SUMMARY
T-boxes are gene-regulatory mRNA elements with which Gram-positive bacteria sense amino acid availability. T-boxes have two functional domains. Stem I recognizes the overall shape and anticodon of tRNA, while a 3′ domain evaluates its aminoacylation status, overcoming an otherwise stable transcriptional terminator if the bound tRNA is uncharged. Although T-boxes are believed to evaluate tRNA charge status without using any proteins, this has not been demonstrated experimentally because of the instability of aminoacyl-tRNA. Using a novel, facile method to prepare homogeneous aminoacyl-tRNA, we show that the Bacillus subtilis GlyQS T-box functions independently of any tRNA-binding protein. Comparison of aminoacyl-tRNA analogs demonstrates that the T-box detects the molecular volume of tRNA 3′-substituents. Calorimetry and fluorescence lifetime analysis of labeled RNAs shows that the tRNA acceptor end coaxially stacks on a helix in the T-box 3′ domain. This intimate intermolecular association, selective for uncharged tRNA, stabilizes the antiterminator conformation of the T-box.
INTRODUCTION
T-box riboswitches are non-coding mRNA elements that directly bind their cognate tRNAs. T-boxes evaluate the aminoacylation status of bound tRNA and respond by terminating transcription of downstream open reading frames when the tRNA 3′ terminus is esterified to an amino acid (Green et al., 2010; Grundy and Henkin, 1993). These riboswitches are widespread in Gram-positive bacteria, where, in response to nutritional status, they control expression of aminoacyl-tRNA synthetases (aaRSs) and other genes involved in the biosynthesis and transport of amino acids.
T-box riboswitches are comprised of two phylogenetically conserved domains: Stem I and the antiterminator (Gutierrez-Preciado et al., 2009; Vitreschak et al., 2008). Stem I is necessary and sufficient for specific, high-affinity binding to tRNA, and is insensitive to its aminoacylation status (Zhang and Ferré-D’Amaré, 2013). Stem I achieves sequence specificity by base pairing with the tRNA anticodon, and structure specificity by recognizing the elbow of tRNA (Grigg and Ke, 2013; Zhang and Ferré-D’Amaré, 2013). The structurally bistable antiterminator domain lies 3′ to Stem I and is responsible for both, evaluating the aminoacylation status of the bound tRNA and controlling transcription (Grundy and Henkin, 1993; Yousef et al., 2005). In isolation in solution, it consists of two short helices, A1 and A2, separated by a seven-nucleotide (nt) bulge (Gerdeman et al., 2003). Biochemical and genetic analyses (Fauzi et al., 2009; Grundy and Henkin, 1993; Yousef et al., 2005) imply that the NCCA-3′ terminus of uncharged tRNA base pairs with the conserved first four residues of the seven-nt bulge. When not bound to uncharged tRNA, and in the context of the full-length T-box, the domain adopts a more thermodynamically stable terminator conformation, consisting of a long stem-loop followed by a run of uridines. Such structures are canonical ρ-independent, intrinsic transcriptional terminators (Peters et al., 2011).
The T-box has been proposed to discriminate directly between charged and uncharged tRNA 3′ termini (Grundy and Henkin, 1993). This implies that the antiterminator domain can detect an amino acid residue as small as 44 Da (glycine) attached to a ~24 kDa tRNA. Alternatively, it is possible that in vivo, the T-box only needs to discriminate between uncharged tRNA and the ternary complex formed between aa-tRNA, EF-Tu and GTP. In the cell, most aa-tRNAs are thought to be bound to EF-Tu, an abundant translation factor (> 5% of cell dry weight) with high affinity for aa-tRNAs (Kd ~ 10−10 M; Buttner et al., 2001; Louie et al., 1984). Thus, it is unclear a priori whether the T-box needed to evolve the ability to discriminate between protein-free charged and uncharged tRNAs. To date, and due to the difficulty in preparing aminoacyl-tRNAs free of uncharged tRNA, this fundamental question regarding T-box function has not been examined experimentally. To definitively establish the selectivity of the T-box, we developed an efficient procedure for preparing highly purified (>95%) aa-tRNA and evaluated its effect on T-box-controlled transcriptional termination in vitro (the essential cellular role of EF-Tu precludes examination of the effect of deleting it in vivo). Remarkably, we find that the Bacillus subtilis glyQS T-box can efficiently distinguish between charged and uncharged tRNAGly in the complete absence of EF-Tu. Further, analyses with tRNAs bearing diverse 3′ substitutions demonstrates that termination efficiency correlates linearly with the molecular volume of the 3′ substituent. Strict steric discrimination suggests intimate packing between the tRNA acceptor end and the T-box antiterminator domain. Chemical modification, fluorescence, and isothermal titration calorimetry (ITC) experiments indicate that the uncharged tRNA binds forming a coaxial stack with the A1 helix of the antiterminator, and that this takes place in a snug binding pocket that is destabilized by tRNA 3′ substituents of even modest size. Coaxial stacking of the tRNA with the antiterminator therefore provides the stabilization energy that allows this structure to form preferentially over the transcriptional terminator.
RESULTS
Facile Preparation of Homogeneous Aminoacyl-tRNAs
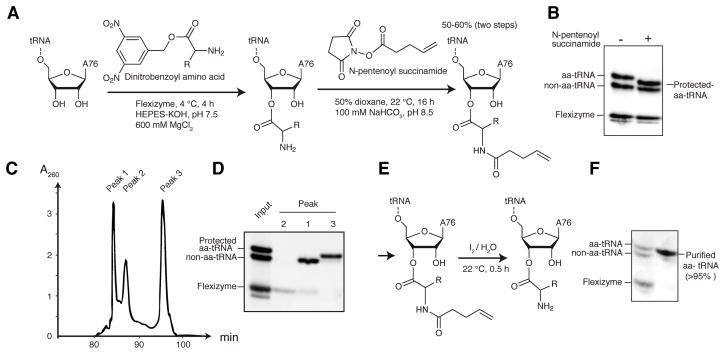
To examine the transcriptional termination activity of T-boxes in vitro, preparations of aa-tRNA essentially free from uncharged tRNA are needed (Putzer et al., 2002). The hydrolytic instability of the aminoacyl bond (Figure S1), and the laborious published procedure for the purification of aa-tRNAs (aminoacylation by cognate aaRSs followed by EF-Tu affinity purification; Ohtsuki et al., 2010) have precluded direct analysis of T-box function. To overcome this challenge, and to definitively establish the mechanism of T-box function, we developed an efficient method to obtain purified aa-tRNAs. This method is also generally compatible with mutant or misacylated tRNAs, that would be rejected by native aaRSs and EF-Tu, respectively. Our method employs a 46-nt catalytic RNA, the flexizyme, which can aminoacylate virtually any single-stranded RNA 3′ terminus with any natural and many unnatural amino acids (Goto et al., 2011; Xiao et al., 2008). A flexizyme (dFX) charged tRNAGly with 50–60% yield (Figure 1A). To stabilize the labile aminoacyl bond and provide a hydrophobic purification handle, we reacted the α-amino group of the esterified glycine with N-pentenoyl succinamide (Figure 1B). Although the aminoacyl bond is unstable at pH 8 (Figure S1), under our reaction conditions, N-pentenoylation is faster than hydrolysis. Pentenoylated aa-tRNAs can be easily separated from uncharged tRNAs using reversed-phase high-performance liquid chromatography (RP-HPLC; Fig 1C–D). Finally, the purified, protected aa-tRNA can be deprotected rapidly under mild conditions with dilute iodine (Lodder et al., 2005) immediately before use (Figure 1E–F; Experimental Procedures).
Figure 1. An Efficient Method to Prepare Homogeneous aa-tRNAs.
(A) Aminoacylation of tRNA and protection of aa-tRNA. The overall yield (on a tRNA basis) for the two steps is 50–60%.
(B) Protection of the α-amino group of aa-tRNA alters its gel mobility.
(C) C18 RP-HPLC chromatogram of the pentenoylation reaction mixture.
(D) Acid-PAGE analysis of fractions from (C).
(E) Mild deprotection of protected aa-tRNA regenerates aa-tRNA.
(F) Acid-PAGE analysis of the purified, deprotected aa-tRNA. See also Figure S1.
T-box Readout of tRNA Aminoacylation Status
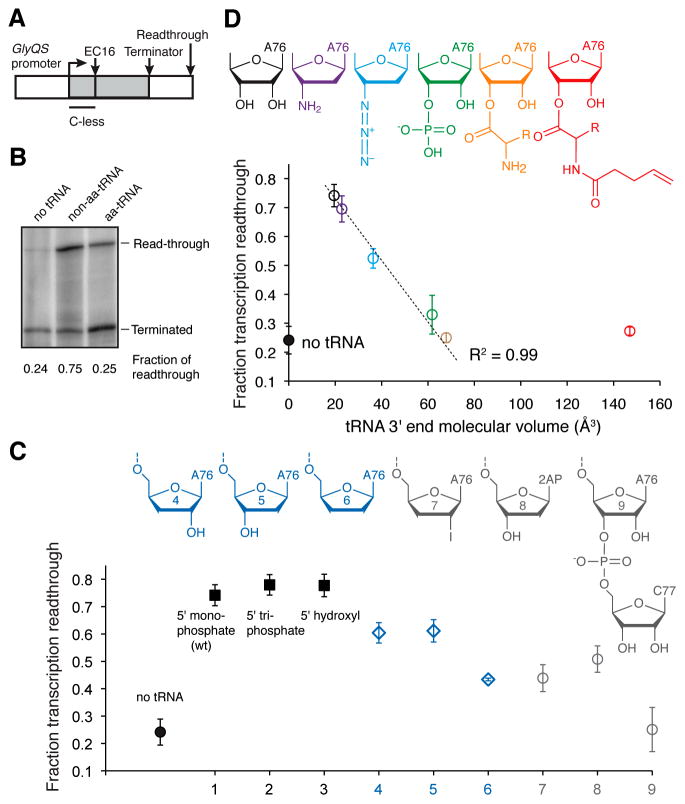
Using purified glycyl-tRNAGly, we examined transcriptional termination by the B. subtilis glyQS T-box riboswitch (Figure 2A–B). Under our experimental conditions, transcription in the absence of tRNA results in 24% readthrough. Addition of uncharged tRNAGly markedly suppresses transcriptional termination, yielding 75% readthrough. Remarkably, addition of glycyl-tRNAGly resulted in just 25% readthrough, which is indistinguishable from the efficiency of intrinsic transcriptional termination in the absence of tRNA. This experiment demonstrates that this T-box riboswitch efficiently distinguishes aa-tRNAs from non-aa-tRNAs in vitro in the absence of EF-Tu, a notable feat for its 33-nt antiterminator domain. The dynamic range of transcriptional antitermination we observe is comparable to that reported previously for a T-box riboswitch (Grundy et al., 2002b), and other riboswitches, such as the B. subtilis metE SAM-I riboswitch, (18 to 85% readthrough; Epshtein et al., 2003).
Figure 2. Mechanisms of tRNA 3′ End Recognition by a T-box Riboswitch.
(A) DNA template used for the in vitro antitermination assay. Transcription elongation complexes (ECs) were synchronized at position 16 by withholding CTP. Purified EC16 was incubated with 3 μM tRNA, before restarting by adding 100 μM of all NTPs.
(B) Representative Urea-PAGE analysis showing tRNA aminoacylation-dependent transcriptional discrimination by the T-box.
(C) Top, a panel of tRNA variants (Figure S2). Bottom, fraction of transcription readthrough induced by nine tRNA variants. Error bars represent s.d. from three independent experimental replicates.
(D) Top, a panel of tRNA variants carrying a range of 3′ substituents. Bottom, plot of fraction of transcription readthrough against the van der Waals volume of each tRNA 3′ substituent. Pearson’s R2 was computed omitting the protected-aa-tRNA. Error bars represent s.d. from three independent experimental replicates.
We next examined the antitermination effect of tRNA analogs with various 5′ and 3′ substituents. Consistent with its function in sensing tRNA 3′ aminoacylation, the T-box is indifferent to the presence of 5′ mono- or tri-phosphate on tRNA (Figure 2C). Uncharged and aa-tRNAs differ only in the absence of an esterified amino acid and the presence of a free 3′-OH (or 2′-OH) group in the former. In principle, discrimination between uncharged tRNA and aa-tRNA could be achieved either through active recognition of the free tRNA 2′ or 3′-OH on the former, or through a passive mechanism in which aa-tRNAs are rejected due to steric clash of the amino acid with the antiterminator structure. To distinguish between these mechanisms, we generated a series of tRNA derivatives with various chemical modifications on the ribose of the 3′ terminal tA76 (tRNA residues are preceded by “t”, Figure S2) and compared antitermination efficiencies. tRNAGly with 2′-deoxy, 3′-deoxy, or 2′,3′-dideoxy modifications on tA76 introduce modest antitermination defects (Figure 2C), suggesting a minor role for 2′-OH or 3′-OH recognition, possibly through direct hydrogen bonding between the tRNA 3′ terminus and the T-box antiterminator.
To directly test for steric readout of the tRNA 3′-terminus by the T-box, we generated a panel of tRNA variants that carry a range of chemical modifications of different molecular sizes (Figure 2D). In vitro antitermination assays using these tRNA variants reveal a strong inverse correlation between the van der Waals volume of the tRNA 3′ end substituent with its ability to induce downstream gene transcription (Pearson’s R2=0.99; Figure 2D). Notably, large tRNA 3′-substitutions, such as an additional nucleotide (Figure 2C; Grundy et al., 2005), or pentenoylated glycine (Figure 2D) do not induce any additional termination compared to glycyl-tRNAGly. Thus, the 44-kD EF-Tu, which envelops the tRNA 3′ terminus (Figure S2), would induce the same level of termination. Our results indicate that the glycyl moiety is sufficient to elicit the full dynamic range of termination. These results imply that steric rejection of the esterified amino acid is the principal mechanism by which T-boxes sense aminoacylation of tRNAs.
T-box Antiterminator Stabilization by tRNA
How does the uncharged tRNA stabilize the antiterminator against formation of the energetically more favorable terminator structure? Transcription of the antiterminator is completed before that of the terminator, conferring a kinetic advantage to the former. However, the large difference in stability (calculated ΔΔG=−16.4 kcal/mol; Lorenz et al., 2011), suggests that interactions with the tRNA in addition to putative hydrogen bonding to its terminal 3′-OH contribute to stabilization of the antiterminator. In isolation, the antiterminator comprises helices A1 and A2 flanking a 7-nt bulge (Gerdeman et al., 2003; Gutierrez-Preciado et al., 2009; Vitreschak et al., 2008). Four of the bulge nucleotides were genetically shown to base pair with the -NCCA-3′ acceptor terminus of tRNA, forming a third, 4-bp intermolecular helix. RNA duplexes are stabilized by coaxial stacking (Hermann and Patel, 1999; Zhang et al., 2010), and there are at least three distinct structural arrangements for these three helices. The intermolecular helix could stack against either A1 or A2, or instead remain unstacked if A1 and A2 stack on each other. We next sought to distinguish among these possibilities.
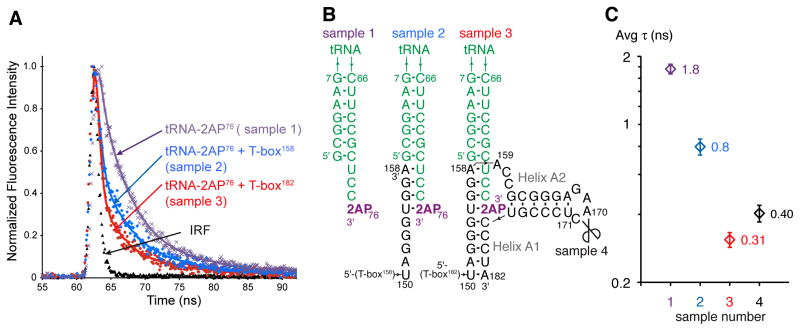
The fluorescent analogue of adenine, 2-aminopurine (2AP) has been extensively used as a structural probe for nucleic acids, because its quantum yield and fluorescence lifetime are sensitive to its chemical environment, with reduction in both typically attributed to stacking (Jean and Hall, 2001). We substituted the 3′-terminal adenosine of tRNA with 2AP (tRNA-2AP76) and used the 2AP fluorescence lifetime as a reporter of tRNA 3′ end interaction with the T-box antiterminator. First, we performed benchmarking experiments using synthetic RNA oligonucleotides of known structure (Figure S3). Free 2AP triphosphate exhibited a lifetime of 9.9±0.1 ns, in line with previously reported lifetimes of ~10 ns (Table S1; Jean and Hall, 2001). 2AP at the tRNA 3′ terminus is quenched, exhibiting a biphasic decay consisting of fast (τ1=0.28±0.1 ns; 68%) and slow components (τ2=4.9±0.1 ns; 32%), with an average lifetime (τavg) of 1.8±0.1 ns (Figure 3). 2AP in the interior of a base-paired, stacked duplex is further quenched, yielding a very short lifetime (τavg=0.33±0.1 ns; τ1=0.13±0.05 ns, 95%; τ2=3.9±0.1 ns, 5%; Figure S3B–D).
Figure 3. 2AP Fluorescence Lifetimes Reveal the Structural Context of the tRNA 3′ End Bound to the T-box Antiterminator.
(A) Normalized traces of 2AP fluorescence decay and best fits for RNAs in (B). IRF: instrument response function.
(B) Secondary structures of RNAs analyzed in (A). Scissors indicate nick position in the split T-box reconstituted with T-box170 and T-box171–182.
(C) Average 2AP fluorescence lifetimes of the RNAs shown in (B), as colored in (A). See also Figure S3. Error bars represent s.d. from three independent experimental replicates.
We assembled T-box-tRNA complexes by in vitro transcription of two T-box constructs in the presence of excess pre-folded tRNA-2AP76. One is truncated after the four nucleotides complementary to the -NCCA-3′ terminus of tRNA (T-box158); the other comprises the complete antiterminator domain (T-box182). The resulting complexes were purified from uncomplexed RNAs under native conditions through size-exclusion chromatography (SEC; Figure S3A). Incubation of purified T-boxes with tRNA results in heterogeneous, polydisperse mixtures (J.Z. and A.R.F., unpublished). Co-transcriptional assembly and native purification overcomes this. Fluorescence analysis of these complexes revealed average lifetimes of 0.80±0.06 ns and 0.31±0.02 ns for complexes containing T-box158 and T-box182 RNAs, respectively (Figure 3). Comparison with the benchmarks suggests that when complexed with T-box158, the 2AP at the tRNA 3′ end is base paired but only stacked on one face, whereas with T-box182, which contains the full antiterminator, it is base paired and stacked on both faces (Figure S3B–D). Thus, when stabilizing the T-box antiterminator, the 3′ terminus of uncharged tRNA is likely to lie at the interface of two coaxially stacked helices.
tRNA Stacks Coaxially with Antiterminator Helix A1
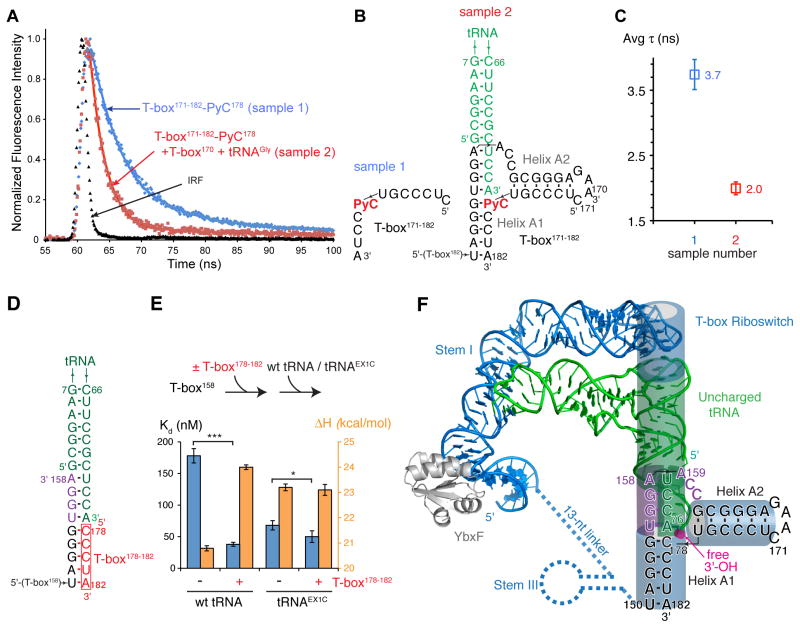
Since the intermolecular helix is invariably immediately adjacent to helix A1, we hypothesize that in the T-box-tRNA complex, tA76 stacks on the near-invariant G154•C178 pair at the top of A1 (Figure 3B), rather than on helix A2. To probe the structural context of C178 in the complex, we employed the fluorescent cytosine analog pyrrolocytosine (PyC). PyC forms a base pair with G that exhibits structural and thermodynamic properties of a normal G•C pair, and its fluorescence is sensitive to its environment (Buskiewicz and Burke, 2012; Tinsley and Walter, 2006; Zhang et al., 2008; Zhang and Wadkins, 2009). To introduce PyC at T-box position 178, we nicked the non-conserved loop of the antiterminator between positions 170 and 171, generating a two-piece variant of T-box182. This nick did not adversely affect interaction of the antiterminator with tRNA, as nearly identical tRNA-2AP76 fluorescence intensities and lifetimes were observed for the nicked and one-piece T-box182 constructs (Figure 3B–C). Additionally, substituting C178 with PyC in the oligonucleotide T-box171–182 does not prevent its association with tRNA and T-box170, as it co-eluted with these two larger RNAs (data not shown).
We benchmarked the effect of base pairing and stacking interactions on PyC fluorescence lifetime using oligonucleotides of known structure. In a single-stranded RNA oligonucleotide, PyC exhibits average fluorescence lifetimes of 3.6–5.7 ns, which are dramatically shortened upon base pairing (Figure S3E–G). When base-paired to a guanine, and stacked only on its 3′ side (on a cytosine), PyC exhibited a fluorescence lifetime of 1.55±0.03 ns. When this base-paired PyC was also stacked on its 5′ face (with an adenosine, yielding a structural context similar to that in our proposed tRNA-antiterminator complex model, Figure 4B), the lifetime increased appreciably, to 1.92±0.04 ns. Overall, our benchmarks are consistent with previous reports that base pairing quenches whereas stacking with cytosine or adenosine enhances PyC fluorescence quantum yield and lifetime through increased solvent shielding (Hardman et al., 2008; Hardman and Thompson, 2006). These benchmarks establish an empirical correlation between the structural context of PyC and its lifetime.
Figure 4. PyC Fluorescence Lifetime and Calorimetric Analyses Suggest Coaxial Stacking between the tRNA 3′ end and Antiterminator Helix A1.
(A) Normalized traces of PyC fluorescence decay and best fits for RNAs in (B).
(B) Secondary structures of RNAs analyzed in (A).
(C) Average PyC lifetimes of the RNAs shown in (B), as colored in (A). Error bars represent s.d. from three independent experimental replicates. See also Figure S3.
(D) Secondary structure schematic of a truncated tRNA-T-box complex without helix A2.
(E) Two-step ITC analysis of the energetic contribution from the proposed stacking between tRNA 3′ end tA76 and T-box C178. The upper panel explains the sequential ITC experiment, and the lower panel shows the Kd (blue) and enthalpy changes (orange) from the ITC titrations (Figure S4). Error bars represent s.d. from three independent replicates. (*, p=0.052; ***, p=0.0011; two-tailed t-test). See also Figure S4.
(F) Model of a full-length T-box in complex with uncharged tRNA based on this work, phylogenetic analysis, and the co-crystal structure of a T-box Stem I bound to its cognate tRNA (Zhang and Ferré-D’Amaré, 2013). tRNA is in green, T-box in blue, 7-nt antiterminator bulge in purple, aminoacylation site in magenta. Stacked cylinders denote coaxially stacked RNA helices.
As expected, the isolated oligonucleotide T-box171–182 containing PyC at position 178 exhibited a lifetime of 3.7±0.2 ns. In contrast, when this same oligonucleotide was assembled into a ternary complex with uncharged tRNA and T-box170, the lifetime decreased to 2.0±0.1 ns. This is consistent with the PyC residue at position 178 being base-paired and stacking with a 5′ adenosine (Figure 4A–C), since model compounds in which PyC is flanked by a 5′ adenosine exhibit lifetimes of 4.7±0.3 ns (single-stranded) or 1.92±0.04 ns (PyC base-paired to G; Figure S3E–G).
We further reasoned that if the tRNA 3′ adenosine stacks directly on C178 of T-box helix A1, there should be a corresponding energetic signature. To measure this stabilization energy, we devised a two-step ITC assay in which a 5-nt RNA oligonucleotide (T-box178–182) is pre-bound to T-box158 (Kd ~ 1 μM; saturation of binding was monitored by ITC, Figure S4A) and tRNA is then titrated into the split T-box construct lacking helix A2. The presence of T-box178–182 increased the affinity with which tRNA binds to T-box158 by ~4.7 fold (p=0.0011; two-tailed t-test), reflecting additional favorable enthalpy and free energy of −3.2 kcal/mol and −0.9 kcal/mol, respectively (Figure 4D–E & S4). These energetic contributions are in line with previous studies of coaxial helical stacking (Discussion). As expected, extending the tRNA 3′ end by one extra cytosine residue (previously used as a surrogate for aa-tRNA by Grundy et al., 2005) prevented stacking with T-box178–182 and thus abolished the affinity enhancement (Figure 4E). Overall, our results support direct coaxial stacking between the tRNA 3′ stem and the antiterminator helix A1, but not with helix A2. We therefore propose that this stacking interaction represents the mechanism by which uncharged tRNAs stabilize the antiterminator structure, thereby dictating the transcription outcome.
DISCUSSION
T-boxes Sense tRNA Aminoacylation Status Independent of EF-Tu
To our knowledge, this is the first demonstration that a T-box riboswitch can evaluate the aminoacylation status of its cognate tRNA in the absence of any protein (other than RNA polymerase). EF-Tu can discriminate between aa-tRNA and free tRNA by more than 2000-fold (Janiak et al., 1990). It is notable that Gram-positive bacteria have evolved a second, independent class of molecular devices sensitive to tRNA aminoacylation. In many bacteria, archaea, and organelles such as mitochondria, initially misacylated Glu-tRNAGln and Asp-tRNAAsn undergo transamidation to produce Gln-tRNAGln and Asn-tRNAAsn. By virtue of having both a tRNA body and an esterified amino acid that exhibit low affinities for EF-Tu, these misacylated tRNAs do not appreciably bind EF-Tu (Asahara and Uhlenbeck, 2002). Another group of tRNAs that do not bind EF-Tu are the isodecoder tRNAs that have evolved to be poor substrates of aaRSs and EF-Tu, to escape the translation machinery and function in roles such as cell wall remodeling or viral packaging (Geslain and Pan, 2011; Roy and Ibba, 2008). The inherent ability of T-boxes to detect tRNA aminoacylation directly prevents interference from these misacylated tRNA intermediates, tRNAs not used for translation, or otherwise aberrant tRNAs that fail to bind EF-Tu. If T-boxes relied on EF-Tu for sensing aminoacylation, the accumulation of such tRNAs (aminoacylated but not bound by EF-Tu) would be misinterpreted as signals for amino acid starvation. Alternatively, T-boxes may have evolved prior to the emergence of EF-Tu, providing amino acid surveillance for a primordial organism that relied on direct binding of aa-tRNAs to the ribosomal A site.
Steric Readout of tRNA Aminoacylation by Riboswitches and Proteins
The strong phylogenetic conservation of the antiterminator domain among more than 1100 annotated T-boxes (Gutierrez-Preciado et al., 2009; Vitreschak et al., 2008) suggests a common structure. Thus, it is likely that the steric discrimination mechanism employed by the GlyQS riboswitch to sense the presence of esterified glycine on tRNAGly is employed by T-boxes specific to other amino acids. Nonetheless, because some aminoacyl moieties (those bearing, for instance aromatic or cationic side chains) might stabilize the antiterminator structure despite being rejected sterically, the intrinsic stability (and structure) of T-box antiterminators that respond to different tRNA types may be subtly different. Such specialization is more readily achieved than in the case of EF-Tu, which has to compensate its affinity for the tRNA acceptor stem with that for the esterified amino acid (LaRiviere et al., 2001). Steric readout is also important in the double-sieve mechanism by which some aaRSs achieve specificity (Moras, 2010; Tsui and Fersht, 1981). In eukaryotes, amino acid availability is monitored by the multi-domain eIF2α kinase GCN2, which is activated by the direct binding of any uncharged tRNA (Hinnebusch, 2005). GCN2 binds tRNAs using a domain that resembles Histidyl-tRNA synthetase, and steric readout has been proposed to be an important mechanism by which that protein senses tRNA aminoacylation (Dong et al., 2000).
tRNA as an Integral Part of the Transcription Antiterminator
The favorable free energy derived from stacking the tRNA 3′ adenosine and T-box C178 (−0.9 kcal/mol, Figure 4E) falls between the favorable energies associated with appending a 3′ single-stranded (dangling) cytosine (−0.5 kcal/mol) or a G•C pair (−2.1 kcal/mol) to a pre-existing U•A base pair (Sugimoto et al., 1987). This is consistent with our structural model. Coaxial stacking between the tRNA and helix A1 would also explain the strong phylogenetic bias for the presence of four consecutive purines in the 5′ strand of helix A1 (Grundy et al., 2002a), reminiscent of the continuous track of stacked purines bridging the K-turn and the specifier elements in the proximal region of the T-box Stem I (Zhang and Ferré-D’Amaré, 2013).
The T-box antiterminator domain, consisting of just 33 nt, simultaneously senses tRNA aminoacylation status and functions as a transcriptional switch. Our model (Figure 4F) indicates that tRNA forms an integral part of this transcriptional antiterminator. Association of the tRNA acceptor end and helix A1 gives rise to a strikingly long helical stack. This would start at the interdigitated T-loops of the Stem I, which pack against the tRNA elbow (Zhang and Ferré-D’Amaré, 2013). The stack would then continue through the fourteen base pairs of the tRNA T- and acceptor stems, the four base-pair intermolecular helix formed between the tRNA 3′-NCCA terminus and the antiterminator bulge, and end with the five base-pairs of helix A1, resulting in a total of 29 layers of base pairs and triples. The T-box sandwiches the top half of tRNA between its two structural and functional domains, thereby exploiting the structural stability of tRNA to stabilize the energetically unfavorable antiterminator conformation.
The T-box is the only known RNA that can directly sense aminoacylation status of a tRNA, a challenging task commonly reserved for elaborate proteins such as aaRSs and EF-Tu. The T-box adds sensing of tRNA charge state to the repertoire of RNA-mediated cellular functions that may enable the operation and regulation of a primordial translation machinery, including free amino acid recognition (Serganov and Patel, 2009), amino acid activation (Kumar and Yarus, 2001), RNA aminoacylation (Illangasekare et al., 1995; Lee et al., 2000; Xiao et al., 2008), and peptidyl transfer (Noller et al., 1992). The central roles of the T-box system in many aspects of amino acid metabolism in most Gram-positive bacteria attest to its efficacy.
EXPERIMENTAL PROCEDURES
Preparation of Homogeneous Aminoacylated-tRNA
In vitro transcribed tRNA was aminoacylated using dinitro-flexizyme (dFx) as described (Goto et al., 2011). The aminoacylation mixture was reacted with N-pentenoyl succinamide, fractionated on a C18 column, and deprotected with aqueous iodine (Lodder et al., 2005). Details are in Supplemental Experimental Procedures.
Preparation of tRNA Variants Carrying 3′ Modifications
An in vitro transcribed tRNA fragment (residues 1–57) was ligated with RNA oligonucleotides corresponding to tRNA residues 58–76 bearing desired modifications.
In Vitro Transcription Antitermination Assay
Antitermination assays were performed essentially as described (Grundy et al., 2002b).
Assembly and Native Purification of T-box-tRNA Complexes by SEC
T-box fragments were transcribed by T7 RNA polymerase in the presence of pre-folded tRNA. The mixture was treated with Proteinase K and DNase I, and then fractionated by SEC. After fluorescence lifetime measurements, samples were re-analyzed on the same column to verify the intactness of the tRNA-T-box complex (Figure S3A).
2AP and PyC Fluorescence Lifetime Measurements
Fluorescence lifetime measurements were performed at 21 °C on an EasyLife-LS (Photon Technology International) system with excitation at 280 nm (2AP) or 340 nm (PyC) and emission at 376 nm (2AP) and 448 nm (PyC).
Isothermal Titration Calorimetry (ITC)
ITC was performed at 20 °C as described (Zhang and Ferré-D’Amaré, 2013).
Supplementary Material
HIGHLIGHTS.
An efficient method to obtain purified aminoacyl-tRNAs with broad applicability
T-box riboswitches directly sense tRNA aminoacylation status without proteins
T-boxes gauge the molecular volume of tRNA 3′ substituents to detect aminoacylation
Intermolecular coaxial stacking with uncharged tRNA stabilizes T-box antiterminator
Acknowledgments
We thank J. Posakony for assistance with chemical synthesis and NMR analysis, Y. Goto and H. Suga for a gift of dinitrobenzylglycine and suggestions on flexizyme use, S. Hecht for suggesting the use of the pentenoyl protecting group, N. Tjandra for access to NMR, G. Piszczek for fluorescence and ITC support, R. Levine and D.- Y. Lee for help with mass spectrometry, and N. Baird, J. Brunelle, K. Fredrick, R. Green, J. Hogg, M. Ibba, M. Lau, J. Lee, P. Nissen, C. Jones, O. Uhlenbeck, A. Roll-Mecak, and K. Warner for discussions. This work employed the Biochemistry and Biophysics core facilities of the National Heart, Lung and Blood Institute (NHLBI) and was supported in part by the intramural program of the NHLBI, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz IA, Burke JM. Folding of the hammerhead ribozyme: pyrrolo-cytosine fluorescence separates core folding from global folding and reveals a pH-dependent conformational change. RNA. 2012;18:434–448. doi: 10.1261/rna.030999.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner K, Bernhardt J, Scharf C, Schmid R, Mader U, Eymann C, Antelmann H, Volker A, Volker U, Hecker M. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis. 2001;22:2908–2935. doi: 10.1002/1522-2683(200108)22:14<2908::AID-ELPS2908>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell. 2000;6:269–279. doi: 10.1016/s1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc Natl Acad Sci USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauzi H, Agyeman A, Hines JV. T box transcription antitermination riboswitch: influence of nucleotide sequence and orientation on tRNA binding by the antiterminator element. Biochim Biophys Acta. 2009;1789:185–191. doi: 10.1016/j.bbagrm.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdeman MS, Henkin TM, Hines JV. Solution structure of the Bacillus subtilis T-box antiterminator RNA: seven nucleotide bulge characterized by stacking and flexibility. J Mol Biol. 2003;326:189–201. doi: 10.1016/s0022-2836(02)01339-6. [DOI] [PubMed] [Google Scholar]
- Geslain R, Pan T. tRNA: Vast reservoir of RNA molecules with unexpected regulatory function. Proc Natl Acad Sci U S A. 2011;108:16489–16490. doi: 10.1073/pnas.1113715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Katoh T, Suga H. Flexizymes for genetic code reprogramming. Nat Protoc. 2011;6:779–790. doi: 10.1038/nprot.2011.331. [DOI] [PubMed] [Google Scholar]
- Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–324. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Ke A. Structural determinants for geometry and information decoding of tRNA by T Box leader RNA. Structure. 2013;21:2025–2032. doi: 10.1016/j.str.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Moir TR, Haldeman MT, Henkin TM. Sequence requirements for terminators and antiterminators in the T box transcription antitermination system: disparity between conservation and functional requirements. Nucleic Acids Res. 2002a;30:1646–1655. doi: 10.1093/nar/30.7.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci USA. 2002b;99:11121–11126. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Yousef MR, Henkin TM. Monitoring uncharged tRNA during transcription of the Bacillus subtilis glyQS gene. J Mol Biol. 2005;346:73–81. doi: 10.1016/j.jmb.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardman SJ, Botchway SW, Thompson KC. Evidence for a nonbase stacking effect for the environment-sensitive fluorescent base pyrrolocytosine--comparison with 2-aminopurine. Photochem Photobiol. 2008;84:1473–1479. doi: 10.1111/j.1751-1097.2008.00368.x. [DOI] [PubMed] [Google Scholar]
- Hardman SJ, Thompson KC. Influence of base stacking and hydrogen bonding on the fluorescence of 2-aminopurine and pyrrolocytosine in nucleic acids. Biochemistry. 2006;45:9145–9155. doi: 10.1021/bi060479t. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Stitching together RNA tertiary architectures. J Mol Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Illangasekare M, Sanchez G, Nickles T, Yarus M. Aminoacyl-RNA synthesis catalyzed by an RNA. Science. 1995;267:643–647. doi: 10.1126/science.7530860. [DOI] [PubMed] [Google Scholar]
- Janiak F, Dell VA, Abrahamson JK, Watson BS, Miller DL, Johnson AE. Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu•GTP: evidence for a new functional role for elongation factor Tu in protein biosynthesis. Biochemistry. 1990;29:4268–4277. doi: 10.1021/bi00470a002. [DOI] [PubMed] [Google Scholar]
- Jean JM, Hall KB. 2-Aminopurine fluorescence quenching and lifetimes: role of base stacking. Proc Natl Acad Sci USA. 2001;98:37–41. doi: 10.1073/pnas.011442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RK, Yarus M. RNA-catalyzed amino acid activation. Biochemistry. 2001;40:6998–7004. doi: 10.1021/bi010710x. [DOI] [PubMed] [Google Scholar]
- LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Lee N, Bessho Y, Wei K, Szostak JW, Suga H. Ribozyme-catalyzed tRNA aminoacylation. Nature Structural Biology. 2000;7:28–33. doi: 10.1038/71225. [DOI] [PubMed] [Google Scholar]
- Lodder M, Wang B, Hecht SM. The N-pentenoyl protecting group for aminoacyl-tRNAs. Methods. 2005;36:245–251. doi: 10.1016/j.ymeth.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie A, Ribeiro NS, Reid BR, Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J Biol Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- Moras D. Proofreading in translation: dynamics of the double-sieve model. Proc Natl Acad Sci USA. 2010;107:21949–21950. doi: 10.1073/pnas.1016083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Yamamoto H, Doi Y, Sisido M. Use of EF-Tu mutants for determining and improving aminoacylation efficiency and for purifying aminoacyl tRNAs with non-natural amino acids. J Biochem. 2010;148:239–246. doi: 10.1093/jb/mvq053. [DOI] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putzer H, Condon C, Brechemier-Baey D, Brito R, Grunberg-Manago M. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 2002;30:3026–3033. doi: 10.1093/nar/gkf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci USA. 2008;105:4667–4672. doi: 10.1073/pnas.0800006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Kierzek R, Turner DH. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- Tinsley RA, Walter NG. Pyrrolo-C as a fluorescent probe for monitoring RNA secondary structure formation. RNA. 2006;12:522–529. doi: 10.1261/rna.2165806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui WC, Fersht AR. Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res. 1981;9:4627–4637. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–735. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Murakami H, Suga H, Ferré-D’Amaré AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNAGly and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–287. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- Zhang CM, Liu C, Christian T, Gamper H, Rozenski J, Pan D, Randolph JB, Wickstrom E, Cooperman BS, Hou YM. Pyrrolo-C as a molecular probe for monitoring conformations of the tRNA 3′ end. RNA. 2008;14:2245–2253. doi: 10.1261/rna.1158508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ferré-D’Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–366. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D’Amaré AR. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry. 2010;49:9123–9131. doi: 10.1021/bi1012645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wadkins RM. DNA hairpins containing the cytidine analog pyrrolo-dC: structural, thermodynamic, and spectroscopic studies. Biophys J. 2009;96:1884–1891. doi: 10.1016/j.bpj.2008.12.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.