [4, 12‡] |
| Y51H1A.3 | NDUFB8 | |||
| Y53G8AL.2 | NDUFA9 | √ | RNAi | [4] |
| Y54F10AM.5 | NDUFA8 | |||
| Y56A3A.19 | NDUFAB1 | √ | RNAi | [4, 5‡] |
| Y63D3A.7 | NDUFA2 | |||
| Y71H2AM.4 | NDUFC2 | √ | RNAi | [4] |
| Y94H6A.8 | NDUFA12 | |||
| ZK809.3 | NDUFB6 | √ | RNAi | [4] |
| COMPLEX II | ||||
| C03G5.1 (sdha-1) | SDHA | no effect | RNAi | [2, 15‡] |
| C34B2.7 (sdha-2) | SDHA | no effect | RNAi | [2, 15‡] |
| F42A8.2 (sdhb-1) | SDHB | no effect | RNAi | [2, 7‡, 15‡] |
| T07C4.7 (mev-1) | SDHC | decrease | genetic & RNAi | [2, 11, 15‡, 16] |
| F33A8.5 (sdhd-1) | SDHD | no effect | RNAi | [2, 15‡] |
| COMPLEX III | ||||
| C54G4.8 (cyc-1) | CYC1 | √ | RNAi | [7‡, 9, 10, 17] |
| E04A4.7 (cyc-2.1) | CYCS (cytochrome c) | √ | RNAi | [4] |
| ZC116.2 (cyc-2.2) | CYCS (cytochrome c) | |||
| F42G8.12 (isp-1) | UQCRFS1 | √ | genetic & RNAi | [11, 17] |
| F56D2.1 (ucr-1) | UQCRC1 | √ | RNAi | [4, 7‡] |
| VW06B3R.1 (ucr-2.1) | UQCR2 | |||
| T10B10.2 (ucr-2.2) | UQCR2 | no effect | RNAi | [7‡] |
| T24C4.1 (ucr-2.3) | UQCR2 | decrease | genetic & RNAi | [7‡, 18] |
| F45H10.2 | UQCRQ | √ | RNAi | [4] |
| F57B10.14 | UQCR11 | |||
| R07E4.3 | UQCRQ | √ | RNAi | [7‡] |
| T02H6.11 | UQCRB | √ | RNAi | [7‡, 14‡] |
| T27E9.2 | UQCRH | √ | RNAi | [7‡] |
| COMPLEX IV | ||||
| F26E4.9 (cco-1) | COX5B | √ | RNAi | [4, 7‡, 9, 10, 11, 12‡, 14‡, 17] |
| Y37D8A.14 (cco-2) | COX5A | √ | RNAi | [7‡, 10, 19] |
| F26E4.6 (cco-4) | COX7C | √ | RNAi | [5‡, 7‡, 12‡, 14‡] |
| F29C4.2 | COX6C | √ | RNAi | [4, 7‡] |
| F40G9.2 | COX17 | |||
| F54D8.2 | COX6A1 | √ | RNAi | [4, 7‡] |
| JC8.5 | COX11 | |||
| T06D8.5 | COX15 | √ | RNAi | [14] |
| W09C5.8 | COX4 | √ | RNAi | [4, 7‡, 12, 14] |
| Y46G5A.2 | COX10 | |||
| Y71H2AM.5 | COX6B | √ | RNAi | [7‡] |
| COMPLEX V | ||||
| F35G12.10 (asb-1) | ATP5F1 (b) | |||
| F02E8.1 (asb-2) | ATP5F1 (b) | √ | RNAi | [10] |
| K07A12.3 (asg-1) | ATP5L (g) | |||
| C53B7.4 (asg-2) | ATP5L (g) | √ | RNAi | [12‡] |
| C34E10.6 (atp-2) | ATP5B (β) | √ | genetic & RNAi | [5, 6‡] |
| F27C1.7 (atp-3) | ATP5O (OSCP) | √ | RNAi | [9, 10, 11] |
| T05H4.12 (atp-4) | ATP5J (F6) | √ | RNAi | [10] |
| C06H2.1 (atp-5) | ATP5H (d) | √ | RNAi | [10] |
| F32D1.2 (hpo-18) | ATP5E (ε) | |||
| F58F12.1 | ATP5D (δ) | |||
| H28O16.1 | ATP5A1 (α) | √ | RNAi | [5] |
| R04F11.2 | ATP5I (e) | |||
| R05D3.6 | ATP5E (ε) | |||
| R53.4 | ATP5J2 (f) | |||
| T26E3.7 | ATP5A1 (α) | |||
| Y69A2AR.18 | ATP5C1 (γ) | |||
| Y82E9BR.3 | ATP5G3 (c) | |||
| ZC262.5 | ATP5E (ε) | |||
| OTHER ± | ||||
| T06D8.6 (cchl-1) | HCCS | √ | RNAi | [4] |
| ZC395.2 (clk-1) | COQ7 | √ | genetic | [20] |
| F59G1.7 (frh-1) | FXN | √ | genetic & RNAi | [11, 21, 22] |
| ZC395.6 (gro-1) | TRIT1 | √ | genetic | [23] |
| C37H5.8 (hsp-6) | mtHSP70 | √ | RNAi | [22] |
| ZK524.3 (lrs-2) | LARS2 | √ | genetic | [14‡] |
| T21C9.1 (mics-1) | OMP25 | √ | genetic & RNAi | [24‡] |
| F56B3.8 (mrpl-2) | MRPL2 | √ | RNAi | [25‡] |
| W09D10.3 (mrpl-12) | MRPL12 | √ | RNAi | [4] |
| Y48E1B.5 (mrpl-37) | MRPL37 | √ | RNAi | [25‡] |
| B0261.4 (mrpl-47) | MRPL47 | √ | RNAi | [14‡] |
| E02A10.1 (mrps-5) | MRPS5 | √ | RNAi | [25‡] |
| F09G8.3 (mrps-9) | MRPS9 | √ | RNAi | [4] |
| Y37D8A.18 (mrps-10) | MRPS10 | √ | RNAi | [4] |
| F21D5.8 (mrps-33) | MRPS33 | √ | RNAi | [4] |
| F43E2.7 (mtch-1) | MTCH1 | √ | RNAi | [5‡] |
| F10D11.1 (sod-2) | SOD2 (MnSOD) | √ | genetic | [26‡] |
| C08A9.1 (sod-3) | SOD2 (MnSOD) | no effect | genetic | [26‡] |
| ZK637.9 (tpk-1) | TPK1 | √ | genetic | [4] |
| K08F11.4 (yars-1) | YARS2 | √ | RNAi | [4] |
| F13G3.7 | SLC25A44 | √ | RNAi | [14‡] |
| K01C8.7 | SLC25A32 | √ | RNAi | [14‡] |
Blank space indicates nothing has been reported to date.
List is not exhaustive.
Numbers refer to references listed below.
FUdR used.
Hartman, P.S., et al., Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mech Ageing Dev, 2001. 122(11): p. 1187–201.
Pujol, C., et al., Succinate dehydrogenase upregulation destabilize complex I and limits the lifespan of gas-1 mutant. PLoS One, 2013. 8(3): p. e59493.
Van Raamsdonk, J.M. and S. Hekimi, FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech Ageing Dev, 2011. 132(10): p. 519–21.
Kim, Y. and H. Sun, Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging Cell, 2007. 6(4): p. 489–503.
Curran, S.P. and G. Ruvkun, Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet, 2007. 3(4): p. e56.
Tsang, W.Y., et al., Mitochondrial respiratory chain deficiency in Caenorhabditis elegans results in developmental arrest and increased life span. J Biol Chem, 2001. 276(34): p. 32240–6.
Zuryn, S., et al., Mitochondrial dysfunction in Caenorhabditis elegans causes metabolic restructuring, but this is not linked to longevity. Mech Ageing Dev, 2010. 131(9): p. 554–61.
Grad, L.I. and B.D. Lemire, Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Hum Mol Genet, 2004. 13(3): p. 303–14.
Dillin, A., et al., Rates of behavior and aging specified by mitochondrial function during development. Science, 2002. 298(5602): p. 2398–401.
Hansen, M., et al., New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet, 2005. 1(1): p. 119–28.
Rea, S.L., N. Ventura, and T.E. Johnson, Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol, 2007. 5(10): p. e259.
Hamilton, B., et al., A systematic RNAi screen for longevity genes in C. elegans. Genes Dev, 2005. 19(13): p. 1544–55.
Yang, W. and S. Hekimi, Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging Cell, 2010. 9(3): p. 433–47.
Lee, S.S., et al., A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet, 2003. 33(1): p. 40–8.
Kuang, J. and P.R. Ebert, The failure to extend lifespan via disruption of complex II is linked to preservation of dynamic control of energy metabolism. Mitochondrion, 2012. 12(2): p. 280–7.
Ishii, N., et al., A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutat Res, 1990. 237(3–4): p. 165–71.
Feng, J., F. Bussiere, and S. Hekimi, Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell, 2001. 1(5): p. 633–44.
Butler, J.A., et al., Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J, 2010. 24(12): p. 4977–88.
Suthammarak, W., et al., Complex I function is defective in complex IV-deficient Caenorhabditis elegans. J Biol Chem, 2009. 284(10): p. 6425–35.
Wong, A., P. Boutis, and S. Hekimi, Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics, 1995. 139(3): p. 1247–59.
Ventura, N., et al., Reduced expression of frataxin extends the lifespan of Caenorhabditis elegans. Aging Cell, 2005. 4(2): p. 109–12.
Ventura, N. and S.L. Rea, Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnol J, 2007. 2(5): p. 584–95.
Lakowski, B. and S. Hekimi, Determination of life-span in Caenorhabditis elegans by four clock genes. Science, 1996. 272(5264): p. 1010–3.
Hoffmann, M., et al., MICS-1 interacts with mitochondrial ATAD-3 and modulates lifespan in C. elegans. Exp Gerontol, 2012. 47(3): p. 270–5.
Houtkooper, R.H., et al., Mitonuclear protein imbalance as a conserved longevity mechanism. Nature, 2013. 497(7450): p. 451–7.
Van Raamsdonk, J.M. and S. Hekimi, Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet, 2009. 5(2): p. e1000361.
Figure 1. Identity and Location of Mitochondrial Respiratory Complex Subunits.
(A) Schematic of Complex I (core subunits only). (B–E) Schematics of complexes II-V, respectively. All complexes are oriented such that their matrix-facing surface is at the top. Large text - human subunit; small text - orthologous C. elegans subunit(s); § mitochondrial DNA encoded; * no known C. elegans ortholog.
In humans, mutations in some of the same genes that produce longevity in Mit mutants paradoxically result in devastating diseases. This observation remained puzzling until it was discovered that both humans and worms are, in fact, much alike in that they both exhibit a threshold response to mitochondrial ETC perturbation. In worms this can be demonstrated very easily using RNAi targeting individual subunits of the ETC, such as atp-3. Starting with dilute doses of RNAi, increasing concentrations produce no detectable phenotype until a threshold is reached. Beyond this point, progressively greater target knockdown monotonically increases lifespan up to a second threshold, beyond which lifespan decreases until animals are eventually short-lived compared to wild-type (Rea and others 2007). In humans, mitochondrial diseases also present with variable phenotypes, and in some cases may even be asymptomatic (Limongelli and others 2004). Clearly, in both organisms, compensatory mechanisms are invoked and much work has gone into studying what these processes might be. While mitochondrial disruption is obviously undesirable in humans, it is notable that even mammals can achieve a longer life by this mechanism. In mice, knockout of the cytochrome c assembly factor Surf1 (Dell'agnello and others 2007), hemizygous knockout of Mclk1 (the mouse ortholog of clk-1) (Liu and others 2005), or reduction of mitochondrial ribosomal subunit S5 (Houtkooper and others 2013), have all been reported to prolong life.
2. The Mit Phenotype
C. elegans Mit mutants display a set of co-clustering phenotypes that may be fully or only partially penetrant. In addition to longevity, these include delayed embryonic and/or larval development, small adult size, slow muscle function, and reduced fertility and fecundity (Rea and others 2007). Using an RNAi dilution series, it has been clearly established that concentrations of RNAi that extend lifespan consistently induce the entire Mit phenotype (Rea and others 2007). When RNAi doses are severe enough, however, animals can arrest and even be short-lived. It is interesting to note that, while an RNAi screen against ETC components reported neither change in developmental rate nor alteration of adult size to be predictive of lifespan, the longest-lived worms in the screen were, in fact, both the smallest in size and the slowest to develop (Zuryn and others 2010).
However, the various co-segregating traits of the Mit phenotype are linked only superficially. Uncoupling of longevity from developmental delay in the robust isp-1(qm150) Mit mutant, for example, was shown following the serendipitous discovery of isp-1(qm150); ctb-1(qm189) double mutants. Cytochrome b (CTB-1) is a mitochondrial DNA (mtDNA)-encoded subunit of complex III. The ctb-1(qm189) mutation almost fully mitigates the delayed development of isp-1(qm150) mutant worms but has very little effect on their lifespan (Feng and others 2001). In a study of the CCO-1 subunit of complex IV, Dillin and colleagues demonstrated that intestinal- or neuronal-specific RNAi knockdown were each sufficient to extend nematode lifespan. Neither treatment, however, resulted in the slow movement characteristic of Mit mutants. Conversely, cco-1 knockdown in the body wall muscles decelerated movement but actually shortened lifespan (Durieux and others 2011). Clearly, in this system, life extension and slowness are traits that can also be uncoupled. Whether one should further conclude that different tissues control different aspects of the Mit phenotype, or that differing degrees of cco-1 knockdown in different tissues are required to elicit life extension, remains an open question. Nonetheless, one important point to consider for all Mit mutants is that their final phenotypic outcome almost certainly represents an integrated response from multiple tissues. Perhaps then, what is most remarkable about the Mit mutants of C. elegans is that there is such significant overlap in their final phenotype – enough to be recognized as the “Mit phenotype”. In line with earlier findings (Rea and others 2007), this indicates the existence of a “sweet spot” for life extension in the face of ETC disruption.
3. Mechanisms of Mit Mutant Life Extension
In the following sections we discuss several areas of investigation that have been pursued as possible causes of life extension in Mit mutants. This section is not meant to be exhaustive and in all likelihood the longevity of these animals reflects the outcome of multiple processes acting in concert. When studying whole organisms, it is vital to remember that we are studying an integrated system that is dynamic and which can respond to various systemic perturbations in multiple ways.
3.1. Reactive Oxygen Species (ROS) Signaling
In 1956 Denham Harman published his seminal paper proposing free radicals as the proximal cause of aging (Harman 1956). This hypothesis posits that free radicals, both from the environment and especially aerobic respiration, cause damage that accumulates over time and eventually manifests as aging and death. Accordingly, it has been proposed that Mit mutants live longer because they have reduced ETC flux, resulting in decreased generation of reactive oxygen species (ROS) (Rea 2005). The fact that the short-lived mev-1(kn21) mutant produces high levels of superoxide radical (Senoo-Matsuda and others 2001) would seem to support a causative role of ROS in aging. However, contrary to expectations, not only Mit mutants, but also the long-lived daf-2(e1370) mutant, actually show higher levels of oxidative damage than wild-type worms at young adulthood (Labuschagne and others 2013). Other measures of oxidative damage have shown no correlation with lifespan (Rea and others 2007). It is notable that the unexpected finding of high levels of oxidative damage in long-lived animals is not peculiar to C. elegans. The exceptionally long-lived naked mole rat has higher levels of oxidatively damaged proteins, lipids, and DNA than physiologically age-matched mice (Andziak and others 2006).
In recent years, it has become clear that ROS are not always damaging and in fact play important roles as signaling molecules (Ray and others 2012). As one example of this, ROS generated by dysfunctional mitochondria inhibit cytosolic translation through GCN-2 (Baker and others 2012). When this kinase is knocked out, Mit mutants show stronger induction of the mitochondrial unfolded protein response – evidence that ROS-induced GCN-2 function plays a role in mitigating mitochondrial stress (Baker and others 2012). ROS may also be required for activation of HIF-1, a transcription factor shown to be required for maximal Mit mutant longevity (Hwang and Lee 2011; Lee and others 2010) and which shall be discussed in section 3.2. Counter intuitively, it has been proposed that Mit mutants and other models of extended lifespan live longer not by lowering ROS, but by increasing it just enough to induce protective stress-response genes that bring about the animal’s subsequent longevity. This hypothesis has been termed hormesis (Masoro 1998) and specifically “mitohormesis” when ROS leak from the mitochondria (Ristow and Zarse 2010). The effects of eliminating the mitochondrial superoxide dismutase SOD-2 seem to support an hormetic role for ROS: While oxidative damage is predictably increased, sod-2 deletion not only prolongs life, but further increases longevity in the clk-1(qm30) Mit mutant (Van Raamsdonk and Hekimi 2009) and extends the lifespan of the otherwise short-lived gas-1(fc21) mutant beyond wild-type, effectively making this particular double-knockout a Mit mutant (Suthammarak and others 2013).
3.2. Transcription Factors
While numerous transcription factors have been tested for a role in the longevity of Mit mutants, others have been identified from large RNAi screens. Some of these transcription factors have been clearly linked to a specific mechanism such as autophagy and are only briefly mentioned here. Others are not so easily categorized or the nature of their role in Mit mutants has yet to be defined.
When looking for mediators of longevity, the usual suspects are DAF-16 and SKN-1, which are the C. elegans orthologs of mammalian FOXO and Nrf2, respectively. Both are required for normal lifespan and have been shown to play a role in extending life via reduced insulin signaling (Lin and others 1997; Tullet and others 2008), TOR inhibition (Robida-Stubbs and others 2012), and at least some forms of dietary restriction (Bishop and Guarente 2007; Greer and others 2007; Honjoh and others 2009). DAF-16, however, has been repeatedly shown to be unnecessary for Mit mutant longevity (Dillin and others 2002; Durieux and others 2011; Feng and others 2001; Hansen and others 2005; Kim and Sun 2007; Lee and others 2003; Ventura and others 2009). Mit mutants might be expected to require SKN-1 since this transcription factor mediates the oxidative stress response (An and Blackwell 2003) and is essential for life extension invoked by both complex I inhibitors (Schmeisser and others 2013a) and ROS inducers (Schmeisser and others 2011; Zarse and others 2012). Surprisingly, however, the short-lived skn-1(zu67) mutant, which is deficient in skn-1 isoforms a and c, achieves life extension equivalent to wild-type worms on atp-3, cco-1, or cyc-1 RNAi (Rea and others 2007; Tullet and others 2008). Even the skn-1(zu135) mutant, wherein all skn-1 isoforms (a, b and c) are knocked out, was similarly long-lived on frh-1 RNAi (Ventura and others 2009).
The first transcription factor shown to modulate longevity in Mit mutants was the C. elegans homolog of p53, CEP-1 (Ventura and others 2009), which contributes to their stress response by upregulating protective genes such as the antioxidant glutathione-S-transferase (Torgovnick and others 2010). While CEP-1 contributes to life extension in response to moderate mitochondrial disturbance, most surprising is its role in the early demise of worms with severe mitochondrial dysfunction. This is most clearly seen in worms on undiluted atp-3 RNAi: Severe knockdown of this complex V subunit resulted in larval arrest and the early death of wild-type worms and yet this same treatment caused dramatic life extension in cep-1 null worms (Ventura and others 2009). This is significant as it might be assumed that the pathology resulting from severe mitochondrial dysfunction would be a direct consequence of the impaired mitochondria themselves, but this study shows that this is not the case. Even in the face of dramatic mitochondrial impairment, pathology and life shortening seem to be the result of maladaptive cellular responses.
The next transcription factor recognized for its role in Mit mutant life extension was even more unexpected. As its name suggests, hypoxia inducible factor-1 (HIF-1) mediates survival under conditions of low oxygen. Under normal conditions, HIF-1 is targeted for degradation (Ivan and others 2001; Jiang and others 2001). Consequently, loss of hif-1 is not detrimental to otherwise wild-type worms under normal conditions (Jiang and others 2001) and can even prolong life (Zhang and others 2009). It was therefore surprising to learn that knockdown of hif-1 or aha-1 – which encodes the binding partner of HIF-1 (Jiang and others 2001), significantly abrogates life extension in several Mit mutants, in spite of normoxic conditions (Khan and others 2013; Lee and others 2010). Life extension is reduced even when hif-1 knockdown is initiated in adulthood (Lee and others 2010), an unexpected finding since the Mit phenotype has been shown to be specified during larval development (Dillin and others 2002; Rea and others 2007). This suggests that adaptation to either hypoxic conditions or dysfunctional mitochondria share overlapping requirements (Rea 2005). Interestingly, HIF-1 also plays a role in life extension mediated by dietary restriction (Chen and others 2009).
Additional transcription factors mediating life extension in Mit mutants also overlap with those utilized by various forms of dietary restriction, despite differences between these two mechanisms (Durieux and others 2011; Wolff and Dillin 2006). An RNAi screen for transcription factors required for the longevity of the isp-1(qm150); ctb-1(qm189) double mutant identified the nuclear hormone receptor nhr-25 (Walter and others 2011). While knockdown of nhr-25 had minimal effect on either wild-type worms or the long-lived insulin-like signaling pathway mutant age-1(hx546), it significantly shortened the lifespan of Mit mutants as well as eat-2(ad1116) mutants (this latter mutation affects the worms’ ability to eat and is thus a dietary restriction model) (Walter and others 2011). As the known functions of nhr-25 all pertain to development, the nature of its specific role in mediating the longevity of these animals is unknown.
In a separate RNAi screen conducted by our own group in search of transcription factors controlling Mit mutant lifespan, we found that knockdown of jun-1 completely abolished life extension in both isp-1(qm150) and tpk-1(qm162) Mit mutants but had no effect on wild-type worms (Khan and others 2013). Previously, this transcription factor was shown to mediate life extension in response to intermittent fasting (Uno and others 2013). Induced JUN-1 targets included genes related to aging and ubiquitin-dependent protein catabolism. Enrichment of these genes led the authors to propose that intermittent fasting may prolong life through increased protein turnover via SCF E3 ligase components under the control of JUN-1 (Uno and others 2013). This may also be the role of JUN-1 in Mit mutant life extension. Consistent with this, several SCF ubiquitin ligases are also upregulated in response to mitochondrial dysfunction (Nargund and others 2012).
One unexpected transcription factor identified by our screen was TAF-4, which forms part of the core transcription factor complex TFIID. A specific role in longevity was surprising, given that TAF-4 is considered a general transcription factor and its removal is embryonic lethal (Walker and others 2001). However, while wild-type worms on taf-4 RNAi did show reduced fecundity, they were otherwise phenotypically normal and had lifespan identical to controls. In contrast, both isp-1(qm150) and tpk-1(qm162) Mit mutants had their life extension completely negated by RNAi knockdown of taf-4 (Khan and others 2013). We have hypothesized previously (Khan and others 2013), that TAF-4 mediates metabolic adaptations through its interaction with the cAMP responsive element-binding protein transcription factor, CRH-1 (C. elegans CREB homolog family member 1) (Altarejos and Montminy 2011).
Two homeobox domain transcription factors, namely, CEH-18 (Khan and others 2013) and CEH-23 (Walter and others 2011), also contribute to Mit mutant longevity. The roles these transcription factors play in life extension is unknown, but given the central role of neurons in modulating the lifespan of the entire worm (Durieux and others 2011), it is interesting that CEH-23 plays a role in neuronal differentiation during development (Altun-Gultekin and others 2001). RNAi knockdown of ceh-23 specifically reduced the life extension of Mit mutants while affecting neither the long-lived age-1(hx546) nor eat-2(ad1116) mutants, nor the short-lived mev-1(kn1) mutant, and actually increased the lifespan of both wild-type and daf-16(mgDf47) worms. Significantly, transgenic overexpression of ceh-23 in either a subset of neurons or in the intestine was sufficient to extend lifespan in otherwise wild-type animals (Walter and others 2011).
Autophagy has been established as contributing to longevity in multiple worm models of life extension and will be discussed in section 3.4 with respect to Mit mutants. However, it is worth noting here that it has recently been shown that all of these models share not only a requirement for autophagy, but also the same transcription factor to induce it, namely, the TFEB ortholog, HLH-30 (Lapierre and others 2013).
Finally, the transcription factors most associated with Mit mutants are the Activating Transcription Factor associated with Stress-1 (ATFS-1), Ubiquitin-Like Protein-5 (UBL-5), and Defective Proventriculus in Drosophila homolog-1 (DVE-1). These are key mediators of the mitochondrial unfolded protein response (UPRmt) (Benedetti and others 2006; Haynes and others 2007; Haynes and others 2010) and will be examined in section 3.3.
In summary of this section, among all the transcription factors discussed, DAF-16, SKN-1, and HLH-30 are known to mediate life extension in multiple classes of long-lived C. elegans, but only HLH-30 is active in Mit mutants. Several transcription factors required by Mit mutants are also essential in various models of dietary restriction. That said, not all Mit mutants have the exact same transcription factor requirements, and neither do all dietary restriction models, but both classes overlap in their requirement for JUN-1, HIF-1, and NHR-25. Other transcription factors seem to modulate lifespan in a manner that is specific to Mit mutants (ATFS-1, UBL-5, DVE-1, and CEH-23) or have not, to our knowledge, been tested for their effects on lifespan in other classes of long-lived C. elegans (TAF-4, CEH-18, and AHA-1).
3.3. Mitochondrial Unfolded Protein Response
One mechanism by which the lifespan of Mit mutants may be extended entails activation of the mitochondrial unfolded protein response (UPRmt). Much like the prototypic UPR induced by the endoplasmic reticulum (UPRER), an accumulation of misfolded proteins in the mitochondria triggers retrograde signaling to the nucleus, whereupon mitochondrial chaperones and proteases are then upregulated to re-establish homeostasis. This process has been described in detail elsewhere (Haynes and others 2013; Pellegrino and others 2013) and here we provide only a summary (Figure 2).
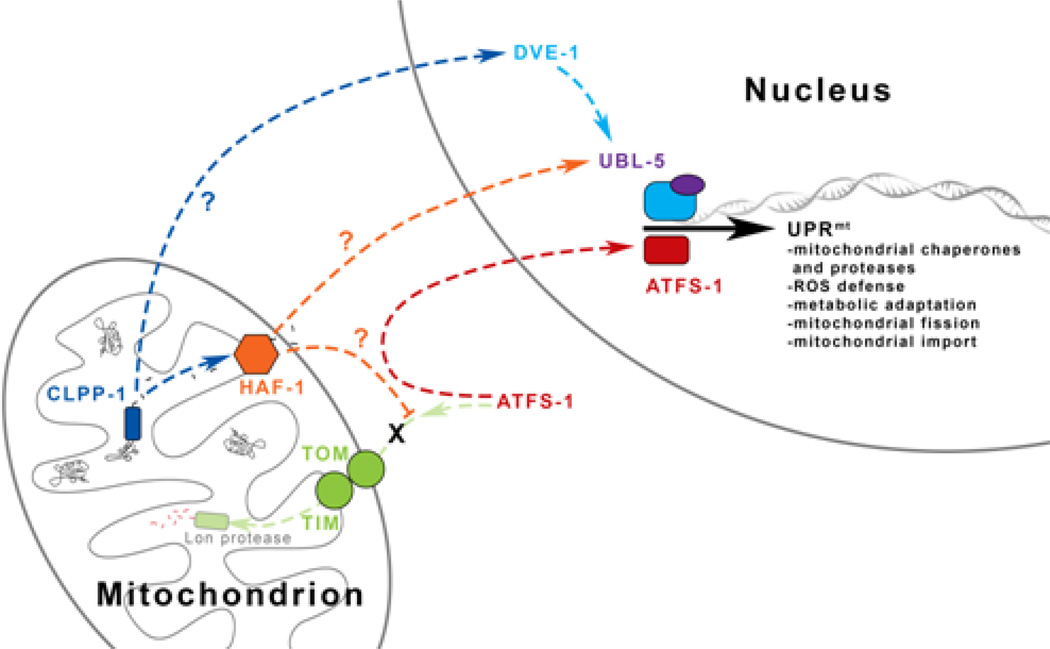
Figure 2. Activation of the Mitochondrial Unfolded Protein Response in C. elegans.
In the mitochondrial matrix, misfolded proteins are degraded to peptides by the protease CLPP-1. These peptide fragments are directed out of the matrix and into the cytosol by HAF-1. This efflux prevents the import of the transcription factor ATFS-1 into the matrix where it is normally degraded and instead redirects it into the nucleus. Two additional transcription factors – DVE-1 and UBL-5 – form a complex which cooperates with ATFS-1 to induce the mitochondrial unfolded protein response. While the response of DVE-1 depends on CLPP-1 activation, and the upregulation of UBL-5 on HAF-1 function, the signaling mechanisms involved that allow each proteins to reach the nucleus, independent of ATFS-1 activation, has not been deciphered.
Much of the work on the UPRmt of C. elegans has been undertaken by the Ron and Haynes labs. It was these investigators who first discovered two mitochondrial chaperones, namely, HSP-6 and HSP-60 – orthologous to mammalian mtHSP70 and HSP60, respectively – that were specifically induced in response to disruption of mitochondrial proteostasis (Yoneda and others 2004). The identification of these two proteins allowed fluorescent reporters to be constructed that, in turn, facilitated RNAi screens to find genes necessary for their induction following mitochondrial dysfunction (Benedetti and others 2006; Haynes and others 2007; Haynes and others 2010).
The proximal mediator of UPRmt appears to be the mitochondrial protease, CLPP-1, which degrades misfolded proteins in the mitochondrial matrix (Haynes and others 2007). The resulting peptides are then exported to the cytosol by the ATP-binding cassette transporter, HAF-1, which also modulates protein import into the mitochondria (Haynes and others 2010). Because mitochondrial protein import utilizes the mitochondrial membrane potential (Δψm) generated by the ETC, monitoring protein import may be an indirect way of monitoring ETC function. Diminished Δψm has been shown to activate retrograde signaling and increase replicative lifespan in yeast (Miceli and others 2011), but whether it plays a role in UPRmt induction in C. elegans is unknown.
ATFS-1 is an unusual transcription factor in that it contains both mitochondrial and nuclear targeting sequences. Under normal conditions, ATFS-1 is imported into the mitochondrial matrix and degraded by the Lon protease. During mitochondrial dysfunction, some ATFS-1 is not imported and is instead redirected to the nucleus. HAF-1 plays a role in this process but the specifics are not yet known (Haynes and others 2010; Nargund and others 2012). Also required for UPRmt activation is the homeodomain-containing transcription factor DVE-1, which undergoes nuclear redistribution downstream of CLPP-1 activation but independent of HAF-1 function (Haynes and others 2007). While HAF-1 does not play a role in the redistribution of DVE-1, it is needed for DVE-1-dependent upregulation of the ubiquitin-like protein UBL-5 (Haynes and others 2007), also required for UPRmt induction (Benedetti and others 2006). Factors mediating signaling from CLPP-1 and HAF-1 to the nucleus independent of ATFS-1 have not yet been identified. In the nucleus, UBL-5 and DVE-1 form a transcription complex (Haynes and others 2007) that cooperates with ATFS-1 to activate a broad transcriptional response that includes induction of mitochondrial chaperones, proteases, and transporters; genes mediating mitochondrial fission; and various metabolic enzymes (Nargund and others 2012).
In an elegant series of experiments, Dillin and colleagues further delineated C. elegans UPRmt induction at the organismal level. Surprisingly, these investigators discovered that the UPRmt could be induced in the worm intestine by mitochondrial disruption that occurred distally in neuron or muscle cells. This non-cell-autonomous UPRmt response was not activated in the canonical manner: While intestine-specific knockdown of ubl-5 blocked UPRmt activated by mitochondrial disruption in the intestine, it did not block UPRmt induced by the non-cell-autonomous response activated by neuronal or muscle mitochondrial disruption (Durieux and others 2011).
Based on the pioneering work of Ron and colleagues, it was suggested that induction of UPRmt may contribute to the extended lifespan of Mit mutants (Ventura and Rea 2007). In support of this hypothesis, RNAi knockdown of ubl-5 was found to abrogate isp-1(qm150) life extension but to not affect wild-type worms nor other long-lived mutants (Durieux and others 2011). Additionally, while atfs-1 RNAi did not impact the growth of wild-type worms, it impaired both isp-1(qm150) and clk-1(qm30) Mit mutant development (Nargund and others 2012). It was recently asserted that induction of UPRmt is the major mechanism for Mit mutant longevity (Houtkooper and others 2013), yet most data suggests that this cannot be the only mechanism required: Knockdown of cco-1 in either neuronal, intestine, or muscle cells induces UPRmt in the intestine, but only neuronal or intestinal knockdown prolongs life (Durieux and others 2011). Likewise, while Phsp-6::GFP is induced in most Mit mutants, it is not induced in all of them (Ventura and Rea 2007), suggesting that UPRmt is not ubiquitously required for Mit mutant longevity.
RNAi knockdown of several nuclear DNA-encoded mitochondrial genes induce UPRmt without extending life (Durieux and others 2011) (see also Figure 3A–B), as do the mev-1(kn1) and gas-1(fc21) mutations (Durieux and others 2011; Pujol and others 2013). Neither the mev-1(kn-1) nor gas-1(fc21) mutation causes pathology incompatible with longevity as both mutants can become long-lived upon additional gene knockdowns. (This short-lived mutants will be discussed further in section 4.)
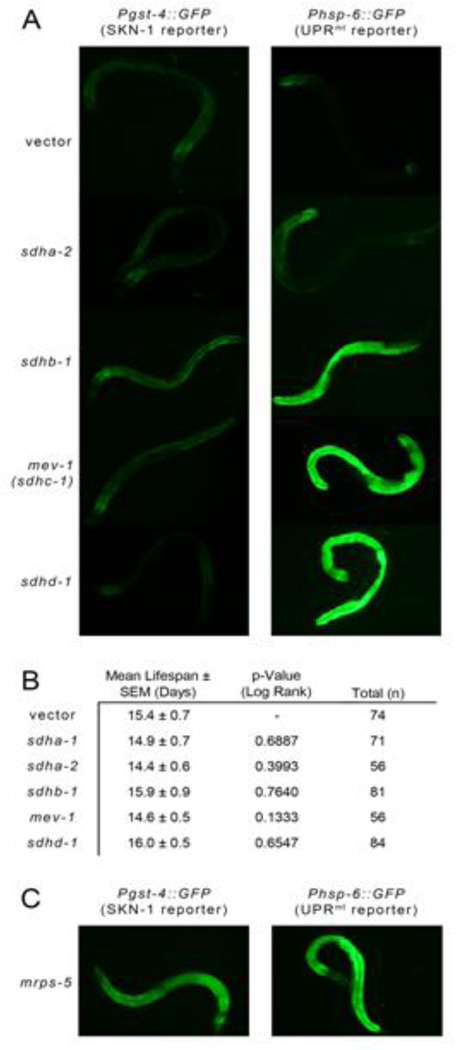
Figure 3. Robust Activation of UPRmt Signaling Following Complex II Disruption is Not Sufficient to Extend C. elegans Lifespan.
(A) RNAi knockdown of three out of four complex II subunits strongly activates UPRmt (marked by a Phsp-6::GFP transcriptional reporter) but the on SKN-1 activation (marked by a Pgst-4::GFP transcriptional reporter) is minimal. Data for sdha-1 was not available. (B) RNAi knockdown of any complex II subunit in wild-type (N2) animals has no effect on lifespan (data from Figure 1b, Kuang et. al. 2012). (C) Unlike disruption of complex II subunits, life-extending RNAi knockdown of mprs-5 strongly activates both SKN-1 and UPRmt signaling pathways (control worms same as in panel A).
If UPRmt induction is responsible for the life extension of Mit mutants, then the prediction would be that constitutive activation of UPRmt in otherwise wild-type worms should cause them to be longlived. This has not been the case. A recent screen for worms resistant to statins found mutations in the mitochondrial-targeting sequence of atfs-1 that caused the transcription factor to be sent into the nucleus regardless of mitochondrial status. As assayed both by transcriptional reporters and increased resistance to mitochondrial stressors such as ethidium bromide, this nuclear-directed ATFS-1 constitutively induced UPRmt in these mutants, yet they were less healthy and had reduced lifespan compared with wild-type worms (Rauthan and others 2013). This atfs-1 gain-of-function mutation suggests that UPRmt is not causative of longevity, yet it is important to consider whether this condition truly recapitulates endogenous UPRmt. While the transcription factors DVE-1 and UBL-5 have been shown to be integral to endogenous UPRmt, they are not downstream of ATFS-1 and are presumably not activated in these mutants. Clearly, the subject of UPRmt and its role in Mit mutants and lifespan specification warrants further study.
3.4. Autophagy
Haynes and colleagues have proposed three broad responses to mitochondrial dysfunction: UPRmt to restore mitochondrial homeostasis, mitophagy to remove defective mitochondria and restore cellular homeostasis, and, finally, apoptosis to eliminate the damaged cell and protect organismal homeostasis. The extent to which each pathway is induced depends on the severity and duration of the problem (Pellegrino and others 2013). We now move from discussion of the first response – UPRmt, to the second – autophagy, which encompasses degradation and turnover of numerous cellular components, including the mitochondria (where the process is called mitophagy).
Recent work by Ventura and colleagues studying frh-1 (the worm ortholog of Frataxin, required for Fe-S cluster assembly), showed that the core autophagy regulating genes bec-1 and unc-51 are upregulated upon frh-1 knockdown (Schiavi and others 2013). Both genes were found to be essential for the increased autophagy and subsequent life extension of this Mit mutant (Schiavi and others 2013). Previously, the Hekimi lab had reported that while autophagy is upregulated in response to isp-1 and nuo-6 RNAi, it is not upregulated in isp-1(qm150) and nuo-6(qm200) mutants, suggesting that increased autophagy is not a universal requirement for Mit mutant life extension (Yang and Hekimi 2010). Another recent study, however, showed that RNAi knockdown of the autophagy genes bec-1 and vps-34 reduced lifespan in isp-1(qm150) and clk-1(qm30) mutants while having no effect on wild-type worms (Lapierre and others 2013). Additionally, several studies have shown autophagy to also be increased in other long-lived C. elegans mutants and required for their life extension as well (Lapierre and others 2013; Toth and others 2008). Hansen and colleagues demonstrated that these long-lived worms not only share increased autophagy, but upregulate it through the same transcription factor, namely, HLH-30, orthologous to mammalian TFEB (Lapierre and others 2013).
Transgenic overexpression of hlh-30 actually increased lifespan of wild-type worms, suggesting that autophagy is not merely differentially required in long-lived worms, but actually plays a causative role in their life extension (Lapierre and others 2013). The role of autophagy in determining lifespan has been reviewed by Tavernarakis and colleagues (Lionaki and others 2013). Broadly, autophagy has been found to decline with age, and upregulating autophagy is believed to favor longevity through removal of damaged molecules and organelles that can otherwise accumulate and contribute to pathology. Autophagy has been shown to be protective by degrading the aggregate-prone proteins that contribute to neurodegenerative disease (Ravikumar and others 2002).
Autophagy likely also contributes to Mit mutant longevity through its role in lipolysis, another process found to be increased in long-lived worms (Lapierre and others 2012; Schiavi and others 2013). The role of lipid metabolism in determining lifespan has been most studied in worms made long-lived by elimination of the germline precursor cells and several lipases are required for this mode of longevity. One of these lipases is LIPL-4 (Wang and others 2008). Significantly, overexpression of LIPL-4 extends lifespan in otherwise wild-type worms (Wang and others 2008). This lipase is also upregulated under conditions of nutrient deprivation and, like increased autophagy, is dependent on the transcription factor HLH-30 (O'Rourke and Ruvkun 2013).
Returning to the three broad responses that can be activated by dysfunctional mitochondria, it is important to note that each response may facilitate the next if it fails to restore homeostasis. Among the myriad genes activated by ATFS-1 are several involved in mitochondrial fission (Nargund and others 2012), a process which mediates the segregation of damaged mitochondria for degradation by mitophagy (Twig and others 2008). Similarly, autophagy can contribute either to cell survival or cell death (Kourtis and Tavernarakis 2009). Thus, while increasing autophagy can be protective and even extend lifespan in multiple model organisms (Lapierre and others 2013; Pyo and others 2013; Simonsen and others 2008), excessive autophagy can result in cellular damage and death (Kang and others 2007) and contribute to premature aging (Marino and others 2008). Autophagy, like mitochondrial dysfunction, seems to exhibit a threshold effect, being protective and extending life within a certain range, beyond which, it becomes pathological and life-shortening.
3.5. Metabolism
While the term “metabolism” encompasses many levels of metabolite processing in multicellular organisms, we use it here to refer specifically to the set of cellular reactions that cooperate to convert food sources into biomass and usable energy. The underlying premise for metabolism playing a role in aging is that a change in metabolic configuration may itself be conducive to long life. Several studies have examined changes in metabolism and its role in potentially controlling Mit mutant longevity. These studies can be grouped into two broad approaches: microarray-based analyses and metabolomics.
Of the microarray-based analyses, one of the most extensive was undertaken by Morgan and colleagues (Falk and others 2008): Utilizing whole genome affimetrix microarrays and Gene Set Enrichment Analysis, these authors analyzed eight worm lines in which an ETC subunit was disrupted either genetically (gas-1(fc21), mev-1(kn1), and isp-1(qm150)) or via RNAi (nuo-5, nduf-6, C34B2.8, D2030.4, and Y56A3A.19). Compared to wild-type worms, the six strains in which complex I was disrupted all showed upregulation of genes involved in oxidative phosphorylation (OXPHOS), pyruvate metabolism, fat metabolism, glycolysis, and the TCA cycle, as well as several genes related to stress protection – including glutathione and P450-requiring stress-response pathways. Additionally, a number of genes related to amino acid catabolism were differentially regulated in complex I disruptants. In a related study (Cristina and others 2009), the global transcriptome of the long-lived clk-1(qm30), isp-1(qm150), and cco-1 RNAi-treated worms, representing disruption of ubiquinone biosynthesis, and complexes III and IV, respectively, were examined by microarray analysis. While there was considerable variation in the expression level of individual genes among the strains, gene ontology (GO) term analysis uncovered several processes that were significantly over-represented and were essentially the same metabolic processes found in the aforementioned complex I disruptants (Falk and others 2008). All three Mit mutants upregulated components of their ETC, presumably as a primary compensation for their defects. Fatty acid β-oxidation was also increased. In addition, several alternate energy-generating pathways that bypass all or part of the ETC were upregulated, including glycolysis, the glyoxylate cycle, and glycerol fermentation (Cristina and others 2009). In summary, both studies demonstrate that disruption of the ETC in C. elegans is compensated for either by increasing ETC activity and feeding more substrates into the TCA cycle, or by bypassing the ETC altogether and essentially turning the worms into fermenters.
One unexpected finding made by Morgan and colleagues in their study was that, relative to wild-type animals, several amino acids were more abundant in the three genetic mutants studied. This was surprising because mRNA data indicated that the catabolic processes that degrade these amino acids were in fact also elevated. Nevertheless, alanine and the three branched-chain amino acids (leucine, isoleucine, and valine) were increased. Interestingly, glutamate was decreased. This configuration, it was suggested, likely reflects glutamate-mediated transamination of the corresponding α-ketoacids of alanine, valine, leucine and isoleucine. Normally, degradation of these α-ketoacids requires a functional ETC and so their removal in mitochondrial mutants may sink glutamate from the cell leading to knock-on consequences for metabolism (Falk and others 2008).
In another study, Ebert and colleagues (Zuryn and others 2010) measured a variety of metabolic parameters for a panel of RNAi-induced Mit mutants. By comparing their findings alongside data collected for two different complex II RNAi disruptants, which showed no life extension, Ebert and colleagues showed that neither reduction of whole body ATP levels, decreased brood size, sensitivity to the uncoupler FCCP, reduction in adult volume, nor reduced oxygen consumption differed between the various disruptants, including those that affected complex II. Intriguingly, they also showed that while genes required for β-oxidation (acs-2), the glyoxylate cycle (gei-7), gluconeogenesis (PEPCK), and glycolysis (gpd-3) were robustly upregulated in RNAi disruptants representing four ETC complexes, their complete abrogation by removal of the transcription factor NHR-49 had no effect on lifespan. These findings argue that despite obvious metabolic readjustments in Mit mutants, some other parameter must be working to extend life.
A clue to what this life-extending parameter might be came from our own recent metabolomics studies. We have described a suite of mass-spectrometry techniques to study the metabolic consequences of mitochondrial dysfunction in C. elegans (Butler and others 2012; Butler and others 2010; Mishur and others 2013). Using these techniques, we identified a collection of compounds, enriched in α-ketoacids and α-hydroxyacids, that were restricted to mitochondrial mutants exhibiting the classic “Mit phenotype” (Butler and others 2013). We posited that these compounds originate with a build-up of NADH in the mitochondria and consequential inhibition of the three α-ketoacid dehydrogenases (namely, α-ketoglutarate dehydrogenase, pyruvate dehydrogenase, and branchedchain ketoacid dehydrogenase). In recent work, we have obtained evidence that the α-ketoacids and α-hydroxyacids that accumulate in Mit mutants contribute to aspects of their phenotype (Mishur and others, manuscript in review). Notably, many of these compounds are structurally related to α-ketoglutarate and some are already known to act as inhibitors of a large family of enzymes called the α-ketoglutarate dependent hydroxylases (Chowdhury and others 2011; Cunliffe and others 1992; Hutchinson and others 2012). One such enzyme is EGL-9, which negatively regulates the hypoxia-inducible transcription factor HIF-1 under normoxic conditions (Epstein and others 2001). As discussed in section 3.2, HIF-1 is active and required for Mit mutant life extension even under normoxic conditions (Khan and others 2013; Lee and others 2010). In light of this, it is significant that exogenous administration of some of the α-ketoacids and α-hydroxyacids that accumulate in Mit mutants stabilize HIF-1 (Mishur and others, manuscript in review).
4. NADH, Ubiquinone, and the Mit Phenotype
We will conclude our review of responses to mitochondrial disruption in Mit mutants with a discussion of two of the short-lived ETC mutants as their exception may prove instructive. Life-shortening mutations have been reported in complex I (Grad and Lemire 2004; Hartman and others 2001), complex II (Ishii and others 1990), and complex III (Butler and others 2010). Since severe RNAi-mediated knockdown of some ETC subunits result in larval arrest and reduced lifespan, these life-shortening ETC mutations may likewise be situated at the far end of the response curve (Rea and others 2007). It might be assumed that these mutations produce dysfunction incompatible with longevity, however, like the role of CEP-1 upon atp-3 knockdown (Ventura and others 2009), a closer look at the two best-studied of these mutants suggest this is not the case.
4.1 Response to Complex II Disruption
As can be seen in Table 1, disruption of almost any ETC subunit, as well as various mitochondrial ribosomal, metabolite carrier, and chaperone proteins, among others, can all extend life. However, disruption of complex II has never been reported to increase lifespan (Ichimiya and others 2002; Kuang and Ebert 2012; Rea and others 2007). When most forms of mitochondrial disruption can extend life, it is puzzling why complex II should be different.
It has been suggested that the mitochondrial unfolded protein response (UPRmt) is responsible for Mit mutant life extension and that this response is triggered by an imbalance between nuclear- and mitochondrial-encoded subunits within respiratory complexes (Houtkooper and others 2013). Since complex II is entirely encoded by nuclear genes, knockdown of its subunits would not be expected to create such an imbalance and this has been proposed as the reason complex II inhibition does not extend life (Houtkooper and others 2013). However, as established by induction of the UPRmt reporter Phsp-6::GFP, this response is, in fact, robustly activated by individual knockdown of three of the four complex II subunits (Figure 3A, see also (Durieux and others 2011; Pujol and others 2013)). A similar response is observed when the Phsp-6::GFP reporter is crossed into the complex II mutant, mev-1(kn1) (unpublished observation).
It is possible that UPRmt would extend the lifespan of these complex II disruptants, except that some pathology is simultaneously induced that is life shortening. There is evidence for this in that the mev-1(kn1) mutant is hypersensitive to oxygen (Ishii and others 1990) and generates excessive ROS from complex II (Senoo-Matsuda and others 2001), leading to pathological apoptosis and aging (Senoo-Matsuda and others 2003). Interestingly, evidence suggests that complex II plays a unique role in signaling apoptosis and that high ROS generation is part of this process (Grimm 2013). It could be that complex II disruption triggers this response in addition to the life extending ones. However, these same short-lived mev-1(kn1) mutants can live even longer than wild-type worms following the additional knockdown of mrps-5, which encodes a mitochondrial ribosomal subunit (Houtkooper and others 2013). This suggests that it is not overt pathology that makes mev-1(kn1) worms short-lived, but a failure to induce longevity mechanisms. Alternatively, mrps-5 knockdown may somehow counter the pathology and thereby allow the UPRmt and other protective mechanisms to extend life. Our group has previously shown Pgst-4::GFP to be a robust reporter for activation of SKN-1 (Kahn and others 2008), the latter of which, among its many roles, protects against oxidative stress (An and Blackwell 2003). While Pgst-4::GFP was only marginally induced by each of the complex II RNAi that we tested (Figure 3A), it was strongly induced by mrps-5 RNAi (Figure 3C). The SKN-1 antioxidant response induced by mrps-5 knockown may act to counter the ROS caused by the mev-1(kn1) mutation.
Ebert and colleagues systematically analyzed the effects of RNAi knockdown of each complex II subunit on lifespan, growth rate, fecundity, oxygen consumption, ATP content, mitochondrial membrane potential (Δψm), FCCP sensitivity, and gene expression. RNAi knockdown was efficacious, with mRNA levels being reduced to an average of 40% of wild-type, yet none had any effect on lifespan (Kuang and Ebert 2012) (Figure 3B). RNAi knockdown of sdhb-1, mev-1, and sdhd-1 was clearly sufficient to impact mitochondrial function, as evinced by FCCP hypersensitivity, reduced ATP, and diminished egg-laying (Kuang and Ebert 2012). These are the same complex II knockdowns that we have found to activate UPRmt (Figure 3A); and reduced egg-viability has been previously reported on both sdhb-1 and mev-1 RNAi (Rea and others 2007). Interestingly, while the reductions in ATP levels were comparable to that observed upon knockdown of subunits of complex I, III, or IV, the impact of complex II disruption on Δψm was much less (Kuang and Ebert 2012). Similarly, the upregulation of genes involved in alternate metabolic pathways was also less than observed in Mit mutants, suggesting that metabolism may be perturbed to a lesser extent, and that the worm is able to adapt without invoking the mechanisms that lead to Mit mutant longevity (Kuang and Ebert 2012).
4.2 Response to Complex I Disruption
Ironically, the one instance wherein knockdown of complex II subunits (specifically sdhb-1, mev-1, or sdhd-1) has been reported to extend life is in the otherwise short-lived complex I mutant, gas-1(fc21) (Pujol and others 2013). Using blue native gel electrophoresis, this study showed that the gas-1(fc21) mutation results in a reduction of assembled complex I and thereby diminishes complex I activity. Knocking down complex II apparently corrects this deficit. This loss of complex I accompanied by an upregulation of complex II is not observed in the long-lived complex I mutant, nuo-6(qm200). Taken together, these observations led the authors to hypothesize that it was the upregulation of complex II in gas-1(fc21) mutants that destabilized complex I and caused these worms to be short-lived (Pujol and others 2013). As the authors pointed out, this could explain the previously reported life extension of gas-1(fc21) worms cultured at 15°C (Hartman and others 2001) as a more fragile complex I may be stabilized at lower temperatures. However, it does not explain how an increase in complex II should destabilize complex I. While most evidence indicates that complex II does not join supercomplex formations (Genova and Lenaz 2013), complex I depends on forming supercomplexes with III and IV to function properly (Acin-Perez and others 2004; Suthammarak and others 2009). As a consequence of this interaction, the complex III mutant isp-1(qm150) also has reduced complex I activity and yet, unlike gas-1(fc21), this mutant is long-lived (Suthammarak and others 2010). Finally, this hypothesis for the gas-1(fc21) mutant’s diminished lifespan offers no clear explanation for why other manipulations such as eliminating sod-2 or daf-16 (Suthammarak and others 2013) or culturing the worms on FUdR (Van Raamsdonk and Hekimi 2011) also increase the lifespan of these worms. Indeed, all of these gene knockdowns would seem to cause further stress in the worm, arguing that, like complex II disruptants, gas-1(fc21) are not long-lived because they are able to adapt without inducing the mechanisms that produce longevity in Mit mutants.
4.3 Rethinking the Mitochondrial Electron Transport Chain
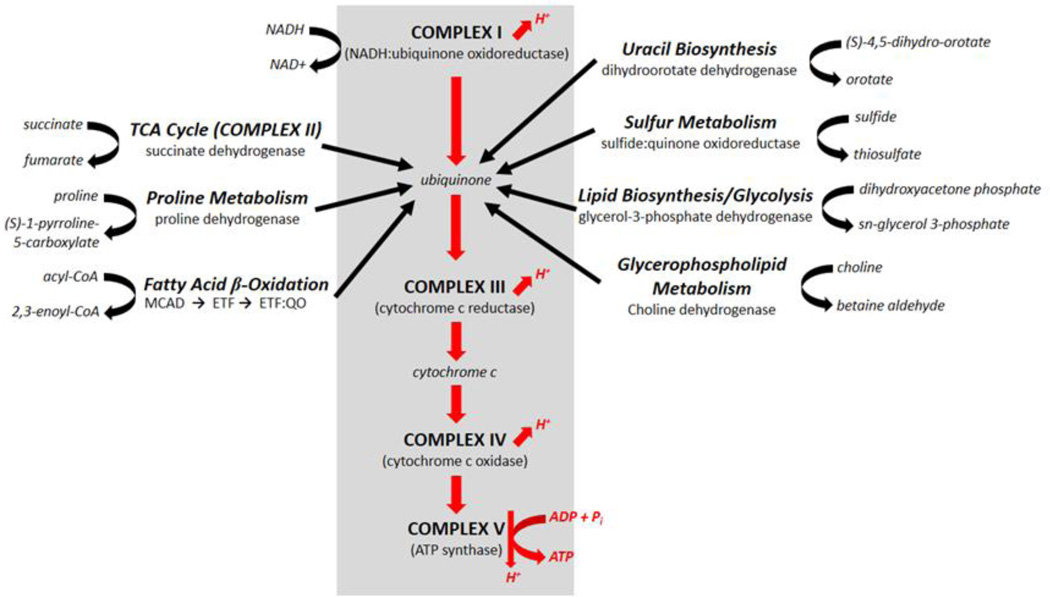
The NADH and succinate dehydrogenases are not unique in contributing electrons to the mitochondrial ubiquinone pool. Numerous other enzymes do so as well, including electron transfer flavoprotein (ETF) ubiquinone oxidoreductase (which transfers electrons gained from β-oxidation of fatty acids to ubiquinone), dihydroorotate dehydrogenase, glycerol-3-phosphate dehydrogenase, and proline hydroxylase, among others (Figure 4). It is, perhaps, an historical artifact that succinate dehydrogenase is counted as part of the ETC while these other enzymes are not. Indeed, succinate dehydrogenase has more in common with these other matrix-localized enzymes than with the other canonical ETC complexes: It does not pump protons, it is comprised exclusively of nuclear-encoded subunits, and it is not integral to supercomplex formation (Genova and Lenaz 2013; Schagger and Pfeiffer 2001). Succinate dehydrogenase is an enzyme of the TCA cycle and it contributes electrons collected from succinate oxidation to the ubiquinone pool. Crippling succinate dehydrogenase does not directly impair generation of the mitochondrial proton motive force, nor its generation from other electron donors. As summarized in sections 3.5 and 4.1, while metabolism is impacted in complex II disruptants, as one would expect upon impairment of the TCA cycle, it does not cause the wholesale metabolic reprogramming seen in Mit mutants (Butler and others 2013; Kuang and Ebert 2012).
Figure 4. Mit Mutant Phenotype Arises from Disruption of the Core Electron Transport Pathway Comprised of Complexes I, III, IV and V.
At least eight pathways contribute electrons to the ubiquinone pool in mitochondria. Of these, only complex I pumps protons and contains subunits encoded by mitochondrial DNA.
As is the case for complex II, no effect on lifespan has been reported following disruption of the other metabolic pathways contributing electrons to the ubiquinone pool. An exception, however, are genes within the ETF pathway: RNAi knockdown is embryonic lethal (Kamath and others 2003; Simmer and others 2003; Sonnichsen and others 2005) and this may reflect the importance of lipid metabolism to C. elegans development. In light of all the above, the question seems to be less, “What is different about complex II among the ETC complexes that its disruption does not result in life extension?” and more, “What is unique about complex I among all the contributors to the ubiquinone pool that its disruption DOES result in life extension?” As has been stated, complex I, like complexes III, IV, and V, pumps protons and contains subunits encoded by mtDNA. What seems key, however, is that all the mitochondrial disruptions that result in life extension are downstream of NADH, suggesting NAD+/NADH ratio as the most proximal cause of Mit mutant longevity, as our recent metabolomics data also suggest (Butler and others 2013) (Mishur and others, manuscript in review). As explained in section 3.5, NADH is normally one of the products generated by the α-ketoacid dehydrogenases. A build-up of NADH will render these enzymes inactive and may be one explanation for the observed accumulation of α-ketoacids and α-hydroxyacids in Mit mutants (Butler and others 2013). These compounds are capable of inhibiting α-ketoglutarate dependent hydroxylases, including EGL-9 which negatively regulates HIF-1, and ultimately may also play an active part in specifying the Mit phenotype (Mishur and others, manuscript in review).
5. Conclusion
While the mechanism controlling life extension in Mit mutants was first viewed as a single phenomenon, after 20 years of study, evidence now increasingly indicates that different Mit mutants induce a collection of overlapping, life-promoting mechanisms. Yang and Hekimi once suggested that Mit mutants should be viewed as two separate classes: genetic and RNAi (Yang and Hekimi 2010). However, even among these two broad classes it appears that different mechanisms control life extension, depending on which gene is being knocked down or mutated. This is illustrated by two of the best studied Mit mutants – clk-1(qm30) and isp-1(qm150) – wherein additional mutations can have very different phenotypic effects: While the lifespan of clk-1(qm30) worms is further increased by either the daf-2(e1370) mutation (Lakowski and Hekimi 1996) or sod-2 knockdown (Van Raamsdonk and Hekimi 2009; Yang and others 2007), isp-1(qm150) worms show either no effect or life-shortening when exposed to these same treatments (Feng and others 2001; Suthammarak and others 2013; Van Raamsdonk and Hekimi 2009). It could be possible that clk-1(qm30) and isp-1(qm150) extend life by the same mechanisms and that the opposite effects of additional knockouts/knockdowns are due to where they sit on the “dilution curve” (Rea and others 2007; Van Raamsdonk and Hekimi 2009). However, the fact that nuo-6 RNAi further extends isp-1(qm150) lifespan argues that this mutant cannot already be at the threshold where further impairment is pathological (Yang and Hekimi 2010). Another peculiarity of the clk-1(qm30) mutant is that life extension is completely negated by whole gonad ablation (Dillin and others 2002), despite being daf-16 independent (Lakowski and Hekimi 1996).
Without knowing the potential differences between various Mit mutants, it seems prudent to include more than the isp-1(qm150) and clk-1(qm30) mutants when testing the role of a particular mechanism in Mit mutant longevity. Especially neglected are the RNAi-induced Mit mutants. A requirement for the various transcription factors is assumed but, with the exception of CEP-1 (Ventura and others 2009), none have been verified in these worms. Even UPRmt, which by reporter induction is seen to be strongly activated in most RNAi-induced Mit mutants (Ventura and Rea 2007), has never been formally tested for its role in their life extension. Taken as a whole, current research indicates that there is not one pathway that is solely responsible for Mit mutant longevity but, rather, a network of interacting responses that cooperate to extend life.
Highlights.
We examine responses to mitochondrial dysfunction in C. elegans that may extend life.
We discuss ROS signaling, UPRmt, autophagy, metabolism, and transcription factors.
The consequences of complexes I versus complex II disruption are explored.
Acknowledgements
Financial support was provided by a fellowship from the Glenn Foundation for Medical Research and a training grant from the NIA (EM). We would also like to thank our reviewers for their thoughtful feedback and helpful recommendations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Molecular cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature reviews Molecular cell biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, Hobert O. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 2001;128:1951–1969. doi: 10.1242/dev.128.11.1951. [DOI] [PubMed] [Google Scholar]
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes & development. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andziak B, O'Connor TP, Qi W, DeWaal EM, Pierce A, Chaudhuri AR, Van Remmen H, Buffenstein R. High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging cell. 2006;5:463–471. doi: 10.1111/j.1474-9726.2006.00237.x. [DOI] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS genetics. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Butler JA, Mishur RJ, Bhaskaran S, Rea SL. A metabolic signature for long life in the Caenorhabditis elegans Mit mutants. Aging cell. 2013;12:130–138. doi: 10.1111/acel.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JA, Mishur RJ, Bokov AF, Hakala KW, Weintraub ST, Rea SL. Profiling the anaerobic response of C. elegans using GC-MS. PloS one. 2012;7:e46140. doi: 10.1371/journal.pone.0046140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JA, Ventura N, Johnson TE, Rea SL. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2010;24:4977–4988. doi: 10.1096/fj.10-162941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS genetics. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, McDonough MA, King ON, Clifton IJ, Klose RJ, Claridge TD, Ratcliffe PJ, Schofield CJ, Kawamura A. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO reports. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS genetics. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe CJ, Franklin TJ, Hales NJ, Hill GB. Novel inhibitors of prolyl 4-hydroxylase. 3. Inhibition by the substrate analogue N-oxaloglycine and its derivatives. Journal of medicinal chemistry. 1992;35:2652–2658. doi: 10.1021/jm00092a016. [DOI] [PubMed] [Google Scholar]
- Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Human molecular genetics. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Nissim I, Sedensky MM, Yudkoff M, Morgan PG. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Molecular genetics and metabolism. 2008;93:388–397. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochimica et biophysica acta. 2013 doi: 10.1016/j.bbabio.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Grad LI, Lemire BD. Mitochondrial complex I mutations in Caenorhabditis elegans produce cytochrome c oxidase deficiency, oxidative stress and vitamin-responsive lactic acidosis. Human molecular genetics. 2004;13:303–314. doi: 10.1093/hmg/ddh027. [DOI] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Current biology : CB. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. Respiratory chain complex II as general sensor for apoptosis. Biochimica et biophysica acta. 2013;1827:565–572. doi: 10.1016/j.bbabio.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS genetics. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. Journal of gerontology. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hartman PS, Ishii N, Kayser EB, Morgan PG, Sedensky MM. Mitochondrial mutations differentially affect aging, mutability and anesthetic sensitivity in Caenorhabditis elegans. Mechanisms of ageing and development. 2001;122:1187–1201. doi: 10.1016/s0047-6374(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Fiorese CJ, Lin YF. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends in cell biology. 2013;23:311–318. doi: 10.1016/j.tcb.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Developmental cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Molecular cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson SE, Leveridge MV, Heathcote ML, Francis P, Williams L, Gee M, Munoz-Muriedas J, Leavens B, Shillings A, Jones E, Homes P, Baddeley S, Chung CW, Bridges A, Argyrou A. Enabling lead discovery for histone lysine demethylases by high-throughput RapidFire mass spectrometry. Journal of biomolecular screening. 2012;17:39–48. doi: 10.1177/1087057111416660. [DOI] [PubMed] [Google Scholar]
- Hwang AB, Lee SJ. Regulation of life span by mitochondrial respiration: the HIF-1 and ROS connection. Aging. 2011;3:304–310. doi: 10.18632/aging.100292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimiya H, Huet RG, Hartman P, Amino H, Kita K, Ishii N. Complex II inactivation is lethal in the nematode Caenorhabditis elegans. Mitochondrion. 2002;2:191–198. doi: 10.1016/s1567-7249(02)00069-7. [DOI] [PubMed] [Google Scholar]
- Ishii N, Takahashi K, Tomita S, Keino T, Honda S, Yoshino K, Suzuki K. A methyl viologen-sensitive mutant of the nematode Caenorhabditis elegans. Mutation research. 1990;237:165–171. doi: 10.1016/0921-8734(90)90022-j. [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidative-stress response in Caenorhabditis elegans. The Biochemical journal. 2008;409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes & development. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MH, Ligon M, Hussey LR, Hufnal B, Farber R, 2nd, Munkacsy E, Rodriguez A, Dillow A, Kahlig E, Rea SL. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging. 2013 doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sun H. Functional genomic approach to identify novel genes involved in the regulation of oxidative stress resistance and animal lifespan. Aging cell. 2007;6:489–503. doi: 10.1111/j.1474-9726.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell death and differentiation. 2009;16:21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- Kuang J, Ebert PR. The failure to extend lifespan via disruption of complex II is linked to preservation of dynamic control of energy metabolism. Mitochondrion. 2012;12:280–287. doi: 10.1016/j.mito.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, Stigter EC, Hendriks MM, Berger R, Rokach J, Korswagen HC, Brenkman AB. Quantification of in vivo oxidative damage in Caenorhabditis elegans during aging by endogenous F3-isoprostane measurement. Aging cell. 2013;12:214–223. doi: 10.1111/acel.12043. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nature communications. 2013;4:2267. doi: 10.1038/ncomms3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Melendez A, Hansen M. Autophagy links lipid metabolism to longevity in C. elegans. Autophagy. 2012;8:144–146. doi: 10.4161/auto.8.1.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Current biology : CB. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nature genetics. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Limongelli A, Schaefer J, Jackson S, Invernizzi F, Kirino Y, Suzuki T, Reichmann H, Zeviani M. Variable penetrance of a familial progressive necrotising encephalopathy due to a novel tRNA(Ile) homoplasmic mutation in the mitochondrial genome. Journal of medical genetics. 2004;41:342–349. doi: 10.1136/jmg.2003.016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lionaki E, Markaki M, Tavernarakis N. Autophagy and ageing: insights from invertebrate model organisms. Ageing research reviews. 2013;12:413–428. doi: 10.1016/j.arr.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes & development. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Ugalde AP, Salvador-Montoliu N, Varela I, Quiros PM, Cadinanos J, van der Pluijm I, Freije JM, Lopez-Otin C. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Human molecular genetics. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Hormesis and the antiaging action of dietary restriction. Experimental gerontology. 1998;33:61–66. doi: 10.1016/s0531-5565(97)00071-5. [DOI] [PubMed] [Google Scholar]
- Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quinones JF, Jazwinski SM. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Frontiers in genetics. 2011;2:102. doi: 10.3389/fgene.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishur RJ, Butler JA, Rea SL. Exometabolomic mapping of Caenorhabditis elegans: a tool to noninvasively investigate aging. Methods in molecular biology. 2013;1048:195–213. doi: 10.1007/978-1-62703-556-9_15. [DOI] [PubMed] [Google Scholar]
- Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, Miyoshi H, Sakamoto K, Ishii N, Hekimi S, Kita K. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. The Journal of biological chemistry. 2001;276:7713–7716. doi: 10.1074/jbc.C000889200. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke EJ, Ruvkun G. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nature cell biology. 2013;15:668–676. doi: 10.1038/ncb2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochimica et biophysica acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol C, Bratic-Hench I, Sumakovic M, Hench J, Mourier A, Baumann L, Pavlenko V, Trifunovic A. Succinate dehydrogenase upregulation destabilize complex I and limits the lifespan of gas-1 mutant. PloS one. 2013;8:e59493. doi: 10.1371/journal.pone.0059493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nature communications. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Human molecular genetics. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cellular signalling. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea SL. Metabolism in the Caenorhabditis elegans Mit mutants. Experimental gerontology. 2005;40:841–849. doi: 10.1016/j.exger.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS biology. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K. How increased oxidative stress promotes longevity and metabolic health: The concept of mitochondrial hormesis (mitohormesis) Experimental gerontology. 2010;45:410–418. doi: 10.1016/j.exger.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell metabolism. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. The Journal of biological chemistry. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- Schiavi A, Torgovnick A, Kell A, Megalou E, Castelein N, Guccini I, Marzocchella L, Gelino S, Hansen M, Malisan F, Condo I, Bei R, Rea SL, Braeckman BP, Tavernarakis N, Testi R, Ventura N. Autophagy induction extends lifespan and reduces lipid content in response to frataxin silencing in C. elegans. Experimental gerontology. 2013;48:191–201. doi: 10.1016/j.exger.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Molecular Metabolism. 2013a;2:92–102. doi: 10.1016/j.molmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Schmeisser K, Weimer S, Groth M, Priebe S, Fazius E, Kuhlow D, Pick D, Einax JW, Guthke R, Platzer M, Zarse K, Ristow M. Mitochondrial hormesis links low-dose arsenite exposure to lifespan extension. Aging cell. 2013b;12:508–517. doi: 10.1111/acel.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Zarse K, Ristow M. Lonidamine extends lifespan of adult Caenorhabditis elegans by increasing the formation of mitochondrial reactive oxygen species. Hormone and metabolic research = Hormon- und Stoffwechselforschung = Hormones et metabolisme. 2011;43:687–692. doi: 10.1055/s-0031-1286308. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Hartman PS, Akatsuka A, Yoshimura S, Ishii N. A complex II defect affects mitochondrial structure, leading to ced-3- and ced-4-dependent apoptosis and aging. The Journal of biological chemistry. 2003;278:22031–22036. doi: 10.1074/jbc.M211377200. [DOI] [PubMed] [Google Scholar]
- Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, Nakazawa H, Hartman PS, Ishii N. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. The Journal of biological chemistry. 2001;276:41553–41558. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS biology. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. The Journal of biological chemistry. 2010;285:40724–40731. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthammarak W, Somerlot BH, Opheim E, Sedensky M, Morgan PG. Novel Interactions between Mitochondrial Superoxide Dismutases and the Electron Transport Chain. Aging cell. 2013 doi: 10.1111/acel.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suthammarak W, Yang YY, Morgan PG, Sedensky MM. Complex I function is defective in complex IV-deficient Caenorhabditis elegans. The Journal of biological chemistry. 2009;284:6425–6435. doi: 10.1074/jbc.M805733200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgovnick A, Schiavi A, Testi R, Ventura N. A role for p53 in mitochondrial stress response control of longevity in C. elegans. Experimental gerontology. 2010;45:550–557. doi: 10.1016/j.exger.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochemical and biophysical research communications. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno M, Honjoh S, Matsuda M, Hoshikawa H, Kishimoto S, Yamamoto T, Ebisuya M, Yamamoto T, Matsumoto K, Nishida E. A fasting-responsive signaling pathway that extends life span in C. elegans. Cell reports. 2013;3:79–91. doi: 10.1016/j.celrep.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS genetics. 2009;5:e1000361. doi: 10.1371/journal.pgen.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S. FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mechanisms of ageing and development. 2011;132:519–521. doi: 10.1016/j.mad.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura N, Rea SL. Caenorhabditis elegans mitochondrial mutants as an investigative tool to study human neurodegenerative diseases associated with mitochondrial dysfunction. Biotechnology journal. 2007;2:584–595. doi: 10.1002/biot.200600248. [DOI] [PubMed] [Google Scholar]
- Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging cell. 2009;8:380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Rothman JH, Shi Y, Blackwell TK. Distinct requirements for C.elegans TAF(II)s in early embryonic transcription. The EMBO journal. 2001;20:5269–5279. doi: 10.1093/emboj/20.18.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS biology. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Experimental gerontology. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139:1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell metabolism. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PloS one. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S, Kuang J, Tuck A, Ebert PR. Mitochondrial dysfunction in Caenorhabditis elegans causes metabolic restructuring, but this is not linked to longevity. Mechanisms of ageing and development. 2010;131:554–561. doi: 10.1016/j.mad.2010.07.004. [DOI] [PubMed] [Google Scholar]