Abstract
The Hexosamine Biosynthetic Pathway leads to elevated post-translation addition of O-linked-βN-acetylglucosamine (O-GlcNAc) on intracellular proteins. Cancer cells elevate total O-GlcNAcylation by increasing O-GlcNAc transferase (OGT) and/or decreasing O-GlcNAcase (OGA) levels. Reducing O-GlcNAcylation in cancer cells inhibits oncogenesis. Here, we demonstrate that O-GlcNAcylation regulates glycolysis in cancer cells via HIF-1α and its transcriptional target GLUT1. Reducing O-GlcNAcylation increases α-ketoglutarate, HIF-1 hydroxylation and interaction with VHL resulting in HIF-1α degradation. Reducing O-GlcNAcylation in cancer cells results in activation of ER stress and apoptosis of cancer cells mediated through CHOP induction of BCL2-family proteins. HIF-1α and GLUT1 are critical for OGT-mediated regulation of metabolic stress as overexpression of stable HIF-1 or GLUT1 rescues metabolic defects and apoptosis. Human basal-like breast cancers with high levels of HIF-1α contain elevated OGT, O-GlcNAcylation and lower OGA levels correlate independently with poor patient outcome. Thus, O-GlcNAcylation regulates cancer cell metabolic reprograming and survival stress signaling via regulation of HIF-1α.
Keywords: O-GlcNAc, OGT, cancer, metabolism, HIF1α, VHL, GLUT1, ER stress, CHOP, BIM, mTOR, hexosamine, epithelial, breast cancer
INTRODUCTION
Deregulating cellular energetics is emerging as a characteristic hallmark of cancer cells (Hanahan and Weinberg, 2011). Within such cells glucose and glutamine are used at an increased rate (DeBerardinis et al., 2008), resulting in the production of ATP in a manner independent of oxygen concentration (Dang and Semenza, 1999). Elevated glucose and glutamine flux are needed not only to serve the energetic demands of cancer cells but also to provide the essential carbon and nitrogen used in macromolecule synthesis, fueling the rapid growth and proliferation seen in tumors (DeBerardinis et al., 2008). This increase in glucose and glutamine uptake can alter multiple metabolic and signaling pathways in cancer cells including, for example, the hexosamine biosynthetic pathway (HBP) (Marshall, 2006), the liver kinase B1 (LKB1)/AMP-activated kinase (AMPK) signaling pathway (Shaw and Cantley, 2012), and the mammalian target of rapamycin (mTOR) (Zoncu et al., 2011).
The HBP relies on glucose and glutamine uptake and approximately 3–5% of the total glucose entering a cell is shunted into this pathway (Marshall et al., 1991). Fructose-6-phosphate, obtained from glucose during glycolysis, is converted through the enzymes of the HBP into the end product UDP-N-Acetylglucosamine (UDP-GlcNAc) (Marshall, 2006). This critical metabolite is required for the biosynthesis of a variety of extracellular glycopolymers, including both N- and O-glycans (Marshall, 2006); however, it also serves as the substrate for O-linked β-N-acetlyglucosamine (O-GlcNAc) transferase (OGT). This enzyme catalyzes the transfer of the GlcNAc moiety onto the free hydroxyl of select serine and threonine residues of target proteins and is the sole known glycosyltransferase responsible for the post-translational modification of a diverse population of nuclear and cytosolic proteins. This modification can be removed by the glycoside hydrolase O-GlcNAcase (OGA) (also referred to as MGEA5) that catalyzes cleavage of O-GlcNAc from proteins (Gao et al., 2001). This modification can alter protein function directly or, in some cases, by competing with phosphorylation sites (Hart et al., 2011). O-GlcNAcylation has been proposed to serve primarily to regulate cellular signaling and transcription regulatory pathways in response to altered nutrients and stress (Hart et al., 2011). Many of these O-GlcNAcylated proteins are known to be associated with oncogenesis and tumor progression and, recently elevated O-GlcNAcylation has been described in various cancers (Lynch and Reginato, 2011). The mechanism by which O-GlcNAcylation becomes elevated within cancer cells is not clear but may be caused by elevated levels of OGT seen in some cancers such as breast (Caldwell et al., 2010), prostate (Lynch et al., 2012), and lung (Mi et al., 2011) and/or decrease in OGA levels as seen in breast (Krzeslak et al., 2012), and colon cancers (Yehezkel et al., 2012). Importantly, OGT is required for tumor growth in vitro (Caldwell et al., 2010) and metastasis in vivo (Gu et al., 2010) (Lynch et al., 2012). Moreover, it has been proposed that aberrant O-GlcNAcylation can contribute to metabolic disorders, such as insulin resistance (Yang et al., 2008) suggesting that it could also play a role in the altered metabolism occurring in cancer cells.
Cancer cells can alter metabolism and energy homeostasis by a number of ways. Oncogenes can directly regulate key pathways and enzymes involved in glycolysis. Specifically, the phosphoinositide-3 kinase (PI-3K)/Akt pathway mediating activation of the mTOR pathway has been shown to play a major role in coordinating cell growth and metabolism (Zoncu et al., 2011). Multiple environmental cues including growth factors and nutrients can regulate mTOR signaling including the tumor suppressor LKB1, which activates AMPK. This activation of AMPK leads to inhibition of mTOR activity and loss of mTOR signaling in turn results in a decreased in the translation of critical cell growth and metabolic regulators, including HIF-1α (Zoncu et al., 2011). The transcription factor HIF-1α promotes the transcription of a set of genes that contribute to aerobic glycolysis and the shuttling of carbons from glucose and nitrogen from glutamine into macromolecule synthesis that is typically seen in cancer cells (Shaw and Cantley, 2012).
The levels of HIF-1α protein are controlled by the availability of oxygen and metabolites, such that during normoxic conditions, HIF-1α is hydroxylated by oxygen and α-ketoglutarate-dependent prolyl hydroxylases (PHDs) (Semenza, 2010).. This modification results in HIF-1α proteasome-dependent degradation through hydroxylation-dependent interactions with the E3 ligase von Hippel-Lindau (pVHL). Cancer cells are capable of stabilizing HIF-1α levels independent of oxygen concentrations in response to growth factor stimulation, oncogenic activation and loss of tumor suppressor function, allowing for the transcriptional upregulation of pro-glycolytic factors (Semenza, 2010). In cancer cells, HIF-1α activates a transcriptional program that facilitates the metabolic shift to aerobic glycolysis through the upregulation of several glycolytic proteins, such as glucose transporter GLUT1, hexokinase 2 (HK2), and lactate dehydrogenase A (LDHA) (Iyer et al., 1998) (Semenza, 2010). Moreover, increased HIF-1α expression predicts poor clinical response and clinical outcome in human breast cancer (Generali et al., 2006) and, consistent with this observation, GLUT1 has also been shown to be overexpressed in breast cancer (Brown and Wahl, 1993).
Cell metabolism is tightly linked to cell death pathways through the mitochondria, which plays a key role in both metabolism and apoptosis. Cancer cells are hypersensitive to metabolic stress such as glucose or glutamine deprivation and will undergo apoptosis if nutrients are limiting (El Mjiyad et al., 2011). Inhibition of metabolism in cancer cells can lead to induction of apoptosis by a number of pathways including activation of ER stress apoptotic response (El Mjiyad et al., 2011). A shortage of glucose in cancer cells can induce ER stress pathway, resulting in the PKR-like ER-localized eIF2α kinase (PERK) phosphorylation of eIF2α and the induction of C/EBP homologous protein (CHOP), which results in the induction of Bcl2-family BH3-only proteins including Bim, Puma and Noxa (El Mjiyad et al., 2011).
Here, we present evidence that O-GlcNAcylation within breast cancer cells regulates cancer cell metabolism via regulation of HIF-1α and its downstream target GLUT1. Mechanistically, we show that OGT regulates HIF-1α proteasomal degradation in a manner that is dependent on regulation of α-ketoglutarate, HIF-1α hydroxylation and the tumor suppressor pVHL. Furthermore, decreasing O-GlcNAcylation leads to ER-mediated apoptosis in breast cancer cells.. In addition, we show that in human breast cancers containing high HIF-1α levels also contain elevated OGT and O-GlcNAcylation. Importantly, in overall breast cancer patients lower OGA expression correlates with poor clinical outcome. Thus, O-GlcNAcylation serves as a critical link between the nutrient sensing and metabolic pathways that are critical for cancer cell survival via regulation of HIF-1α hydroxylation.
RESULTS
O-GlcNAcylation Regulates Metabolic Reprogramming and Signaling in Cancer Cells
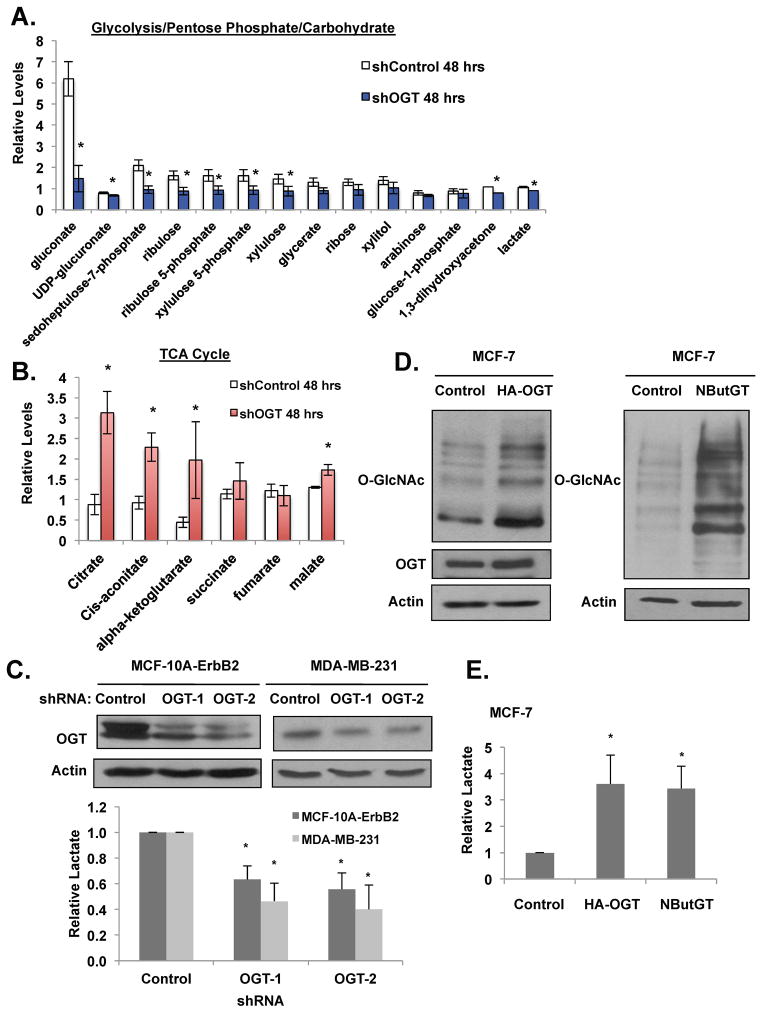
Since OGT and O-GlcNAc has been associated with regulation of metabolic diseases such as insulin resistance (Yang et al., 2008), we hypothesized that OGT could serve as important regulator of glycolytic metabolism to regulate cancer cell growth. To test this idea we initially examined the effect of OGT reduction on metabolites from a triple negative human breast cancer cells MDA-MB-231 using liquid chromatography-mass spectrometry (LC-MS). The metabolic profile of MDA-MB-231 cells containing OGT knockdown with RNAi demonstrated a general decrease in glycolytic and pentose phosphate pathways (PPP) metabolites (Figures 1A) and increase in TCA cycle metabolites (Figure 1B) consistent with a reversal of the Warburg effect and inhibition of cancer cell growth under these conditions that we (Caldwell et al., 2010) and others have previously shown (Itkonen et al., 2013).
Figure 1. OGT regulates glycolytic metabolism in breast cancer cells.
(A) MDA-MB-231 cells expressing control or OGT-1 shRNA were collected and levels of glycolytic, pentose phosphate pathway, and carbohydrate metabolites and (B) TCA cycle intermediates were measured using LC-MS analysis. (C) Cell lysates from MDA-MB-231 and MCF-10A-ErbB2 expressing control, OGT-1 or OGT-2 shRNA were collected for immunoblot analysis with indicated antibodies (top). Lactate levels were measured and normalized to control shRNA treated cells (bottom). (D) Cell lysates from MCF-7 cells stably overexpressing control or HA-OGT (left) or MCF-7 cells were treated with DMSO control or 100 μM NButGT for 48 hours (right) were collected for immunoblot analysis with indicated antibodies. (E) Changes in lactate levels were measured from cells in (D) and normalized to control treated cells. (*p-value <0.05). See also Figure S1, S2.
To confirm that the metabolite patterns reflected decrease in glycolysis, we measured specific glycolytic parameters in MDA-MB-231 cells and in the human immortalized mammary epithelial cells transformed with oncogene ErbB2/HER2 (MCF-10A-ErbB2) stably expressing OGT RNAi. Both transformed and breast cancer cells stably expressing OGT RNAi contained decreased levels of lactate (Figure 1C), glucose flux (Figure S1A), and ATP levels (Figure S1B). Conversely, elevation of total O-GlcNAcylation levels in the breast cancer cells MCF-7 either by overexpression of HA-tagged OGT or treatment with the OGA inhibitor NButGT, which elevates total O-GlcNAcylation (Figures 1D) (Macauley & Vocadlo 2010), caused an increase in glucose flux (Figures S1F) and lactate levels (Figures 1E). In agreement with previous reports, which indicated that targeting OGT has minimal effects on non-transformed epithelial cell types (Caldwell et al., 2010), reducing OGT levels in the human immortalized mammary epithelial cell line MCF-10A resulted in negligible effects on glucose or lactate levels (data not shown). These data suggest that in transformed and cancer cells OGT and O-GlcNAc levels redirect cell metabolism in favor of glycolysis and reducing OGT can reverse the Warburg effect.
Since cellular metabolism can alter specific signaling pathways such as mTOR, we next examined the effects of altered O-GlcNAcylation on the mTOR pathway. Decreased OGT expression in MDA-MB-231 or MCF-10A-ErbB2 cells via stable expression of shRNA decreased the phosphorylation of the mTOR downstream effectors S6 kinase and 4EBP1 at Thr389 and Thr70, respectively (Figure S1C). A number of pathways are known to regulate mTOR activity including the PI3-kinase-AKT pathway. However, since we previously showed that reducing OGT levels in breast cancer cells does not decrease AKT activity nor MEK-Erk pathway (Caldwell et al., 2010), we examined additional regulators of mTOR. Consistent with O-GlcNAcylation regulating energy metabolism, decreasing OGT expression in MCF-10A-ErbB2 or MDA-MB-231 cells via stable expression of shRNA resulted in the activation of the metabolic sensor kinase LKB1, as indicated by its increased phosphorylation at Ser428 (Figures S1C). We also detected an increase in phosphorylation of the downstream effector, AMPK (Figures S1C). Activated AMP kinase is known to decrease phosphorylation of S6 kinase and 4EBP1 at Thr389 and Thr70, respectively (Zoncu et al., 2011). In addition to genetic approaches to repress OGT mRNA expression, we also pharmacologically inhibited the catalytic activity of OGT in MDA-MB-231 cells through use of a known OGT inhibitor, Ac5SGlcNAc (Gloster et al., 2011). Treatment of MDA-MB-231 cells with Ac-5SGlcNAc diminished O-GlcNAc levels and increased LKB1 and AMP kinase activation and also decreased phosphorylation of mTOR effectors (Figures S2A). OGT inhibitor also decreased MDA-MB-231 cell growth in basement membrane 3-dimensional (3D) cultures as compared to vehicle treated cells (Figures S2B) and reduced metabolic outputs, including glucose uptake (Figures S2C). Conversely, elevation of O-GlcNAcylation by either overexpressing OGT (Figures S1D) or treating cells with the OGA inhibitor NButGT (Figures S1E), decreased LKB1 and AMP kinase activation and elevated levels of phosphorylated S6 kinase. Thus, consistent with role of O-GlcNAcylation in regulating glycolysis, changes in O-GlcNAcylation can also regulate metabolic signaling associated with energy status in cancer cells.
O-GlcNAcylation Regulates HIF-1α Proteasomal Degradation in a VHL-dependent Manner
HIF-1α is a critical regulator of aerobic glycolysis in cancer cells (Semenza, 2010). Therefore, we examined HIF-1α expression in the context of altered O-GlcNAcylation. MDA-MB-231 cells stably expressing OGT shRNA contained lower levels of HIF-1α protein at either normoxic or hypoxic (1% O2) conditions (Figures S3A), whereas there was no significant change in HIF-1α RNA levels as measured by QRT-PCR (Figures S3B). Consistent with a mechanism in which OGT mediates regulation of HIF-1α protein stability or activity, decreased expression levels of several known HIF-1 target genes were observed upon OGT knockdown, including adrenomedullin (ADM), BNIP3L, GLUT1, and LDHA (Figures S3B). Likewise, similar reductions in HIF-1α protein levels were observed in MCF-10A-ErbB2 (data not shown) and in MCF-7 cells (Figures S3C) in which OGT expression was reduced. Finally, by using MEFs containing an Ogt exon flanked by the loxP recombination sites (MEFs-OGTF/Y) (O’Donnell et al., 2004), we found that post-Cre recombinase transduction, OGT protein and O-GlcNAc levels were reduced, which inhibited HIF-1α protein expression under hypoxic conditions (Figures S3D). Conversely, we observe stabilization of HIF-1α protein when MCF-7 cells stably express OGT (Figures S1D) or are treated with the OGA inhibitor NButGT (Figures S1E). However, HIF-1α protein does not appear to be directly O-GlcNAc modified as immunoprecipitated HIF-1α protein contained no detectable O-GlcNAcylation when compared to the positive control, the transcription factor Sp1 (Han and Kudlow, 1997) (data not shown). Thus, O-GlcNAcylation likely regulates HIF-1α protein stability via an indirect mechanism.
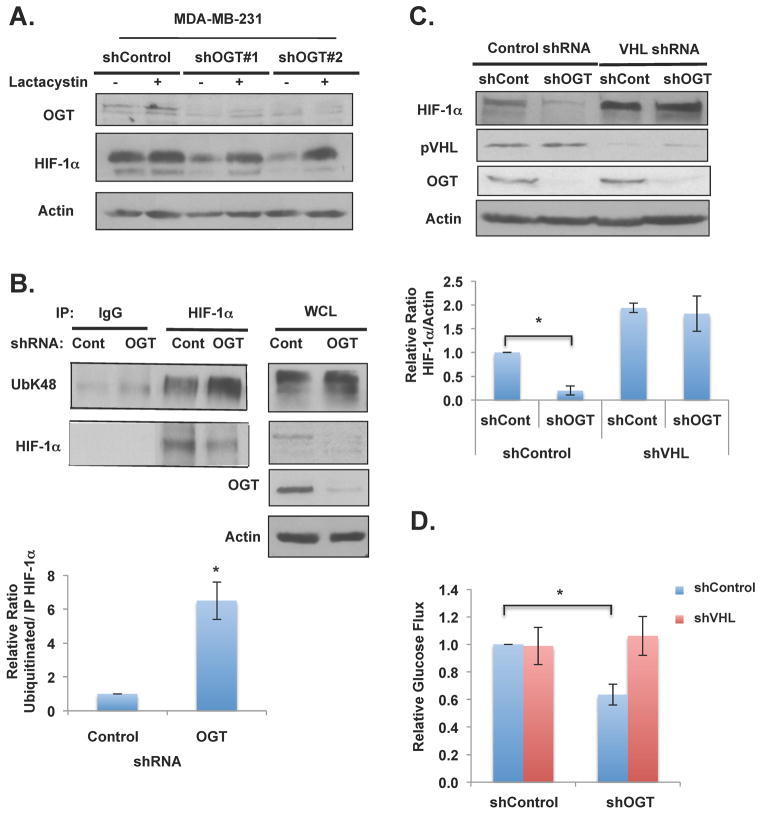
To test whether O-GlcNAcylation regulates HIF-1α degradation, MDA-MB-231 cells stably expressing control shRNA or OGT shRNA were treated with the proteasome inhibitor lactacystin. OGT shRNA-mediated inhibition of HIF-1α protein levels could be reversed by treatment of lactacystin (Figure 2A) suggesting that OGT regulation of HIF-1α was proteasome-dependent. Consistent with idea that OGT regulates HIF-1α via proteasomal pathway, we detected a six-fold increase in HIF-1α ubiquitination under conditions of decreased OGT levels in MDA-MB-231 cells compared to controls (Figures 2B). Since pVHL is well know to regulate HIF-1α degradation pathway we examined whether OGT regulation of HIF-1α was dependent on pVHL-mediated degradation. We compared the effects of OGT knockdown on HIF-1α levels in MDA-MB-231 cells stably expressing pVHL RNAi and as expected, reducing pVHL levels in MDA-MB-231 cells increased the basal expression of HIF-1α protein (Figure 2C). However, HIF-1α levels were not altered in pVHL shRNA treated cells in which OGT expression was also reduced compared to four-fold decrease of HIF-1α levels in OGT knockdown cells (Figure 2C). Consistent with the idea that OGT-mediated regulation of glycolysis is HIF-1α/VHL dependent, we were able to rescue glucose uptake defect in OGT knockdown cells by reducing VHL levels in MDA-MB-231 cells (Figure 2D). In addition, reducing VHL in MDA-MB-231 cells elevated lactate levels compared to controls and abrogated OGT knockdown mediated glycolytic effects (Figure S3E). To ensure this effect was VHL-dependent and occurred in other cell types, we examined kidney epithelial (T132) cells that are wildtype for VHL and renal cell carcinoma (RCC4) cells that are known to be VHL-deficient. While wildtype VHL cells exhibited a twofold decrease in HIF-1α levels in response to OGT depletion, no significant change in HIF-1α expression was observed in RCC4 cells (Figures S3F). In addition, re-expression of VHL in the RCC4 cells, restored OGT knockdown-mediated regulation of HIF-1α (Figure S4A) suggesting that O-GlcNAc regulation of HIF-1α is pVHL-dependent in multiple cell types. We tested whether VHL protein itself may be O-GlcNAcylated by mass spectometry analysis and we did not identify any pVHL ser/thr residues as being O-GlcNAcylated (data not shown). Together, these data suggest that O-GlcNAcylation regulates HIF-1α proteasomal degradation in a pVHL-dependent manner.
Figure 2. O-GlcNAc Regulation of HIF-1α via Protein Degradation Requires VHL.
(A) MDA-MB-231 cells expressing control or OGT shRNA were treated with DMSO control or 10mM lactacystin for 6 hours and lysates collected for immunoblot analysis. (B) Protein lysates from MDA-MB-231 cells expressing control or OGT shRNA were collected and subjected to immunoprecipitation with the indicated antibodies. Data are quantified and presented as an average from three or more independent experiments. (C) MDA-MB-231 cells expressing control or VHL shRNA and 48 hours later infected with control or OGT-2 shRNA. Protein lysates were collected for immunoblot analysis. Data are quantified and presented as an average from three or more independent experiments. (D) Glucose flux from cells described in (C) were measured and normalized to control shRNA treated cells. * p-value<0.05. See also Figure S3, S4.
O-GlcNAcylation Regulates HIF-1α Hydroxylation
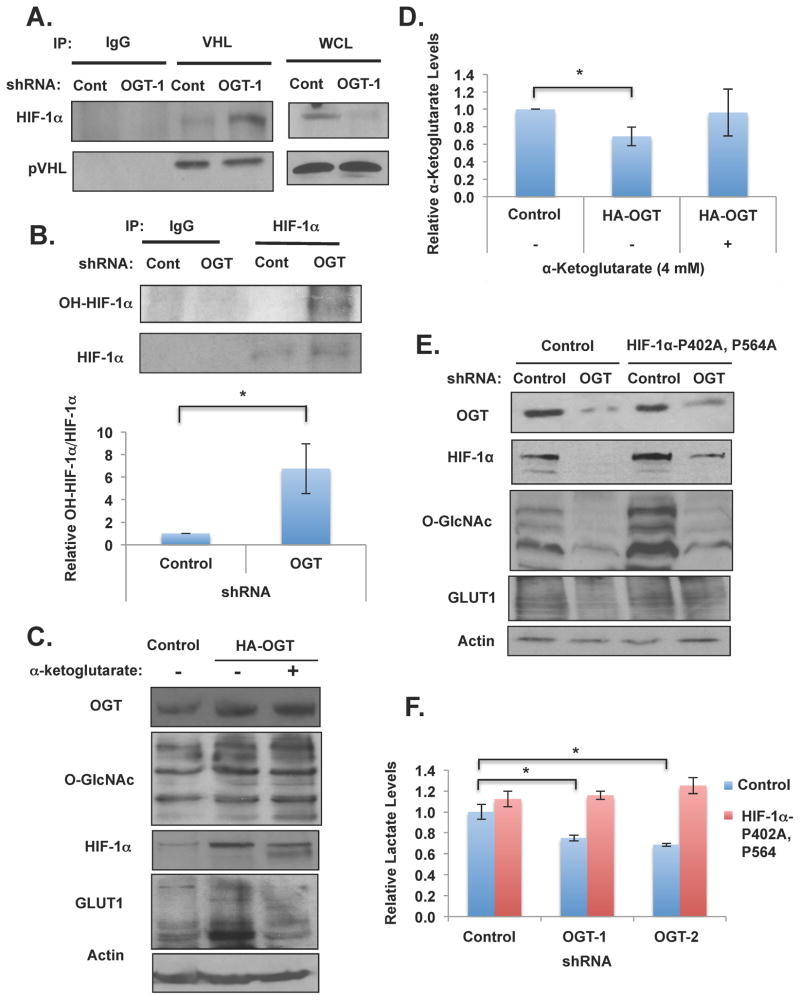
Under aerobic conditions HIF-1α is hydroxylated by specific prolyl hydroxylases (PHDs) at two conserved proline residues (Pro402 and Pro564) in a reaction requiring oxygen, α-ketoglutarate and ascorbate and hydroxylation of HIF-1α facilitates pVHL binding and degradation (Jaakkola et al., 2001). To examine whether OGT regulation of HIF-1α was dependent on PHD activity, we treated MDA-MB-231 cells with dimethyloxaloylglycine (DMOG), a prolyl-4-hydroxylase inhibitor. MDA-MB-231 cells containing OGT RNAi that were treated with DMOG completely rescued the decrease in HIF-1α expression observed in response to OGT RNAi as compared to vehicle treated cells (Figure S4B). Consistent with the observations that pVHL is required for HIF-1α degradation and requires PHD activity under conditions of decreased O-GlcNAcylation, reducing OGT expression in breast cancer cells led to increased interaction between HIF-1α and pVHL as indicated by co-immunoprecipitation experiments (Figures 3A). Conversely, increasing O-GlcNAcylation in cancer cells by treating cells with NButGT reduced the interaction between HIF-1α and pVHL (Figures S4C) consistent with increased HIF-1α protein levels observed under these conditions (Figure S4C). We next examined whether O-GlcNAcylation regulates HIF-1α stability by modulating PHD activity by measuring the extent of HIF-1α hydroxylation. We assessed PHD activity in control and OGT knockdown MDA-MB-231 cells by pretreating with proteasomal inhibitor lactacystin (to prevent hydroxylated HIF-1α from being degraded). Lactacystin treatment of OGT knockdown cells contained similar levels of HIF-1α compared to control cells (Figure 2A), however these cells contained six-fold increase in hydroxylated HIF-1α indicating that PHD activity is increased in OGT knockdown cells (Figure 3B). Since HIF-1α and VHL were not directly O-GlcNAcylated, we examined other mechanisms by which O-GlcNAcylation may regulate HIF-1α hydroxylation. Results from LC-MS analysis showed that α-ketoglutarate, a metabolite required for HIF-1 hydroxylation, was significantly elevated in OGT knockdown MDA-MB-231 cells (Figure 1B) consistent with increased HIF-1α hydroxylation (Figure 3B) and degradation under these conditions (Figure 2B). This data suggests that O-GlcNAcylation may regulate HIF-1α levels via regulation of α-ketoglutarate levels. To test this idea further, we measured α-ketoglutarate levels in MDA-MB-231 cells stably overexpressing HA-OGT which increased total O-GlcNAcylation levels and increased HIF-1α levels compared to control cells (Figure 3C). Overexpression of OGT significantly reduced α-ketoglutarate levels in MDA-MB-231 cells compared to controls (Figure 3D). Importantly, treatment of HA-OGT MDA-MB-231 cells with exogenous α-ketoglutarate to similar levels seen in control cells (Figure 3D) was able to reverse OGT-mediated induction of HIF-1α and HIF1-target GLUT1 (Figure 3C). These results show that O-GlcNAcylation regulation of HIF-1α is PHD-, hydroxylation-dependent and suggests that O-GlcNAcylation regulates HIF-1α protein, in part, via regulation of α-ketoglutarate levels.
Figure 3. O-GlcNAc Regulation of HIF-1α Hydroxylation is Required for OGT-mediated Regulation of Metabolism and Metabolic Signaling.
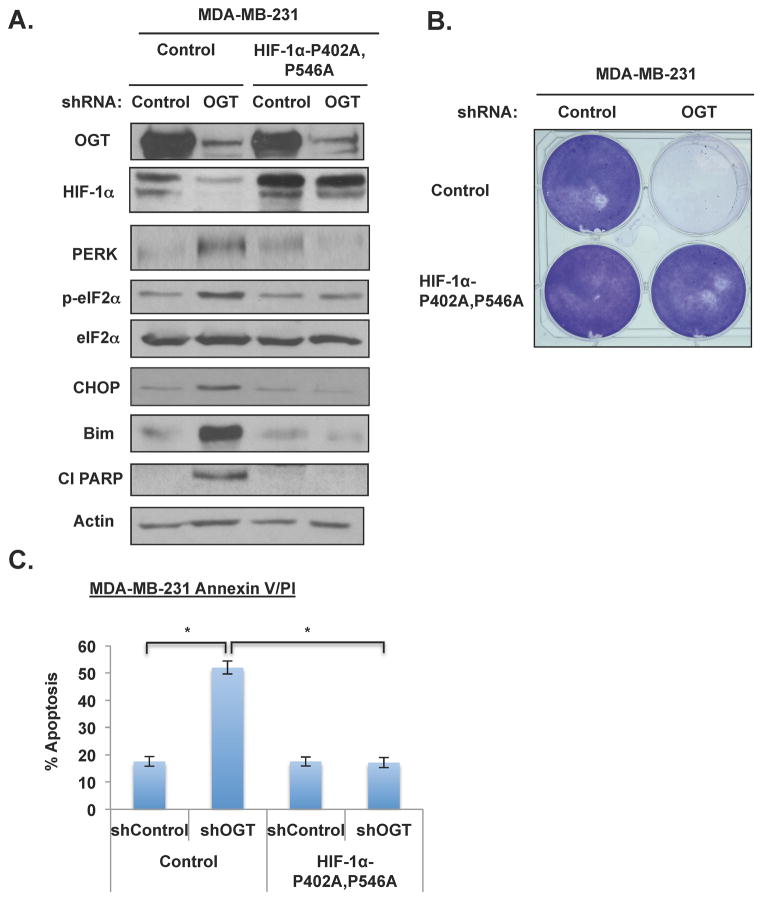
(A) (B) Immunoprecipitation was performed with indicated antibodies from MDA-MB-231 cell lysates expressing control or OGT-1 shRNA. Data are quantified and presented as an average from three or more independent experiments. (C) MDA-MB-231 cells expressing control or HA-OGT plasmids and then treated with control or α-ketoglutarate (4 mM) for 16 hrs and lysates were collected for immunoblot analysis. (D) Cells in (C) were measured for changes in α-ketoglutarate levels. (E) Cell lysates were collected from MDA-MB-231 cells stably expressing control or HIF-1αP402A, P546A and either infected with control or OGT-1 shRNA and expression of proteins was analyzed by immunobloting. (F) Cells expressing control vector or HIF-1α (P402A-P546A) as in (E) were measured for changes in lactate. Levels were normalized to control shRNA treated cells. * p-value<0.05. See also Figure S5.
HIF-1α Hydroxylation is Required for O-GlcNAcylation-mediated Regulation of Metabolic Flux
Given the glycolytic defects observed with decreased O-GlcNAcylation, including decreased glycolytic/PPP metabolites, the activation of LKB1/AMPK and corresponding loss of mTOR signaling, and the reduction of HIF-1α protein, we next sought to characterize the role of HIF-1α in OGT-mediated regulation of cancer metabolism. A constitutively expressed mutant version of HIF-1α was utilized, in which the hydroxylation sites at P402 and P564 are mutated, making it resistant to PHD-mediated hydroxylation and subsequent VHL-dependent ubiquitination (Semenza, 2010). Consistent with our data that O-GlcNAcylation, in part, regulates HIF-1α via hydroxylation and VHL-mediated degradation, stable expression of HIF-1α mutant (P402/P564) restored HIF-1α levels from 72% reduction in OGT knockdown cells to 23% reduction compared to control (Figures 3E and S4D). Importantly, overexpression of HIF-1α stabilized mutant did not alter OGT levels, although mutant HIF-1α slightly increased O-GlcNAcylation in cell expressing OGT RNAi (Figures 3E), most likely by increasing glycolysis and flux through the HBP. However, upon OGT knockdown in MDA-MB-231 cells expressing HIF-1α-P402A, P564A, we did not detect activation of LKB1/AMP kinase phosphorylation and the phosphorylation state of the mTOR effectors S6 kinase and 4EBP1 was not reduced (Figure S5A). These results suggested that expression of HIF-1α hydroxylation mutant reversed OGT-knockdown mediated metabolic stress and signaling. We observed that the glycolytic defect mediated by the expression of OGT shRNA in breast cancer cells was abrogated in cells stably expressing HIF-1α-mutant since lactate levels are restored to near control level in both MDA-MB-231 (Figures 3F) and MCF7 (data not shown) cells. Overexpression of HIF-1α stable mutant in MDA-MB-231 cells lowered glucose flux compared to control cells but nevertheless reducing OGT in these cells had no effect on glucose flux (Figure S4E). These results, along with reducing VHL levels in OGT knockdown cells, further suggested that O-GlcNAcylation-mediated changes in metabolic flux requires regulation of HIF-1α. Consistent with this idea, GLUT1 levels are decreased in MDA-MB-231 cells expressing OGT shRNA as compared to control shRNA cells, yet GLUT1 levels are restored in cells with expression of the stable HIF-1α-P402A, P564A mutant (Figure 3E), consistent with the reversal of glycolytic parameters (Figure 3F). In addition, cells expressing stable HIF-1α also reversed OGT shRNA-mediated induction of α-ketoglutarate levels (Figure S4F) suggesting that HIF-1α can reverse GLUT1 levels, glycolytic parameters and can also regulate key metabolites associated with HIF-1α’s own regulation.
HIF-1α regulates many glycolytic enzymes including phosphofructokinase-1 (PFK1) (Iyer et al., 1998) and PFK2 isoform 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) (Semenza, 2010) both key enzymes regulating glycolytic flux. Reducing OGT expression in three different breast cancer cell lines representative of the major breast cancer subtypes including MDA-MB-231 (triple negative) (Figure S5B), MCF-7, (luminal), & MCF10A-ErbB2 (HER2+) (data not shown) decreased PFK1 and PFKFB3 protein levels. Importantly, both PFK1 and PFKFB3 levels were rescued in OGT knockdown MDA-MB-231 cells overexpressing stable HIF-1α mutant (Figure S5A). OGT regulation of PFK1 occurs at the level of RNA transcription, as OGT knockdown cells contained significant decrease in muscle- and liver-type PFK-1 (PFK-M and PFK-L) isoforms (Figure S5C). Conversely, elevating O-GlcNAcylation in cancer cells causes increased expression of PFK1 in MCF7 cells overexpressing OGT (Figure S1D) or NButGT treated cells (Figures S1E) consistent with increased glycolytic flux (Figure 1E and S1F). These data suggest that OGT regulation of key glycolytic proteins, including GLUT1, PFK1 and PFKFB3, in breast cancer cells is dependent on O-GlcNAc regulation of HIF-1α pathway.
O-GlcNAcylation Selectively Regulates Tumor Cell Survival via an ER Stress CHOP-dependent Mechanism
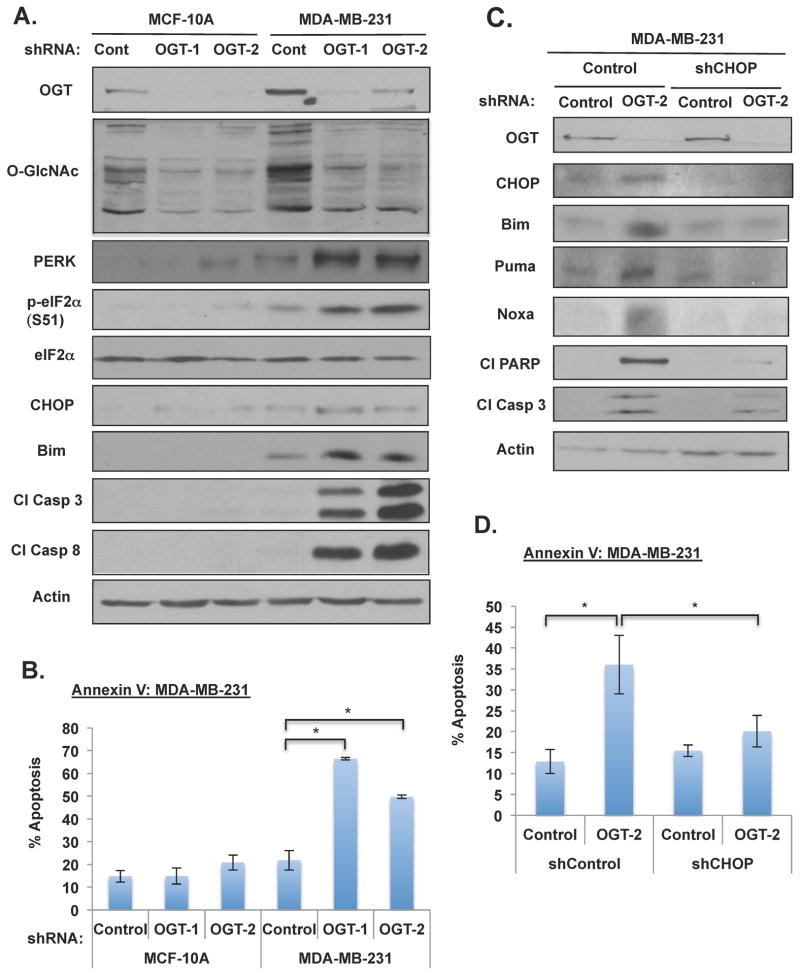
Glucose deprivation and antiglycolytic drugs can selectively induce tumor cell death (El Mjiyad et al., 2011), thus we examined the effect of reducing O-GlcNAcylation on apoptosis in non-transformed immortalized mammary epithelial cells compared to breast cancer cells. In MCF-10A and MDA-MB-231 cells stably expressing control shRNA or OGT shRNA, at day eight post-infection we observed cell rounding and detachment of MDA-MB-231 cells containing OGT knockdown while MCF-10A cells appeared healthy and attached to the plate (Figure S5D). The breast cancer cells, but not immortalized human mammary epithelial cells, that expressed decreased OGT levels had elevated cleaved caspase-3 and caspase-8 (Figure 4A) and up to 50–70% of cells were positive for Annexin-V (Figure 4B), indicating high level of apoptosis that was selective to cancer cells. A similar level of induction in apoptosis was observed in MDA-MB-231 cells in which OGT was knocked down when cells were cultured in 3D conditions (Figure S5E), as well as in MCF7 and MCF10A-ErbB2 cells (Figure S6A). Treatment of MDA-MD-231 cells cultured in 3D with the OGT inhibitor also activated apoptosis to a similar extent observed with genetic deletion (Figure S2D and S2E).
Figure 4. Targeting OGT Selectively Induces ER Stress-mediated Apoptosis in Cancer Cells.
(A) Cell lysates were collected from MCF-10A and MDA-MB-231 cells after 7 days of lentiviral infection with control, OGT-1, or OGT-2 shRNA. Cells were lysed and expression of proteins were analyzed by immunobloting. (B) Cells expressing control, OGT-1 or OGT-2 shRNA were stained with Annexin V/PI and analyzed by flow cytometry. (C) Cell lysates were collected from cells expressing either control or CHOP shRNA after lentiviral infection with control or OGT-2 shRNA. Cells were lysed and proteins were analyzed by immunobloting. (D) Cells as in (C) were collected, stained with Annexin V/PI and analyzed by flow cytometry. * p-value<0.05. See also Figure S2, S5, S6.
Decrease in bioenergetics may impair protein translational modifications in the ER that can trigger the unfolded protein response (UPR) (El Mjiyad et al., 2011). Therefore, we next examined whether reducing O-GlcNAcylation in breast cancer cells led to activation of ER stress response. Consistent with apoptotic effects in cancer cells, we detected activation of ER stress sensor PERK and the phosphorylation of its downstream target eIF2α in MDA-MB-231 breast cancer cells, but not in non-transformed epithelial cells MCF-10A depleted of OGT (Figure 4A). Activation of UPR interfaces with apoptotic pathways by inducing the expression of BCL2 family BH3-only proteins, including Bim in a CHOP-dependent manner (Shore et al., 2011), and Puma and Noxa (El Mjiyad et al., 2011). Reducing OGT expression in MDA-MB-231 cells led to the induction of transcription factor CHOP, Bim, Puma and Noxa (Figure 4A and 4C). Similar activation of ER stress, BH3-only proteins and apoptosis was observed in multiple breast including MDA-MB-231, MCF7 and MCF10A-ErbB2 (Figure S6A) and lung cancer (H2199) cells (Figure S6B) depleted of OGT, or in MDA-MB-231 cells treated with OGT inhibitor (Figure S2E).
To test whether CHOP expression was required for OGT depletion-mediated apoptosis in cancer cells, we reduced CHOP levels via shRNA, which blocked OGT depletion-mediated induction of Bim, Puma, Noxa, as well as reduced caspase-3, PARP cleavage (Figure 4C) and significantly reduced apoptosis (Figure 4D). Thus, reducing O-GlcNAcylation in cancer cells leads to metabolic stress and the induction of ER stress, resulting in apoptosis in a CHOP-dependent manner.
To test whether OGT-depletion-induced ER stress and apoptosis was related to defects in metabolic flux mediated by HIF-1α regulation, we examined apoptosis in MDA-MB-231 cells overexpressing HIF-1α hydroxylation mutant (Figure 5A). Expression of HIF-1α-P402A, P564A in MDA-MB-231 cells rescues the OGT-knockdown mediated growth defect, as indicated by crystal violet staining (Figure 5B). Cells overexpressing HIF-1α mutant were resistant to OGT-depletion mediated ER stress signaling including activation of eIF2α and induction of CHOP, Bim, and PARP cleavage (Figure 5A) and apoptosis (Figure 5C). Thus, O-GlcNAcylation regulation of ER stress-mediated survival pathways acts through the effects of O-GlcNAc on HIF-1α hydroxylation.
Figure 5. Overexpression of Non-degradable HIF-1α Mutant Rescues ER Stress-mediated Apoptosis in Cancer Cells with Reduced OGT In Vitro.
(A) Cell lysates were collected from MDA-MB-231 cells overexpressing control or HIF-1αP402A, P546A and either infected with control or OGT-1 shRNA. Cells were lysed and proteins were analyzed by immunobloting. (B) MDA-MB-231 cells were treated with control or OGT shRNA after stable infection with control or HIF-1αP402A, P546A. Cell colonies were stained with crystal violet. (C) MDA-MB-231 cells as in (B) were analyzed for apoptosis after lentiviral infection with control or OGT shRNA. Cells were stained with Annexin V/PI and analyzed by flow cytometry. * p-value<0.05.
O-GlcNAcylation Regulates Tumor Cell Metabolism and Stress Survival via GLUT1 In Vitro and In Vivo
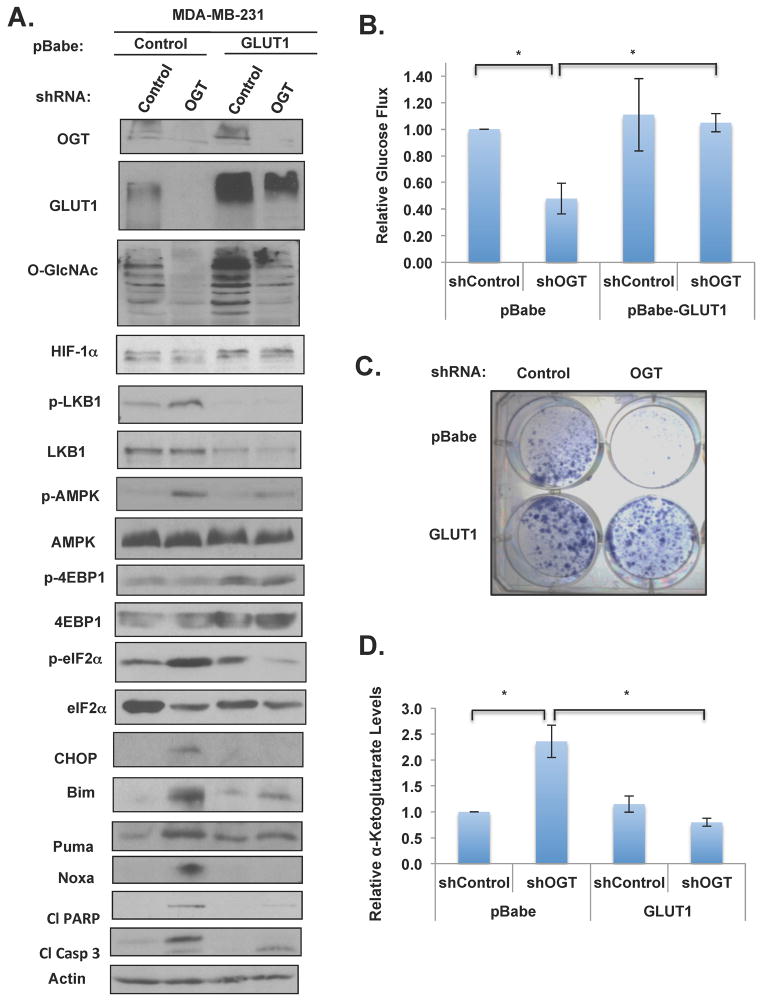
Consistent with OGT knockdown-mediated decrease in HIF-1α levels, we also detected a decrease in both GLUT1 RNA (Figure S3B) and protein levels (Figure 3E). To test whether the bioenergetics and survival stress effects we observed following the OGT-mediated decrease in HIF-1α was due to glucose flux, we tested whether overexpression of GLUT1 would rescue the metabolic and apoptotic defects observed in OGT-knockdown cells. Ectopic expression of GLUT1 in MDA-MB-231 cells elevated O-GlcNAcylation in control cells, but only slightly in OGT knockdown cells (Figure 6A). Nevertheless, cells overexpressing GLUT1 were able to rescue all of the metabolic and ER stress signaling effects that result from decreasing OGT expression that we have described herein, including effects on the LKB1/AMPK/mTOR pathway effects and the induction of eIF2α/CHOP/Bim pathway (Figure 6A), glucose uptake (Figure 6B), growth (Figure 6C) and apoptosis (Figure S6C). Importantly, overexpression of GLUT1 alone in MDA-MB-231 cells was able to reverse HIF-1α and PFK1 levels in an O-GlcNAcylation-independent manner, as overexpression of GLUT1 in control RNAi cells elevated basal levels of HIF-1α and PFK-1 and reducing OGT levels in GLUT1 overexpressing cells had no effect on HIF-1α and PFK-1 levels (Figure 6A). Consistent with the idea that O-GlcNAcylation regulates HIF-1α protein via regulation of α-ketoglutarate levels, overexpression of GLUT1 in MDA-MB-231 cells was able to reverse OGT shRNA-mediated induction of α-ketoglutarate levels (Figure 6D).
Figure 6. Overexpression of HIF-1α Transcriptional Target, GLUT1 is Sufficient to Rescue Stress Signaling, Glucose Uptake, α-ketoglutarate levels and ER Stress-induced Apoptosis in Breast Cancer Cells Caused by Targeting OGT.
(A) Cell lysates were collected from cells expressing either control or GLUT1 and either infected with control or OGT shRNA. Cells were lysed and proteins were analyzed by immunobloting. (B) Glucose uptake levels in cells as in (A) were measured and normalized to control shRNA treated cells. (C) MDA-MB-231 cells were infected with control or OGT shRNA after stable expression of control or GLUT1. Cell colonies were stained with crystal violet. (D) α-ketoglutarate levels were measured from MDA-MB-231 cells stably overexpressing either control or GLUT1 and either infected with control or OGT shRNA. Levels were normalized to control shRNA treated cells. * p-value<0.05. See also Figure S6.
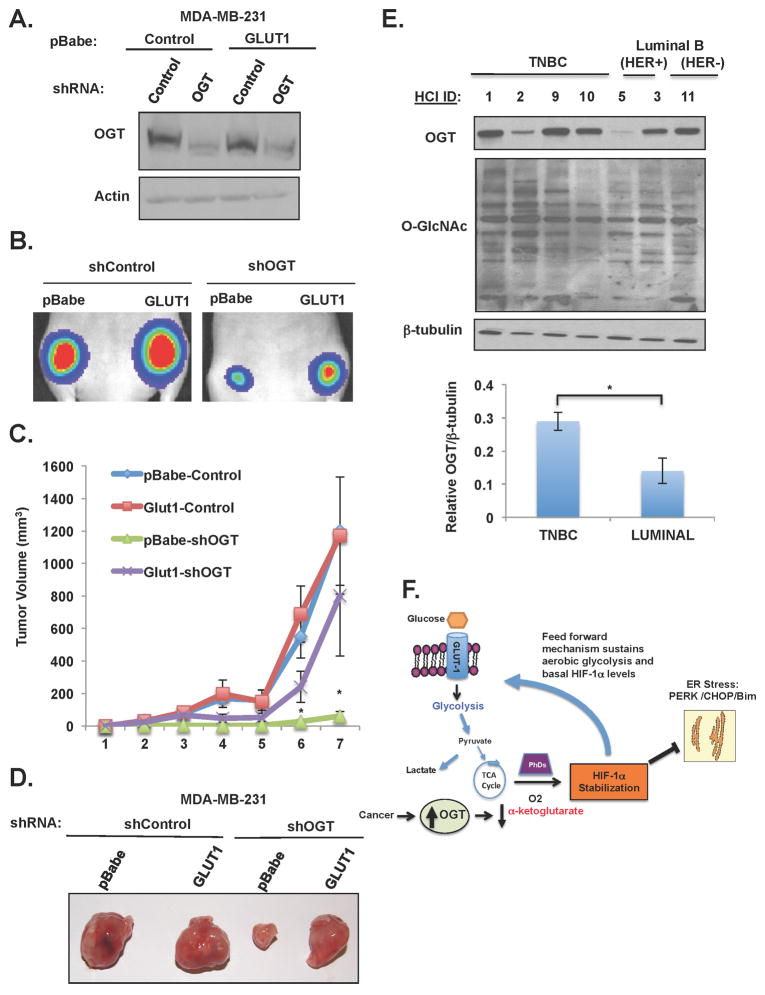
To test whether OGT regulation of GLUT1 was required for tumor growth in vivo, MDA-MB-231 cells stably expressing luciferase and either control shRNA or OGT shRNA in presence or absence of GLUT1 overexpression (Figure 7A) were injected orthotopically into the mammary fat pads of Nude (Nu/Nu) mice. Tumor growth was visualized over time utilizing an IVIS Spectrum bio-imaging system. As previously reported (Caldwell et al., 2010), reducing OGT in MDA-MB-231 cells causes decreased tumor growth in vivo. However, cells overexpressing GLUT1 were able to partially rescue the effect of OGT depletion in cancer cells in vivo (Figure 7B), since GLUT1 overexpression in the context of OGT depletion increased tumor volume (Figure 7C,D) and tumor weight (Figure S6D) compared to control OGT-depleted cells. Thus, OGT regulation of tumor growth in vivo is partly dependent on GLUT1 expression.
Figure 7. Overexpressing GLUT1 Partially Rescues Tumor Regression Induced by Targeting OGT In Vivo.
(A) Cell lysates from indicated groups were collected before injected into mice and immunoblotted with the indicated antibodies. (B) Representative bioluminescent images from week 3 post-injection. (C) Mean tumor volume (mm3) of MDA-MB-231 cells with indicated treatment (n=13/group) shown at indicated week. (D) Representative tumors in mice 8 weeks after injection. (E) OGT/O-GlcNAc Levels in TNBC lysates. Cell lysates prepared from PDX tumor tissues representing various breast cancer subtypes and profiled for OGT and O-GlcNAc expression by immunoblotting; β-tubulin is included as a loading control. *p ≤0.05. (F) A schematic illustration of proposed model depicting OGT regulation of HIF-1α stabilization via regulation of α-ketoglutarate that promotes a feed forward mechanism increasing glycolytic flux and blocking ER stress-mediated apoptosis in cancer cells. See also Figure S7.
O-GlcNAc Cycling Enzyme Expression in Human Breast Cancer Patients
The over-expression of HIF-1α in human solid cancer biopsies, including breast cancers, correlates with metastatic potential and decreased survival (Generali et al., 2006). Since we have previously shown that breast cancer cells express elevated OGT levels, we examined whether conditions that would be expected to elevate O-GlcNAcylation, such as an increase in OGT or a decrease in OGA/MGEA5 expression, would be associated with breast cancer subtype or with poor survival in breast cancer patients. Data mining from the Cancer Genome Atlas Research Network (TCGA) repository of a total of 804 breast cancer patients (Network, 2012) did not reveal a significant difference in OGT expression among subtypes, except for a small, but significant increase in basal like tumors relative to HER2-enriched tumors (Figure S7A). In contrast, OGA/MGEA5 expression was highest in luminal A and B tumors, followed by HER2-enriched tumors, and lowest in basal-like and normal-like breast tumors and most comparison among subtypes were statistically significant (Figure S7B). We also mined the NKI van’t Veer dataset (van ‘t Veer et al., 2002) to determine if expression of either OGT or OGA/MGEA5 correlates with survival from breast cancer. Although no significant correlation between OGT expression and survival was observed (Figure S7C), relatively low-level expression of OGA/MGEA5 was independently correlated with both decreased metastasis-free (Figure S7D) and overall survival (Figure S7E) in breast cancer patients.
Data from the TCGA dataset previously identified the HIF-1α network hub to be hyperactivated in basal/triple-negative breast cancers (TNBC) tumors as compared to luminal cancers (Network, 2012). Thus, we next examined levels of OGT, and O-GlcNAc in patient-derived breast cancer mouse xenograft (PDX) samples. These xenograft tumors were shown to replicate the original tumor pathology and clinical metastasis patterns when xenografted and serially-passaged in recipient mice (DeRose et al., 2011). Consistent with the idea that TNBCs have an enriched HIF-1-dependent gene network, we recently reported that these TNBCs xenograft tumors contain high levels of HIF-1α protein (Regan Anderson et al., 2013) compared to other subtypes represented in the HCI tumor bank. Consistent with idea that increased O-GlcANcylation/OGT are associated with HIF-1α levels, we find that TNBC lysates contain elevated total O-GlcNAcylation and significant increase of OGT levels compared to luminal breast cancers (Figure 7E). Thus, clinical samples are consistent with idea that cancer cells alter expression of O-GlcNAc cycling enzymes that lead to elevation of O-GlcNAcylation and is significantly associated with human mortality in breast cancers. These data also support the idea that OGT may serve as a novel therapeutic target in HIF-1 overexpressing breast cancers, including TNBCs.
DISCUSSION
Taken together, the data presented here demonstrate for the first time that the hexosamine biosynthetic pathway, via O-GlcNAcylation, regulates metabolic reprogramming in cancer cells via regulation of HIF-1α hydroxylation and directly links key nutrient sensing pathways, including the LKB1/AMP kinase and mTOR. The upregulation of OGT and elevated O- GlcNAcylation is documented in several cancer types (Caldwell et al., 2010) (Mi et al., 2011) (Lynch et al., 2012) and our data strongly suggest a model (Figure 7F) in which OGT and O- GlcNAcylation play an important role in contributing to the Warburg effect by reducing α-ketoglutarate levels thereby increasing HIF-1α stabilization and enhancing aerobic glycolysis in a feed forward mechanism and contributing to the glycolytic phenotype consistently observed in rapidly dividing tumors (DeBerardinis et al., 2008). Elevated O-GlcNAcylation helps to maintain aerobic glycolysis and cancer HIF-1α levels that protect from ER stress-mediated apoptosis. Using a combination of genetic and chemical inhibitor approaches, we show that O- GlcNAcylation regulates HIF-1α protein degradation in a α-ketoglutarate-, PHD- and VHL- dependent manner since decreasing O-GlcNAcylation in cancer cells increases α-ketoglutarate, HIF-1α hydroxylation, increases HIF-1α’s association with VHL, leading to enhanced proteasomal degradation. The loss of HIF-1α expression achieved by reducing O-GlcNAcylation leads to a decrease in expression of multiple HIF-1 transcriptional targets that directly regulate glucose metabolism including GLUT1, LDHA, PFKFB3 and PFK-1, resulting in decreased glycolysis and the induction of ER stress. Additionally, we find that induction of the ER stress- regulated protein CHOP is required for this O-GlcNAcylation-dependent survival pathway via regulation of the BH3-only proteins. Finally, and more importantly in terms of clinical utility, we show that O-GlcNAcylation depletion in immortalized mammary epithelial cells does not activate this stress pathway, suggesting that targeting OGT in cancer cells may selectively induce metabolic stress and apoptosis.
The hexosamine biosynthetic pathway is emerging as a critical metabolic pathway in cancer. Recently, other groups have shown that pancreatic tumors derived from inducible expression of KrasG12DA in mice contain elevated flux through hexosamine pathway and have increased O-GlcNAcylation. Moreover, reduction of KrasG12DA expression in these tumors leads to reduction of glucosamine-6-phosphate (GlcN-6P) levels, the product of committed state allowing entry into HBP, and downregulation of RNA and protein levels of rate-limiting enzyme glutamine fructose-6-phosphate amidotransferase 1 (Gfpt1). Importantly, RNAi knockdown of Gfpt1 reduced overall O-GlcNAcylation and blocked KrasG12DA-mediated tumor growth in vitro and in vivo (Ying et al., 2012). Another recent study showed that hypoxic pancreatic cancer cells compared to normoxic cells, contain elevated glycolytic markers and increased gene expression of key HBP genes including Gfpt2 as well as elevated O-GlcNAcylated protein levels (Guillaumond et al., 2013). Consistent with our results showing that O-GlcNAcylation regulates HIF-1α levels and function, this study also showed that inhibition of O-GlcNAcylation blocked cell survival of hypoxic pancreatic cancer cells. In addition to being required for O-GlcNAcylation of nuclear and cytoplasmic proteins, the HBP is also required for initiation of N-glycosylation in the ER and N-glycan branching in the Golgi apparatus (Dennis et al., 2009). Although these studies implicate HBP in cancer regulation none distinguish between requirements of different branches of HBP pathway. Our results provide direct evidence that the HBP via O-GlcNAcylation can directly link to glucose uptake and glycolysis in cancer cells via regulation of HIF-1α pathway that is required for cancer growth in vitro and in vivo.
Recent data has shown that phosphofructokinase 1 (PFK1), a key enzyme of glycolytic flux that can be activated by fructose-2,6-bisphosphate (F2,6BP), is O-GlcNAcylated and that this modification is responsible for its inactivation that redirects glucose flux through pathways critically involved in tumor growth such as the oxidative pentose phosphate pathway (PPP) (Yi et al., 2012). Consistent with this data, metabolite analysis of cancer cells with reduced O-GlcNAcylation had significant effect on flux thru PPP as a number of PPP metabolites including ribulose-5-phosphate, xylulose-5-phosphate and sedoheptulose-7-phosphate were decreased under these conditions. It is possible that decreasing PFK1 O-GlcNAcylation reduced flux through PPP. However, key enzymes of the PPP such as glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase are known to be regulated by HIF-1α (Guo et al., 2009). Moreover, our results show that PFK-1 protein and RNA levels (including both PFK-L and PFK-M isoforms) are very sensitive to alterations in O-GlcNAcylation in breast cancer cells. Consistent with role of O-GlcNAcylation enhancing glycolysis, under conditions of elevated O-GlcNAcylation, we detect increased levels of PFK-1. Interestingly, decreasing O-GlcNAc in cancer cells reduced protein levels of PFK-1, which could be restored in cells that expressed a hydroxylation HIF-1α mutant, which is consistent with previous report that PFK-1 is transcriptionally regulated by HIF-1α (Iyer et al., 1998). Thus, we propose that O-GlcNAcylation-mediated regulation of HIF-1α may be a dominant pathway regulating cancer bioenergetics since HIF-1α directly transcriptionally regulates nearly all glycolytic enzymes (Semenza, 2010). Thus, cancer cells may co-opt the nutrient sensor O-GlcNAcylation pathway that can contribute to cancer metabolism pathways by regulating key metabolic factors such as HIF-1α and c-Myc (Itkonen et al., 2013) which are necessary to maintain glycolytic flux, rapid cell growth and survival.
In summary, we have described a novel role for O-GlcNAcylation in driving breast cancer glycolysis, and have identified that these changes are mediated through a HIF-1/GLUT1-dependent mechanism. As global elevations in O-GlcNAcylation are recognized as a common feature of cancer cells, OGT is positioned as a novel therapeutic target with potent anti-glycolytic activity. In addition, since both HIF-1α and OGT are elevated in chemoresistant basal-like/triple-negative breast cancers, our data suggests that targeting OGT in these tumors may have potential therapeutic benefit.
EXPERIMENTAL PROCEDURES
Detailed Experimental Procedures are available online in the Supplemental Experimental Procedures including materials, mouse xenografts, metabolic assays, viral transductions, immunoprecipitations, 3D culture, qRT-PCR, immunoblotting, metabolic and bioinformatics analysis.
Cell Culture
MCF-10A-ErbB2 cells were cultured in DMEM/F12 supplemented with 5% horse serum, 100 μg/ml EGF, 1 mg/ml hydrocortisone, 1 mg/ml cholera toxin, and 10 mg/ml insulin. MDA-MB-231 and HEK-293T cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. MCF-7 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, 1% penicillin/streptomycin and 10 mg/ml insulin. T132 (VHL +/+) and RCC4 (VHL −/−) cells (kindly provided by C. Simon, University of Pennsylvania) were cultured in DMEM supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% penicillin/streptomycin. MEFs-OGTF/Y cells (kindly provided by N. Zachara, John Hopkins University) were cultured as previously described (O’Donnell et al., 2004).
Statistics
All results shown as averages are presented as mean + SE from three or more independent experiments. Unless otherwise noted, p-values were calculated using a Student’s two-tailed test: (*p-value <0.05).
Supplementary Material
HIGHLIGHTS.
Reducing OGT/O-GlcNAcylation in cancer cells decreases cancer glycolysis
OGT regulates stability of HIF-1α via regulation of α-ketoglutarate levels
Reducing OGT levels or activity induces ER-stress and apoptosis in cancer cells
Stable HIF-1α mutant or GLUT1 overexpression rescues OGT-mediated phenotypes
Acknowledgments
We thank G. Jones, S. R. Jackson, and K. Shahriari for technical support. We thank Mignon Keaton for help in analyzing LC-MS metabolite data. This work is supported by NCI grants CA183574 (to C.M.F.), CA138488 (to T.N.S.) and CA155413 (to M.J.R.). D.J.V. thanks the Natural Sciences and Engineering Research Council of Canada for support through an E.W.R Steacie Memorial Fellowship and Dr. Lehua Deng for the synthesis of compounds.
Footnotes
Supplemental information includes seven figures and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brown RS, Wahl RL. Overexpression of Glut-1 glucose transporter in human breast cancer. An immunohistochemical study. Cancer. 1993;72:2979–2985. doi: 10.1002/1097-0142(19931115)72:10<2979::aid-cncr2820721020>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, Vosseller K, Reginato MJ. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends Biochem Sci. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, Factor R, Matsen C, Milash BA, Nelson E, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mjiyad N, Caro-Maldonado A, Ramirez-Peinado S, Munoz-Pinedo C. Sugar-free approaches to cancer cell killing. Oncogene. 2011;30:253–264. doi: 10.1038/onc.2010.466. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, Bersiga A, Allevi G, Milani M, Aguggini S, et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006;12:4562–4568. doi: 10.1158/1078-0432.CCR-05-2690. [DOI] [PubMed] [Google Scholar]
- Gloster TM, Zandberg WF, Heinonen JE, Shen DL, Deng L, Vocadlo DJ. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7:174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C, Yang J, Han F, Lu X, Yu W. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70:6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- Guillaumond F, Leca J, Olivares O, Lavaut MN, Vidal N, Berthezene P, Dusetti NJ, Loncle C, Calvo E, Turrini O, et al. Strengthened glycolysis under hypoxia supports tumor symbiosis and hexosamine biosynthesis in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2013;110:3919–3924. doi: 10.1073/pnas.1219555110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Miyake M, Liu KJ, Shi H. Specific inhibition of hypoxia inducible factor 1 exaggerates cell injury induced by in vitro ischemia through deteriorating cellular redox environment. J Neurochem. 2009;108:1309–1321. doi: 10.1111/j.1471-4159.2009.05877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I, Kudlow JE. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Molecular and cellular biology. 1997;17:2550–2558. doi: 10.1128/mcb.17.5.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkonen HM, Minner S, Guldvik IJ, Sandmann MJ, Tsourlakis MC, Berge V, Svindland A, Schlomm T, Mills IG. O-GlcNAc Transferase Integrates Metabolic Pathways to Regulate the Stability of c-MYC in Human Prostate Cancer Cells. Cancer Res. 2013;73:5277–5287. doi: 10.1158/0008-5472.CAN-13-0549. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Krzeslak A, Forma E, Bernaciak M, Romanowicz H, Brys M. Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med. 2012;12:61–65. doi: 10.1007/s10238-011-0138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TP, Ferrer CM, Jackson SR, Shahriari KS, Vosseller K, Reginato MJ. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287:11070–11081. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TP, Reginato MJ. O-GlcNAc transferase: a sweet new cancer target. Cell Cycle. 2011;10:1712–1713. doi: 10.4161/cc.10.11.15561. [DOI] [PubMed] [Google Scholar]
- Marshall S. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci STKE. 2006:re7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, Cong Q, Yu W. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011;1812:514–519. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Molecular and cellular biology. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan Anderson TM, Peacock DL, Daniel AR, Hubbard GK, Lofgren KA, Girard BJ, Schorg A, Hoogewijs D, Wenger RH, Seagroves TN, et al. Breast Tumor Kinase (Brk/PTK6) Is a Mediator of Hypoxia-Associated Breast Cancer Progression. Cancer Res. 2013;73:5810–5820. doi: 10.1158/0008-5472.CAN-13-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Current opinion in genetics & development. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Cantley LC. Decoding key nodes in the metabolism of cancer cells: sugar & spice and all things nice. F1000 Biol Rep. 2012;4:2. doi: 10.3410/B4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, et al. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- Yehezkel G, Cohen L, Kliger A, Manor E, Khalaila I. O-linked beta-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-beta-D-glucosaminidase silencing on cell phenotype and transcriptome. J Biol Chem. 2012;287:28755–28769. doi: 10.1074/jbc.M112.345546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, 3rd, Peters EC, Driggers EM, Hsieh-Wilson LC. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 2012;337:975–980. doi: 10.1126/science.1222278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.