Abstract
Ion channels have played a substantial role in the evolution of novel traits across all of the domains of life. A fascinating example of a novel adaptation is the convergent evolution of electric organs in the Mormyroid and Gymnotiform electric fishes. The regulated currents that flow through ion channels directly generate the electrical signals which have evolved in these fish. Here, we investigated how the expression evolution of two sodium channel paralogs (Scn4aa and Scn4ab) influenced their convergent molecular evolution following the teleost-specific whole-genome duplication. We developed a reliable assay to accurately measure the expression stoichiometry of these genes and used this technique to analyze relative expression of the duplicate genes in a phylogenetic context. We found that before a major shift in expression from skeletal muscle and neofunctionalization in the muscle-derived electric organ, Scn4aa was first downregulated in the ancestors of both electric lineages. This indicates that underlying the convergent evolution of this gene, there was a greater propensity toward neofunctionalization due to its decreased expression relative to its paralog Scn4ab. We investigated another derived muscle tissue, the sonic organ of Porichthys notatus, and show that, as in the electric fishes, Scn4aa again shows a radical shift in expression away from the ancestral muscle cells into the evolutionarily novel muscle-derived tissue. This study presents evidence that expression downregulation facilitates neofunctionalization after gene duplication, a pattern that may often set the stage for novel trait evolution after gene duplication.
Keywords: gene duplication, gene expression, evolution, neofunctionalization, dosage balance, quantitative PCR
Introduction and Background
Many complex and novel phenotypes evolve through the duplication of genes. The long-term maintenance of gene duplicates by natural selection is generally explained by two processes: neofunctionalization and subfunctionalization. Duplicate genes neofunctionalize when they gain a new function and are maintained by purifying selection for that new function (Ohno 1970). Genes subfunctionalize when the expression patterns and pleiotropic functions of the singleton ancestral gene are partitioned among the descendant duplicate genes (Force et al. 1999). Initially, it was assumed that gene duplicates either obtained new functions through the accumulation of neutral mutations or rapidly degraded into pseudogenes. This “neutral trajectory” would thus only afford duplicated genes a brief window of opportunity to obtain new functions (Lynch and Conery 2000; Bershtein and Tawfik 2008). However, new data suggest that duplicate genes often persist for long periods of time before becoming pseudogenes or gaining novel functions (He and Zhang 2005; Rastogi and Liberles 2005; Conant and Wolfe 2008; Sato et al. 2009). This means that many stably maintained duplicate genes are either under selection immediately after duplication for increased gene dosage (Papp et al. 2003; Kondrashov 2012; Nasvall et al. 2012) or soon come under selection following the neutral process of subfunctionalization (Force et al. 1999). How so many duplicate genes coevolve for long spans of time with apparently redundant function before they obtain novel ones, and how this process facilitates the evolution of novel adaptations in nature is a complex and interesting problem which is unresolved. To fully investigate this problem, we must follow the evolutionary trajectory of paralogs in large, diversifying clades.
When novel organ systems evolve, numerous genes must be co-opted (exapted) and optimized for the new phenotype. The convergent evolution of electric organs derived from muscle cells provides an elegant example of the tinkering nature of phenotypic evolution. Between 100 and 200 Ma, two different lineages of teleost fish independently evolved electrical communication and navigation systems (Lavoue et al. 2012). In the separate ancestors of the Mormyroids in Africa and the Gymnotiforms in South America, subpopulations of muscle cells transformed into noncontractile electrocytes where the action potential was repurposed for generating electrical signals which they broadcast through the water (Ferrari and Zakon 1993; Hopkins 1995); this put the ion channels that generate sodium currents under a novel, diversifying selective regime.
One of the genetic changes allowing the convergent evolution of the electric fishes’ unique communication system occurred over 100 My before electric fish existed, when the ancestor of all teleost fish underwent a whole-genome duplication (WGD). In that duplication event, the voltage-gated sodium channel of vertebrate skeletal muscle, Scn4a, duplicated to Scn4aa and Scn4ab (Novak et al. 2006). When myogenic electric organs evolved over 100 My later in Mormyroids and Gymnotiforms, the same paralog, Scn4aa, lost expression in skeletal muscle and was compartmentalized into the electric organ (Zakon et al. 2006) where its protein sequence diversified in a pattern mirroring the diversity of electric signals found in the numerous species of the two electric clades (Arnegard et al. 2010). This substantial convergent change in expression and function of the Scn4aa paralog occurred despite the fact that it and its paralog have been broadly conserved in the skeletal muscle of all nonelectric teleost fishes for ∼300 My. This observation elicits two interesting questions. Can more than chance explain the convergent neofunctionalization of Scn4aa? And, what selective force is maintaining both paralogs in nonelectric fish when one sodium channel is sufficient in both electric lineages’ muscle? This system allows us to apply years of gene duplication theory and research to the simultaneous investigation of the role gene duplication plays in the evolution of novelty as well as the role convergent molecular evolution plays in convergent phenotypic evolution.
Following the teleost WGD, Scn4aa and Scn4ab likely originated with identical expression levels in the same cells. We hypothesize that the relative expression of the two genes gradually diverged under continual selection until Scn4aa expression in the ancestors of both electric lineages was substantially downregulated relative to Scn4ab. Thus, in the electric fish ancestors, Scn4ab contributed more to the abundance of sodium channels than Scn4aa in muscle, and mutations that co-opted Scn4aa expression for the new cell type would have been relatively less deleterious to muscle function. Under this hypothesis, there was a change in expression in teleosts after genome duplication that facilitated neofunctionalization in electric fish.
Here, we investigate the role gene expression evolution played in the convergent molecular evolution of Scn4aa in Mormyroid and Gymnotiform electric fish. Real-time quantitative PCR (qPCR) is a well-developed assay for measuring gene expression; however, to our knowledge, qPCR has not yet been used to study the evolution of expression stoichiometry. We developed and validated a simple qPCR assay that accurately measures the relative expression of multiple genes to each other, and analyzed the relative expression of both paralogs in widely divergent taxa in a phylogenetic context. Our analysis supports the hypothesis that Scn4aa was downregulated relative to its paralog in the ancestors of both electric lineages before its expression was lost in skeletal muscle and it neofunctionalized in electric organs. We also found that Scn4aa shows a similar pattern of expression in the sound-producing teleost the Midshipman (Porichthys notatus), being downregulated in the skeletal muscle and up-regulated in the novel myogenic sonic organ. Despite the apparent propensity for neofunctionalization, analysis of branch-specific dN/dS (nonsynonymous mutations per nonsynonymous site over synonymous mutations per sysnonymous site) indicates that Scn4aa and Scn4ab had similar levels of purifying selection throughout their shared evolutionary history even when Scn4aa had significantly lower expression than Scn4ab. This supports the conclusion that Scn4aa evolved a new function under continual purifying selective pressure leading up to both electric lineages.
Results
Validation of qPCR to Measure the Expression Stoichiometry of Scn4aa and Scn4ab
To our knowledge, the accuracy of qPCR in comparing the expression of two genes has not been empirically determined. If hydrolysis probes such as TaqMan are used to avoid amplicon length effects and the primers and probes have a high enough concentration to efficiently bind all amplicon copies, then theoretically a simple model can be used for relative gene expression estimates. We assessed the accuracy of using an equation based on a simple model of PCR kinetics: Expression ratio Scn4aa/Scn4ab = 2CqB−CqA (“2ΔCq” method) for estimating the expression stoichiometry, where ΔCq is the difference in number of cycles (Cq) each gene (A and B) takes to pass a fluorescence threshold. This method was tested in two ways (see supplementary information, Supplementary Material online). First, samples with known relative abundances for the two paralogs (1:1) were created by ligating fragments of both paralogs into the same plasmid. Four different plasmid samples were created from Gasterosteus aculeatus (two plasmid samples for two primer-probe sets), Osteoglossum bicirrhosum, and Ictalurus punctatus. qPCR analysis demonstrated that the assumptions of the 2ΔCq method introduce less than a 1.5-fold bias (ΔCq < 0.5 cycles, where ΔCq = 0 was the expectation) in the Scn4aa/Scn4ab expression ratio, since both paralogs passed the threshold within half a cycle of each other (fig. 1). Relative abundance estimates were measured and averaged over multiple concentrations of plasmids covering a dilution range of 100-fold to 50,000-fold (supplementary fig. S1, Supplementary Material online). For the final analysis, the relative expression measured from Ga. aculeatus, O. bicirrhosum, and I. punctatus was corrected for bias by subtracting from ΔCq the value of the bias, that is, Scn4aa/Scn4ab = 2(Cb−Ca + bias), where bias = Ca − Cb as measured from the plasmid experiment.
Fig. 1.
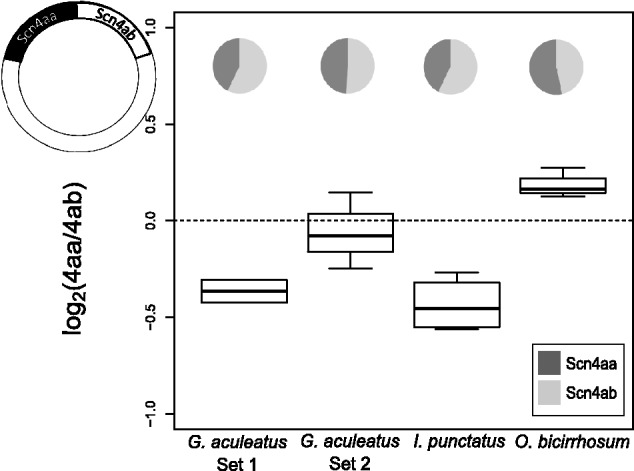
Box and whisker plots of relative expression of Scn4aa and Scn4ab estimates from RT qPCR reactions on plasmid samples containing 1:1 relative abundance. Dashed line denotes the expected log2 ratio and the boundaries (−1 to 1) denote a full cycle’s difference (2×). In all four plasmid samples, the two paralogs crossed the threshold within half a cycle of each other. The bold line represents the median value, the box spans the two interquartiles, and the whiskers represent the minimum and maximum values of the data, excepting outliers, which are represented as dots. Pie charts represent the proportion of total sodium channel expression contributed by each gene.
Another validation technique used two primer-probe sets targeting the same paralog and comparing the relative estimated expression of those two to each other, with the expectation of equal expression since they target the same gene. This study yielded similar results to the plasmid samples with bias being slightly higher, probably due to the added bias of variable reverse transcription (RT) efficiency in addition to the PCR bias (supplementary fig. S2, Supplementary Material online). Together, these results suggest that 2ΔCq accurately detects relatively small (e.g., 1.5-fold) differences in expression between genes.
We also investigated expression variability among multiple locations along the trunk musculature in four fish species to determine the extent we can generalize the results of this study beyond the muscle sections we sample for our comparative analysis. In three of the species, Ga. aculeatus, Xenomystus nigri, and I. punctatus, relative expression was highly similar in two different locations (supplementary fig. S3, Supplementary Material online). In O. bicirrhosum, there was greater variability between individuals in the mid-trunk section, this variation was accounted for in our Bayesian analysis and appears to be anomalous. Relative expression measured from RNA purified from zebrafish whole embryos also shows very similar relative expression to that which we measured from muscle samples in zebrafish adults (see supplementary information, Supplementary Material online). In summary, these results indicate expression measured at the mid-trunk is likely an adequate representation of expression for the entire musculature.
Comparative Expression and Ancestral State Reconstruction Results
We used qPCR to compare the expression of the two paralogs in several nonelectric and electric teleost fish. The two electric fishes, Eigenmannia virescens and Gnathanemus petersii, showed almost no detectable expression of Scn4aa in skeletal muscle, confirming previous qualitative findings (Arnegard et al. 2010). Among eight nonelectric fish, the distribution of relative expression (Scn4aa:Scn4ab) shows Scn4aa having lower expression than Scn4ab in all but one species, varying between a >6-fold lower expression to a slight 0.2-fold higher expression. The closest relatives to the electric fishes tended to have the lowest expression of Scn4aa (fig. 2).
Fig. 2.
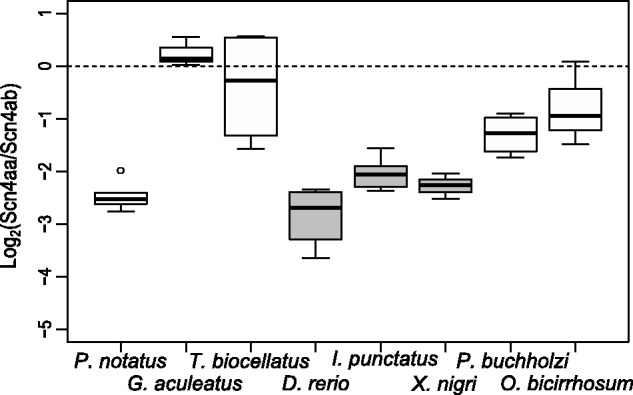
Log2 relative expression of each nonelectric species. Dotted line represents equal expression of the paralogs (1:1). The three closest relatives of the electric lineages are shaded in gray. Species tend to have a lower expression of Scn4aa relative to Scn4ab, especially among the closest relatives of electric fish. Species and sample sizes included were: Porichthys notatus (N = 5), Gasterosteus aculeatus (N = 3), Tetraodon biocellatus (N = 4), Danio rerio (N = 4), Ictalurus punctatus (N = 6), Xenomystus nigri (N = 4), Pantodon buchholzi (N = 4), and Osteoglossum bicirrhosum (N = 3). Dot represents an outlier that was included in the analysis since its inclusion would make statistical tests more conservative.
Model Selection and Maximum-Likelihood Estimation of Ancestral States
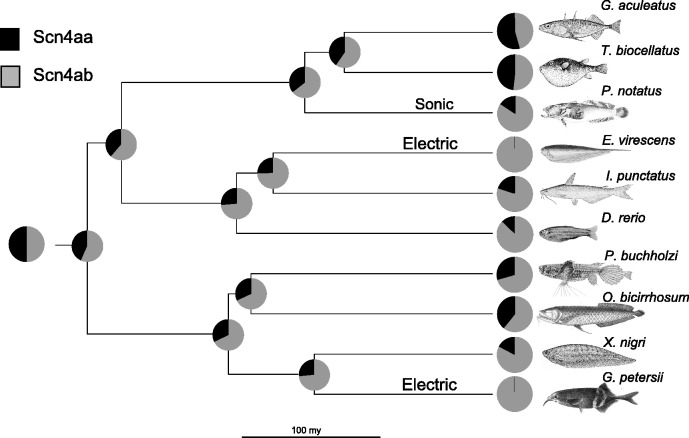
To investigate the possibility that a downregulation of Scn4aa expression preceded the parallel loss of expression in muscle and neofunctionalization in electric organ, we performed ancestral state reconstruction on the expression ratios of the two genes. Neutral genetic drift and randomly fluctuating natural selection can be described by Brownian motion (Hansen and Martins 1996; O’Meara et al. 2006; Losos 2011). Evolution under stabilizing selection for an optimal value can be modeled as an Ornstein–Uhlenbeck (OU) process (Hansen 1997). Stabilizing selection for long periods of time with punctuated shifts in selected optima can also be modeled as an OU process (Butler and King 2004). To select the best evolutionary model for estimating ancestral expression pattern, expression data were fit to the tips of a highly comprehensive and recently published time-calibrated phylogeny (Betancur-R et al. 2013), and a Brownian motion model of neutral evolution was compared with several OU models of evolution under purifying selection. The OU models ranged in complexity from a single optimal Scn4aa:Scn4ab expression for the whole tree to three different optima mapped onto different subclades of the tree according to the largest shifts in relative expression pattern (supplementary table S1, Supplementary Material online). An OU model with two optima, one shared by the convergent electric fishes (little to no detectable expression of Scn4aa) and the other optima for nonelectric fish, had the lowest AICc value, though it was only marginally better (2 < ΔAICc < 4) than a more complex model with a third separate optimum for the clade containing Ga. aculeatus and Tetraodon biocellatus, which both show the highest expression ratio between the paralogs. When the electric fish lineages were removed from the tree, a Brownian motion model (no selection/randomly fluctuating selection) showed a much better fit than all OU models tested (ΔAICc > 4) for the remaining nonelectric fish (supplementary table S2, Supplementary Material online). This pattern indicates the relative expression was evolving under a neutral evolutionary process until a shift toward selection for a significant decrease in Scn4aa expression occurred along the branches leading to the Mormyroids and Gymnotiforms.
The initial relative expression immediately after duplication was likely a 1:1 expression ratio, since these genes duplicated during a WGD and therefore their regulatory sequence was likely identical. This information should make ancestral state estimation much more accurate than in cases where the state at the root of the tree is much more ambiguous. The WGD could have occurred anywhere along the teleost root branch, which spans 39 My (Betancur-R et al. 2013). Model testing and ancestral states were estimated using three different time points along that branch for the WGD event. The earliest possible duplication event timing was set to the time of divergence of teleosts and their closest relatives, the holostei (322 Ma). The most recent possible duplication scenario was placed at the most basal bifurcation in the teleost tree which produced the elopomorpha clade (283 Ma) (Betancur-R et al. 2013). The third, intermediate, scenario placed the WGD at the midpoint of the tree root (302 Ma). The results of the three ancestral state reconstruction scenarios (early, intermediate, and late duplication times) predict very similar ancestral expressions (supplementary table S1, Supplementary Material online).
The OU model that fit the full data and tree best showed all the nonelectric fish under selection for a ratio of 0.39. Under the model with a separate optimum for the pufferfish/stickleback clade, the rest of the nonelectric fish were predicted to be evolving around an optimal value of 0.305 and the pufferfish/stickleback clade evolving under the optimum of 1.34. Both models support the hypothesis that the two electric fish lineages evolved from an ancestor that had Scn4aa downregulated relative to Scn4ab after the teleost WGD.
Ancestral state reconstruction using a Brownian motion model was performed on a tree with the electric fish data removed. Reconstruction under different timings of the WGD (283, 302, and 322 Ma) consistently predicted that the ancestor of both electric fish lineages had Scn4aa greatly downregulated relative to Scn4ab in skeletal muscle with the most recent nonelectric Mormyrid ancestor having a 2- to 2.3-fold reduction of Scn4aa expression relative to Scn4ab and the Gymnotiform ancestor having a 2.7- to 3.1-fold reduction (fig. 3; WGD at 302 My). A ratio of 1:1 fell outside the 95% confidence interval (Schluter et al. 1997) for the most recent estimated ancestor’s relative expression of Scn4aa to Scn4ab for the two electric clades under all three WGD timings. In every evolutionary scenario analyzed, Scn4aa is predicted to have been downregulated in the ancestor of both electric lineages before its expression was lost in muscle and neofunctionalized in the electric organ.
Fig. 3.
Maximum likelihood ancestral reconstruction of relative expression of Scn4aa and Scn4ab. Earliest point at the root of the tree corresponds to 302 Ma. The scale bar signifies 100 My. The tree topology and branch lengths were derived from Betancur-R et al. (2013). Danio rerio and Pantodon buchholzi images by Pogrebnoj-Alexandroff and Robert Back, respectively.
Bayesian Estimation of Ancestral States
To relax several of the assumptions behind our maximum-likelihood estimation, we developed a hierarchical Bayesian framework to infer ancestral states (see Materials and Methods). It is possible that the teleost ancestor was an allotetraploid and thus potentially had unequal expression between paralogs (depending upon how genetically divergent the two parental species were; Rapp et al. 2009). However, even in the case of allopolyploidy, we would not expect the expression between paralogs to differ substantially, as the longer two hybridizing species have diverged, the more Dobzhansky–Muller incompatibilities are expected to accrue. This accumulation of mutations is expected to be more rapid in more complex genomes and developmental programs, putting a narrow limit on how genetically diverged hybridizing species can be. The conserved synteny of these paralogs among teleosts likely precludes the possibility of any large-scale structural regulatory changes happening very soon after duplication (Novak et al. 2006; Widmark et al. 2011). In light of this, we argue that equal expression is the most likely state after genome duplication, but we weaken our certainty about this by modeling the value at the root as a highly dispersed random normal variable. Our maximum-likelihood estimates are also built on the assumption that our measured mean log expression ratios for each species are the true means for those species. Although we demonstrated that our assay introduced little bias, there were still different amounts of variation between individuals within the different species and we did not get an individual measure of PCR bias effects for every species in the study, so a more explicit treatment of the uncertainty of our measurements would provide more confidence in interpreting the results of our analysis. Therefore, we treated the measured mean log expression ratio for each species as a probability function of the sampling distribution and the signal introduced by the qPCR assay (see Materials and Methods).
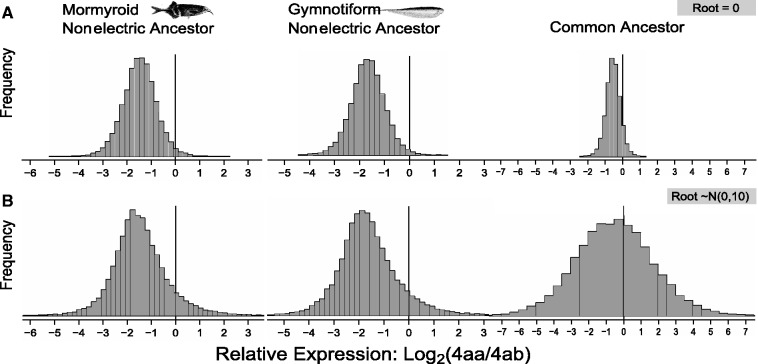
If we are certain that the ancestral state at the WGD is 1:1 like in the maximum-likelihood analysis then the probability that Scn4aa had lower expression than Scn4ab for both electric fish ancestors is 0.98 and 0.99 for the most recent nonelectric ancestor of the Mormyroids and the Gymnotiforms, respectively. The probability Scn4aa < Scn4ab is 0.91 for their common ancestor (fig. 4A). Next, instead of treating the state after duplication at the root of the tree as known (1:1), we modeled it as a random normal variable with mean log2(1). We made a conservative assumption that the expression difference upon duplication likely did not exceed twice the largest expression difference observed in our data set, which is a log2(ratio) of −2.85 observed in zebrafish. We therefore set the variance of the root at 10, reflecting our assumption that the probability that the relative expression ratio at the root was greater than zebrafish (P[Ancestor > |−2.85|]) is ∼0.33 and greater than twice that of zebrafish is 0.22. We view this as a highly conservative assumption since a 1:1 ratio biases values near the root away from the phylogenetic mean of the data set, which is less than 1:1. With these models, and the assumptions of the evolutionary model selected, the marginal posterior probability that expression of Scn4aa was less than Scn4ab (4aa < 4ab) in the most recent nonelectric ancestor of the Mormyroids is 0.91, that of the most recent nonelectric ancestor of the Gymnotiforms is also 0.91, whereas that of the common ancestor of both was 0.60 (fig. 4B). Thus, the stronger the prior belief that these genes had equal expression after the genome duplication, the more probable that Scn4aa had lower expression than Scn4ab in the common ancestor of the two electric lineages. Not surprisingly, the means of the posterior distributions are robust to uncertainty about the state at the WGD. The mean of the posterior distribution for the ancestral expression ratio Scn4ab:Scn4aa is 2.8:1, 3.1:1, and 1.4:1 for Mormyroids, Gymnotiforms, and their common ancestor, respectively.
Fig. 4.
Bayesian inference of ancestral states. Vertical lines at zero in each of the three distributions indicate equal expression between the paralogs. Frequency shows the relative proportion of samples (20,000) falling within a bin of 0.2. The distribution represents the marginal posterior distribution of each ancestral state given the measured expression in each species. (A) Panel of posterior distributions for the three ancestral states assuming µroot = 0. (B) Panel of posterior distributions for the three ancestral states assuming a conservative prior of P(µroot) = N(0,10) for the state after the genome duplication.
We interpret these overall results to mean that if the relative expression is evolving by a largely neutral evolutionary process and if the tree we used to do the analysis accurately models the relationships of the species in our data set, then even with some highly conservative assumptions (see Materials and Methods) the probability Scn4aa was downregulated before neofunctionalizing in a novel tissue is high for both lineages of electric fish. We outline a possible mechanism for this slow neutral rate of expression divergence in the discussion.
Porichthys notatus and Sonic Muscle
Like the convergent evolution of myogenic electric organs, sonic muscles have evolved multiple times in teleost fishes (Bass and Zakon 2005; Boyle et al. 2013). In the Midshipman, Por. notatus, where vocalizations are used by males to attract females, highly derived sonic muscle fibers vibrate the swim bladder at frequencies in the hundreds of hertz. These are frequencies in excess of 10 times what is possible in the fastest known skeletal muscle fibers used for locomotion (Rome et al. 1996). The muscle fibers comprising the sonic muscle have highly derived structural and molecular features (Bass and Marchaterre 1989; Rome et al. 1996; Lewis et al. 2003). It is possible that the sodium channel expression pattern in the sonic muscle also evolved to optimize the high frequency contraction rate of these cells. If Scn4aa was expressed at lower levels in the ancestor of midshipmen then perhaps it was more evolvable in the derived sonic muscle fibers.
Along with measuring the relative expression of the duplicate sodium channel genes in trunk muscle, we measured their expression in the sonic muscle of these fish. In contrast to the skeletal muscle in these species, where Scn4aa has over a 5-fold reduction in expression relative to Scn4ab, the sonic muscle on average shows 80% higher expression of Scn4aa than Scn4ab (fig. 5). Although not as extreme as the electric fish, this is another example of a fish with a derived muscle cell type up-regulating Scn4aa relative to Scn4ab; in no other species measured for this study does Scn4aa have a significantly higher expression than Scn4ab (fig. 2). This presents the exciting possibility that Scn4aa shows a recurrent tendency to gain new functions when novel muscle-derived organs evolve in teleost fish. Under the assumption that the relative expression of the two paralogs experienced positive selection for a radical shift in expression in Por. notatus, maximum-likelihood ancestral state reconstruction was performed without Por. notatus. The results were highly similar and were still statistically significant as before.
Fig. 5.
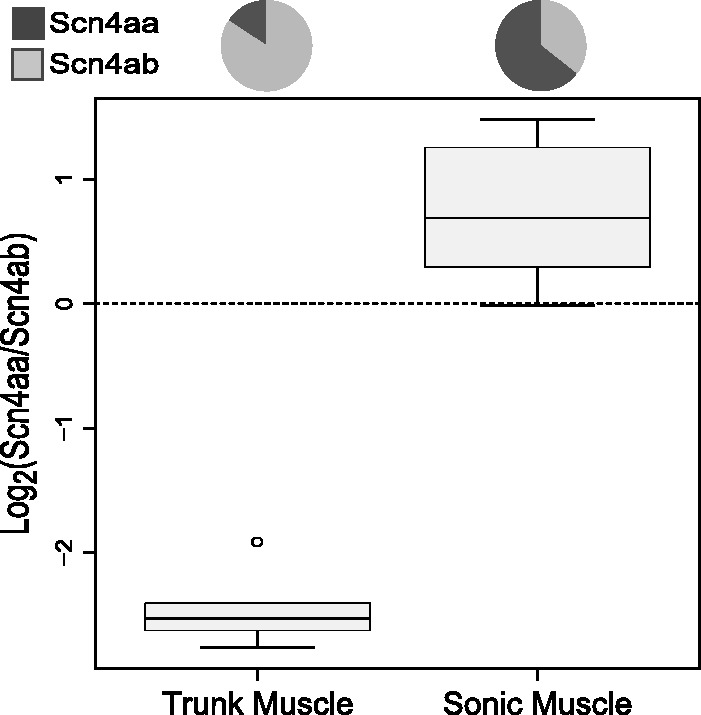
Expression divergence between derived muscle tissue and trunk muscle in the midshipman Porichthys notatus. Sonic muscle, which is attached to the swim bladder, shows greater than a 9-fold increase in expression relative to Scn4ab compared with trunk muscle (N = 5).
Patterns of Selection
Genes with low expression may be less essential to fitness and evolve at faster rates than highly expressed genes (Krylov et al. 2003; Drummond et al. 2005). To explore the possibility that Scn4aa was under weaker selection than Scn4ab prior to neofunctionalization, we compared the dN/dS of each branch in the Scn4aa clade with the same branch in the Scn4ab clade (fig. 6). Both genes had identical evolutionary histories since they coexisted in the same genomes; this means the Scn4aa clade is identical in topology to the Scn4ab clade. We estimated the mean dN/dS (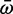 ) ratio for each branch using a random effects likelihood branch sites model (Kosakovsky Pond et al. 2011; see Materials and Methods for tree construction) (supplementary fig. S4, Supplementary Material online). There was no significant difference in
) ratio for each branch using a random effects likelihood branch sites model (Kosakovsky Pond et al. 2011; see Materials and Methods for tree construction) (supplementary fig. S4, Supplementary Material online). There was no significant difference in 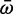 between the two paralogs (P = 0.24 Wilcoxon-signed rank test) and no significant relationship between relative expression and relative
between the two paralogs (P = 0.24 Wilcoxon-signed rank test) and no significant relationship between relative expression and relative 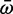 between the two paralogs (P = 0.45). Scn4aa appears to have been under similar levels of selective pressure as Scn4ab in the most recent nonelectric ancestors of the electric fishes.
between the two paralogs (P = 0.45). Scn4aa appears to have been under similar levels of selective pressure as Scn4ab in the most recent nonelectric ancestors of the electric fishes.
Fig. 6.
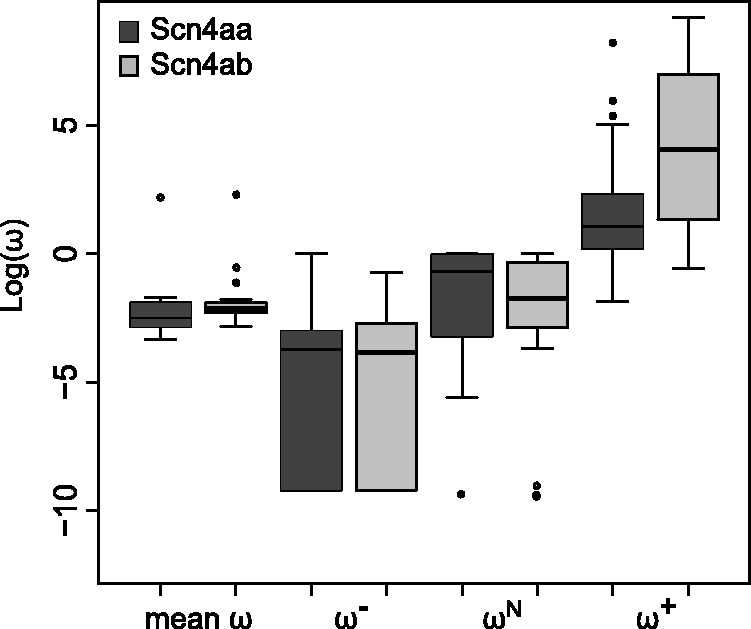
Comparison of dN/dS (ω) between the two paralogs. Up to three different ω rates were fit to each branch of a phylogeny containing both paralogs: Purifying selection (ω−), nearly neutral selection (ωN), and positive selection (ω+). Box and whisker plots compare the distribution of each of the rate classes between the two paralogs Scn4aa and Scn4ab. Because some estimates in the purifying selection class were zero, a constant of 0.0001 was added to each value. This had no effect on the results of statistical tests but prevented box plots from stretching to negative infinity in the purifying selection rate class. The bold line represents the median value, the box spans the two interquartiles, and the whiskers represent the minimum and maximum values of the data, excepting outliers, which are represented as dots.
Discussion
How a gene’s expression evolves after duplication has an impact on how the function of that gene evolves. A change in magnitude, timing, and spatial pattern of expression can change the cellular context of the gene and thus the fitness consequences of mutations in that gene. Therefore, determining the evolutionary changes in gene expression can greatly illuminate the selective causes behind evolutionary changes in protein sequence and function.
Validating qPCR to Measure the Relative Expression of Duplicate Genes
We measured the accuracy of qPCR using hydrolysis probes to quantify the relative expression of two paralogous genes. We have shown that a simple model of PCR kinetics introduces little bias and the bias appears not to come from the assumption of equal and 100% efficiency (supplementary fig. S1, Supplementary Material online). Having ascertained that qPCR can be accurate in detecting a 1.5-fold difference in expression between genes, we studied the comparative relative expression of a duplicate pair of sodium channel genes to gain insight into the possible role expression evolution played in their parallel neofunctionalization at the base of the Gymnotiform and Mormyroid clades. This study demonstrates that qPCR using hydrolysis probes can be used to investigate the expression stoichiometry of genes.
Parallel Loss of Scn4aa Expression from Muscle in Electric Fish
The evolutionary potential of a gene is determined by its current function and expression. The unique accumulation of mutations experienced by paralogs following gene duplication can alter their evolutionary potential relative to one another, such that one paralog of a pair might be more easily co-opted for a given novel phenotype than its counterpart. The fact that the closest relatives of the two electric lineages show the lowest expression of Scn4aa whereas more distant relatives show higher expression supports a convergent downregulation before convergent neofunctionalization. However, given the widespread pattern of lower expression of Scn4aa (fig. 1), downregulation could have happened soon after duplication, indicating that both electric lineages may have inherited this pattern from a common ancestor. In either case, the convergent neofunctionalization of Scn4aa appears to have been facilitated by a downregulation in relative expression.
Interestingly, electric fish have no apparent muscle deficiency despite being the only teleost fish expressing just one of the paralogs in their muscle (Arnegard et al. 2010). Large- and small-scale studies on protein interaction networks and knockout phenotypes indicate that duplicate genes often have a high degree of functional redundancy for a much longer period of time than theoretical studies first predicted (Lynch and Conery 2000; Solomon and Fritz 2002; Gu et al. 2003; Tvrdik and Cappecchi 2006). In fact, duplicate genes often maintain considerable functional overlap for tens of millions of years (Vavouri et al. 2008), even in rapidly reproducing lineages (yeast: Gu et al. 2003; Dean et al. 2008; DeLuna et al. 2008). In Caenorhabditis elegans, numerous paralogous pairs have been shown to be highly redundant in function through RNAi knockout experiments, even among paralogs that duplicated over 80 Ma (Tischler et al. 2006). In vertebrates, there are numerous examples of single knockouts of one duplicate gene with little phenotypic consequence, whereas a double knockout of both paralogs proves highly deleterious (Solomon and Fritz 2002; Kafri et al. 2008, 2009).
Another line of evidence suggesting high functional overlap between ancient duplicates comes from the nonfunctionalization of paralogs long after duplication. Paralogs produced by the teleost WGD were often maintained over 100 My before becoming pseudogenes (Sato et al. 2009). This is much longer than expected for genes evolving under neutral selection (Lynch and Conery 2000). If duplicate genes can become lost from the genome tens to hundreds of millions of years after duplication, then genes can also neofunctionalize tens to hundreds of millions of years after duplication. There may be myriad selective mechanisms maintaining the apparent redundancy of genes and the type of gene would likely be an important factor in post-duplication evolutionary dynamics (He and Zhang 2005; Kafri et al. 2006; Makino and McLysagt 2010). The fact that all teleost fish observed to date express both sodium channel paralogs in muscle while electric fish exhibit no apparent phenotypic consequence by only expressing one implies a large degree of functional similarity between the duplicates in skeletal muscle.
Genome Duplication and the Dosage Balance Hypothesis
These observations of functionally redundant yet essential gene duplicates may be explained by the interaction of gene expression evolution and protein sequence evolution (Nuzhdin et al. 2004; Anderson and Evans 2009). A change in relative gene dosage (copy number) can have a profound effect on the evolutionary dynamics of genes after duplication. Therefore, it is expected that the type and scale of duplication (e.g., tandem vs. whole genome) will greatly impact the subsequent evolutionary potential of the duplicate genes involved (Papp et al. 2003; Gu et al. 2005; Aury et al. 2006; Henrichsen et al. 2009; Qian et al. 2010; Kondrashov 2012) and thus determine the time scale of functional divergence.
Many pathologies, such as Down syndrome, arise from an increase in gene dosage which increases transcript abundance (Kahlem et al. 2004; Arron et al. 2006; Stranger et al. 2007). The relative expression of genes typically has a large impact on the stoichiometric ratios of interacting proteins and, thus, on gene duplicability (Papp et al. 2003; Makino and McLysaght 2010; Schwanhauser et al. 2011). Generally, if a gene is dosage-balanced (in stoichiometric balance with other genes), then it cannot duplicate through small-scale duplications. The dosage balance hypothesis states that dosage-balanced genes can only duplicate during WGDs, where relative expression is not altered (Papp et al. 2003; Kondrashov and Koonin 2004; Aury et al. 2006). This hypothesis predicts that following WGD, both paralogs will be maintained by purifying selection to preserve correct stoichiometry among interacting proteins produced by the sum of the two paralogs’ expression (Aury et al. 2006; Blomme et al. 2006; Chain and Evens 2006; Sato et al. 2009; Birchler and Veitia 2012). Instead of paralogs going through a neutral phase before neofunctionalization, they diverge under continuous selection throughout their evolutionary history. Selection on expression could facilitate the maintenance of the important structural properties that all proteins share such as solubility and fold stability (Bloom et al. 2006) for long enough periods of time to facilitate neofunctionalization within those constraints. This suggests that WGD, or polyploidization, is a source of selectively maintained new genetic material that can contribute to phenotypic evolution long after WGD (Freeling and Thomas 2006).
The numerous polyploidizations in several vertebrate lineages of varying ages provide insight into the time scale of divergence of WGD genes (“ohnologs”; sensu Wolfe 2000). The African clawed frogs (Xenopus and Silurana) have experienced multiple recent polyploidizations. Xenopus laevis, which is descended from a tetraploid frog ∼40 Ma, exhibits strong purifying selection in branches immediately following duplication as well as little asymmetry in sequence evolution between ohnologs, supporting the dosage balance prediction that ohnologs should be maintained by purifying selection to maintain an optimal level of protein abundance (Chain and Evans 2006; Chain et al. 2008). Although different Xenopus species descend from ancestors of varying levels of polyploidization they show little apparent phenotypic diversity (Hughes and Friedman 2010), though subtle molecular changes in expression have been documented (Chain and Evans 2006). Thus, there seems to be little evolutionary divergence of ohnologs in Xenopus tens of millions of years later.
Conversely, ohnologs from the more ancient teleost WGDs show much more widespread asymmetric sequence divergence from the inferred ancestral state (Brunet et al. 2006). Around half of all duplicates generated by the teleost WGD have been retained in at least one extant lineage (Jaillon et al. 2004; Sato et al. 2009) with many showing lineage-specific loss from the genome (Sato et al. 2009). A comprehensive study of teleost ohnologs found that a majority of the genes underwent subfunctionalization in expression, with a smaller subset of genes neofunctionalizing (Kassahn et al. 2009). These patterns of gene duplicate evolution observed in the more recent Xenopus and the ancient teleost polyploidizations support the interpretation that dosage balance is not a permanently stable state maintaining functional redundancy, but a force that considerably slows divergence of dosage-balanced ohnologs. Therefore, these gradual changes in expression of ohnologs could account for both the long time course of divergence after WGD and also the long-term maintenance of the two Scn4aa sodium channels in teleost muscle.
A number of characteristics specific to ion channels, and particularly the muscle-specific sodium channels, suggest that these channels are dosage-balanced and were/are being maintained by selection for stoichiometric balance with other ion channels. Teleost fish have eight voltage-gated sodium channels in their genomes, all of which were produced by a series of WGDs from a single gene: two at the base of vertebrates and one at the base of teleosts (Lopreato et al. 2001; Novak et al. 2006; Zakon 2012). Scn4a has not duplicated in any vertebrates except teleosts (Zakon 2012). This pattern suggests that Scn4a is dosage sensitive and therefore produces a deleterious phenotype when its expression changes relative to interacting genes such as the beta subunits as well as other ion channels, such as potassium channels.
Electrophysiological studies also indicate that excitable cells are highly sensitive to changes in expression of ion channels (Waxman 2001). Theoretical and empirical studies demonstrate that intrinsic electrical properties of neurons are fine-tuned through the control of ion channel expression (Marder and Goaillard 2006). Often, if one ion channel’s expression is perturbed, other sets of channels are up- or downregulated to maintain the correct electrical properties of the cell (Marder and Goaillard 2006; Schulz et al. 2006; O’leary et al. 2013). This illustrates the importance of expression levels in ion channels and also a widespread overlap in function between ion channels. These patterns support our hypothesis that Scn4a was dosage sensitive before the teleost WGD and therefore Scn4aa and Scn4ab were under purifying selection for dosage balance following duplication.
Neofunctionalization of Dosage-Balanced Genes
Gene dosage is a compelling reason for the maintenance of redundant genes following WGD, but elicits the question of why certain ohnologs are able to deviate greatly from their original expression levels or why selection shifted from dosage maintenance to a form promoting a novel function. Expression is often an evolutionarily labile trait for genes (Rifkin et al. 2003; Chain and Evans 2006; Brawand et al. 2011). It is therefore possible for a necessary range of absolute protein abundance to be maintained even if the relative expression of dosage-balanced duplicate genes changes through time, through either transcriptional or translational compensation by paralogs (fig. 7). This pattern of “expression drift” may be common, where the rate of drift is a function of the strength of selection on correct stoichiometry. A dosage-balanced state could then be a stable, though transitory, state until more pronounced changes like neofunctionalization eventually occur (Hughes et al. 2007). Ancient genome duplications can therefore seed evolutionary change long after the duplication event occurred.
Fig. 7.
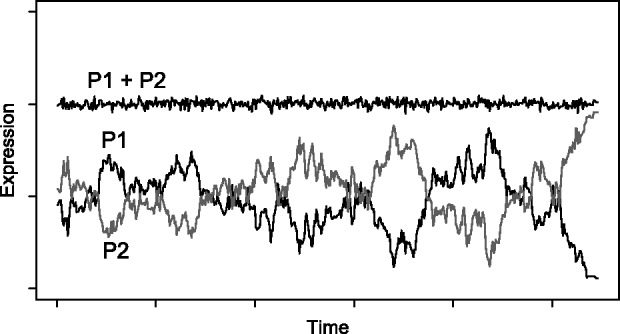
Schematic of expression drift under dosage balance selection. The summed expression of the two paralogs (P1 + P2) evolves under stabilizing selection and remains constant through time. Each individual paralog P1 and P2 evolves through a series of nearly neutral deleterious and compensatory changes in expression level until one paralog makes up most of the stoichiometrically required expression freeing the other one to neofunctionalize or become a pseudogene.
Scn4aa and Scn4ab likely originated with identical expression patterns and identical levels of purifying selection. The force of selection on dosage balance may have reduced the rate of expression divergence. Our results support this possibility; although relative expression has diversified to an extent in teleosts, Scn4aa exhibits a general tendency of reduced expression relative to Scn4ab, and there is a pronounced phylogenetic signal in the pattern of expression on the tree (fig. 3). We argue that Scn4aa has two unique features that explain its continued maintenance in nonelectric teleost muscle: 1) stoichiometric relationships with other ion channels in the muscle membrane likely have maintained both paralogs and under that selective regime each paralog’s expression diverged through a series of nearly neutral expression changes where some changes were slightly deleterious and other compensatory changes were slightly advantageous. This occurred until Scn4aa was at low but significant levels in the ancestors of electric fish, and 2) by virtue of its reduced relative expression, it was the more evolvable of the two paralogs for novel muscle-derived organ function. In summary, we propose that both ohnologs were maintained by selection for their combined expression, while their relative expression evolved by nearly neutral dynamics (Ohta 1992) until one of the ohnologs, Scn4aa, became relatively freed from selective constraints in muscle and neofunctionalized. The fact that Brownian motion best models the evolution of relative expression between the paralogs in nonelectric fish (supplementary table S2, Supplementary Material online) supports this hypothesis. It could either be that Scn4aa drifted to lower expression or originally had lower expression in the event that the WGD is due to an ancient allotetraploidy.
The relative downregulation of Scn4aa measured by qPCR could be due to either a global downregulation of expression in all muscle cells or to a decrease in the number of muscle cells in which Scn4aa is expressed compared with Scn4ab. In the Gymnotiform Sternopygus macrurus the electric organ has been shown to develop from a subset of fast muscle fibers (Unguez and Zakon 1998). The genome duplication could have facilitated the diversification of muscle fibers in teleost fish. Thus, it is possible that Scn4aa was compartmentalized into a subset of muscle fibers before co-option into the electric organ while Scn4ab was expressed in all muscle fibers and thus more constrained in its evolution. In this scenario, gene duplication facilitated the elaboration of muscle fiber types which facilitated the evolution of electrocytes. Higher resolution methods such as in situ hybridization performed in a phylogenetic context would be needed to explore this possibility.
Transcription is an important process for regulating protein abundance but there is substantial regulation complexity of protein levels beyond transcriptional regulation (Vogel and Marcotte 2012), because of this, the decreased transcription of Scn4aa relative to Scn4ab does not necessarily mean there is a lower relative density of Nav1.4a channels in the membrane. The rate of translation and the stability of the mRNA and protein can compensate for a decrease in transcription. It is also possible that the long-term maintenance of the ohnologs is due to a divergence of function to some degree. Although dN/dS studies here and previous (Arnegard et al. 2010) show high conservation between these two ohnologs in nonelectric fish, the evolutionary time scales are large enough that these analyses are expected to have low power. If there was some functional divergence, it was either nonessential to the electric fish ancestor, or the gain of electric organs more than compensated for the functional deficit of losing a unique protein expressed in muscle. Interestingly, there is some indication of a slight up-tick in evolutionary rate in Scn4ab when Scn4aa was lost from muscle in the two electric lineages (Arnegard et al. 2010). This question could be investigated through electrophysiology comparing sodium currents between the ohnologs from a species which shows constrained evolutionary rates, that is, nonelectric fish or from a sequence based on ancestral state reconstruction if possible. Gene knockout studies through morpholinos can also be informative about differences in function, though the effects can be confounded if the expression is different between the two paralogs, so a species where expression is highly similar such as Ga. aculeatus would be optimal.
Sodium Channels in Sonic Muscles
There are numerous examples of novel cell types and organ systems evolving from muscle cells in teleosts, such as heater organs, electric organs, and sonic organs (Block 1991; Rome et al. 1996) yet no study has investigated whether Scn4a paralog expression ratios vary in tissues other than electric organs. We found that midshipman sonic organ muscle has the highest expression of Scn4aa relative to Scn4ab of any muscle type measured, while having highly downregulated expression in skeletal muscle. The difference in expression pattern between the two muscle types is over a factor of 9. These patterns are highly reminiscent of the electric organs and skeletal muscle of Gymnotiforms and Mormyrids (Arnegard et al. 2010), indicating that this sodium channel paralog may be more easily co-opted into novel muscle-derived organ systems. Future investigations can explore whether Scn4aa is consistently upregulated in these novel organs (and downregulated in skeletal muscle). Seeking out the roles of these duplicate channels in the evolution and function of these other muscle-derived excitable tissues would be particularly exciting, and could help elucidate general patterns of expression evolution following gene duplication.
Conclusion
By studying the comparative genetic changes associated with the convergent evolution of electric organs, we can investigate the role that gene duplication plays in the evolution of complexity. More broadly, studies such as these can reveal the diversity (and constraints) of genetic solutions to adaptive problems. By measuring relative gene expression and analyzing it in a phylogenetic context, we have demonstrated a post-WGD evolutionary process where downregulation precedes neofunctionalization. Our findings are in agreement with a previous study that found a gradual downregulation before convergent nonfunctionalization (Anderson and Evans 2009), implying that this phenomenon of “expression drift” in paralogs over exceptionally long time scales may be more common than previously considered. The two-step process of expression drift during dosage maintenance and co-option of the downregulated paralog for novel function may be important in the evolution of novel organ systems, particularly those involving ion channels and other proteins involved in complex functional networks. The gradual downregulation of a dosage-balanced ohnolog can be an evolutionary preamble to drastic functional changes. Under the expression drift model, we expect the expression of dosage-balanced duplicates as well as quantitatively subfunctionalized duplicates (Force et al. 1999; Qian et al. 2010) to evolve by nearly neutral dynamics until one becomes more susceptible to neofunctionalization or nonfunctionalization. We predict that for clades where the expression tends to be lower there should be many more instances of neofunctioanlization or nonfunctionalization of the lower expressed ohnolog in that clade. With the growing accumulation of expression data, these results may emerge as a common pattern across broad taxonomic samples.
Materials and Methods
RNA Extraction and Preparation
Tissues were collected from 10 species (Por. notatus [N = 5], I. punctatus [N = 6], O. bicirrhosum [N = 3], Danio rerio [N = 4], Pantodon buchholzi [N = 4], T. biocellatus [N = 4], Ga. aculeatus [N = 3], Gn. petersii [N = 3], E. virescens [N = 4], and Xenom. nigri [N = 4]). All fish except for Por. notatus and Ga. aculeatus were purchased from commercial aquariums. Porichthys notatus samples from trunk muscle and sonic organ muscle were kindly provided by the laboratory of Andrew Bass and Ga. aculeatus was provided by the laboratory of Daniel Bolnick. In a subset of species (I. punctatus, O. bicirrhosum, Xenom. nigri, and Ga. aculeatus), separate muscle samples from two or three locations in the hypaxial rostral-caudal axis were analyzed using qPCR to determine whether expression stoichiometry varied among different locations in the body. Muscle tissue was flash frozen in liquid nitrogen immediately after excision and stored at −80 °C until RNA extraction.
RNA extraction was conducted by homogenizing frozen tissue in STAT60 (TEL-TEST, Friendswood, TX) using a powerGen 150 homogenizer (Fisher). Homogenized tissue was then column purified using Direct-zol RNA miniprep (Zymo, Orange, CA) according to the manufacturer’s protocol. Genomic DNA was removed using the Turbo DNase kit following the manufacturer’s protocol (Life Technologies, Grand Island, NY). Final RNA concentrations and purity were measured using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). All RNA samples had A260/A280 > 1.7. RNA quality was assessed by visualization on an agarose ethidium bromide gel.
Reverse Transcription
RT was performed on total RNA (30–700 ng) using the Super Script III kit (Life Technologies) and random hexamers (700 nM) and oligo-dT20 (1.25 µM) primers. Primer annealing occurred during a 5-min incubation on ice followed by a 5-min incubation at 25 °C. The subsequent extension time ran at 50 °C for 1.5 h followed by a 15-min enzyme inactivation step at 70 °C. Large fragments of both paralogs in Pa. buchholzi, Por. notatus, T. biocellatus, and the single ortholog in a Bichir (Polypterus senegalensis) were sequenced using an AB 3730 Analyzer (Applied Biosystems, Branchburg, NJ) on PCR samples and homology was confirmed using a BLAST alignment search.
Real-Time qPCR
Since this investigation requires the measurement of expression across several species and between genes, hydrolysis probes were used in all qPCR reactions both to confer specificity between the two paralogs and because hydrolysis probes create a fluorescent signal that is independent of amplicon length. cDNA from RT reactions were diluted to 2 ng/µl to guarantee no PCR inhibitor effects (Ramakers et al. 2003). Hydrolysis probes (IDT, Coralville, IA) were used to analyze all species’ relative expression. Primers and probes were designed to span exon splice junctions using IDT online software. To guarantee paralog specificity both primers and the probe were targeted to sequences that showed at least 20% difference between paralogs and were evenly spaced along the primer-/probe-binding site. Paralog specificity of primers was confirmed by PCR (Takara Bio Inc., Japan) followed by direct sequencing of purified bands cut from agarose gel. Primer-targeted exons were then tested for alternative splicing by matching the forward and reverse primers with primers in upstream and downstream exons. The resulting PCR products were then visualized using electrophoresis on an agarose gel. If a band pattern appeared which indicated the primers and probes were targeted to exons involved in alternative splicing then new primers were designed targeting different exons.
Ten microliters qPCR reactions contained: 2 ng cDNA, 2× TaqMan Universal Master Mix No UNG (Applied Biosystems), 250 nM of each sense and anti-sense primers, and 250 nM hydrolysis probes. The reactions were run in a Viia7 Real-Time PCR System (Applied Biosystems) and run for 40 cycles at 60 °C annealing and 1 min extension at 60 °C with fluorescence reading integrated over the extension phase. RT reactions with no reverse transcriptase added were used as a negative control for all samples analyzed. All negative controls showed either no amplification or amplification more than eight cycles later than the sample.
qPCR Data Analysis
Baseline corrected raw amplification readings were analyzed using LinRegPCR software (Ramakers et al. 2003; Ruijter et al. 2009). A common threshold fluorescence that crosses the exponential phase of both genes’ amplification curves was used to measure and compare the Cq value for both genes in each species. The exponential phase was determined by fitting a regression to the log-linear phase of each reaction’s raw baseline-corrected amplification readings. If the R2 > 0.98, and there were at least four data points, then the slope of the regression was used to estimate doubling efficiency for that reaction. The doubling efficiency was taken as the average of all the reactions: Six technical replicates and three to six biological replicates, the software did not include values that were >10% away from the median in this calculation. If no amplification was shown then a conservative estimate of the Cq value was set to 40 since the qPCR reaction was run for 40 cycles. If the doubling efficiency was within 5% of 1 then relative expression was computed using a 2ΔCq, where ΔCq = Cq(Scn4ab) − Cq(Scn4aa), since two different genes are being compared in the same sample (rather than the more common comparison of the same gene in two different samples normalized to a reference gene). Finally, the equation Scn4aa/Scn4ab = 2ΔCq computes the paralogs’ relative expression by measuring how many cycles it takes both genes to reach the same threshold in a sample and assuming both genes are amplifying with 100% efficiency.
Calibration Plasmid Preparation
Amplicons were created which contained qPCR primer and probe target sites with a buffer of 50–200 bases upstream and downstream. In order to produce amplicons which could be ligated together, the reverse primer for Scn4aa had the ECOR1 target sequence added to the 5′-end with an extra ATA or ATT added to provide sufficient space for the restriction enzyme to bind: 5′-ATAGAATTC—3′. The same method was used for Scn4ab but in the forward primer. PCR produced ∼400–800 base amplicons, containing the ∼150 base qPCR amplicon near the center and an ECOR1 cut site (located at the 3′-end of the Scn4aa amplicon and at the 5′-end of the Scn4ab amplicon) which would allow ligation of the two amplicons.
PCR reactions were run through a 2% agarose gel and bands cut according to their expected sizes. Gel cuts were purified using PureLink quick gel extraction kits (Invitrogen, Carlesbad, CA). The purified amplicons were then restriction digested with ECOR1 HF (New England Biolabs (NEB), Ipswitch, MA). In brief; a 33 µl mixture of both amplicons were incubated with 5 µl 10× buffer and 40 units ECOR1-HF (NEB) for 2 h. Following the digestion reaction, ECOR1 was inactivated at 65 °C for 30 min. The digested fragments were then mixed in the presence of 800 units of T4 ligase (Quick Ligation kit, NEB) in the following reaction mixture: 19µl restriction-treated amplicon mixture, 19 µl 2× buffer, and 2 µl T4 ligase. This mixture was incubated for 10 min followed by inactivation at 65 °C for 10 min. The ligated aa/ab amplicon was then PCR amplified by using the forward primer of Scn4aa and the reverse primer of Scn4ab. The PCR product was then run through an agarose gel where the expected hybrid band was cut and purified. The new amplicon containing both genes was then ligated into the pCR 4-TOPO plasmid using the TOPO TA cloning kit (Invitrogen) and transformed into One Shot Top Ten chemically competent cells (Invitrogen). Plasmids were purified using the Gene Elute Plasmid Miniprep kit (Sigma-Aldrich, St Louis, MO). Plasmids were confirmed through sequencing to contain a single amplicon containing both genes with no mutations at any of the primer- or probe-binding sites.
Dilution series of the plasmids were created to cover a range of concentration of 100- to 50,000-fold. RT qPCR was performed on 10 µl reaction mixtures as described above. Efficiency was also measured by fitting a linear regression to the log-linear phase of each reaction’s amplification plot using the software LinRegPCR (Ramakers et al. 2003; Ruijter et al. 2009).
Maximum-Likelihood Ancestral State Reconstruction
Relative gene expression was measured in a total of ten species from several divergent lineages within teleosts, including one electric fish from the Gymnotiforms (E. virescens) and one from the Mormyroids (Gn. petersii). The mean log2 expression ratio Scn4aa/Scn4ab among the biological replicates from each species was used along with a chronogram derived from a recent comprehensive maximum-likelihood tree of the bony fishes (Betancur-R et al. 2013), with the branch lengths in millions of years. The tree was pruned to the taxa with expression data using Dendroscope (Huson and Scornavacca 2012). Since the relative expression of both paralogs immediately after the WGD was presumably 1:1, ancestral state estimates were determined for trees with a ratio of 1 set at the root of the chronogram.
Evolutionary model selection was done using the R package “OUCH” (Butler and King 2004). The model with the lowest AICc was selected as the model that best fit the data. Ancestral state reconstruction was performed using the “ace” function in the R package “ape” (Schluter et al. 1997) under several phylogenetic scenarios involving assumptions about the character state at the root of the tree. The ace function was modified to provide estimates at specified points along a branch as well as all nodes (code available on request). This function uses a geometric Brownian motion model of evolution for a continuous trait since the trait data is log transformed.
Bayesian Ancestral State Reconstruction
To account for uncertainty at the root as well as uncertainty about our measurements of relative expression, we developed a hierarchical Bayesian expression for the probability distribution of ancestral states. To estimate the posterior probability of relative expression of the most recent nonelectric ancestors of the electric fish as well as their most recent common ancestor, we estimated the marginal posterior probability of the vector of these ancestral states given the data by simulating the joint posterior distribution: P(x, y, β, µroot|data) = P(y|x, β, µroot)P(β|x, µroot)P(x|data)P(µroot). Where y is the vector containing the three ancestral states of interest and x is the vector containing the true mean relative expression of each species in this study, β is the Brownian rate parameter, and µroot is the mean log expression at the root of the tree. The probability of the root is assumed to be N(µroot=0, 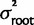 =10), this is highly conservative with respect to the data and the hypothesis and still assumes that equal expression is the most likely state of the relative expression after WGD.
=10), this is highly conservative with respect to the data and the hypothesis and still assumes that equal expression is the most likely state of the relative expression after WGD.
Rather than assume that mean relative expression is known, and is equal to the measured mean 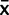 , for each species, we modeled x as a random variable to account for variation within species as well as use the information we gained from our plasmid calibration experiments to model PCR bias effects (fig. 1). We model the mean of our qPCR measurements, φ, in each species as the sum of two random effects; the true mean of the species and the effect of the PCR assay used, θ; φ = x + θ. Since the true mean x is what we are interested in to make evolutionary inferences, we use the equation x = φ – θ. We model the mean expression from our measurements as a Student’s t distribution: φ ∼ tn-1(
, for each species, we modeled x as a random variable to account for variation within species as well as use the information we gained from our plasmid calibration experiments to model PCR bias effects (fig. 1). We model the mean of our qPCR measurements, φ, in each species as the sum of two random effects; the true mean of the species and the effect of the PCR assay used, θ; φ = x + θ. Since the true mean x is what we are interested in to make evolutionary inferences, we use the equation x = φ – θ. We model the mean expression from our measurements as a Student’s t distribution: φ ∼ tn-1(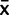 ,
, 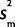 /n), where
/n), where 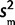 is the sample variance. To model our uncertainty about the PCR effects in species for which we did not perform a calibration experiment and correction, we assumed that our calibration data are normally distributed with known mean, 0, and unknown variance
is the sample variance. To model our uncertainty about the PCR effects in species for which we did not perform a calibration experiment and correction, we assumed that our calibration data are normally distributed with known mean, 0, and unknown variance 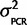 . The mean is known since there is no reason to believe that qPCR systematically favors one ohnolog over the other in every species, but since we have observed some small amount of PCR bias effect we expect some nonzero chance there will be a small percentage of the total signal emanating from artifacts of the PCR reaction favoring one ohnolog over the other. Therefore, the joint probability, P(θ,
. The mean is known since there is no reason to believe that qPCR systematically favors one ohnolog over the other in every species, but since we have observed some small amount of PCR bias effect we expect some nonzero chance there will be a small percentage of the total signal emanating from artifacts of the PCR reaction favoring one ohnolog over the other. Therefore, the joint probability, P(θ, 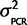 |Calibration data) = P(θ |
|Calibration data) = P(θ | 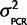 ) P(
) P(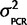 |Calibration data) = N(0,
|Calibration data) = N(0, 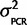 )P(
)P(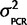 |Calibration data). The posterior distribution for the variance given the calibration data, P(
|Calibration data). The posterior distribution for the variance given the calibration data, P(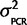 |Calibration data), can be attained if P(Calibration data|
|Calibration data), can be attained if P(Calibration data| 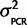 ) = N(0,
) = N(0, 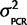 ) and P(
) and P(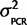 ) = scaled inverse-χ2 with υ degrees of freedom. Therefore, if we have a conservative prior belief that
) = scaled inverse-χ2 with υ degrees of freedom. Therefore, if we have a conservative prior belief that 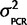 is twice that observed in the data and we weight that belief equally to the data (set υ = sample size from the data; 4) then the posterior distribution P(
is twice that observed in the data and we weight that belief equally to the data (set υ = sample size from the data; 4) then the posterior distribution P(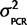 | Calibration data) is a scaled inverse-χ2 distribution with degrees of freedom as the number of independent primer/probe sets we investigated with plasmids (n =4) multiplied by 2 and the scale is the weighted average of the observed mean-squared deviates from the expected value (
| Calibration data) is a scaled inverse-χ2 distribution with degrees of freedom as the number of independent primer/probe sets we investigated with plasmids (n =4) multiplied by 2 and the scale is the weighted average of the observed mean-squared deviates from the expected value (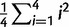 ) = 0.0883 (Gelman et al. 2004), where i is the mean of each of the four calibration experiments, and the prior belief that the scale is actually twice that observed. In other words P(
) = 0.0883 (Gelman et al. 2004), where i is the mean of each of the four calibration experiments, and the prior belief that the scale is actually twice that observed. In other words P(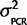 |Calibration data) = scaled inverse-χ2 (df = 8, scale = 0.0883/2 + 0.0883) =
|Calibration data) = scaled inverse-χ2 (df = 8, scale = 0.0883/2 + 0.0883) = 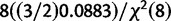 . In summary, we model x = tn−1 (
. In summary, we model x = tn−1 (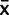 ,
, 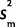 /n) − N(0,
/n) − N(0,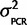 )
) 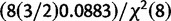 . We compared analyses using this model with results using a much simpler approximation where we instead assume φ ∼ N(
. We compared analyses using this model with results using a much simpler approximation where we instead assume φ ∼ N(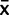 ,
, 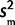 /n) and
/n) and 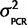 is known with θ ∼ N(0,
is known with θ ∼ N(0, 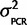 ) and
) and 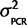 =
= 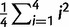 and thus x ∼ N(
and thus x ∼ N(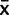 ,
, 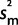 /n +
/n + 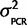 ) the results were very similar. If we ignore uncertainty about PCR effects, our certainty about inferred ancestral states only changes very slightly (data not shown) confirming that we can assume our measured relative expression robustly supports our hypothesis. The results reported are from the full model of uncertainty.
) the results were very similar. If we ignore uncertainty about PCR effects, our certainty about inferred ancestral states only changes very slightly (data not shown) confirming that we can assume our measured relative expression robustly supports our hypothesis. The results reported are from the full model of uncertainty.
If the data are generated by a Brownian motion process over a tree, then the probability of any set of points on that tree is multivariate normal with covariance matrix determined by the branch lengths of the tree multiplied by the Brownian rate parameter β. According to Schluter et al. (1997), the marginal posterior distribution of β given a set of values on the tree and a value at the root and uniform priors for the mean and variance is a scaled inverse-χ2 distribution with N degrees of freedom, where N is the number of internal node estimates plus tips, and Q is the sum of squared differences of all branches between the maximum-likelihood estimate of the ancestor, i, and that of its descendants, i′, divided by the branch length ti,i′ connecting the two: 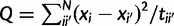 and so β = Q/ χ2(N) (Schluter et al. 1997). We used the ace function from the package “ape” to calculate Q.
and so β = Q/ χ2(N) (Schluter et al. 1997). We used the ace function from the package “ape” to calculate Q.
Since the probability of a subset of a vector from a multivariate normal distribution given another subset is also a multivariate normal, we modeled P(y|x, β, µroot) as multivariate normal N(µy, Σ) with µy being the mean y|x and Σ is the 3 × 3 variance/covariance matrix of y|x (Gelman et al. 2004), similar to the approach taken in Gu (2004). To compute the distribution of P(y|x, β, µroot), we set the variance/covariance matrix of all tips and ancestors of interest, V = βT, where T is the matrix of branch lengths with diagonals being the distance from root to either tip or ancestor of interest and the off-diagonals being the length of all branches shared by two species on the tree, either extant or ancestral. The submatrix Y is the (3 × 3) variance/covariance matrix of the three ancestors of interest, X is the (8 × 8) variance/covariance matrix of extant nonelectric species with relative expression data, and H is the (3 × 8) covariance matrix of y with x.
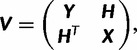 |
(1) |
| (2) |
| (3) |
| (4) |
We sampled y (n = 20,000) from the simulated joint posterior distribution P(x, y, β, µroot |data) using the software package R to measure statistics from the marginal posterior distribution P(y|data) (see supplementary material for code, Supplementary Material online). Marginal posterior distributions are displayed in figure 4.
Selection Analysis
Nucleotide sequence was gathered from published sequences deposited on GenBank as well as through our own sequencing effort. Codon alignments containing at least two domains of both Scn4a paralogs in several teleost species and several nonteleost orthologs were produced using the MAFFT algorithm on the GUIDANCE server (Penn et al. 2010). Codon sites with scores below the arbitrary 60 threshold were removed. Tree from Betancur-R et al. (2013) was used and pruned using Dendroscope as above. We took the teleost subclade of the resultant tree and duplicated it to produce two identical subtrees, one for each paralog. The new duplication node was placed at the midpoint of the root branch for the teleost clade. Branch-specific dN/dS (ω) was estimated using a random effects likelihood branch-site model (Kosakovsky Pond et al. 2011) on the Datamonkey server. This method avoids partitioning branches into “foreground” positive selection and “background” neutral or negative selection. This method estimates, using maximum-likelihood, the proportion of sites in an alignment at each branch which falls into three ω ratios corresponding to negative, neutral, and positive selection (ω−, ωN, ω+). These three selection classes are also estimated for each individual branch in the phylogeny. The  for each branch was calculated as a weighted sum of the three ω rates, where the weight was the proportion of sites that fell into that selection class. The log ratio of
for each branch was calculated as a weighted sum of the three ω rates, where the weight was the proportion of sites that fell into that selection class. The log ratio of  for each branch in the Scn4aa clade to its corresponding branch in the Scn4ab clade was estimated. To test for a relationship between relative expression and relative selection constraint, the same tree was paired down to the taxa for which we measured relative expression and dN/dS was again measured using the same likelihood method. This ratio was regressed against the estimated relative expression of the two paralogs at the terminal node of each branch in the ancestral state reconstruction. See supplementary table 4 (Supplementary Material) online for accession numbers.
for each branch in the Scn4aa clade to its corresponding branch in the Scn4ab clade was estimated. To test for a relationship between relative expression and relative selection constraint, the same tree was paired down to the taxa for which we measured relative expression and dN/dS was again measured using the same likelihood method. This ratio was regressed against the estimated relative expression of the two paralogs at the terminal node of each branch in the ancestral state reconstruction. See supplementary table 4 (Supplementary Material) online for accession numbers.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Laura Crothers and Ben Liebeskind for helpful feedback on the manuscript and the laboratory of Andrew Bass for kindly providing tissue samples from Porichthys notatus and the lab of Daniel Bolnick for providing Gasterasteus aculeatus tissue. We also thank Drs Eric Peatman and Chao Li for providing sequence for Scn4ab in Catfish. This work was funded by NIH grant GM084879, NSF IOS 1122115 (HHZ), the EEB program at the University of Texas, NSF DEB 1311521 to A.M.T., and an Undergraduate Research Fellowship from the University of Texas to D.V.
References
- Anderson DW, Evans BJ. Regulatory evolution of a duplicated heterodimer across species and tissues of allopolyploid clawed frogs (Xenopus) J Mol Evol. 2009;68:236–247. doi: 10.1007/s00239-009-9197-6. [DOI] [PubMed] [Google Scholar]
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. Proc Natl Acad Sci U S A. 2010;107:22172–22177. doi: 10.1073/pnas.1011803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang C, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Aury J, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurents B, Daubin V, Anthouard V, Aiach N, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Bass AH, Marchaterre MA. Sound-generating (sonic) motor system in a teleost fish (Porichthys notatus): sexual polymorphisms and general synaptology of a sonic motor nucleus. J Comp Neurol. 1989;286:154–169. doi: 10.1002/cne.902860203. [DOI] [PubMed] [Google Scholar]
- Bass AH, Zakon HH. Sonic and electric fish: at the crossroads of neuroethology and behavioral neurobiology. Horm Behav. 2005;48:360–372. doi: 10.1016/j.yhbeh.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Tawfik DS. Ohno’s model revisited: measuring the frequency of potentially adaptive mutations under various mutational drifts. Mol Biol Evol. 2008;25:2311–2318. doi: 10.1093/molbev/msn174. [DOI] [PubMed] [Google Scholar]
- Betancur-R R, Broughton RE, Wiley EO, Carpenter K, López JA, Li C, Holcroft NI, Arcila D, Sanciangco M, Cureton II JC, et al. The tree of life and a new classification of bony fishes. PLoS Curr. 2013;5 doi: 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288. : ecurrents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block BA. Evolutionary novelties: how fish have built a heater out of muscle. Am Zool. 1991;31:726–742. [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci U S A. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle KS, Dewan AK, Tricas TC. Fast drum strokes: novel and convergent features of sonic muscle ultrastructure, innervation, and motor neuron organization in the pyramid butterflyfish (Hemitaurichthys polylepis) J Morph. 2013;274:377–394. doi: 10.1002/jmor.20096. [DOI] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–350. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brunet FG, Crollius HR, Paris M, Aury J, Gilbert P, Jaillon O, Laudet V, Robinson-Rechavi M. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol Biol Evol. 2006;23:1808–1816. doi: 10.1093/molbev/msl049. [DOI] [PubMed] [Google Scholar]
- Butler MA, King AA. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am Nat. 2004;164:683–695. doi: 10.1086/426002. [DOI] [PubMed] [Google Scholar]
- Chain FJJ, Evans BJ. Multiple mechanisms promote the retained expression of gene duplicates in the tetraploid frog Xenopus laevis. PLoS Genet. 2006;2:0478–0490. doi: 10.1371/journal.pgen.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain FJJ, Ilieva D, Evans BJ. Duplicate gene evolution and expression in the wake of vertebrate allopolyploidization. BMC Evol Biol. 2008;8:43. doi: 10.1186/1471-2148-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Dean EJ, Davis JC, Davis RW, Petrov DA. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008;4:e1000113. doi: 10.1371/journal.pgen.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, Chao S, Kishony R. Exposing the fitness contribution of duplicated genes. Nat Genet. 2008;40:676–681. doi: 10.1038/ng.123. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari MB, Zakon HH. Conductances contributing to the action potential of Sternopygus electrocytes. J Comp Physiol A. 1993;173:281–292. doi: 10.1007/BF00212692. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian data analysis. 2nd ed. Boca Raton (FL): Chapman & Hall/CRC; 2004. [Google Scholar]
- Gu X. Statistical framework for phylogenomic analysis of gene family expression profiles. Genetics. 2004;167:531–542. doi: 10.1534/genetics.167.1.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zhang Z, Huang W. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc Natl Acad Sci U S A. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZG, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Hansen TF. Stabilizing selection and the comparative analysis of adaptation. Evolution. 1997;51:1341–1351. doi: 10.1111/j.1558-5646.1997.tb01457.x. [DOI] [PubMed] [Google Scholar]
- Hansen TF, Martins EP. Translating between microevolutionary process and macroevolutionary patterns: the correlation structure of interspecific data. Evolution. 1996;50:1404–1417. doi: 10.1111/j.1558-5646.1996.tb03914.x. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrichsen CN, Chaignat E, Reymond A. Copy number variants diseases and gene expression. Hum Mol Genet. 2009;18:R1–R8. doi: 10.1093/hmg/ddp011. [DOI] [PubMed] [Google Scholar]
- Hopkins CD. Convergent designs for electrogenesis and electroreception. Curr Opin Neurobiol. 1995;5:769–777. doi: 10.1016/0959-4388(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Myths and realities of gene duplication. In: Dittmar K, Liberles D, editors. Evolution after gene duplication. Hoboken (NJ): Wiley-Blackwell; 2010. pp. 77–103. [Google Scholar]
- Hughes T, Ekman D, Ardawatia H, Elofsson A, Liberles DA. Evaluating dosage compensation as a cause of duplicate gene retention in Paramecium tetraurelia. Genome Biol. 2007;8:213. doi: 10.1186/gb-2007-8-5-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury J, Brunet F, Petit J, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kafri R, Dahan O, Levy J, Pilpel Y. Preferential protection of protein interaction network hubs in yeast: evolved functionality of genetic redundancy. Proc Natl Acad Sci U S A. 2008;105:1243–1248. doi: 10.1073/pnas.0711043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Springer M, Pilpel Y. Genetic redundancy: new tricks for old genes. Cell. 2009;136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Kahlem P, Sultan M, Herwig R, Steinfath M, Balzereit D, Eppens B, Saran NG, Pletcher T, South ST, Stetten G, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of Down syndrome. Genome Res. 2004;14:1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B Biol Sci. 2012;279:5048–5057. doi: 10.1098/rspb.2012.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–291. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. A random effects branch-site model for detecting episodic diversifying selection. Mol Bio Evol. 2011;28:3033–3043. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoue S, Miya M, Arnegard ME, Sullivan JP, Hopkins CD, Nishida M. Comparable ages for the independent origins of electrogenesis in African and south American weakly electric fishes. PLoS One. 2012;7:e36287. doi: 10.1371/journal.pone.0036287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MK, Nahirney PC, Chen V, Adhikari BB, Wright J, Reedy MK, Bass AH, Wang K. Concentric intermediate filament lattice links to specialized Z-band junctional complexes in sonic muscle fibers of the type I male midshipman fish. J Struct Biol. 2003;143:56–71. doi: 10.1016/s1047-8477(03)00121-7. [DOI] [PubMed] [Google Scholar]
- Lopreato GF, Lu Y, Southwell A, Atkinson NS, Hillis DM, Wilcox TP, Zakon HH. Evolution and divergence of sodium channel genes in vertebrates. Proc Natl Acad Sci U S A. 2001;98:7588–7592. doi: 10.1073/pnas.131171798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB. Seeing the forest for the trees: the limitations of phylogenies in comparative biology. Am Nat. 2011;177:709–727. doi: 10.1086/660020. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Goaillard J. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Nasvall J, Sun L, Roth JR, Andersson DI. Real-time evolution of new genes by innovation, amplification, and divergence. Science. 2012;338:384–387. doi: 10.1126/science.1226521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Jost M, Lu Y, Taylor A, Zakon H, Ribera A. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Common pattern of evolution of gene expression level and protein sequence in Drosophila. Mol Biol Evol. 2004;21:1308–1317. doi: 10.1093/molbev/msh128. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer; 1970. [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol. 1992;23:263–286. [Google Scholar]
- O’Leary T, Williams AH, Caplan JS, Marder E. Correlations in ion channel expression emerge from homeostatic tuning rules. Proc Natl Acad Sci U S A. 2013;110:E2645–E2654. doi: 10.1073/pnas.1309966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara BC, Ane C, Sanderson MJ, Wainwright PC. Testing for different rates of continuous trait evolution using likelihood. Evolution. 2006;60:922–933. [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 2010;38:W23–W28. doi: 10.1093/nar/gkq443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Liao B, Chang AY, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Cell. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA. Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol. 2005;5:28. doi: 10.1186/1471-2148-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nature. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rome LC, Syme DA, Hollingworth S, Lindstedt SL, Baylor SM. The whistle and the rattle: the design of sound producing muscles. Proc Natl Acad Sci U S A. 1996;93:8095–8100. doi: 10.1073/pnas.93.15.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hashiguchi Y, Nishida M. Temporal pattern of loss/persistence of duplicate genes involved in signal transduction and metabolic pathways after teleost-specific genome duplication. BMC Evol Biol. 2009;9:127. doi: 10.1186/1471-2148-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Price T, Mooers AO, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.1111/j.1558-5646.1997.tb05095.x. [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard J, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Rev Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- Schwanhauser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Solomon KS, Fritz A. Concerted action of two dlx paralogs in sensory placode formation. Development. 2002;129:3127–3138. doi: 10.1242/dev.129.13.3127. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de Grassi A, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler J, Lehner B, Chen N, Fraser A. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 2006;7:R69. doi: 10.1186/gb-2006-7-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tvrdik P, Capecchi MR. Reversal of Hox1 gene subfunctionalization in the mouse. Dev Cell. 2006;11:239–250. doi: 10.1016/j.devcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Unguez GA, Zakon HH. Phenotypic conversion of distinct muscle fiber populations to electrocytes in a weakly electric fish. J Comp Neurol. 1998;399:20–34. [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24:485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman SG. Transcriptional channelopathies: an emerging class of disorders. Nature. 2001;2:652–659. doi: 10.1038/35090026. [DOI] [PubMed] [Google Scholar]
- Widmark J, Sundstrom G, Ocampo Daza D, Larhammar D. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;28:859–871. doi: 10.1093/molbev/msq257. [DOI] [PubMed] [Google Scholar]
- Wolfe K. Robustness it’s not where you think it is. Nat Genet. 2000;25:3–4. doi: 10.1038/75560. [DOI] [PubMed] [Google Scholar]
- Zakon HH. Adaptive evolution of voltage-gated sodium channels: the first 800 years. Proc Natl Acad Sci U S A. 2012;109:10619–10625. doi: 10.1073/pnas.1201884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Lu Y, Zwickl DJ, Hillis DM. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc Natl Acad Sci U S A. 2006;103:3675–3680. doi: 10.1073/pnas.0600160103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.