Abstract
Denervation is known to induce skeletal muscle atrophy and fiber-type transitions, the molecular mechanisms of which are poorly understood. To investigate the effect of denervation on skeletal muscle, proteomic analysis was performed to compare denervated soleus muscle with normal soleus muscle. The muscles were fractionated to myofibrillar and sarcoplasmic fractions, which were analysed using two-dimensional gel electrophoresis (2-DE), followed by MALDI-TOF-MS. At least 30 differentially regulated proteins were identified in the sarcoplasmic fractions of normal and denervated soleus muscles. This group included metabolic enzymes, signaling molecules, chaperones, and contractile proteins. We also found two proteins, APOBEC-2 (RNA-editing enzyme) and Gamma-synuclein (breast cancer related protein), which have not been recognized as denervation-induced proteins to date. Our results might prove to be beneficial in elucidating the molecular mechanisms of denervation-induced muscle atrophy.
Keywords: denervation, skeletal muscle, proteome, sarcoplasmic protein, myofibrillar protein
Skeletal muscle is a specialized organ that performs the dynamic work of force development, and it brings about structural and functional changes in response to the amount of mechanical load and activity. Skeletal muscle atrophy is easily caused by load decreases. For example, long-term lying down, plaster fixing due to treatment of fracture, microgravity exposure, and hind limb suspension of the rat lead to immobilized atrophy. Muscle atrophy is also observed with aging and the diseases of the nervous system. From the standpoint of muscular atrophy prevention, it is important to elucidate the molecular mechanisms of atrophy, but our understanding of this process remains unclear.
Sciatic nerve denervation is a typical experimental model for muscle atrophy. In fact, sciatic nerve denervation induces a dramatic shift of protein metabolism from protein synthesis toward protein degradation and reduction in the sizes of muscle fibers. Skeletal muscle atrophy is caused by both a decrease in protein synthesis and an increase in protein degradation.1,2) It has been found that denervation induces degradation of muscle proteins, especially contractile protein, by the activation of proteolytic systems, including ubiquitin-proteasome and some specific proteases.1,3–5) Selective loss of contractile proteins relative to all cellular proteins results in smaller myofibers, but is not accompanied by a reduction in myofiber numbers.6) Sciatic nerve denervation is also known to induce a shift of muscle fiber-type from slow toward last in the rat soleus muscle.7,8) The loss of slow-type specific nerve stimuli in slow-type muscle causes changes in protein expression, a decrease of slow-type specific proteins, and a concomitant increase of fast-type specific proteins, but the molecular mechanisms of skeletal muscle atrophy and fiber-type transition are poorly understood.
To achieve a better understanding of skeletal muscle atrophy and fiber-type transition, we compared normal soleus muscle with denervated soleus muscle by two-dimensional gel electrophoresis (2-DE), followed by MALDI-TOF-MS analysis. Some researchers have reported on proteomic analysis of denervated muscle. Their studies provide powerful evidence that might elucidate the molecular mechanism of muscle atrophy and fiber-type transition.9–11) However, all such studies were performed on the whole proteome, including both sarcoplasmic and myofibrillar fractions.9–11) Most of the proteins identified in these reports are metabolic enzymes and contractile proteins, which are the most abundant proteins in muscles.12) In addition, because contractile proteins have been demonstrated to be selectively lost during muscle atrophy,1,3–5) one should separate whole muscle into myofibrillar and sarcoplasmic fractions to apply 2-DE to investigate the less-expressed proteins in atrophying muscle. Recent reports on proteomic analysis also emphasize that fractionation methods help to detect and identify more lowly expressed proteins.13,14)
The objective of this study was to investigate changes of skeletal muscle proteome following denervation in order to understand the molecular mechanism of nerve-dependent muscle atrophy, and shift in muscle fiber-type. For the reasons mentioned above, we analyzed the proteomes of both normal and denervated slow rat muscles that were separated into myofibrillar and sarcoplasmic fractions. We identified some unique proteins, which have not been reported as markers of denervated skeletal muscles. Further detailed characterization of the role of these newly identified proteins in muscle might improve diagnostic procedures and the design of future strategies to treat muscle pathologies that involve fiber transformation.
Materials and Methods
Animals and treatments
Experiments were performed on young Wistar rats weighing 200–220 g. The rats had free access to food and water, and were subjected to standard conditions of humidity, temperature, and a 12-h light cycle. For sciatic nerve denervation, the rats were anesthetized with pentobarbital (1 mg/kg). The right hind limb was prepared for surgery, a 1-cm incision was made in the skin along with the axis of the femur, and the sciatic nerve was isolated. To prevent reinnervation, 3–5 mm section of sciatic nerve was cut and removed. The left hind limb was sham-operated. Denervated rats were anesthetized and killed by decapitation. Five rats were used as control and, five as the denervation group. The soleus and EDL muscles, were carefully excised from the hind limb, cleaned of tendons and connective tissues, weighed, and immediately frozen in liquid nitrogen. The muscles were stored at −80°C until processed. This procedure was carried out according to Kyusyu University’s rules for animal welfare.
Sample preparation
The soleus and EDL muscles were prepared for 2-DE as follows: Frozen muscles were ground to a fine powder in liquid nitrogen using a pestle. The powder (50 mg) was resuspended in ice-cold extraction buffer (1 ml) containing 20 mm Tris–HCl (pH 6.8), 2 mm EDTA, 175 mm KC1, and 0.5% Triton X. The lysate was centrifuged at 10,000 × g for 15 min al 4°C to obtain supernatant and pellets, containing sarcoplasmic and myofibrillar proteins respectively. The supernatant (sarcoplasmic fraction) was ultracentrifuged at 100,000 × g for 60 min at 4°C. To precipitate proteins, 4 volumes of acetone (−20°C) were added to the supernatant and the mixture was incubated at −20°C for 40 min. After centrifugation (8,000 × g for 10 min), the precipitate was washed with cold acetone (−20°C) and the pellet was solubilized in urea buffer containing 7 m urea, 2 m thiourea, 4% CHAPS, 1% DTT, 0.5% IPG buffer (pH 4–7), and a trace of bromophenol blue. The myofibrillar fraction (10,000 × g pellet) was also solubilized in urea buffer. The sample was incubated in the urea buffer for 1 h at room temperature. For separation of proteins, in the pH 6–11 range, a half volume of DeStreak Rehydration Solution (Amersham Pharmacia Biotech, Piscataway, NJ) was added to the sample.
Two-dimensional gel electrophoresis (2-DE)
Proteins from control and denervated soleus muscle fractions were run in parallel. Isoelectric focusing was carried out with an IPGphor system (Amersham Pharmacia Biotech, Piscataway, NJ). Immobilized pH gradient strips (pH 4–7, 6–11, 18 cm) rehydrated for 12 h (300 μg for the pH 4–7 dry strip, 200 μg for the pH 6–11 dry strip) in urea buffer was focused in 4 steps at 500 V (1 h), 500–1,000 V (1 h), 1,000–8,000 V (1 h), and 8,000 V (8 h). After completion of focusing, the strips were equilibrated with a buffer containing 6 m urea, 130 mm DTT, 30% glycerol, 2% SDS, and a trace of bromophenol blue, and then with a buffer containing 6 m urea, 135 mm iodoacetamido, 30% glycerol, 2% SDS, and a trace of bromophenol blue. They were loaded onto 11% polyacrylamide gels (20 × 20 cm). The system was run at 1,000 V at 24 mA per gel. The gels were fixed in 10% acetic acid and 40% methanol for 2 h and stained for 3 h using Flamingo gel stain solution (Bio-Rad Laboratories, Hercules, CA). The stained gels were washed in diluted water for 10 min. A molecular Imager FX (Bio-Rad, UK) was utilized to visualize protein spots with excitation at 488 nm. The spots were indexed using a PDQUEST.
In-gel tryptic digestion
In-gel tryptic digestion was performed as described by Rosenfeld et al.,15) with a slight modification. The target spot was excised with a clean scalpel and cut into 2-mm cubes. The gel pieces were transferred into a clean 0.2-ml microcentrifuge tube and washed with 40% 1-propanol at room temperature for 15 min. After removal of the 1-propanol solution, 200 mm ammonium bicarbonate in 50% ACN was added, and the sample was incubated at room temperature for 15 min. The gel pieces were dried and covered with 20 ng/μl of modified trypsin (Promega, Madison, W1) in a minimal volume of 100 mm ammonium bicarbonate for rehydration. Each gel piece was chopped into 4 to 5 smaller pieces and incubated 37°C. Then the supernatant was collected, and the gel pieces were extracted once with 100 mm ammonium bicarbonate, followed by two extractions with 80% ACN containing 0.05% TFA. The supernatant and extracts were combined and concentrated to the required concentrations.
MALDI-TOF-MS analysis
The resulting peptide mixtures were desalted using Zip-Tips C18 (Millipore, Billerica, MA), and eluted onto a 96-well MALDI target plate. Then 2-ml samples on the plate were mixed with 1-ml 10 mg/ml CHCA solution in 0.1% TFA in H2O/ACN (1:1). They were then dried at room temperature. MS data were obtained using a Voyager DE mass spectrometer equipped with a 337-nm N2 laser in positive ion reflectron mode (Applied Biosystems). Spectral data were obtained by averaging 64 spectra, each of which was the composite of 64 laser firings. Internal mass calibration was performed using bradykinin (904.45 Da) and ACTH (2465.75 Da).
Identification of protein
PMF was utilized for protein identification by analyzing the sizes of tryptic fragments via the MASCOT (Matrix Science, Boston, MA) search engines using the entire NCBI protein database. For effective PMF analysis, it was assumed that the peptides were monoisotopic, and the possibility that the methionine residues were oxidized was considered. The fingerprinting method allowed for a maximum of one missed tryptic cleavage per protein. The maximum deviation permitted in matching the peptide mass values was 100 ppm. Scores greater than 71 were considered to be significant (p < 0.005).
Immunoblot analysis
Muscle tissue lysates were subjected to SDS–PAGE under reducing conditions. The separated proteins were transferred to nitrocellulose membranes, which were then blocked with 10% powdered milk in 0.1% polyoxyethylene sorbitan monolaurate (Tween 20) in Tris-buffered saline (TTBS) prior to incubation with a 1:1,000 dilution of polyclonal anti-gamma-synuclein (SK23 antibody and E-20 Santa Cruz Biotechnology, CA) or anti-APOBEC-2 antibody (Abcam, Cambridge, MA) overnight at room temperature. The membranes were subsequently treated with biotinylated goat anti-rabbit (for anti-APOBEC-2 antibodies) or rabbit anti-goat (for anti-gamma-synuclein antibody E-20) IgG secondary antibody (Vector Laboratories, Burlingame, UK) at a 1:5,000 dilution in 1% powdered milk in TTBS for 1 h at room temperature, then with horseradish peroxidase-labeled avidin (Vector Laboratories, Burlingame, CA) at a 1:500 dilution in TTBS for 30 min at room temperature, followed by enhanced chemiluminescence (ECL) detection onto Kodak BioMax XAR films according to the manufacturer’s recommendations (Amersham Pharmacia Biotech, Piscataway, NJ).
Real-time RT-PCR
Total RNA was isolated from normal and denervated muscle (50 mg) using Trizol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized from 3 μg of total RNA by a reverse transcriptase SuperScriptII (Invitrogen) using oligo-dT primer. Real-time quantitative RT-PCR was performed in triplicate with Lyghtcycler Systems (Roche Diagnostics, Penzberg, Germany) according to the manufacturer’s protocol. The expression levels of mRNA of APOBEC-2 and gamma-synuclein were determined by normalizing them relative to HPRT expression. The forward and reverse primers were as follows: for HPRT, gaccggttctgtcatgtcg acctggttcatcatcactaatcac (probe no. 95), for gamma-synuclein, caccaccggggtagtacg cgccctcctcttgctctt (probe no. 79), and for APOBEC-2, tcctgaagtaggcaacagagc gccatcctggtcattgct (probe no. 40).
Results
Soleus muscle mass and protein concentrations of myofibrillar and sarcoplasmic fractions after denevation
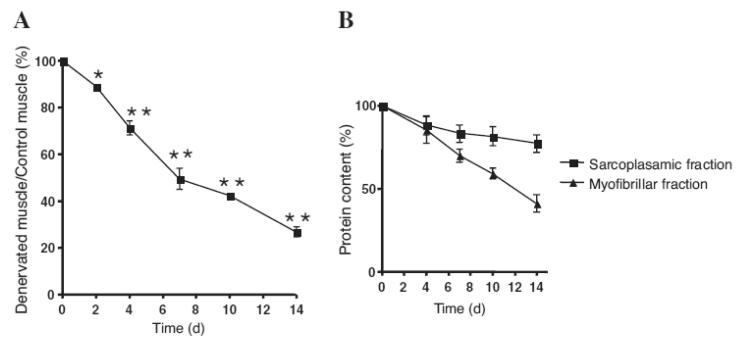
The change in soleus muscle mass after sciatic nerve denervation is shown in Fig. 1A. Soleus muscle mass decreased time-dependently post-denervation. Similarly, the content of protein in the soleus muscle decreased significantly (Fig. 1B). More than a 50% decrease in protein content was observed in the myofibrillar fraction by 14 d post-denervation, whereas only a 20% decrease was observed in the sarcoplasmic fraction. The different degradation rates of muscle protein during denervation atrophy might have been due to selective degradation of muscle proteins by the ubiquitin-proteasome system, because this system is known to degrade myofibrillar proteins more aggressively than sarcoplasmic proteins.1,3–5) This result also suggests the importance of employing the method separating whole muscle protein into myofibrillar and sarcoplasmic fractions for proteomic analysis of muscle. Furthermore, this separation procedure appeared to be useful for better detecting the low-expressed proteins in sarcoplasm, because myofibrillar proteins comprise more than 60% of whole protein in skeletal muscle. All previous proteomic studies on denervation atrophy have been performed on whole proteome.9–11) Considering these advantages, myofibrillar and sarcoplasmic fractions were used separately in our proteomic analysis.
Fig. 1. Denervation Effect on Rat Soleus Muscle.
A, Changes in soleus muscle mass after denervation. Values are shown as the ratio of the right soleus muscle (denervated) weight to the left soleus muscle (control, innervated) weight 2, 4, 7, 10, and 14 d post-denervation. Values are means ± SE, n = 5. Significantly different from sham-operated (left) soleus, * p < 0.05, ** p < 0.01. B, Changes in protein content of sarcoplasmic and myofibrillar fractions prepared from rat soleus muscle after denervation. Values are shown as the ratio of the right soleus muscle (denervated) weight to the left soleus muscle (control, innervated) weight 2, 4, 7, 10, and 14 d post-denervation. Values are means ± SE, n = 5.
Differential proteomic analysis
Proteomic differential display experiments were repeated at least 3 times, and the average fluorescence intensity for each spot was utilized to determine the expression level. More than about 800 and 200 spots were visualized in the sarcoplasmic and myofibrillar fractions respectively. Protein spots from denervated soleus muscle samples, which were more than double or less than half the fluorescent intensity (level of expression) compared to the control muscles, were designated up-regulated and down-regulated proteins respectively.
Proteomic analysis of myofibrillar fraction
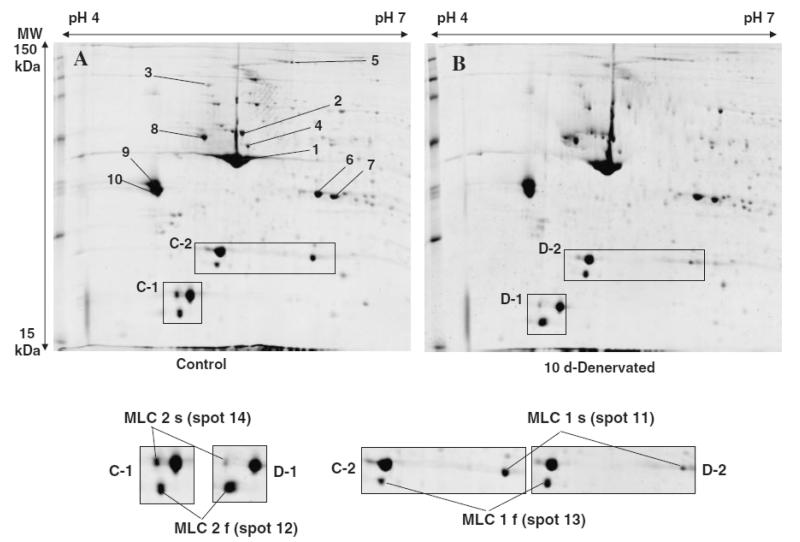
Myofibrillar and sarcoplasmic fractions prepared from normal and denervated soleus muscles were applied to 2-DE at the same level of protein (pH 4–7 for 300 μg and pH 6–11 for 200 μg respectively). The 2-DE at pH 4–7 and pH 6–11 (data not shown) of myofibrillar proteins of normal and denervated soleus muscles (10 d) exhibited almost same patterns of protein expression (Fig. 2). This means that the myofibrillar proteins were not specifically degraded, but rather decreased almost simultaneously in the same manner during denervation atrophy (Figs. 1B and 2). Regardless of the similarity of the expression pattern, denervation affected the expression level of a certain type of myofibrillar proteins. Particularly, slow-type myosin light chain (MLC1) was significantly reduced and fast-type MLC1 was increased as compared to control (Fig. 2, spots 11, 13). A similar change was also observed in MCL2 (Fig. 2, spots 12, 14). Our proteomic results are consistent with previous reports that denervation caused a change in the distribution of both alkali and regulatory MLC isoforms, with a decrease in slow-type isoforms (Fig. 2, spots 11, 14) and an increase in fast-type isoforms (Fig. 2, spots 12, 13)7,8,16) MALDI-TOF-MS analysis led to the identification of 14 myofibrillar proteins (Table 1).
Fig. 2. Proteome Map of Myofibrillar Fractions of (A) Normal Soleus Muscle (B) and 10 d Denervated Soleus Muscle.
Myofibrillar protein of normal and denervated soleus muscle applied equally (300 μg) to 2-DE. Proteomic differential display experiments were repeated at least 3 times, and the average fluorescent intensity (stained with SYPRO Ruby) for each spot was utilized to determine the expression level. The pH-values of the first dimension gel system and molecular mass standards (in kDa) of the second dimension are indicated at the top and the left of the panels respectively. Protein spots in myofibrillar fraction are marked by circles and are numbered 1–14. See Table 1 for a detailed listing of proteins in the myofibrillar fraction. Shown below is an expanded view of the C-1, D-1, C-2, D-2 of the original gel images. The expression levels of the MLCs show the shift of fiber-type transition from slow to fast.
Table 1. Identification of Myofibrillar Proteins.
| Spot no. | Accesion no. | Protein name | tpI | tMW | Cov. | Peptides matched |
|---|---|---|---|---|---|---|
| 1 | gi∣61217738 | Actin, alpha skeletal muscle | 5.23 | 42,051 | 47 | 16 |
| 2 | gi∣1352241 | Desmin | 5.21 | 53,325 | 45 | 12 |
| 3 | gi∣1142640 | Alpha actinin | 5.31 | 102,564 | 34 | 12 |
| 4 | gi∣11560133 | Tubulin, alpha 1 | 4.9 | 50,135 | 23 | 10 |
| 5 | gi∣109481744 | Myosin binding protein C, slow type | 5.5 | 132,040 | 25 | 11 |
| 6 | gi∣1256719 | Troponin T class Ia alpha-1 | 6.1 | 30,748 | 36 | 11 |
| 7 | gi∣207384 | Troponin T class IIa beta-1 | 6.5 | 30,356 | 33 | 12 |
| 8 | gi∣110347600 | Tubulin, beta 2b | 4.8 | 49,953 | 45 | 13 |
| 9 | gi∣20178269 | Tropomyosin beta chain | 4.66 | 32,836 | 50 | 10 |
| 10 | gi∣148840439 | Tropomyosin gamma chain | 4.69 | 29,006 | 63 | 16 |
| 11 | gi∣6981240 | Myosin, light polypeptide 3 (MLC1s) | 5.03 | 22,156 | 63 | 14 |
| 12 | gi∣127177 | Myosin regulatory light chain 2, skeletal muscle isoform (MLC 2f) | 4.82 | 18,838 | 89 | 15 |
| 13 | gi∣127131 | Myosin light chain 1, skeletal muscle isoform (MLC 1f) | 4.99 | 20,548 | 45 | 15 |
| 14 | gi∣127167 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform (MLC-2v) | 4.86 | 18,749 | 95 | 17 |
Theoretical pI.
Theoretical mass.
Sequence coverage (%) in PMF.
Proteomic analysis of sarcoplasmic fraction
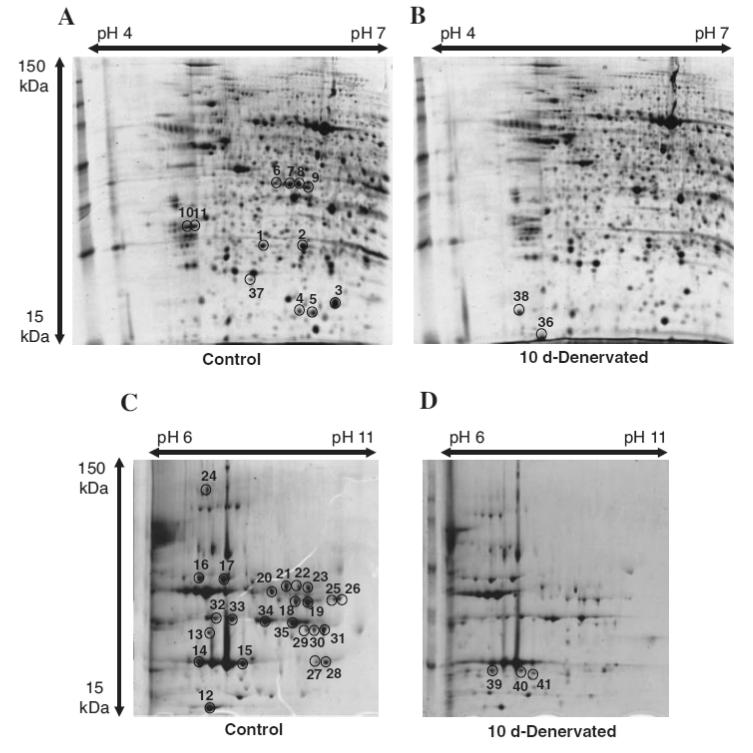
To detect changes in less abundant proteins in atrophying muscle more clearly, we prepared a sarcoplasmic fraction from whole soleus muscle. The 2-DE gel images of sarcoplasmic fraction of both normal and denervated (10 d) soleus muscle are shown in Fig. 3. The sarcoplasmic fraction potentially includes membrane and nucleus proteins and a small amount of myofibrillar proteins. The top layer of the supernatant after centrifugation was carefully siphoned off to prevent contamination with them, but we could not exclude this completely.
Fig. 3. Proteome Map of Sarcoplasmic Fraction of (A, pH 4–7), (C, pH 6–11) Normal Soleus Muscle (B, pH 4–7). (D, pH 6–11) and Denervated (10 d) Soleus Muscle.
Sarcoplasmic protein of normal and denervated soleus muscle applied equally (300 μg for pH 4–7, 200 μg for pH 6–11) to 2-DE. The pH-values of the first dimension gel system and the molecular mass standards (in kDa) of the second dimension are indicated at the top and the left of the panel respectively. Muscle proteins with drastically different expression levels are marked by circles, and are numbered 1–41. See Table 2 for a detailed listing of proteins in the sarcoplasmic fraction with changed abundances in the denervated soleus muscle.
2-DE gel image analysis revealed about 800 spots, of which 75 spots were differentially regulated during denervation atrophy. Subsequent MALDI-TOF-MS analysis led to the identification of 41 differentially expressed spots, corresponding to 21 different proteins (Table 2). Some of these corresponded to previously reported proteins (for example: myoglobin, spot 12;17) parvalbumin, spot 36;18) Hsp27, spots 1 and 2;19) and alpha-crystalline B, spot 320)).
Table 2. Identification of Defferentially Regulated Sarcoplasmic Proteins.
| Spot no. | Accesion no. | Protein name | tpI | tMW | Cov. | Peptides matched |
|---|---|---|---|---|---|---|
| Down-regulated proteins | ||||||
| 1 | gi|94400790 | Heat shock 27 kDa protein 1 | 6.12 | 22,821 | 65 | 12 |
| 2 | gi|94400790 | Heat shock 27 kDa protein 1 | 6.12 | 22,821 | 54 | 7 |
| 3 | gi|57580 | Alpha B-crystallin | 6.84 | 19,957 | 48 | 7 |
| 4 | gi|109477617 | Heat shock protein family, member 7 (cardiovascular) | 5.96 | 18,618 | 42 | 8 |
| 5 | gi|6671746 | Cofilin 2, muscle | 7.66 | 18,709 | 68 | 9 |
| 6 | gi|109463889 | Ankyrin repeat domain protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp) | 5.76 | 45,100 | 44 | 12 |
| 7 | gi|109463889 | Ankyrin repeat domain protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp) | 5.76 | 45,100 | 40 | 17 |
| 8 | gi|109463889 | Ankyrin repeat domain protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp) | 5.76 | 45,100 | 42 | 14 |
| 9 | gi|109463889 | Ankyrin repeat domain protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp) | 5.76 | 45,100 | 45 | 19 |
| 10 | gi|27681627 | Probable C → U editing enzyme APOBEC-2 | 4.57 | 25,660 | 44 | 10 |
| 11 | gi|27681627 | Probable C → U editing enzyme APOBEC-2 | 4.57 | 25,660 | 55 | 11 |
| 12 | gi|47682699 | Myoglobin | 7.84 | 17,157 | 51 | 7 |
| 13 | gi|57527204 | Alpha-ETF | 8.62 | 34,951 | 43 | 11 |
| 14 | gi|31377484 | Carbonic anhydrase 3 | 6.89 | 29,431 | 32 | 8 |
| 15 | gi|31377484 | Carbonic anhydrase 3 | 6.89 | 29,431 | 30 | 6 |
| 16 | gi|126723393 | Enolase 3, beta | 7.08 | 47,013 | 38 | 18 |
| 17 | gi|126723393 | Enolase 3, beta | 7.08 | 47,013 | 29 | 17 |
| 18 | gi|202837 | Aldolase A | 8.31 | 39,259 | 40 | 12 |
| 19 | gi|202837 | Aldolase A | 8.31 | 39,259 | 41 | 14 |
| 20 | gi|206113 | Phosphoglycerate kinase | 7.53 | 44,554 | 33 | 9 |
| 21 | gi|206113 | Phosphoglycerate kinase | 7.53 | 44,554 | 38 | 10 |
| 22 | gi|206113 | Phosphoglycerate kinase | 7.53 | 44,554 | 38 | 10 |
| 23 | gi|206113 | Phosphoglycerate kinase | 7.53 | 44,554 | 38 | 10 |
| 24 | gi|34861509 | Glycogen phosphorylase, muscle form (Myophosphorylase) | 6.65 | 97,287 | 39 | 24 |
| 25 | gi|6980972 | Glutamate oxaloacetate transaminase 2 | 9.13 | 47,314 | 28 | 8 |
| 26 | gi|6980972 | Glutamate oxaloacetate transaminase 2 | 9.13 | 47,314 | 28 | 8 |
| 27 | gi|42476181 | Malate dehydrogenase, mitochondrial | 8.93 | 35,683 | 43 | 12 |
| 28 | gi|42476181 | Malate dehydrogenase, mitochondrial | 8.93 | 35,683 | 41 | 11 |
| 29 | gi|8393948 | Phosphoglycerate mutase 2 | 8.85 | 28,755 | 38 | 8 |
| 30 | gi|8393948 | Phosphoglycerate mutase 2 | 8.85 | 28,755 | 38 | 10 |
| 31 | gi|8393948 | Phosphoglycerate mutase 2 | 8.85 | 28,755 | 46 | 12 |
| 32 | gi|8393418 | Glyceraldehyde-3-phosphate dehydrogenase | 8.14 | 35,828 | 39 | 11 |
| 33 | gi|8393418 | Glyceraldehyde-3-phosphate dehydrogenase | 8.14 | 35,828 | 48 | 10 |
| 34 | gi|8393418 | Glyceraldehyde-3-phosphate dehydrogenase | 8.14 | 35,828 | 48 | 10 |
| 35 | gi|8393418 | Glyceraldehyde-3-phosphate dehydrogenase | 8.14 | 35,828 | 39 | 9 |
| Up-regulated proteins | ||||||
| 36 | gi|11968064 | Parvalbumin | 5 | 11,925 | 60 | 7 |
| 37 | gi|18426864 | Heat shock 27 kDa protein 2 | 5.27 | 20,346 | 67 | 9 |
| 38 | gi|41059678 | Gamma-synuclein | 4.81 | 12,976 | 63 | 8 |
| 39 | gi|117935064 | Triosephosphate isomerase 1 | 6.45 | 26,921 | 39 | 8 |
| 40 | gi|117935064 | Triosephosphate isomerase 1 | 6.89 | 26,849 | 46 | 12 |
| 41 | gi|117935064 | Triosephosphate isomerase 1 | 6.89 | 26,849 | 46 | 8 |
Theoretical pI.
Theoretical mass.
Sequence coverage (%) in PMF.
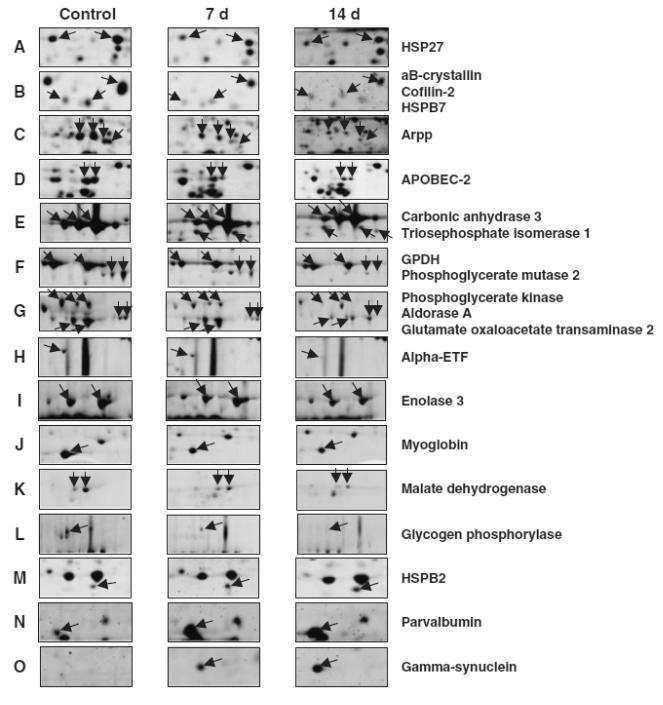
Major proteins that up- or down-regulated are listed in Table 2. Arpp, myoglobin, alpha-crystalline B, alpha-ETF, and hsp27 are normally more abundant in slow-type muscle than in fast-type (data not shown). The expression levels of these proteins were down-regulated post-denervation in soleus muscle (Fig. 4). In contrast, parvalbumin and triosephosphate isomerase, which express at a low level in slow-type muscle, as compared to fast-type muscle, were up-regulated post-denervation (Fig. 4). These results provide the evidence that denervation caused the fiber-type transition from slow to fast in conjunction with the results of MLC shown in Fig. 2. Oppositely to denervation, chronic low frequently stimuli against fast-type muscle induces a shift in protein expression towards the pattern typical of slow-type muscle.21)
Fig. 4. Differential Expression Patterns of Skeletal Muscle Proteins after Denervation.
An expanded view of 2-DE protein spots, depicted in their original position in representative gels shown in Fig. 4, which show a significant change in abundance. Lanes 1 to 3 show gels of normal condition (0 d), 7-d denervated (7 d) and 14-d denervated (14 d) soleus muscles, respectively. Panels A illustrate the gradual changes in the expression levels of HSP27, aB-crystallin, Cofilin-2, HSPB7, Arpp, APOBEC-2, Carbonic anhydrase 3, Triosephosphate isomerase 1, GPDH (Glyceraldehyde-3-phosphate dehydrogenase), Phosphoglycerate mutase, Phosphoglycerate kinase, Ardorase A, Glutamate oxaloacetate transaminase, Alpha-ETF, Enolase 3, Myoglobin, Malate dehydrogenase, Glycogen phosphorylase, HSPB2, Parvalbumin, and Gamma-synuclein.
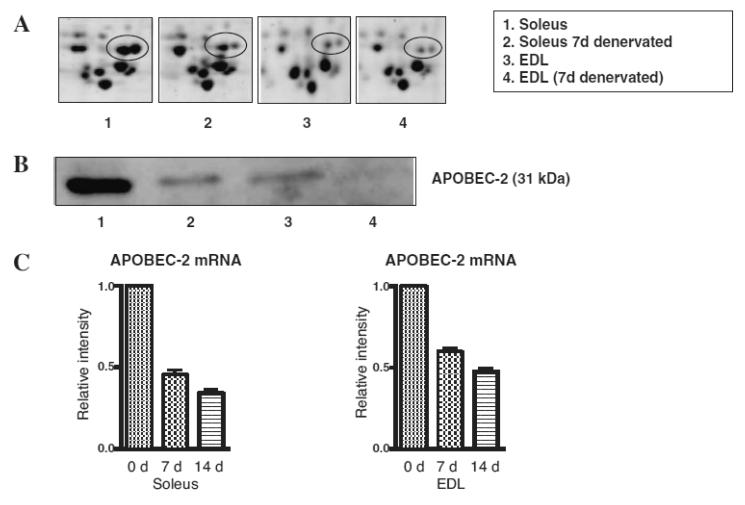
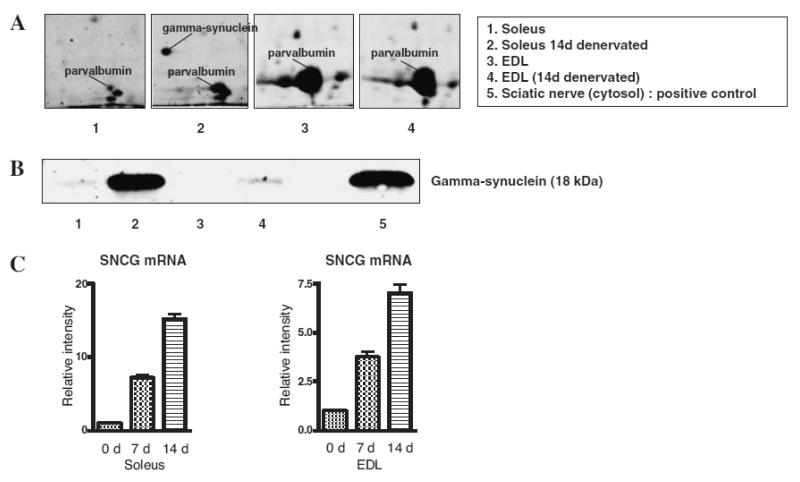
As shown in Fig. 4, we detected some unique proteins in the soleus muscle, APOBEC-2 and gamma-synuclein, which were markedly down- and up-regulated post-denervation respectively. The abundance of the two proteins was also evidently different between soleus (slow-type) muscle and extensor digitorum longus (EDL, fast-type) muscle (Figs. 5A and 6A). Western blotting analysis of muscle extracts using specific antibodies against these proteins confirmed differential expression of APOBEC-2 and gamma-synuclein (Figs. 5B and 6B). RT-PCR analysis demonstrated that APOBEC-2 mRNA expression down-regulated and gamma-synuclein mRNA expression was up-regulated in denervated soleus muscle (Figs. 5C and 6C).
Fig. 5. Differential Expression of APOBEC-2 in Soleus and EDL Muscles.
(A) is an expanded view of APOBEC-2 protein spots. Lanes 1 to 4 show gels of normal condition (0 d), 10-d denervated (10 d) soleus muscles and normal condition (0 d), 10-d denervated (10 d) EDL muscles respectively. Immunoblot analysis (B) and real-time RT-PCR analysis (C) of APOBEC-2 after denervation.
Fig. 6. Differential Expression of Gamma-Synuclein in Soleus and EDL Muscles.
(A) is an expanded view of gamma-synuclein protein spots. Lanes 1 to 4 show gels of normal condition (0 d), 14-d denervated (14 d) soleus muscles, and normal condition (0 d), 14-d denervated (14 d) EDL muscles respectively. Immunoblot analysis (B) and real-time RT-PCR analysis (C) of gamma-synuclein after denervation.
Discussion
In accord with previous studies,7,8,16) our differential proteomic analysis found a fiber-type shift from slow to fast during atrophy (Figs. 2, 3). Skeletal muscle myosin is composed of two heavy chains, two essential light chains (MLC1), and two regulatory light chains (MLC2). MLCs are responsible for force and velocity in muscle fibers. Therefore, the denervation-related shift of MLC toward the fast-type isoform is expected to happen naturally as a consequent of the lowering of mechanical load.
Twenty-three spots were differentially regulated proteins related to metabolic enzymes during denervation atrophy. These were identified as nine proteins, two of oxidative metabolism related enzymes (spots 27, 28, malate dehydrogenase mitochondrial, spots 14, 15, carbonic anhydrase 3) and seven of glycolytic enzymes (glycogen phosphorylase muscle form, aldolase A, enolase 3 beta, triosephosphate isomerase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate mutase 2). Typically, oxidative enzymes are more abundant in slow-type muscle than in fast-type.12) In this study, most of the metabolic enzymes differentially down-regulated during muscle atrophy were glycolytic enzymes, and only two of the oxidative enzymes were down-regulated. This result also gives collateral evidence that slow-type muscle (soleus) is capable of transformation into fast-type muscle following denervation.
Furthermore, by proteomic analysis, we also identified differentially regulated muscle proteins (spots 4, 5, 10–12, 37, 38) that have not been reported. These proteins have crucial roles in denervation-induced atrophy or muscle fiber-type transitions.
APOBEC-2
APOBEC-2 is expressed exclusively in skeletal and cardiac muscles and is considered to mediate the cytidine-to-uridine transcriptional editing of mRNA.22) However, its substrate has not been identified and its physiological role is unknown. APOBEC-2 deficient mice have been already generated, but they showed no phenotypical changes.23) On proteomic analysis of fast-type muscle (data not shown), we found that the expression level of APOBEC-2 was higher in soleus muscle (slow-type) than that of EDL muscle (fast-type). As shown in Fig. 5, APOBEC-2 decreased in soleus and EDL muscles during the atrophy process. Furthermore, we investigated the expression level of APOBEC-2 in soleus and EDL muscles (fast-type) by immunoblot and RT-PCR. The expression level of APOBEC-2 was higher in slow-type soleus muscle than fast-type EDL muscle (Fig. 5). These results suggest that APOBEC-2 may exhibit its RNA editing function to switch muscle fiber-type. We also investigated APOBEC-2 expression in C2C12 myoblast and differentiated myotube. APOBEC-2 was expressed only in differentiated myotube (data not shown).
Ankyrin repeat domain protein 2 (Arpp)
Arpp is a member of the CARP family, a stretch responsive protein, which is more highly expressed in slow-type muscle than in fast-type.24,25) Previous study indicates that Arpp is up-regulated in 4-week denervated gastrocnemius muscle as compared to control muscle,26) in conflict with a recent report that 37 kDa Arpp was down-regulated.27) Our proteomic analysis of soleus muscle showed that Arpp (spot 6–9) was down-regulated at 1-week post-denervation, and rapidly disappeared after that. Since denervation brings about a fiber-type transition from slow to fast, our results are consistent with that of Mckoy et al.27)
Gamma-synuclein
Gamma-synuclein was identified as a developmentally regulated gene in certain neuronal populations,28,29) and independently as breast cancer specific gene 1.30) Gamma-synuclein expression has been demonstrated in the nervous system,28,29) in advanced and metastatic breast carcinomas,30) in the skin,31) and in adipose tissue.32) No gross phenotypic change was observed in gamma-synuclein deficit mice.33) In skeletal muscle, gamma-synuclein expression is very low under normal conditions, and its physiological role remains unclear. Our finding on proteomic analysis of soleus (slow-type) muscle was overexpression of gamma-synuclein during the atrophy process (Fig. 6). To our surprise, it was barely detected before or after denervation in EDL (fast-type) muscle on immunoblot analysis (Fig. 6). Further studies are required to understand the role of gamma-synuclein in skeletal muscle physiology and pathology.
Hspb2
Hspb2 (also called MKBP) is a member of the small heat shock protein family. It is expressed in skeletal and cardiac muscle.34,35) It is more abundantly expressed in slow-type muscle than fast-type.36) Our proteomic analysis showed that Hspb2 was up-regulated in soleus muscle post-denervation, while other Hsp proteins (Hsp27, alpha B-crystalline, hsp family, member 7) were down-regulated. Induction of expression of Hspb2 was observed in both slow and fast-type muscles post-denervation (data not shown).
The proteomic analysis on denervated soleus muscle in this study provides information that might improve our understanding of the mechanisms of denervation-induced atrophy and fiber-type transition. We identified some differentially expressing proteins, especially APOBEC-2 and gamma-synuclein, whose roles in the atrophy process remain unclear. We are studying the roles of these proteins in normal and denervated muscles using APOBEC-2 and gamma-synuclein knock-out mice, as described previously.23,33) We feel that our research approach raises new possibilities of developing targets for therapeutic intervention in skeletal muscle atrophy after nerve injury.
Acknowledgments
We wish to thank Dr. Neuberger, Medical Research Council Laboratory of Molecular Biology, Cambridge, for valuable suggestions regarding APOBEC-2. This work was partly supported by ITO Memorial Foundation, Tokyo, Japan to YI and the Wellcome Trust Grand to VLB.
Abbreviations
- 2-DE
two-dimensional gel electrophoresis
- MALDI-TOF-MS
matrix assisted laser desorption ionization-time of flight-MS
- MLC
myosin light chain
- HSP
heat shock protein
- ACN
acetonitrile
- PMF
peptide mass fingerprint
- TFA
trifluoroacetic acid
References
- 1).Furuno K, Goodman MN, Goldberg AL. J. Biol. Chem. 1990;265:8550–8557. [PubMed] [Google Scholar]
- 2).Taillandier D, Aurousseau E, Meynial-Denis D, Bechet D, Ferrara M, Cottin P, Ducastaing A, Bigard X, Guezennec CY, Schmid HP. Biochem. J. 1996;316:65–72. doi: 10.1042/bj3160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Goldberg AL. J. Biol. Chem. 1969;244:3223–3229. [PubMed] [Google Scholar]
- 4).Goldberg AL, Goodman HM. Am. J. Physiol. 1969;216:1116–1119. doi: 10.1152/ajplegacy.1969.216.5.1116. [DOI] [PubMed] [Google Scholar]
- 5).Goldspink DF. Biochem. J. 1976;156:71–80. doi: 10.1042/bj1560071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Moschella MC, Ontell M. J. Neurosci. 1987;7:2145–2152. doi: 10.1523/JNEUROSCI.07-07-02145.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Nwoye L, Mommaerts WF, Simpson DR, Seraydarian K, Marusich M. Am. J. Physiol. 1982;242:R401–408. doi: 10.1152/ajpregu.1982.242.3.R401. [DOI] [PubMed] [Google Scholar]
- 8).Midrio M, Danieli-Betto D, Megighian A, Velussi C, Catani C, Carraro U. Pflugers Arch. 1992;420:446–450. doi: 10.1007/BF00374618. [DOI] [PubMed] [Google Scholar]
- 9).Isfort RJ, Hinkle RT, Jones MB, Wang F, Greis KD, Sun Y, Keough TW, Anderson NL, Sheldon RJ. Electrophoresis. 2000;21:2228–2234. doi: 10.1002/1522-2683(20000601)21:11<2228::AID-ELPS2228>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 10).Sun H, Liu J, Ding F, Wang X, Liu M, Gu X. J. Muscle Res. Cell Motil. 2006;27:241–250. doi: 10.1007/s10974-006-9067-4. [DOI] [PubMed] [Google Scholar]
- 11).Isfort RJ. J. Chromatogr. B Analyt. Technol. Biomed Life Sci. 2002;771:155–165. doi: 10.1016/s1570-0232(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 12).Okumura N, Hashida-Okumura A, Kita K, Matsubae M, Matsubara T, Takao T, Nagai K. Proteomics. 2005;5:2896–2906. doi: 10.1002/pmic.200401181. [DOI] [PubMed] [Google Scholar]
- 13).Hamelin M, Sayd T, Chambon C, Bouix J. Proteomics. 2007;7:271–280. doi: 10.1002/pmic.200600309. [DOI] [PubMed] [Google Scholar]
- 14).Vitorino R, Ferreira R, Neuparth M, Guedes S, Williams J, Tomer KB, Domingues PM, Appell HJ, Duarte JA, Amado FM. Anal. Biochem. 2007;366:156–169. doi: 10.1016/j.ab.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. Anal. Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 16).Bozzo C, Stevens L, Toniolo L, Mounier Y, Reggiani C. Am. J. Physiol. Cell Physiol. 2003;285:C575–583. doi: 10.1152/ajpcell.00441.2002. [DOI] [PubMed] [Google Scholar]
- 17).Askmark H, Carlson M, Roxin LE. Muscle Nerve. 1984;7:656–661. doi: 10.1002/mus.880070811. [DOI] [PubMed] [Google Scholar]
- 18).Olive M, Ferrer I. Neuropathol. Appl. Neurobiol. 1994;20:495–500. doi: 10.1111/j.1365-2990.1994.tb01001.x. [DOI] [PubMed] [Google Scholar]
- 19).Inaguma Y, Goto S, Shinohara H, Hasegawa K, Ohshima K, Kato K. J. Biochem. (Tokyo) 1993;114:378–384. doi: 10.1093/oxfordjournals.jbchem.a124184. [DOI] [PubMed] [Google Scholar]
- 20).Atomi Y, Yamada S, Nishida T. Bochem. Biophys. Res. Commun. 1991;181:1323–1330. doi: 10.1016/0006-291x(91)92083-v. [DOI] [PubMed] [Google Scholar]
- 21).Donoghue P, Doran P, Wynne K, Pedersen K, Dunn MJ, Ohlendieck K. Proteomics. 2007;7:3417–3430. doi: 10.1002/pmic.200700262. [DOI] [PubMed] [Google Scholar]
- 22).Liao W, Hong SH, Chan BH, Rudolph FB, Clark SC, Chan L. Biochem. Biophys. Res. Commun. 1999;260:398–404. doi: 10.1006/bbrc.1999.0925. [DOI] [PubMed] [Google Scholar]
- 23).Mikl MC, Watt IN, Lu M, Reik W, Davies SL, Neuberger MS, Rada C. Mol. Cell. Biol. 2005;25:7270–7277. doi: 10.1128/MCB.25.16.7270-7277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Kemp TJ, Sadusky TJ, Saltisi F, Carey N, Moss J, Yang SY, Sassoon DA, Goldspink G, Coulton GR. Genomics. 2000;66:229–241. doi: 10.1006/geno.2000.6213. [DOI] [PubMed] [Google Scholar]
- 25).Pallavicini A, Kojic S, Bean C, Vainzof M, Salamon M, Ievolella C, Bortoletto G, Pacchioni B, Zatz M, Lanfranchi G, Faulkner G, Valle G. Biochem. Biophys. Res. Commun. 2001;285:378–386. doi: 10.1006/bbrc.2001.5131. [DOI] [PubMed] [Google Scholar]
- 26).Tsukamoto Y, Senda T, Nakano T, Nakada C, Hida T, Ishiguro N, Kondo G, Baba T, Sato K, Osaki M, Mori S, Ito H, Moriyama M. Lab. Invest. 2002;82:645–655. doi: 10.1038/labinvest.3780459. [DOI] [PubMed] [Google Scholar]
- 27).McKoy G, Hou Y, Yang SY, Vega Avelaira D, Degens H, Goldspink G, Coulton GR. J. Appl. Physiol. 2005;98:2337–2343. doi: 10.1152/japplphysiol.01046.2004. Discussion 2320. [DOI] [PubMed] [Google Scholar]
- 28).Buchman VL, Hunter HJ, Pinon LG, Thompson J, Privalova EM, Ninkina NN, Davies AM. J. Neurosci. 1998;18:9335–9341. doi: 10.1523/JNEUROSCI.18-22-09335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Lavedan C, Leroy E, Dehejia A, Buchholtz S, Dutra A, Nussbaum RL, Polymeropoulos MH. Hum. Genet. 1998;103:106–112. doi: 10.1007/s004390050792. [DOI] [PubMed] [Google Scholar]
- 30).Ji H, Liu YE, Jia T, Wang M, Liu J, Xiao G, Joseph BK, Rosen C, Shi YE. Cancer Res. 1997;57:759–764. [PubMed] [Google Scholar]
- 31).Ninkina NN, Privalova EM, Pinon LG, Davies AM, Buchman VL. Exp. Cell Res. 1999;246:308–311. doi: 10.1006/excr.1998.4292. [DOI] [PubMed] [Google Scholar]
- 32).Oort PJ, Knotts TA, Grino M, Naour N, Bastard JP, Clement K, Ninkina N, Buchman VL, Permana PA, Luo X, Pan G, Dunn TN, Adams SH. J. Nutr. 2008;138:841–848. doi: 10.1093/jn/138.5.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Ninkina N, Papachroni K, Robertson DC, Schmidt O, Delaney L, O’Neill F, Court F, Rosenthal A, Fleetwood-Walker SM, Davies AM, Buchman VL. Mol. Cell. Biol. 2003;23:8233–8245. doi: 10.1128/MCB.23.22.8233-8245.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Iwaki A, Nagano T, Nakagawa M, Iwaki T, Fukumaki Y. Genomics. 1997;45:386–394. doi: 10.1006/geno.1997.4956. [DOI] [PubMed] [Google Scholar]
- 35).Suzuki A, Sugiyama Y, Hayashi Y, Nyu-i N, Yoshida M, Nonaka I, Ishiura S, Arahata K, Ohno S. J. Cell Biol. 1998;140:1113–1124. doi: 10.1083/jcb.140.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Nakagawa M, Tsujimoto N, Nakagawa H, Iwaki T, Fukumaki Y, Iwaki A. Exp. Cell Res. 2001;271:161–168. doi: 10.1006/excr.2001.5362. [DOI] [PubMed] [Google Scholar]