Abstract
Arthropoda is the largest of all animal phyla and includes about 90% of extant species. Our knowledge about regulation of apoptosis in this phylum is largely based on findings for the fruit fly Drosophila melanogaster. Recent work with crustaceans shows that apoptotic proteins, and presumably mechanisms of cell death regulation, are more diverse in arthropods than appreciated based solely on the excellent work with fruit flies. Crustacean homologs exist for many major proteins in the apoptotic networks of mammals and D. melanogaster, but integration of these proteins into the physiology and pathophysiology of crustaceans is far from complete. Whether apoptosis in crustaceans is mainly transcriptionally regulated as in D. melanogaster (e.g., RHG ‘killer’ proteins), or rather is controlled by pro- and anti-apoptotic Bcl-2 family proteins as in vertebrates needs to be clarified. Some phenomena like the calcium-induced opening of the mitochondrial permeability transition pore (MPTP) are apparently lacking in crustaceans and may represent a vertebrate invention. We speculate that differences in regulation of the intrinsic pathway of crustacean apoptosis might represent a prerequisite for some species to survive harsh environmental insults. Pro-apoptotic stimuli described for crustaceans include UV radiation, environmental toxins, and a diatom-produced chemical that promotes apoptosis in offspring of a copepod. Mechanisms that serve to depress apoptosis include the inhibition of caspase activity by high potassium in energetically healthy cells, alterations in nucleotide abundance during energy-limited states like diapause and anoxia, resistance to opening of the calcium-induced MPTP, and viral accommodation during persistent viral infection. Characterization of the players, pathways, and their significance in the core machinery of crustacean apoptosis is revealing new insights for the field of cell death.
Keywords: Metabolic depression, Diapause, Anoxia, Artemia franciscana, Caspase, Bcl-2 family proteins, Apaf-1, Apip, IAP, Radiation, Toxins, Viral infection, EST, Phylogeny
Introduction
Metazoan life forms require mechanisms that ensure the safe and economical disposal of superfluous and potentially harmful cells. Apoptosis is a carefully regulated mechanism of cellular suicide that does not result in loss of plasma membrane integrity or in an inflammatory response. The proper execution of apoptosis is essential for successful organogenesis during embryonic development and for maintenance of tissue homeostasis in adults [1]. Although apoptotic pathways have been well studied in Drosophila melanogaster, insects are only one clade within the subphylum Hexapoda, among three other extant subphyla (Crustacea, Chelicerata, and Myriapoda) in the enormous phylum Arthropoda. Arthropoda includes about 90% of all animal life and is the most diverse group of animals on earth with more than one million described species [2]. To generalize findings from D. melanogaster to all arthropods is prone to erroneous conclusions. The picture that is emerging from the comparison of apoptotic proteins from vertebrates with those found in Cnidaria, Echinodermata, Cephalochordata, Urochordata, and Ecdysozoa, is one of massive lineage-specific expansions and losses of apoptotic genes [3–6]. Regulation of apoptosis may well be more diverse than previously anticipated, even between more closely related groups such as the major arthropod subphyla.
Extrinsic and intrinsic pathways
Two initiation pathways are triggered by distinct events to promote apoptosis in mammals (e.g., [7]). The extrinsic pathway is activated through ligation of death receptors located in the plasma membrane that transmits the death signal into the cell’s interior [8]. This pathway frequently serves as a mechanism to eliminate cells during development, differentiation and tissue remodeling. The intrinsic pathway, which involves mitochondrial signaling, occurs as a response to moderate perturbation of intracellular homeostasis by various cellular stresses (e.g., hypoxia/anoxia, viral or bacterial proteins, reactive oxygen species (ROS), xenobiotics, radiation, and accumulation of mis-folded proteins) [9]. The mitochondrion is an important integrator for the two pathways (although not essential for all modes of extrinsic cell death), and plays a key role in the amplification of the death signal [7].
Extrinsic and intrinsic pathways promote the activation of a set of highly specific cellular proteases that is a hallmark of apoptosis. Caspases are cysteine proteases that cleave substrates after aspartic acid residues, and they are primarily involved in the execution of the cellular demise [10]. Caspases are synthesized as zymogens and can be either activated through proteolytic cleavage by an upstream caspase or by oligomerization at an adapter platform such as the apoptotic peptidase activating factor 1 (APAF-1) in mammals or Dark (Drosophila Apaf-1 related killer) in flies. Caspase activation can also be mediated by interaction between a family member of the tumor necrosis factor receptors (TNFR) and its respective ligand (e.g. TNF-α), as seen during initiation via the extrinsic pathway. This ligand binding promotes the oligomerization of caspases at the intracellular domain of the membrane-spanning TNFR [11]. Independent of the activation mechanism, caspases are strongly inhibited by the inhibitor of apoptosis proteins (IAPs) [12], which are the only family of proteins known to directly regulate the activity of both initiator (caspase 9) and effector (caspase 3, caspase 7) caspases [13].
It is a common convention to classify mammalian cells as Type I or Type II [14]. For Type I cells, receptor-mediated caspase activation via the extrinsic pathway is sufficient to execute the apoptotic program. For Type II cells, receptor-mediated caspase activation is not sufficient to induce cell death. To execute the apoptotic program in these latter cells, the death receptor signal must be amplified by the release of pro-apoptotic proteins from the intermembrane space of mitochondria into the cytoplasm [14]. The use of a highly expanded family of death receptors (e.g., TNFr1, Fas, DR4, etc.) to initiate apoptosis through the extrinsic pathway is likely a vertebrate invention [5], and until now, only one TNFR/TNF-α-like system has been described in any protostome to our knowledge. In D. melanogaster the Type I membrane protein Wengen contains a conserved TNFR/NGFR (nerve growth factor receptor) cysteine-rich region. Overexpression of the TNF homolog Eiger induces Wengen dependent cell death, which signals through the c-Jun N-terminal kinases (JNK) pathway and not by direct activation of the D. melanogastor caspase 8 homolog Dredd [15–17]. There is no experimental evidence for the existence of an extrinsic apoptotic pathway in crustaceans. However, we will discuss some possible scenarios based on the findings in D. melanogaster and putative protein homologs that we have found in expressed sequence tag (EST) libraries of crustaceans in the database of the National Center for Biotechnology Information (NCBI).
Upon induction of apoptosis via the intrinsic pathway in mammals, the permeability of the outer mitochondrial membrane (OMM) is always increased. OMM permeability can be controlled by pro- and anti-apoptotic proteins of the B-cell lymphoma 2 (Bcl-2) family of proteins [18]. In addition, opening of the mitochondrial permeability transition pore can lead to mitochondrial swelling and rupture of the OMM. With either mechanism, the permeabilized OMM facilitates the release of several pro-apoptotic proteins: cytochrome c (cyt-c), apoptosis inducing factor (AIF), endonuclease G (EndoG), second mitochondria-derived activator of caspases/direct IAP binding protein with low pI (SMAC/DIABLO), and high temperature requirement protein A2 (Omi/HtrA2). These proteins promote apoptosis by different mechanisms such as inhibition of IAPs, activation of APAF-1, and caspase-independent nuclear DNA condensation and degradation [19]. Some information is available on the intrinsic pathway in crustaceans [20, 21], and a few downstream apoptotic key players such as caspases and inhibitors of apoptosis proteins (IAPs) have been recently cloned and characterized [22–25]. The role that mitochondria play in the integration and amplification of extrinsic and intrinsic pathways in crustaceans is not clear. Nevertheless, we are starting to appreciate a role of mitochondria in apoptosis in a variety of invertebrate species including insects [7, 26–28]. Whether apoptosis in crustaceans is similar to the situation in D. melanogaster, i.e., largely regulated by transcription of a specific set of “killer” proteins, or is under more complex regulation with Bcl-2 family proteins as in vertebrates [29] needs to be investigated. As with putative proteins for the extrinsic pathway above, we will discuss the possible involvement of Bcl-2-like proteins in crustacean apoptosis based on the findings in D. melanogaster and putative protein homologs that we have found in EST libraries of crustaceans. Finally in this commentary/review, we will illustrate environmental situations when apoptosis is readily triggered and others when it is suppressed and evaluate the molecular mechanisms operative in these situations.
Molecular players for apoptosis in crustaceans
In order to identify proteins with potential apoptotic functions in crustaceans, a broad range of protein sequences with well known apoptotic functions that are listed in the protein database of NCBI for D. melanogaster and Homo sapiens were blasted against the NCBI database for non-redundant protein sequences (blast-p) and non-human, non-mouse expressed sequence tags (dbEST; tblast-n). Matches obtained for crustacean ESTs were translated into the corresponding protein sequence and again blasted against the NCBI protein database for D. melanogaster and H. sapiens (blast-p). All groups of proteins for which crustacean homologues were identified are highlighted by red print and boxed in Fig. 1. The probabilities that the matches between the crustacean sequences and the proteins from D. melanogaster and H. sapiens occurred purely by random chance are given as e-values in Table 1.
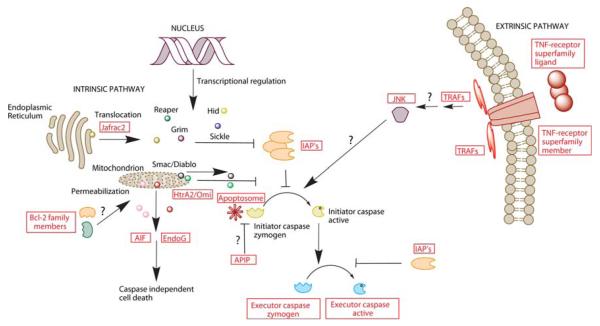
Fig. 1.
Overview of proteins that are possibly conserved in the apoptotic machineries of arthropods. Potential apoptotic players in crustaceans were identified by blasting a wide range of protein sequences with well known apoptotic functions found in the protein database of NCBI for D. melanogaster and H. sapiens against the NCBI database for non-redundant protein sequences (blast-p) and non-human, non-mouse EST’s (tblast-n). All proteins for which homologues in crustaceans were identified are printed and boxed in red. Crustacean homologues could be found for all proteins investigated with the exceptions of Smac/Diablo and the D. melanogaster RHG-proteins. The questions marks indicate apoptotic mechanisms for which data are completely lacking in crustaceans. For example, does Traf signaling activate JNK? Does JNK activate crustacean caspases, and if so, how (i.e., mediated through IAP inhibition, apoptosome activation, or direct interaction with caspases)? Does Apip inhibit recruitment of caspases to the apoptosome? Is mitochondrial outer membrane permeabilization (MOMP), as regulated by homologues of Bcl-2 family proteins, involved in signaling and amplifying apoptosis in crustaceans? For abbreviations please see text
Table 1.
Apoptotic proteins in crustaceans
| GenBank | Species | Source |
E-value Drosophila melanogaster |
NCBI reference | Annotation |
E-value Homo sapiens |
NCBI reference | Annotation |
|---|---|---|---|---|---|---|---|---|
| Mitochondrial factors | ||||||||
| AC015198.1 | Caligus clemensi | Protein | 1.00E-103 | NP_610737.1 | EndoG | 1.00E-71 | NP_004426.2 | EndoG |
| AC010855.1 |
Caligus
rogercresseyi |
Protein | 3.00E-43 | NP_477176.1 | Cyt-c | 4.00E-44 | NP_061820.1 | Cyt-c |
| FE371263.1 | Daphnia pulex | EST | 1.00E-57 | NPJ508649.2 | AIF | 1.00E-78 | NP_004199.1 | Programmed cell death 8 (AIF) |
| FE773261.1 |
Petrolisthes
cinctipes |
EST | 6.00E-37 | NPJ550366.1 | HtrA2 | 6.00E-23 | NP_037379.1 | HtrA2 |
| Apafl | ||||||||
| FE416154.1 | Daphnia pulex | EST | No match | No match | 1.00E-13 | NP_001151.1 | Apoptotic peptidase activating factor 1 isoform b |
|
| IAPS | ||||||||
| AB038431 | Penaeus monodon | Protein | 2.00E-44 | NP_477127.1 | IAP2 | 9.00E-37 | NP_001158.2 | X-linked IAP |
| AC011057 |
Caligus
rogercresseyi |
Protein | 7.00E-15 | NP_650608.1 | Deterin | 2.00E-17 | NP_001159 | Survivin |
| Caspases | ||||||||
| ABK62771.1 |
Marsupenaeus
japonicus |
Protein | 7.00E-05 | NP_524551.2 | DrICE | 4.00E-07 | NP_001073594.1 | Caspase 8 isoform G precursor |
| ABI34434.1 | Penaeus monodon | Protein | 9.00E-38 | NP_524551.2 | DrICE | 1.00E-28 | NP_203124.1 | Caspase 7 isoform delta |
| ABM47410.1 |
Litopenaeus
vannamei |
Protein | 6.00E-32 | NP_524551.2 | DrICE | 3.00E-25 | NP_001217.2 | Caspase 6/Mch2 |
| AAX77407.1 |
Fenneropenaeus
merguiensis |
Protein | 2.00E-39 | NP_524551.2 | DrICE | 3.00E-28 | NP_203124.1 | Caspase 7 isoform delta |
| AB038430.1 | Penaeus monodon | Protein | 2.00E-56 | NP_524551.2 | DrICE | 4.00E-51 | NP_203124.1 | Caspase 7 isoform delta |
| ACA00158.1 |
Litopenaeus
vannamei |
Protein | 5.00E-37 | NP_524551.2 | DrICE | 1.00E-26 | NP_203124.1 | Caspase 7 isoform delta |
| ACO 14647.1 | Caligus clemensi | Protein | 3.00E-48 | NP_524551.2 | DrICE | 1.00E-86 | NP_116786.1 | Caspase 3 preproprotein |
| FE376029.1 | Daphnia pulex | EST | 2.00E-36 | NP_524551.2 | DrICE | 4.00E-31 | NP_203124.1 | Caspase 7 isoform delta |
| G0416273.1 | Triops cancriformis | EST | 1.00E-05 | NP_524551.2 | DrICE | 2.00E-05 | NP_116786.1 | Caspase 3 preproprotein |
| ES518008.1 | Artemia franciscana | EST | 4.00E-53 | NP_524551.2 | DrICE | 1.00E-35 | NP_203124.1 | Caspase 7 isoform delta |
| FE746352.1 |
Petrolisthes
cinctipes |
EST | 2.00E-23 | NP_524551.2 | DrICE | 1.00E-13 | NP_203124.1 | Caspase 7 isoform delta |
| EH666840.1 |
Calanus
finmarchicus |
EST | 2.00E-17 | NP_524551.2 | DrICE | 9.00E-20 | NP_001221.2 | Caspase 10 isoform a |
| FK895557.1 |
Caligu
rogercresseyi |
EST | 1.00E-61 | NP_524551.2 | DrICE | 2.00E-42 | NP_203124.1 | Caspase 7 isoform delta |
| G0416273.1 |
Lernaeocera
branchialis |
EST | 9.00E-06 | NP_524551.2 | DrICE | 9.00E-10 | NP_001220.2 | Caspase 9 isoform alpha preproprotein |
| Bcl-2 Family Members | ||||||||
| ACOl 1568.1 |
Caligus
rogercresseyi |
Protein | 3.00E-51 | NP_523702.1 | Buffy | 4.00E-26 | NP_115904.1 | Bok |
| FK882747.1 |
Caligus
rogercresseyi |
EST | 0.072 | NP_523702.1 | Buffy | 2.00E-10 | NP_004315.1 | Bax |
| CX994504.1 | Carcinus maenas | EST | 3.00E-06 | NP_523702.1 | Buffy | 3.00E-06 | NP_612815.1 | Bcl-xL |
| CV161085.1 | Callinectes sapidus | EST | 4.00E-21 | NP_523702.1 | Buffy | 2.00E-18 | NP_115904.1 | Bok |
| FE824303.1 |
Petrolisthes
cinctipes |
EST | 3.00E-22 | NP_523702.1 | Buffy | 5.00E-19 | NP_115904.1 | Bok |
| FE294837.1 | Daphnia pulex | EST | 3.00E-04 | NPJ788278.1 | Drob-1 | 5.00E-14 | NP_004315.1 | Bax |
| FE790955.1 |
Petrolisthes
cinctipes |
EST | 0.31 | NPJ788278.1 | Drob-1 | 9.00E-10 | NP_612815.1 | Bcl-xL |
| Eiger/TNFa | ||||||||
| FE337796.1 | Daphnia pulex | EST | 1.00E-11 | NPJ724878.2 | Eiger | 5.00E-09 | NP_001005609.1 | Ectodysplasin A isoform EDA-A2 |
| Wengen/TNFR | ||||||||
| FE131999.1 |
Litopenaeus
vannamei |
EST | 0.003 | NPJ728186.1 | Wengen | 7.6 | NP_002498 | Nerve growth factor receptor precursor |
| TRAF | ||||||||
| BAI40013.1 |
Marsupenaeus
japonicus |
Protein | 4.00E-19 | NP_511080.2 | TNF-receptor- associated factor 6 |
5.00E-45 | NP_004611.1 | TNF receptor-associated factor 6 |
| FK896127 |
Caligus
rogercresseyi |
EST | 8.00E-55 | NP_477416.1 | TNF-receptor- associated factor 4 |
2.00E-46 | NP_004286.2 | TNF receptor-associated factor 4 |
| EG949159.1 |
Homarus
americanus |
EST | 2.00E-21 | NP_511080.2 | TNF-receptor- associated factor 6 |
1.00E-31 | NP_004611.1 | TNF receptor-associated factor 6 |
| FK868215.1 |
Calanus
finmarchicus |
EST | 5.00E-29 | NP_477416.1 | TNF-receptor- associated factor 4 |
6.00E-22 | NP_004286.2 | TNF receptor-associated factor 4 |
| APAF-1 Interacting Protein | ||||||||
| FE278296.1 | Daphnia pulex | EST | 6.00E-50 | NP_572916.1 | APIP | 7.00E-47 | NP_057041.2 | APIP |
| FE143002.1 |
Litopenaeus
vannamei |
EST | 3.00E-47 | NP_572916.1 | APIP | 5.00E-48 | NP_057041.2 | APIP |
| FE765580.1 |
Petrolisthes
cinctipes |
EST | 9.00E-55 | NP_572916.1 | APIP | 3.00E-55 | NP_057041.2 | APIP |
| ES508309.1 | Artemia franciscana | EST | 4.00E-39 | NP_572916.1 | APIP | 5.00E-41 | NP_057041.2 | APIP |
| C0157578.1 | Eurydice pulchra | EST | 2.00E-34 | NP_572916.1 | APIP | 4.00E-38 | NP_057041.2 | APIP |
| Jafrac2 | ||||||||
| ABZ80828.1 | Penaeus monodon | Protein | 2.00E-99 | NPJ728793.1 | Jafrac2 | 5.00E-101 | NP_006397.1 | Peroxiredoxin 4 |
| AC012581 |
Lepeophtheirus
salmonis |
Protein | 3.00E-95 | NPJ728793.1 | Jafrac2 | 2.00E-95 | NP_006397.1 | |
| ABN13585 | Artemia franciscana | Protein | 3.00E-91 | NP_728793.1 | Jafrac2 | 6.00E-95 | NP_006397.1 | |
| Death-Inducer Obliterator | ||||||||
| DV467342.1 | Carcinus maenas | EST | 3.00E-23 | NP_J550193 | Protein partner of snf | 9.00E-19 | NP_542987.2 | Death-inducer Obliterator |
| FK897290.1 |
Caligus
rogercresseyi |
EST | 1.00E-22 | NP_J550193 | Protein partner of snf | 2.00E-16 | NP_542987.2 | Death-inducer Obliterator |
| AATF | ||||||||
| FK868966 |
Caligus
rogercresseyi |
EST | 3.00E-30 | NP_J509066.1 | n/d | 3.00E-40 | NP_036270 | Apoptosis antagonizing transcription factor |
| FE398472.1 | Daphnia pulex | EST | 9.00E-14 | NP_J509066.1 | n/d | 3.00E-21 | NP_036270 | Apoptosis antagonizing transcription factor |
| DAD1 | ||||||||
| ABU54835.1 | Penaeus monodon | Protein | 1.00E-45 | NP_J509222.1 | n/d | 2.00E-38 | NP_001335.1 | DAD1 |
| TCTP/Fortilin | ||||||||
| ABZ81535 | Artemia franciscana | Protein | 1.00E-67 | NP_650048.1 | Translational controlled tumor protein |
4.00E-35 | NP_003286.1 | TCCP |
In order to identify proteins with potential apoptotic functions in crustaceans, a broad range of protein sequences with well known apoptotic functions that are listed in the protein database of NCBI for D. melanogaster and H. sapiens were blasted against the NCBI database for non-redundant protein sequences (blast-p) and non-human, non-mouse expressed sequence tags (dbEST; tblast-n). Matching crustacean ESTs were translated into the corresponding protein sequence and again blasted against the NCBI protein database for D. melanogaster and H. sapiens (blast-p)
IAPs and IAP binding proteins
IAPs are a group of proteins that contain at least one baculovirus IAP repeat (BIR) domain through which they interact with caspases and suppress caspase activity [30]. Apoptosis in D. melanogaster is controlled predominantly by transcription of genes that encode the RHG killer proteins—reaper, hid, grim, and sickle. The mechanism by which RHG proteins induce apoptosis is through interaction with the BIR1 and/or BIR2 domains of D. melanogaster IAP 1 (dIAP1). In the fruit fly, binding of RHG proteins liberates caspases that are bound to dIAP1 and induces ubiquitination as well as degradation of dIAP1 [31]. Homologous proteins have not been reported for any of the RHG proteins in crustaceans, or for that matter, outside the Drosophilidae [32]. Thus these proteins are likely restricted to D. melanogaster and closely related species. An IAP containing three BIR domains recently has been cloned from Penaeus monodon, and when expressed in SF-9 cells from an insect, this protein was found to inhibit reaper-induced apoptosis [23]. The finding suggests that caspase activity in the tiger prawn P. monodon is negatively regulated by IAPs but does not resolve the question of whether the inhibition of IAPs is sufficient to induce apoptosis in crustaceans.
The thioredoxin peroxidase Jafrac2 and the serine protease HtrA2/Omi are both proteins that are sequestered in intracellular compartments in healthy cells [33, 34]. In response to induction of apoptosis in D. melanogaster by UV irradiation, Jafrac2 translocates from the ER to the cytoplasm where it promotes cell death by liberating the initiator caspase Dronc from dIAP1 [33]. A second protein known to release caspases from inhibition by IAPs is the mitochondrial serine protease HtrA2/Omi. After HtrA2/Omi is released from the mitochondrion into the cytoplasm, it stimulates apoptosis through cleavage of IAPs in vertebrates [35] and D. melanogaster [34]. The question of whether Jafrac2 plays a significant role in crustacean apoptosis, or whether it functions in protection against oxidative stress similar to the mammalian homolog peroxiredoxin 4 [36], is currently unresolved, as is a role for HtrA2/Omi in crustacean apoptosis. However, two crustacean ESTs found in the NCBI database putatively encode protein fragments projected to increase caspase activity by inhibiting IAPs. ESTs for Jafrac2 and HtrA2/Omi are reported for P. monodon, Petrolisthes cinctipes and other crustaceans (Table 1). Thus, mechanisms to induce apoptosis through inhibition of IAPs could potentially be conserved across multiple arthropod groups, but functional data are not yet available.
Caspases
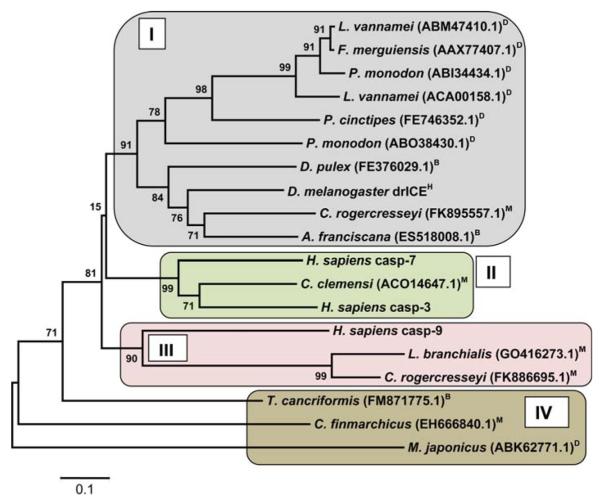
To date, 14 caspase proteins or EST sequences from three major crustacean clades (Maxillopoda, Branchiopoda, and Malacostraca) are present in the NCBI database (Table 1). Neighbor-joining indicates that the proteins likely belong to several different caspase families (Fig. 2), because the grouping does not strictly depend on the known phylogeny of the species analyzed. For example, branch II groups a caspase from the parasitic copepod Caligus clemensi with human caspases, and branch IV contains caspases for species from all three major crustacean clades (Fig. 2). This pattern may not be surprising if one considers there are seven different caspases in the D. melanogaster genome [31] and eleven caspases in the human genome [10, 37]. Remarkably, crustacean caspases only display strong homologies to the D. melanogaster caspase DRICE and not to any of the other six caspases found in the fruit fly. Yet the crustacean caspases do show differing degrees of homology with several human caspases (Table 1). None of the crustacean caspase sequences show any protein–protein interaction motifs such as a caspase recruitment domain (CARD) or a death effector domain (DED) when scanned with ScanProsite (http://ca.expasy.org/) or SMART (http://smart.embl-heidelberg.de). Thus, they are likely executer caspases (cf. [10]). However, this tentative conclusion may be influenced by the fact that half of the investigated sequences are derived from ESTs instead of full-length protein sequences.
Fig. 2.
Phylogenetic tree obtained by neighbor-joining from multiple sequence alignments of the p20 and p10 subunit of crustacean, D. melanogaster (drICE), and H. sapiens caspases (casp-3, 7, and-9). The bootstrap values are added at each branch point and represent percentages of replicate trees in which the associated protein sequences clustered together (500 replicates). The tree is drawn to scale, with branch lengths displayed in the same units (i.e., number of amino acid substitutions per site) as the evolutionary distances used to infer the tree. Similar phylogenetic relationships were obtained based on maximum parsimony analysis. The tree resolves four major branches (I–IV) of which three are related. Analyses of phylogeny and molecular evolution were conducted using MEGA version 4.1 [149]. H arthropod subphylum Hexapoda; D Decapoda, M Maxillopoda, B Branchiopoda, all within arthropod subphylum Crustacea
Bcl-2 family proteins
Several ESTs encoding for protein fragments that show homologies to the Bcl-2 family of proteins can be found for crustaceans in the NCBI database. Neighbor-joining places the deduced protein sequences into three major clades of which two are related to each other (Fig. 3). Only two proteins (Drob-1 and Buffy) that belong to the Bcl-2 protein family are known in D. melanogaster, but twenty different Bcl-2-like proteins are described for vertebrates, where the interplay of pro- and anti-apoptotic members controls the permeability of the outer mitochondrial membrane and thereby impacts the intrinsic apoptotic pathway [18, 38]. Drob-1 and Buffy are closely related to each other and group within the same clade (Fig. 3). We could identify seven different Bcl-2 family proteins from species representing the three major crustacean clades (Maxillopoda, Branchiopoda, and Malacostraca). Two different Bcl-2 family proteins could be identified for the sea louse Caligus rogercresseyi as well as for the porcelain crab P. cinctipes. The fact that the Bcl-2 family proteins found in both species group closer to Bcl-2 family proteins found in other crustaceans, as opposed to the alternative one from their own species, suggests different ancestral proteins (Fig. 3). For example the two different Bcl-2 proteins found for C. rogercresseyi (Maxillopoda) group in one case with a protein found in Malacostraca and in the other case with a Bcl-2 protein found in the branchiopod Daphnia pulex (Fig. 3). These results strongly suggest a higher amount of diversity in crustacean Bcl-2 proteins than one might have anticipated based on studies in D. melanogaster.
Fig. 3.
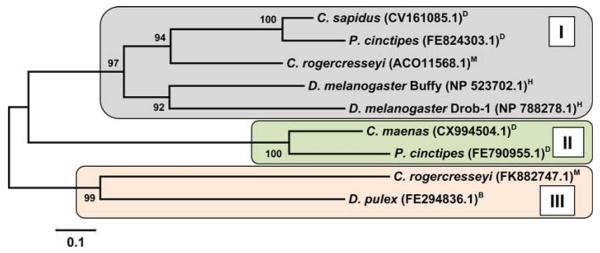
Phylogenetic tree obtained by neighbor-joining from multiple sequence alignments of Bcl-2 proteins for crustaceans and D. melanogaster (Drob-1 and Buffy). The bootstrap values are added at each branch point and represent percentages of replicate trees in which the associated protein sequences clustered together (500 replicates). The tree is drawn to scale, with branch lengths displayed in the same units (i.e., number of amino acid substitutions per site) as the evolutionary distances used to infer the tree. The same phylogenetic relationships were obtained based on maximum parsimony analysis of the corresponding cDNA sequences. The tree resolves three branches of which two are related. Analyses of phylogeny and molecular evolution were conducted using MEGA version 4.1 [149]. H arthropod subphylum Hexapoda; D Decapoda, M Maxillopoda, B Branchiopoda, all within arthropod subphylum Crustacea
Apaf-1 and Apip
Although initiator caspases such as casp-9 or Dronc can be cleaved, they show full activity in their uncleaved form and are activated through dimerization at adaptor platforms [11]. This process is mediated by Apaf-1 in vertebrates and by Dark in D. melanogaster [39, 40]. Despite the similar functions of Apaf-1 and Dark, recent studies revealed that both proteins belong to separate subtypes that were already present at the cnidarian-bilaterian split, leaving insects without orthologs of human Apaf-1 [4]. One EST from the water flea D. pulex (FE416154.1) that encodes for a deduced protein with high similarity to human Apaf-1 (1e−13) was found at NCBI (Table 1). Alignment of both proteins reveals a conserved CARD region and a highly conserved NACHT-NTPase domain (Fig. 4). What seems remarkable to us is that the branchiopod Apaf-1 is not at all similar to Dark or any sequence from D. melanogaster (Table 1), which underscores the potential pitfalls of extrapolating results from D. melanogaster to other arthropods. Unfortunately, a full length sequence is not available for D. pulex Apaf-1 (FE416154.1), which would allow speculation about functional aspects such as the interaction with cyt-c based on the presence or absence of WD40 regions.
Fig. 4.

Sequence alignment of a deduced protein (Apaf-1) from D. pulex (FE416154.1) with H. sapiens Apaf-1 (NP_001151.1). The translated EST from D. pulex includes the Caspase Recruitment and Activation Domain (CARD, shaded in light), the highly conserved NACHT-NTPase domain (shaded in dark), and is highly conserved with the human protein. Importantly, no significant homologies to Dark, the Apaf-1 analog from D. melanogaster, were found (c.f., Table 1). ‘*’ = identical. ‘:’ = conserved substitutions. ‘.’ = semi-conserved substitution
The activity of Apaf-1 can also be regulated by direct interaction with the Apaf-1-interacting protein (Apip). Apip was found to be highly upregulated in mouse skeletal muscle after ischemia and to attenuate hypoxia induced damage by reducing the severity of the apoptotic response [41, 42]. The protein sequence of Apip is conserved between Caenorhabditis elegans and H. sapiens, and the protein inhibits apoptosis by competing with caspase-9 for the CARD region in Apaf-1 [41]. We found several crustacean EST sequences in the NCBI database with high similarities to Apip from D. melanogaster and H. sapiens (Fig. 5). The possibility that apoptosis under hypoxia in crustaceans (see ‘Putting the brakes on apoptosis: negative regulators’) is avoided by direct inhibition of Apaf-1 upstream of the initiator caspase deserves exploration.
Fig. 5.
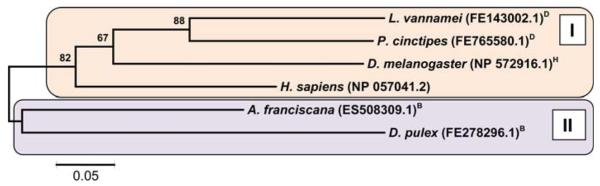
Evolutionary relationships between Apaf-1 Interacting Proteins (Apips) from H. sapiens, D. melanogaster and deduced proteins from crustacean ESTs. The phylogenetic tree was obtained by the neighbor-joining method. The bootstrap values are added at each branch point and represent percentages of replicate trees in which the associated protein sequences clustered together (500 replicates). The tree is drawn to scale, with branch lengths displayed in the same units (i.e., number of amino acid substitutions per site) as the evolutionary distances used to infer the tree. The tree resolves two major branches. Analyses of phylogeny and molecular evolution were conducted using MEGA version 4.1 [149]. H arthropod subphylum Hexapoda; D Decapoda, M Maxillopoda, B Branchiopoda, all within arthropod subphylum Crustacea
Mitochondrial apoptotic factors
At least five proteins that reside in the mammalian mitochondrion have known functions in caspase-dependent and independent programmed cell death when released into the cytoplasm [19, 43]. Crustacean homologs to four of these (EndoG, cyt-c, AIF, and HtrA2/Omi) could be found in the NCBI database (Fig. 1; Table 1). A potential role of cyt-c in crustacean apoptosis cannot be excluded based on the fact that cyt-c is not incorporated in the Dark apoptosome from D. melanogaster [44], but seems unlikely based on the observation that cyt-c fails to increase casp-3 and -9-like activities in cytoplasmic extracts from the branchiopod Artemia franciscana [20]. However, until the molecular structure for a crustacean apoptosome is resolved, a possible role for cyt-c (or another factor) in Apaf-1 activation cannot be completely ruled out. EndoG and AIF are known to contribute to caspase-independent apoptosis in a variety of species including C. elegans and D. melanogaster [45–49]. Based on their conserved role in apoptosis, it is plausible that EndoG and AIF are also playing roles in crustacean apoptosis (Fig. 1; Table 1). No crustacean proteins with homologies to the RHG analog Smac/Diablo [50] are reported in the NCBI database (Table 1) but an EST encoding HtrA2/Omi is reported for P. centipedes (Table 1). Based on the findings that HtrA2/Omi cleaves IAPs after release into the cytoplasm in D. melanogaster [34] and vertebrates [35], it is possible that this function might also be conserved in crustaceans.
Receptor mediated apoptosis
A protein with strong homology to Eiger [a tumor necrosis factor (TNF) homolog from D. melanogaster] is reported in the NCBI database for D. pulex, and a protein from the shrimp Litopenaeus vannamei with moderate homology to the TNF-receptor Wengen is also available (Table 1). In D. melanogaster Eiger and Wengen physically interact with each other through their TNFR/TNF homology domains and induce cell death by signaling through tumor necrosis factor receptor-associated factor (TRAF) and also further downstream through the Jun-N-terminal Kinase-(JNK) signaling pathway [16, 17, 51]. TRAF-like proteins can be found in several crustaceans (Table 1). However, based on the fact that TRAF proteins play roles in a broad range of cellular signaling pathways [52], a specific role in crustacean apoptosis needs to be confirmed.
Mitochondrial permeability transition: a vertebrate phenomenon?
It is well established that when mammalian mitochondria are exposed to Ca++ at approximately 100 μM in the presence of Pi, extensive swelling occurs that results in uncoupled respiration [7, 19, 53–56]. These phenomena are due to an acute increase in permeability of the inner mitochondrial membrane known as the permeability transition, which can be blocked by cyclosporine A [57–59]. The probability of opening of the so-called mitochondrial permeability transition pore (MPTP) is increased when adenine nucleotides and mitochondrial ΔΨ drop [60], but the molecular composition of the MPTP is still not established and is a matter of ongoing research. Of the components traditionally viewed as essential for the pore, the voltage-dependent anion channel has been convincingly eliminated as a candidate based on knock-out experiments [61, 62]. If calcium and other activators of opening are sufficiently high, the pore can be induced even after knockout of the adenine nucleotide transporter (ANT) and cyclophilin D (CyP-D), although these two molecules clearly have important roles in the functioning and regulation of the pore [63–65]. More recently, a strong case has been made that the phosphate carrier may be a key player in the MPTP [66].
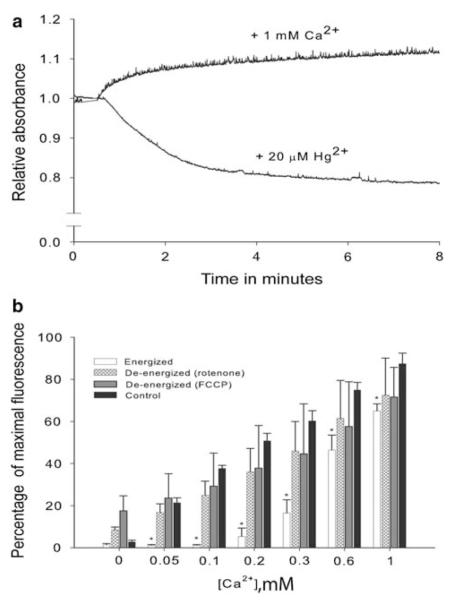
In the open state, matrix swelling then causes the rupture of the outer mitochondrial membrane and release of numerous pro-apoptotic factors from the intermembrane space [43, 59, 67–69]. A number of physiological conditions can trigger the opening of the MPTP, including oxidative stress, elevated intracellular calcium, and cellular energy limitation as a result of hypoxia and anoxia [7, 59,70–73]. Consequently, it was surprising when Menze et al. [21] found for mitochondria from the anoxia-tolerant crustacean, Artemia franciscana, that the MPTP did not open in response to various physiological and pharmacological inducers. We subsequently investigated properties of the permeability transition in mitochondria from another crustacean, the ghost shrimp, Lepidophthalmus louisianensis [74]. Our data demonstrated that calcium concentrations up to 1 mM did not induce swelling in mitochondria from L. louisianensis (Fig. 6a). The increase in absorbance (i.e., apparent decrease in mitochondrial volume) was likely not due to shrinkage but rather to an increase in refractive index because of the formation of Ca2+-phosphate complexes in the matrix during high calcium uptake [75–77]. As an important positive control to document the capacity of ghost shrimp mitochondria to swell, addition of high concentrations of mercury (20 μM) caused substantial swelling of mitochondria of L. louisianensis (Fig. 6a). This level of mercury has been shown to induce mitochondrial permeabilization by unspecific “damage” of membrane proteins that leads to opening of an un-regulated pore, which is not inhibited by cyclosporine A [78]. Experiments on calcium-induced calcium release shown in Fig. 6b confirmed the absence of a MPTP; specifically, exogenously added calcium did not promote release of matrix calcium (MPTP opening) across the entire range of concentrations tested with mitochondria of L. louisianensis. Indirect evidence suggests that a regulated MPTP may be absent in mitochondria from the oyster Crassostrea virginica [79]. For these reasons, we have previously suggested that the lack of a calcium-induced permeability transition may be wide-spread in invertebrates [7].
Fig. 6.
Swelling assay upon addition of Ca2+ and HgCl2 (in the presence of 1 mM phosphate) for mitochondria isolated from hepatopancreas of the ghost shrimp Lepidophthalmus louisianensis. Mitochondrial swelling (i.e., a decrease in absorbance) is not observed after addition of 1 mM CaCl2 to energized mitochondria (succinate plus rotenone). Mitochondrial volume does increase after addition of 20 μM HgCl2. b The absence of Ca2+-induced release of Ca2+ by mitochondria isolated from hepatopancreas of L. louisianensis. Mitochondria were incubated in exogenously added Ca2+ in the presence of the Ca2+ probe fluo-5 N, which reports the extra-mitochondrial Ca2+. Fluorescence expressed as a percentage of the total fluorescence when the dye is saturated with Ca2+ (% maximal fluorescence). Control samples contained no mitochondria. Energized mitochondria (succinate plus rotenone) reduced the concentration of exogenously added Ca2+ at all concentrations investigated, relative to controls. De-energized mitochondria (no succinate or rotenone, but with FCCP) were less effective in calcium uptake than energized mitochondria. Importantly, no release of Ca2+ was observed across the entire range of exogenously added Ca2+ by either the energized or de-energized mitochondria. Each bar represents mean ± SD of n = 4 experiments. * Significantly different from control (ANOVA, Tukey’s pair-wise comparison, P < 0.001) (Modified after [74])
Among non-mammalian vertebrates, liver mitochondria from a goby (Zosterisessor ophiocephallas) show calcium-induced swelling when incubated in the presence of Pi, but the amount of calcium required to induce the permeability transition was substantially higher than that required for liver mitochondria from rat [80]. A mitochondrial permeability transition induced by Cu and inhibited by cyclosporine A has been documented in primary hepatocytes from rainbow trout [81]. While data are not fully conclusive due to methodological issues, a functional MPTP may exist in liver mitochondria from the Baltic lamprey Lampetra fluviatilis [82]. Finally, liver mitochondria isolated from the bob white quail Colinus virginianus also displayed a functional MPTP in response to calcium and phosphate (M. Menze and S. Hand, unpublished observations). A phylogenetic distribution for the MPTP is far from complete. Existing evidence of which we are aware indicates that the MPTP is present in all vertebrates studied to date, yet a regulated MPTP that can be inhibited by cyclosporine A has not been demonstrated thus far in crustaceans or other invertebrates. In parallel with the evolution of diversified pathways for cell death in eukaryotic organisms, perhaps functional trade-offs in the predisposition to tolerance of environmental stresses have occurred [7]. For example, a short-term anoxia may trigger calcium imbalance and apoptosis in mammals, whereas anoxia over months may not initiate cell death in certain crustaceans, as discussed below.
Caspase regulation
The contribution of caspases to the apoptotic process is evolutionarily conserved [10]. Not surprisingly, activation of these cysteine proteases must be tightly regulated in order to avoid cell degradation under physiological conditions when it is unwarranted or maladaptive. Reports characterizing the biochemical regulation of caspases in crustaceans are few (e.g., [20]), as are studies on their cloning and molecular characterization [22, 24, 25].
Allosteric regulation of caspase activity by KCl and Ca2+
Some insight into regulation of caspase activities in crustaceans comes from work on caspase-9-like activity in cell extracts isolated from embryos of the brine shrimp A. franciscana. Casp-9-like activity was measured by following cleavage of LEHD-R110 and was found to be sensitive to Ca2+ and KCl. The inhibition of caspase activity by [K+] has been proposed to serve as a safeguard to inhibit unwarranted activation of caspases in healthy cells [83–85]. An increase in casp-9-like activity of about 14-fold was observed upon reducing the KCl concentration in the enzyme assay from 150 to 0 mM (Fig. 7; [20]). In mammals, inhibition of caspases by KCl is believed to occur by blocking cyt-c binding to Apaf-1, perhaps by competing with cyt-c at the WD40 site, and in turn preventing the processing of procaspase-9 [86, 87]. A. franciscana lacks a cyt-c effect [20], and in this species KCl likely inhibits the processing and/or recruitment of casp-9 to the apoptosome. At low KCl concentration a pronounced effect of calcium on caspase activity was found. [20] The addition of 5 mM calcium leads to a fivefold increase in LEHD-R110-cleavage activity (Fig. 7; [20]), an effect that is likely allosterically mediated. Whether or not the direct effect of calcium on caspase activity is physiologically meaningful obviously requires investigation at lower calcium concentrations. Regulation of A. franciscana caspases by intracellular nucleotides may be important in diapause and anoxia (see “Energy-limited states”, below).
Fig. 7.
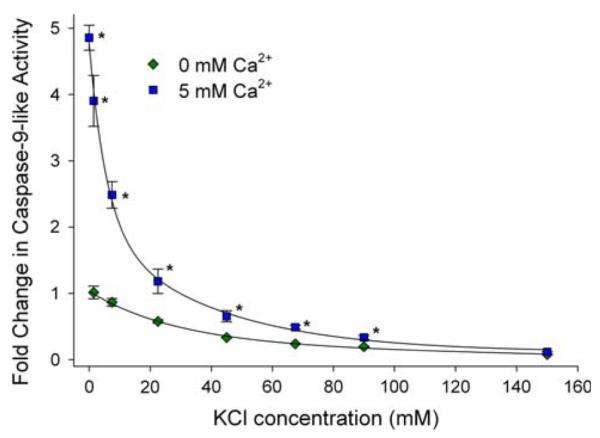
Influence of Ca2+ and KCl on caspase 9 (casp-9)-like activity in cytosolic extracts from diapause embryos of Artemia franciscana. Casp-9-like activity was measured as the increase in fluorescence due to cleavage of the fluorogenic substrate Z-LEHD-R110. As KCl concentrations are decreased, the addition of 5 mM calcium promotes an increase in activity of casp-9. At 0 mM KCl, the addition of 5 mM calcium leads to a 4.86 ± 0.19 (SD)-fold (n = 3) increase in activity. The activation is even more dramatic (67-fold) when expressed relative to the depressed activity at 150 mM KCl. The influence of Ca2+ and KCl was expressed as fold changes relative to control. Each value is expressed as the mean (SD) of n = 3–5 experiments. * Statistically different activity after calcium addition (P < 0.05) (Modified after [20])
Positive stimuli for apoptosis
Radiation
Ultraviolet radiation can be a powerful initiator of apoptosis. UV-B light, comprised of wavelengths between 290 and 320 nm, readily causes genetic damage [88], owing in large part to the fact that its spectrum lies close to the DNA molecule’s maximum absorbance [89]. The energy thus absorbed may then result in DNA lesions, like the formation of cyclobutane pyrimidine dimmers, that block transcription [90]. Cyclobutane pyrimidine dimers result in a proportional upregulation of p53 that facilitates genome repair by arresting the cell in G1 [91]. UV radiation also results in p53 stabilization by p38 kinase mediated phosphorylation [92, 93]. If DNA repair mechanisms fail to correct the damage, p53 mediates downstream inhibition of anti-apoptotic members of the Bcl-2 family of proteins and concurrent activation of pro-apoptotic members, such as Bax, resulting in apoptosis in vertebrates [94]. Miguel et al. [95] demonstrated that UV-B apparently triggered apoptosis in the mangrove crab Ucides cordatus as judged by TdT-mediated dUTP-biotin nick end-labeling (TUNEL) of selected tissues; however, the mechanisms underlying the apoptotic signaling are unclear at this point. It is appropriate to note that Miguel et al. [95] used antibodies directed against mammalian Bax and p53 without regard as to whether these antibodies cross-react with the correct target proteins in crustaceans. For example, a functional Bax pathway or mitochondrial outer membrane permeabilization (MOMP) has never been reported in Crustacea. Two ESTs have been reported (Table 1) that bear marginal resemblance to Buffy and Drob-1 of D. melanogaster and some similarity to human Bax. Obviously, the use of mammalian antibodies for crustacean studies with little concern as to whether the target proteins exist, and if so, whether they are sufficiently conserved to cross-react with a mammalian antibody, should be avoided. In our experiences the majority of mammalian antibodies do not react specifically with the desired target protein in crustaceans.
Whether or not a cell undergoes necrosis or apoptosis after exposure to radiation appears to vary depending primarily on the cell type, and secondarily, with exposure level [96, 97]. Although some necrosis is seen, apoptosis is the primary mode of cell death correlated with UV exposure in murine embryonic fibroblasts [96]. A study conducted by de Oliveira Miguel et al. [97] has shown a similar propensity for apoptosis in retinula and laminar ganglionaris cells of the crab U. cordatus when the crabs were exposed to 254–312 nm UV sources.
Other forms of ionizing radiation, such as γ-rays and α-particles can have even greater oxidizing and genotoxic effects [98]. Ionizing radiation may produce the so-called bystander effect when absorbed in low doses. Although the mechanism underlying this phenomenon is still unclear, irradiated cells have been shown to communicate with adjacent cells resulting in the spread of death signals and the consequent induction of apoptosis in the un-irradiated cells [98, 99]. Cells cultured from hematopoietic tissue of the Norway lobster, Nephrops norvegicus, were exposed to a low dose of 60Co γ-radiation [99], which caused cellular abnormalities and apoptosis. These irradiated cells released a factor into the medium that induced apoptosis in fish and human cells that had not been irradiated [99]. Thus, a bystander-like effect is observable with irradiated cells from a crustacean. The dose dependent experiments showed N. norvegicus cells to be far more sensitive to irradiation than previous reports for other invertebrates [99]. Based on prototypical morphological criteria, apoptosis was the predominant mode of cell death seen at doses below 0.5 Gy and at doses above 0.5 Gy necrosis was seen. This result is in contrast to those observed for UV exposure in U. cordatus [95] and also for mammalian or fish cells treated with higher doses of γ-radiation [99, 100]. In the latter two reports, apoptosis was the primary mode of cell death. No increases in necrotic rates above 0.5 Gy were observed in the case of c-irradiated mammal and fish cell lines [99].
Organic toxins and heavy metals
Organic compounds like phthalate esters [101, 102] have become common contaminants in soil and water and have been shown to induce apoptosis in crustaceans. Benthic invertebrates are commonly affected, since many of these organic pollutants have a substantial affinity for sedimentary absorption [103]. Similarly, heavy metals like manganese (liberated from MnO2 during hypoxia; [104]) and cadmium [105] are frequent contaminants in marine sediments. Cadmium is perhaps the most common apoptosis-inducing environmental pollutant [105]. We offer here a brief synopsis of representative work that illustrates induction of apoptosis by these agents in crustaceans, but unfortunately there is paucity of information addressing the underlying mechanisms of signal transduction.
Sung et al. [102] found that the majority of hydrophobic dialkyl phthalate esters (PAEs) tested on haemocytes of the giant freshwater prawn Macrobrachium rosenbergii induced necrotic cell death with a low incidence of apoptosis. However, two of the PAEs tested, dihexyl phthalate (DHP) and dipropyl phthalate (DPrP), produced primarily apoptotic cell death as confirmed by annexin assay, propidium iodide (PI) staining, and transmission electron microscopy of DNA fragmentation and cytosolic vacuole formation. The frequency of apoptotic cell death was directly correlated with exposure time [102]. An increase in superoxide ion formation (O2−) was also reported, which suggests that oxidative stress could be an important contributor to the induction of apoptosis.
Manganese is a potent neurotoxin and induces apoptosis in neurons and Parkinson’s disease-like symptoms in humans and other animals [106]. Manganese promotes apoptosis by inhibiting mitochondrial complexes I and II, which results in disruption of energy metabolism, increased ROS production, depolarization of the mitochondrial membrane, and cytochrome c release [107]. In hematopoietic cells and circulating hemocytes of N. norvegicus manganese induces apoptosis resulting in hemocytopenia. The frequency of apoptosis, as assessed with the TUNEL assay and DNA ladder assay for DNA fragmentation, was correlated with increasing manganese concentrations, but no such relation was seen in regard to exposure time [104]. Hematopoietic cells were found to be more sensitive to manganese toxicity than hematocytes [104].
To investigate the loss of motility and survivorship, Cheng, et al. [105] examined the apoptotic effects of cadmium exposure on stage II nauplius larvae of the barnacle, Balanus amphitrie. Using the TUNEL assay, these authors determined that the rate of apoptosis increased as the concentration of cadmium was raised to 15 μM, compared to untreated controls that lacked detectable apoptotic cells. Above 15 μM, the incidence of apoptosis was associated with a decrease in larval survivorship.
Diatom-induced apoptosis in copepods
Some diatom species (e.g. Skeletonema costatum and Thalassiosira rotula) possess a chemical defense strategy that is based on activation of apoptotic pathways in their main grazers, the calanoid copepods. In response to cellular damage S. costatum and T. rotula produce unsaturated aldehydes such as 2-trans-4-trans-decadienal (DD) [108,109]. Romano et al. [110] demonstrated that adult females of Calanus helgolandicus reared on a diet consisting of S. costatum or T. rotula produced embryos with reduced viability compared to controls that were fed the aldehyde-free algae Prorocentrum minimum. Embryos from mothers reared on T. rotula also showed increased numbers of TUNEL positive cells, and hatched nauplii exhibited a high frequency of deformations [110]. When mothers were fed S. costatum for 5 days, the spawned embryos later developed teratogenic birth defects such as deformed limbs in 45–65% of the hatched nauplii. After 10 days feeding on S. costatum the egg viability decreased by about 75%, the degree of teratogenesis increased, and hatched embryos were strongly deformed [111]. TUNEL staining and DNA laddering assays clearly demonstrated that the observed birth defects were due to increased apoptosis in the embryos. Increased apoptosis could be contributed to the maternal uptake of DD, because apoptosis also could be induced in healthy embryos by direct incubation with 5 μg/ml DD [110]. However, no increase in enzymatic activity for cleaving Ac-DEVD-FMK (a substrate for human caspase-3) was found in extracts of homogenized C. helgolandicus embryos after exposure to DD [110]. The authors concluded that diatom-derived aldehydes either trigger caspase-independent apoptotic pathways in C. helgolandicus or that activated caspases had no significant specificity for the human caspase-3 substrate [110]. This marine diatom-herbivore system is a remarkable example in which the alga reduces grazing pressure by impairing the reproductive success of the herbivore through induction of apoptosis, instead of repelling or poisoning the predator [108, 109].
Putting the breaks on apoptosis: negative regulators
There are environmental challenges during which apoptotic signaling in crustaceans is blunted [7]. Many of these stresses are manifested at the cellular level as a drop in energy availability. ATP limitation can compromise ion homeostasis, cause dissipation of ion gradients and membrane potential leading to release of pro-apoptotic factors. Initiation of apoptosis under such commonly encountered environmental stresses in crustacean would be maladaptive. Thus, it appears that mechanisms are in place to restrict apoptosis during energy limitation, in striking contrast to the positive stimulation of cell death observed under similar conditions in mammals.
Energy-limited states
Diapause
This latent state is an ontogenetic, programmed arrest of development, and sometimes metabolism, during life cycles of many invertebrates that can lead to restricted ATP levels in cells (e.g., see [112–114]). In some species like embryos of the brine shrimp, Artemia franciscana, aerobic metabolism is depressed to as little as 2% of the control, pre-diapause rate [112]. Considering that proton leak respiration has been estimated to comprise 20% or more of basal metabolic rate in vivo [115], it seems unlikely that mitochondrial membrane potential could be maintained in a physiological state where activity of the electron transport system (and associated proton pumping) is below that necessary for leak compensation. The restriction in ATP production during diapause would presumably disrupt intracellular calcium homeostasis as well. Yet these embryos survive diapause for months, indicative of a lack of accumulated cell death.
Our studies of the regulation of caspase 9-like activity by nucleotides in extracts of A. franciscana embryos [20] revealed the novel finding that ADP is a powerful inhibitor (Fig. 8). A direct effect of ADP on regulation of caspase 9 has not been described for mammals [116]. Binding of various nucleotides (particularly triphosphates) to cyt-c, thereby interfering with Apaf-1 interaction and depressing apoptosome formation has been reported [117]. However, cyt-c does not stimulate caspase activation in A. franciscana embryos [20], so such an indirect mechanism via cyt-c is an unlikely explanation for the strong ADP inhibition of caspase 9. Particularly noteworthy in the context of avoiding apoptosis during diapause, we found that ADP inhibition is about 70% greater in diapause extracts at the approximate physiological concentration of ADP (15–150 μM) than in post-diapause extracts [20], and analysis of IC50 values (Table 2) showed that inhibition by Mg-ADP is far greater in diapause embryos (IC50 = 0.7 μmol l−1) than in post-diapause embryos (IC50 = 44.4 μmol l−1). In A. franciscana embryos, ADP concentrations do not change appreciably between post-diapause and diapause states (J. A. Covi, J. Reynolds and S. Hand, unpublished observations). Consequently, the differential ADP inhibition could represent a new mechanism to prevent caspase activation during diapause when ATP levels fall. As in mammalian systems, high concentrations of ATP inhibit caspases and are prosurvival, while low concentrations activate caspases [116, 117].
Fig. 8.
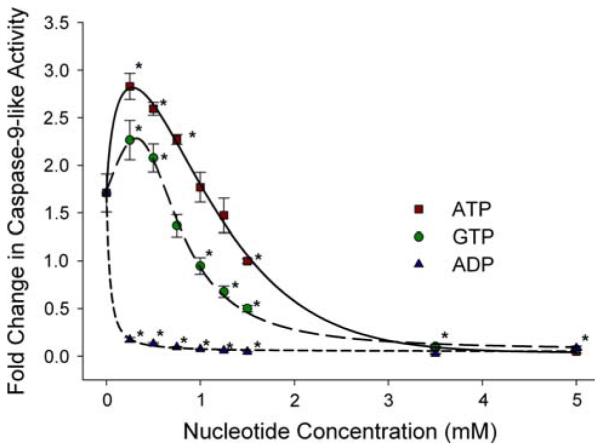
Impact of ATP, GTP, and ADP (in the presence of MgCl2) on casp-9-like activity in cytosolic extracts from diapause embryos of Artemia franciscana. Casp-9-like activity was measured as the increase in fluorescence due to cleavage of the fluorogenic substrate Z-LEHD-R110. Addition of 2.5 mM MgCl2 in the absence of nucleotides leads to a 1.7-fold increase in casp-9 activity in diapause extract. A biphasic pattern of activation followed by inhibition is observed for ATP and GTP. ADP is highly inhibitory at all concentrations investigated. Each value is expressed as the mean (SD) of n = 3–9 experiments. * Statistically different to 0 mM added nucleotides (P < 0.05) (Modified after [20])
Table 2.
Differential inhibition of caspase-9-like activity by Mg-ADP in extracts from diapause versus post-diapause embryos of A. franciscana
| Conditions | IC50 |
|---|---|
| Mg-ADP, diapause | 0.7 ± 0.1 |
| ADP, diapause | 1.5 ± 0.1 |
| ATP, diapause | 1.0 ± 0.1 |
| Mg-ADP, post-diapause | 44.4 ± 12 |
| ADP, post-diapause | 3.1 ± 0.4 |
| ATP, post-diapause | 2.6 ± 0.2 |
IC50 = chemical concentration (μM) that promotes 50% inhibition of enzyme activity. Caspase-9-like activity was followed by measuring cleavage of the fluorogenic substrate LEHD-R110. When added, total Mg2+ concentration was held constant at 2.5 mM (after [20])
Anoxia
The complete absence of oxygen can be tolerated by A. franciscana embryos for years at room temperature [118]. Under this condition ATP levels plummet to almost immeasurable levels as judged by measurement in acid extracts [119, 120] or by NMR [121]. As discussed above, cell stress in mammals leads to a drop in ATP that relieves the inhibition of caspases and aids in fostering apoptosis. However, another novel feature of A. francsicana caspase 9-like activity is its strong inhibition by GTP (Fig. 8); the regulatory pattern for GTP on caspase 9 is very similar to that for ATP [20]. GTP apparently promotes a direct inhibition by binding to caspase 9 or perhaps to the adaptor platform Apaf-1. Because intracellular GTP remains high (>2 mM) in embryos exposed to anoxia [119], it could maintain a high degree of inhibitory control over caspases. The effect could serve as a means to prevent maladaptive apoptosis during oxygen deprivation. Soluble guanylate concentrations are high in A. franciscana embryos because of large stores of P1, P4-diguanosine 5-tetraphosphate (Gp4G) and its complex metabolic interconversions [122]. Multiple mechanisms are in place that apparently serve to suppress apoptosis during energy-limited states in A. franciscana.
Viral challenges and accommodation
Viral infection in crustaceans is a serious problem in aquaculture and consequently has been investigated in a number of shrimp species. This issue is most appropriately cast in the larger setting of viral manipulation of cell death, a field which continues to reveal molecular mechanisms by which viral pathogens evade host cell defense mechanisms that limit viral replication [123]. Immune and inflammatory responses are induced by viral infection, but particularly germane for this review, successful viral replication relies on the ability of viral products to block or delay apoptosis until sufficient progeny have been produced [123]. Thus, from the perspective of the virus, the invader must escape host-induced apoptosis [124–128].
As pointed out by Hilleman [126], blockage of the apoptotic system must have been a positive strategy for viral survival or else it would neither have been evolved nor retained. The mechanisms known for viral evasion of apoptosis are overwhelmingly numerous and diverse [126]. In addition to encoding homologues of anti-apoptotic proteins like Bcl-2 [125] or preventing p53-mediated apoptosis [124], viruses also produce proteins like M11L, which is targeted to the mitochondrion, associates with the peripheral benzodiazepine receptor, and prevents opening of the MPTP and cyt-c release [128]. Viral mitochondrial inhibitor of apoptosis (vMIA) can form a complex with the adenine nucleotide translocator (ANT) [129], and more recently has been shown to bind to Bax and prevent Bax-dependent mitochondrial outer membrane permeabilization (MOMP) by sequestering Bax at mitochondrion as a vMIA–Bax complex [129, 130].
Crustacean species are known for their ability to tolerate viral infection for prolonged periods. Undoubtedly, viral evasion of the host’s apoptotic response, as discussed above, is a substantial contributor to the long-term tolerance of crustaceans to viral infection, but additional factors may also be involved. This phenomenon in arthropods has been termed ‘viral accommodation’, which Flegel [131] defines as the process whereby “crustaceans and other arthropods actively accommodate viral pathogens as persistent infections that act as a kind of memory that functions to specifically reduce the severity of disease and to dampen viral triggered apoptosis.” It is appropriate to emphasize that crustaceans depend on, as do other invertebrates, innate immunity and do not display adaptive immunity [132]. The principle difference is that innate immunity relies upon predetermined molecular pattern recognition of microbial substances, so called pattern recognition proteins [126, 131, 133]. Adaptive immunity relies on specific recognition and response to antigens, which includes the capacity for immunological memory and the presence of antibodies in the serum [126]. Thus, an antibody-based immune response is not present in crustaceans. Consequently, it remains unclear what the ‘memory’ component in viral accommodation actually is, but Flegel [131] suggests that perhaps it is the persistent presence of the viruses themselves that constitute the memory element. A key point here is that crustaceans do not clear the viruses between episodes of infection, but rather viruses are continuously present at low levels, which is a pattern that contrasts to the general theme of viral clearance observed in vertebrates. The low-level, persistent infection is not analogous to ‘latent’ infections in vertebrates when viruses are inserted into the host genome and are not undergoing active replication; rather in crustacean infections, the viruses are replicating continuously and producing infectious virions [131].
So, what factors contribute to depressing apoptosis during persistent viral infection in crustaceans? Three of the most studied viral infections in shrimp are the infectious hypodermal and hematopoietic necrosis virus (IHHNV), the white spot syndrome virus (WSSV) and the yellow head virus (YHV). For these viral infections evidence suggests that when apoptosis rises to high frequency in cells/tissues of the host shrimp, the fate of the host animal is death, i.e., frequency of apoptosis is associated with mortality [134]. Thus, if apoptosis is serving as a host defense mechanism, it is not working well in crustaceans. Rather it seems that avoidance of rampant apoptosis is the successful strategy that correlates with keeping the host alive during viral infection.
At the molecular level, details relating to control of apoptosis during the above viral infections are few. Bangrak et al. [135] have shown that transcript levels for translationally controlled tumor protein (TCTP, or fortilin), which is described as an anti-apoptotic protein [136, 137], show little change early in the infection of Penaeus monodon with WSSV, but decrease substantially late in the episode when shrimp are becoming moribund. This pattern is consistent with apoptosis only becoming prominent when the battle is already lost. Transcript levels of defender against apoptotic death 1 (DAD1) have also been quantified during YHV infection of tiger shrimp [138]. While the frequency of apoptosis was unfortunately not measured in this study, the DAD1 mRNA profile is not consistent with the notion that apoptosis only becomes significant late in the infectious episode: mRNA for DAD1 dropped markedly very early (6 h) after YHV injection and well before the copy number of YHV reached high levels when the shrimp approached death (sometime after 36 h). DAD1 was statistically unchanged after 6 h post-injection. Finally, transcript levels for ribophorin 1, another component of the oligosaccharyltransferase complex (OST) along with DAD1 (cf. [139]), purportedly increase before the onset of DNA fragmentation during apoptosis. Molthathong et al. [140] reported a transcript profile that shows ribophorin 1 upregulated early after infection of tiger shrimp and well before the moribund stage of infection; again, one would predict that strong upregulation of a presumptive proapoptotic protein would occur in late stages of the infection, in order for the pattern to be consistent with apoptosis linked to death of the host.
Ignoring the conflicting patterns above, what is equally disconcerting about the transcripts thus far studied is that they encode proteins whose roles have not at all been fully defined. Linkage between the expression level of a gene product and apoptosis may simply be due to a disruption of cell homeostasis, not because the protein in question has anything to do with regulation of the apoptosis cascade per se. As beautifully articulated by Spierings et al. [141], apoptosis can be triggered by a bewildering array of conditions, including virtually anything that contributes to cellular stress or loss of housekeeping functions; so simply showing that a molecule influences apoptosis does not prove that it directly acts on any of the key players in the pathway. More work on crustacean apoptosis is needed before studies like those on TCTP, DAD1, and ribophorin 1 can be interpreted with confidence.
In a broader context, could it be that the persistent presence of metabolically active viruses in crustacean tissues (i.e., the ‘memory’ component of Flegel, [131]) promotes an altered cellular energy status as reflected by ATP/ADP ratio or energy charge? If so, perhaps a contributor to avoiding apoptosis is the inhibition of caspases by high ADP. Both ADP and GTP are known to strongly inhibit caspases in brine shrimp, as discussed above [20]. Alternatively, it could actually be that cellular ATP may be upregulated during persistent infection, as demonstrated by Chang et al. [142] for vaccinia virus infection of HeLa cells (epithelial cervical cancer cell line); two genes of the electron transport system in HeLa cells were elevated in response to the infection. It would be of interest to quantify adenine and guanine nucleotides in viral-infected and noninfected tissues from crustaceans to investigate whether or not these compounds contribute to the ‘memory’ component of viral accommodation.
Conclusions and future directions
We have summarized recent findings and highlighted emerging issues that provide insights into the regulation of apoptosis in crustaceans. A variety of sequences for crustacean homologues of apoptotic proteins from D. melanogaster and H. sapiens (Table 1) have been deposited at NCBI, but for most of these proteins the significance in regulating apoptosis in this subphylum has not been rigorously evaluated. Our knowledge about the molecular basis of apoptosis in arthropods rests predominantly on the work for D. melanogaster. By emphasizing key points that, in our opinion, need further attention we hope to stimulate more work in the field, because mechanisms that regulate apoptosis in crustaceans may differ significantly from the principles that govern life and death decisions in flies. The diversity of Bcl-2 type proteins in crustaceans appears to be greater than in D. melanogaster (Fig. 3). Interestingly, an EST with high homology to H. sapiens Apaf-1, but with no significant similarity to the D. melanogaster analog Dark, was found for the water flea D. pulex (Fig. 4). The degree to which apoptosis in crustaceans is regulated by proteins of the Bcl-2 family needs to be clarified. The calcium-induced MPTP appears to be absent in crustacean mitochondria [21, 74], but information is needed on the regulation of MOMP via mechanisms similar to Bax/Bak-mediated permeabilization in mammals. Despite a few evaluations of cell death during brain ontogenesis in the European lobster Homarus vulgaris, [143, 144], amazingly little is known about the role of apoptosis in crustacean development. There are several unresolved questions. How is viral accommodation mediated in shrimp? What are the metabolic requirements to survive persistent viral infections? What is the role of IAPs in controlling caspase activity? What additional features place constraints on mitochondrial-based pathways of apoptosis in species that survive severe environmental insults? Expanding our knowledge of how apoptotic pathways are blunted during states of severe metabolic arrest may point to new avenues for preventing cell death in human disease states and for the long-term stabilization of mammalian cells and tissues [145–148].
Acknowledgments
This work was supported by NIH Grants 1-RO1-GM071345-01 and 2-RO1-DK046270-14A1, and NSF Grant IOS-0920254.
Contributor Information
Michael A. Menze, Division of Cellular, Developmental and Integrative Biology, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Bldg, Baton Rouge, LA 70803, USA
Grady Fortner, Division of Cellular, Developmental and Integrative Biology, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Bldg, Baton Rouge, LA 70803, USA.
Suman Nag, Division of Cellular, Developmental and Integrative Biology, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Bldg, Baton Rouge, LA 70803, USA.
Steven C. Hand, Division of Cellular, Developmental and Integrative Biology, Department of Biological Sciences, Louisiana State University, 202 Life Sciences Bldg, Baton Rouge, LA 70803, USA
References
- 1.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Hassanin A. Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol. 2006;38:100–116. doi: 10.1016/j.ympev.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Bottger A, Alexandrova O. Programmed cell death in Hydra. Semin Cancer Biol. 2007;17:134–146. doi: 10.1016/j.semcancer.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Zmasek CM, Zhang Q, Ye Y, Godzik A. Surprising complexity of the ancestral apoptosis network. Genome Biol. 2007;8:R226. doi: 10.1186/gb-2007-8-10-r226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson AJ, Croce J, Carbonneau S, et al. The genomic underpinnings of apoptosis in Strongylocentrotus purpuratus. Dev Biol. 2006;300:321–334. doi: 10.1016/j.ydbio.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 6.Oberst A, Bender C, Green DR. Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ. 2008;15:1139–1146. doi: 10.1038/cdd.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hand SC, Menze MA. Mitochondria in energy-limited states: mechanisms that blunt the signaling of cell death. J Exp Biol. 2008;211:1829–1840. doi: 10.1242/jeb.000299. [DOI] [PubMed] [Google Scholar]
- 8.Tran SE, Meinander A, Eriksson JE. Instant decisions: transcription-independent control of death-receptor-mediated apoptosis. Trends Biochem Sci. 2004;29:601–608. doi: 10.1016/j.tibs.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Ferri KF, Kroemer G. Organelle-specific initiation of cell death pathways. Nat Cell Biol. 2001;3:E255–E263. doi: 10.1038/ncb1101-e255. [DOI] [PubMed] [Google Scholar]
- 10.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 11.Salvesen GS, Riedl SJ. Caspase mechanisms. In: Khosravi-Far R, White E, editors. Programmed cell death in cancer progression and therapy. Springer; Netherlands: 2008. pp. 13–23. [Google Scholar]
- 12.Salvesen GS, Abrams JM. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- 13.Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- 14.Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22:8543–8567. doi: 10.1038/sj.onc.1207107. [DOI] [PubMed] [Google Scholar]
- 15.Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 16.Kanda H, Igaki T, Kanuka H, Yagi T, Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J Biol Chem. 2002;277:28372–28375. doi: 10.1074/jbc.C200324200. [DOI] [PubMed] [Google Scholar]
- 17.Kauppila S, Maaty WS, Chen P, et al. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene. 2003;22:4860–4867. doi: 10.1038/sj.onc.1206715. [DOI] [PubMed] [Google Scholar]
- 18.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 19.Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiol Rev. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 20.Menze MA, Hand SC. Caspase activity during cell stasis: avoidance of apoptosis in an invertebrate extremophile, Artemia franciscana. Am J Physiol Regul Integr Comp Physiol. 2007;292:R2039–R2047. doi: 10.1152/ajpregu.00659.2006. [DOI] [PubMed] [Google Scholar]
- 21.Menze MA, Hutchinson K, Laborde SM, Hand SC. Mitochondrial permeability transition in the crustacean Artemia franciscana: absence of a calcium-regulated pore in the face of profound calcium storage. Am J Physiol Regul Integr Comp Physiol. 2005;289:R68–R76. doi: 10.1152/ajpregu.00844.2004. [DOI] [PubMed] [Google Scholar]
- 22.Leu JH, Wang HC, Kou GH, Lo CF. Penaeus monodon caspase is targeted by a white spot syndrome virus anti-apoptosis protein. Dev Comp Immunol. 2008;32:476–486. doi: 10.1016/j.dci.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Leu JH, Kuo YC, Kou GH, Lo CH. Molecular cloning and characterization of an inhibitor of apoptosis protein (IAP) from the tiger shrimp, Penaeus monodon. Dev Comp Immunol. 2008;32:121–133. doi: 10.1016/j.dci.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 24.Phongdara A, Wanna W, Chotigeat W. Molecular cloning and expression of caspase from white shrimp Penaeus merguiensis. Aquaculture. 2006;252:114–120. [Google Scholar]
- 25.Wongprasert K, Sangsuriya P, Phongdara A, Senapin S. Cloning and characterization of a caspase gene from black tiger shrimp (Penaeus monodon)-infected with white spot syndrome virus (WSSV) J Biotechnol. 2007;131:9–19. doi: 10.1016/j.jbiotec.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009 doi: 10.1146/annurev-genet-102108-134850. 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Krieser RJ, White K. Inside an enigma: do mitochondria contribute to cell death in Drosophila? Apoptosis. 2009;14:961–968. doi: 10.1007/s10495-009-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudgeon C, Qiu W, Sun S, Zhang L, Yu J. Transcriptional regulation of apoptosis. In: Yin XM, Dong Z, editors. Essentials of apoptosis. Humana Press; New York: 2009. pp. 239–260. [Google Scholar]
- 30.Srinivasula SM, Ashwell JD. IAPs: what’s in a name? Mol Cell. 2008;30:123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- 32.Clark AG, Eisen MB, Smith DR, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 33.Tenev T, Zachariou A, Wilson R, Paul A, Meier P. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 2002;21:5118–5129. doi: 10.1093/emboj/cdf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Igaki T, Suzuki Y, Tokushige N, Aonuma H, Takahashi R, Miura M. Evolution of mitochondrial cell death pathway: proapoptotic role of HtrA2/Omi in Drosophila. Biochem Biophys Res Commun. 2007;356:993–997. doi: 10.1016/j.bbrc.2007.03.079. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasula SM, Gupta S, Datta P, et al. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J Biol Chem. 2003;278:31469–31472. doi: 10.1074/jbc.C300240200. [DOI] [PubMed] [Google Scholar]
- 36.Iuchi Y, Okada F, Tsunoda S, et al. Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem J. 2009;419:149–158. doi: 10.1042/BJ20081526. [DOI] [PubMed] [Google Scholar]
- 37.Creagh EM, Conroy H, Martin SJ. Caspase-activation pathways in apoptosis and immunity. Immunol Rev. 2003;193:10–21. doi: 10.1034/j.1600-065x.2003.00048.x. [DOI] [PubMed] [Google Scholar]
- 38.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 39.Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006;18:677. doi: 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Cho DH, Hong YM, Lee HJ, et al. Induced inhibition of ischemic/hypoxic injury by APIP, a novel Apaf-1-interacting protein. J Biol Chem. 2004;279:39942–39950. doi: 10.1074/jbc.M405747200. [DOI] [PubMed] [Google Scholar]
- 42.Cho DH, Lee HJ, Kim HJ, et al. Suppression of hypoxic cell death by APIP-induced sustained activation of AKT and ERK1/2. Oncogene. 2007;26:2809–2814. doi: 10.1038/sj.onc.1210080. [DOI] [PubMed] [Google Scholar]
- 43.Saelens X, Festjens N, Vande Walle L, van Gurp M, van Loo G, Vandenabeele P. Toxic proteins released from mitochondria in cell death. Oncogene. 2004;23:2861–2874. doi: 10.1038/sj.onc.1207523. [DOI] [PubMed] [Google Scholar]
- 44.Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Wang J, Gengyo-Ando K, et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol. 2007;9:541–549. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- 46.Joza N, Pospisilik JA, Hangen E, et al. AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci. 2009;1171:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- 47.Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 48.Temme C, Weissbach R, Lilie H, et al. The Drosophila melanogaster gene cg4930 encodes a high affinity inhibitor for endonuclease G. J Biol Chem. 2009;284:8337–8348. doi: 10.1074/jbc.M808319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joza N, Galindo K, Pospisilik JA, et al. The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ. 2008;15:1009–1018. doi: 10.1038/cdd.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 51.Xue L, Igaki T, Kuranaga E, Kanda H, Miura M, Xu T. Tumor suppressor CYLD regulates JNK-induced cell death in Drosophila. Dev Cell. 2007;13:446–454. doi: 10.1016/j.devcel.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Lee NK, Lee SY. Modulation of life and death by the tumor necrosis factor receptor-associated factors (TRAFs) J Biochem Mol Biol. 2002;35:61–66. doi: 10.5483/bmbrep.2002.35.1.061. [DOI] [PubMed] [Google Scholar]
- 53.Haworth RA, Hunter DR. The Ca2+ -induced membrane transition in mitochondria. II. Nature of the Ca2+ trigger site. Arch Biochem Biophys. 1979;195:460–467. doi: 10.1016/0003-9861(79)90372-2. [DOI] [PubMed] [Google Scholar]
- 54.Hunter DR, Haworth RA. The Ca2+ -induced membrane transition in mitochondria. I. The protective mechanisms. Arch Biochem Biophys. 1979;195:453–459. doi: 10.1016/0003-9861(79)90371-0. [DOI] [PubMed] [Google Scholar]
- 55.Gunter TE, Pfeiffer DR. Mechanisms by which mitochondria transport calcium. Am J Physiol. 1990;258:C755–C786. doi: 10.1152/ajpcell.1990.258.5.C755. [DOI] [PubMed] [Google Scholar]
- 56.Halestrap AP, Doran E, Gillespie JP, O’Toole A. Mitochondria and cell death. Biochem Soc Trans. 2000;28:170–177. doi: 10.1042/bst0280170. [DOI] [PubMed] [Google Scholar]
- 57.Hunter DR, Haworth RA, Southard JH. Relationship between configuration, function, and permeability in calcium-treated mitochondria. J Biol Chem. 1976;251:5069–5077. [PubMed] [Google Scholar]
- 58.Bernardi P. The permeability transition pore. Control points of a cyclosporin A-sensitive mitochondrial channel involved in cell death. Biochim Biophys Acta. 1996;1275:5–9. doi: 10.1016/0005-2728(96)00041-2. [DOI] [PubMed] [Google Scholar]
- 59.Bernardi P, Krauskopf A, Basso E, et al. The mitochondrial permeability transition from in vitro artifact to disease target. Febs J. 2006;273:2077–2099. doi: 10.1111/j.1742-4658.2006.05213.x. [DOI] [PubMed] [Google Scholar]
- 60.Petronilli V, Cola C, Bernardi P. Modulation of the mitochondrial cyclosporin A-sensitive permeability transition pore. II. The minimal requirements for pore induction underscore a key role for transmembrane electrical potential, matrix pH, and matrix Ca2+ J Biol Chem. 1993;268:1011–1016. [PubMed] [Google Scholar]
- 61.Baines CP, Kaiser RA, Sheiko T, Craigen WJ, Molkentin JD. Voltage-dependent anion channels are dispensable for mitochondrial-dependent cell death. Nat Cell Biol. 2007;9:550–555. doi: 10.1038/ncb1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krauskopf A, Eriksson O, Craigen WJ, Forte MA, Bernardi P. Properties of the permeability transition in VDAC1(−/−) mitochondria. Biochim Biophys Acta. 2006;1757:590–595. doi: 10.1016/j.bbabio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 63.Kokoszka JE, Waymire KG, Levy SE, et al. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–465. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baines CP, Kaiser RA, Purcell NH, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 65.Basso E, Fante L, Fowlkes J, Petronilli V, Forte MA, Bernardi P. Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J Biol Chem. 2005;280:18558–18561. doi: 10.1074/jbc.C500089200. [DOI] [PubMed] [Google Scholar]
- 66.Leung AW, Varanyuwatana P, Halestrap AP. The mitochondrial phosphate carrier interacts with cyclophilin D and may play a key role in the permeability transition. J Biol Chem. 2008;283:26312–26323. doi: 10.1074/jbc.M805235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 68.Zamzami N, Kroemer G. The mitochondrion in apoptosis: how Pandora’s box opens. Nat Rev Mol Cell Biol. 2001;2:67–71. doi: 10.1038/35048073. [DOI] [PubMed] [Google Scholar]
- 69.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 70.Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- 71.Kakkar P, Singh BK. Mitochondria: a hub of redox activities and cellular distress control. Mol Cell Biochem. 2007;305:235–253. doi: 10.1007/s11010-007-9520-8. [DOI] [PubMed] [Google Scholar]
- 72.Orrenius S, Gogvadze V, Zhivotovsky B. Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol. 2007;47:143–183. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 73.Ryter SW, Kim HP, Hoetzel A, et al. Mechanisms of cell death in oxidative stress. Antioxid Redox Signal. 2007;9:49–89. doi: 10.1089/ars.2007.9.49. [DOI] [PubMed] [Google Scholar]
- 74.Holmann JD, Hand SC. Metabolic depression is delayed and mitochondrial impairment averted during prolonged anoxia in the ghost shrimp, Lepidophthalmus louisianensis (Schmitt, 1935) J Exp Mar Biol Ecol. 2009;376:85–93. doi: 10.1016/j.jembe.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andreyev AY, Fahy B, Fiskum G. Cytochrome c release from brain mitochondria is independent of the mitochondrial permeability transition. FEBS Lett. 1998;439:373–376. doi: 10.1016/s0014-5793(98)01394-5. [DOI] [PubMed] [Google Scholar]
- 76.Chalmers S, Nicholls DG. The relationship between free and total calcium concentrations in the matrix of liver and brain mitochondria. J Biol Chem. 2003;278:19062–19070. doi: 10.1074/jbc.M212661200. [DOI] [PubMed] [Google Scholar]
- 77.Nicholls DG, Chalmers S. The integration of mitochondrial calcium transport and storage. J Bioenerg Biomembr. 2004;36:277–281. doi: 10.1023/B:JOBB.0000041753.52832.f3. [DOI] [PubMed] [Google Scholar]
- 78.He L, Lemasters JJ. Regulated and unregulated mitochondrial permeability transition pores: a new paradigm of pore structure and function? FEBS Lett. 2002;512:1–7. doi: 10.1016/s0014-5793(01)03314-2. [DOI] [PubMed] [Google Scholar]
- 79.Sokolova IM, Evans S, Hughes FM. Cadmium-induced apoptosis in oyster hemocytes involves disturbance of cellular energy balance but no mitochondrial permeability transition. J Exp Biol. 2004;207:3369–3380. doi: 10.1242/jeb.01152. [DOI] [PubMed] [Google Scholar]
- 80.Toninello A, Salvi M, Colombo L. The membrane permeability transition in liver mitochondria of the great green goby Zosterisessor ophiocephalus (Pallas) J Exp Biol. 2000;203(Pt 22):3425–3434. doi: 10.1242/jeb.203.22.3425. [DOI] [PubMed] [Google Scholar]
- 81.Krumschnabel G, Manzl C, Berger C, Hofer B. Oxidative stress, mitochondrial permeability transition, and cell death in Cu-exposed trout hepatocytes. Toxicol Appl Pharmacol. 2005;209:62–73. doi: 10.1016/j.taap.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 82.Savina MV, Emelyanova LV, Belyaeva EA. Bioenergetic parameters of lamprey and frog liver mitochondria during metabolic depression and activity. Comp Biochem Physiol B Biochem Mol Biol. 2006;145:296–305. doi: 10.1016/j.cbpb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 83.Hughes FM, Jr, Cidlowski JA. Potassium is a critical regulator of apoptotic enzymes in vitro and in vivo. Adv Enzyme Regul. 1999;39:157–171. doi: 10.1016/s0065-2571(98)00010-7. [DOI] [PubMed] [Google Scholar]
- 84.Bortner CD, Cidlowski JA. Caspase independent/dependent regulation of K(+), cell shrinkage, and mitochondrial membrane potential during lymphocyte apoptosis. J Biol Chem. 1999;274:21953–21962. doi: 10.1074/jbc.274.31.21953. [DOI] [PubMed] [Google Scholar]
- 85.Hughes FM, Jr, Bortner CD, Purdy GD, Cidlowski JA. Intracellular K+ suppresses the activation of apoptosis in lymphocytes. J Biol Chem. 1997;272:30567–30576. doi: 10.1074/jbc.272.48.30567. [DOI] [PubMed] [Google Scholar]
- 86.Beem E, Holliday LS, Segal MS. The 1.4-MDa apoptosome is a critical intermediate in apoptosome maturation. Am J Physiol Cell Physiol. 2004;287:C664–C672. doi: 10.1152/ajpcell.00232.2003. [DOI] [PubMed] [Google Scholar]
- 87.Cain K, Langlais C, Sun XM, Brown DG, Cohen GM. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J Biol Chem. 2001;276:41985–41990. doi: 10.1074/jbc.M107419200. [DOI] [PubMed] [Google Scholar]
- 88.Kulms D, Schwarz T. Independent contribution of three different pathways to ultraviolet-B-induced apoptosis. Biochem Pharmacol. 2002;64:837–841. doi: 10.1016/s0006-2952(02)01146-2. [DOI] [PubMed] [Google Scholar]
- 89.Petit-Frere C, Capulas E, Lowe JE, et al. Ultraviolet-B-induced apoptosis and cytokine release in xeroderma pigmentosum keratinocytes. J Invest Dermatol. 2000;115:687–693. doi: 10.1046/j.1523-1747.2000.00093.x. [DOI] [PubMed] [Google Scholar]
- 90.Patrick MH. Studies on thymine-derived UV photoproducts in DNA–I. Formation and biological role of pyrimidine adducts in DNA. Photochem Photobiol. 1977;25:357–372. doi: 10.1111/j.1751-1097.1977.tb07355.x. [DOI] [PubMed] [Google Scholar]
- 91.Murphy G, Young AR, Wulf HC, Kulms D, Schwarz T. The molecular determinants of sunburn cell formation. Exp Dermatol. 2001;10:155–160. doi: 10.1034/j.1600-0625.2001.010003155.x. [DOI] [PubMed] [Google Scholar]
- 92.Chouinard N, Valerie K, Rouabhia M, Huot J. UVB-mediated activation of p38 mitogen-activated protein kinase enhances resistance of normal human keratinocytes to apoptosis by stabilizing cytoplasmic p53. Biochem J. 2002;365:133–145. doi: 10.1042/BJ20020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.She QB, Chen N, Dong Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J Biol Chem. 2000;275:20444–20449. doi: 10.1074/jbc.M001020200. [DOI] [PubMed] [Google Scholar]
- 94.Reed JC. Double identity for proteins of the Bcl-2 family. Nature. 1997;387:773–776. doi: 10.1038/42867. [DOI] [PubMed] [Google Scholar]
- 95.Miguel NC, Wajsenzon IJ, Takiya CM, et al. Catalase, Bax and p53 expression in the visual system of the crab Ucides cordatus following exposure to ultraviolet radiation. Cell Tissue Res. 2007;329:159–168. doi: 10.1007/s00441-007-0410-x. [DOI] [PubMed] [Google Scholar]
- 96.Tomicic MT, Christmann M, Kaina B. Apoptosis in UVC light irradiated p53 wild-type, apaf-1 and p53 knockout mouse embryonic fibroblasts: interplay of receptor and mitochondrial pathway. Apoptosis. 2005;10:1295–1304. doi: 10.1007/s10495-005-1392-3. [DOI] [PubMed] [Google Scholar]
- 97.de Oliveira Miguel NC, Meyer-Rochow VB, Allodi S. Ultrastructural study of first and second order neurons in the visual system of the crab Ucides cordatus following exposure to ultraviolet radiation. Micron. 2002;33:627–637. doi: 10.1016/s0968-4328(02)00030-6. [DOI] [PubMed] [Google Scholar]
- 98.Bonner WM. Low-dose radiation: thresholds, bystander effects, and adaptive responses. P Natl Acad Sci USA. 2003;100:4973–4975. doi: 10.1073/pnas.1031538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mothersill C, Lyng F, Mulford A, et al. Effect of low doses of ionizing radiation on cells cultured from the hematopoietic tissue of the Dublin Bay prawn, Nephrops norvegicus. Radiat Res. 2001;156:241–250. doi: 10.1667/0033-7587(2001)156[0241:eoldoi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 100.Lyng FM, Reilly S, Seymour CB, Mothersill CE. Morphological evidence of radiation induced delayed apoptosis in the distant progeny of irradiated cells. Radiat Environ Biophys. 1996;35:237–283. doi: 10.1007/s004110050040. [DOI] [PubMed] [Google Scholar]
- 101.Peakall DB. Phthalate esters: occurrence and biological effects. Residue Rev. 1975;54:1–41. doi: 10.1007/978-1-4612-9857-1_1. [DOI] [PubMed] [Google Scholar]
- 102.Sung HH, Kao WY, Su YJ. Effects and toxicity of phthalate esters to hemocytes of giant freshwater prawn, Macrobrachium rosenbergii. Aquat Toxicol. 2003;64:25–37. doi: 10.1016/s0166-445x(03)00011-0. [DOI] [PubMed] [Google Scholar]
- 103.Lotufo GR, Farrar JD, Inouye LS, Bridges TS, Ringelberg DB. Toxicity of sediment-associated nitroaromatic and cyclonitramine compounds to benthic invertebrates. Environ Toxicol Chem. 2001;20:1762–1771. [PubMed] [Google Scholar]
- 104.Oweson CA, Baden SP, Hernroth BE. Manganese induced apoptosis in haematopoietic cells of Nephrops norvegicus (L.) Aquat Toxicol. 2006;77:322–328. doi: 10.1016/j.aquatox.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 105.Cheng SH, Chan KW, Chan PK, So CH, Lam PKS, Wu RSS. Whole-mount in situ TUNEL method revealed ectopic pattern of apoptosis in cadmium treated naupliar larvae of barnacle (Balanus amphitrite, Darwin) Chemosphere. 2004;55:1387–1394. doi: 10.1016/j.chemosphere.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 106.Prabhakaran K, Ghosh D, Chapman GD, Gunasekar PG. Molecular mechanism of manganese exposure-induced dopaminergic toxicity. Brain Res Bull. 2008;76:361–367. doi: 10.1016/j.brainresbull.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 107.Kitazawa M, Anantharam V, Yang Y, Hirata Y, Kanthasamy A, Kanthasamy AG. Activation of protein kinase C delta by proteolytic cleavage contributes to manganese-induced apoptosis in dopaminergic cells: protective role of Bcl-2. Biochem Pharmacol. 2005;69:133–146. doi: 10.1016/j.bcp.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 108.Miralto A, Barone G, Romano G, et al. The insidious effect of diatoms on copepod reproduction. Nature. 1999;402:173–176. [Google Scholar]
- 109.Paffenhofer GA, Ianora A, Miralto A, et al. Colloquium on diatom-copepod interactions. Mar Ecol-Prog Ser. 2005;286:293–305. [Google Scholar]
- 110.Romano G, Russo GL, Buttino I, Ianora A, Miralto A. A marine diatom-derived aldehyde induces apoptosis in copepod and sea urchin embryos. J Exp Biol. 2003;206:3487–3494. doi: 10.1242/jeb.00580. [DOI] [PubMed] [Google Scholar]
- 111.Ianora A, Miralto A, Poulet SA, et al. Aldehyde suppression of copepod recruitment in blooms of a ubiquitous planktonic diatom. Nature. 2004;429:403–407. doi: 10.1038/nature02526. [DOI] [PubMed] [Google Scholar]
- 112.Clegg JS, Drinkwater LE, Sorgeloos P. The metabolic status of diapause embryos of Artemia franciscana (SFB) Physiol Zool. 1996;69:49–66. [Google Scholar]
- 113.Reynolds JA, Hand SC. Embryonic diapause highlighted by differential expression of mRNAs for ecdysteroidogenesis, transcription and lipid sparing in the cricket Allonemobius socius. J Exp Biol. 2009;212:2075–2084. doi: 10.1242/jeb.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reynolds JA, Hand SC. Decoupling development and energy flow during embryonic diapause in the cricket, Allonemobius socius. J Exp Biol. 2009;212:2065–2074. doi: 10.1242/jeb.027359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brand MD, Chien LF, Ainscow EK, Rolfe DF, Porter RK. The causes and functions of mitochondrial proton leak. Biochim Biophys Acta. 1994;1187:132–139. doi: 10.1016/0005-2728(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 116.Chereau D, Zou H, Spada AP, Wu JC. A nucleotide binding site in caspase-9 regulates apoptosome activation. Biochemistry. 2005;44:4971–4976. doi: 10.1021/bi047360+. [DOI] [PubMed] [Google Scholar]
- 117.Chandra D, Bratton SB, Person MD, et al. Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome C and inhibiting apoptosome. Cell. 2006;125:1333–1346. doi: 10.1016/j.cell.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 118.Clegg J. Embryos of Artemia franciscana survive four years of continuous anoxia: the case for complete metabolic rate depression. J Exp Biol. 1997;200:467–475. doi: 10.1242/jeb.200.3.467. [DOI] [PubMed] [Google Scholar]
- 119.Stocco DM, Beers PC, Warner AH. Effect of anoxia on nucleotide metabolism in encysted embryos of the brine shrimp. Dev Biol. 1972;27:479–493. doi: 10.1016/0012-1606(72)90187-x. [DOI] [PubMed] [Google Scholar]
- 120.Carpenter JF, Hand SC. Arrestment of carbohydrate-metabolism during anaerobic dormancy and aerobic acidosis in Artemia embryos—determination of Ph-sensitive control points. J Comp Physiol B. 1986;156:451–459. [Google Scholar]
- 121.Busa WB, Crowe JH, Matson GB. Intracellular Ph and the metabolic status of dormant and developing Artemia embryos. Arch Biochem Biophys. 1982;216:711–718. doi: 10.1016/0003-9861(82)90261-2. [DOI] [PubMed] [Google Scholar]
- 122.Finamore FJ, Warner AH. The occurrence of P1, P4-diguanosine 5′-tetraphosphate in brine shrimp eggs. J Biol Chem. 1963;238:344–348. [PubMed] [Google Scholar]
- 123.Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 124.McLean JE, Ruck A, Shirazian A, Pooyaei-Mehr F, Zakeri ZF. Viral manipulation of cell death. Curr Pharm Des. 2008;14:198–220. doi: 10.2174/138161208783413329. [DOI] [PubMed] [Google Scholar]
- 125.Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. Viral control of mitochondrial apoptosis. PLoS Pathog. 2008;4:e1000018. doi: 10.1371/journal.ppat.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hilleman MR. Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14560–14566. doi: 10.1073/pnas.0404758101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Irusta PM, Chen YB, Hardwick JM. Viral modulators of cell death provide new links to old pathways. Curr Opin Cell Biol. 2003;15:700–705. doi: 10.1016/j.ceb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 128.Everett H, Barry M, Sun X, et al. The myxoma poxvirus protein, M11L, prevents apoptosis by direct interaction with the mitochondrial permeability transition pore. J Exp Med. 2002;196:1127–1139. doi: 10.1084/jem.20011247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goldmacher VS. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie. 2002;84:177–185. doi: 10.1016/s0300-9084(02)01367-6. [DOI] [PubMed] [Google Scholar]
- 130.Arnoult D, Bartle LM, Skaletskaya A, et al. Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc Natl Acad Sci U S A. 2004;101:7988–7993. doi: 10.1073/pnas.0401897101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flegel TW. Update on viral accommodation, a model for host-viral interaction in shrimp and other arthropods. Dev Comp Immunol. 2007;31:217–231. doi: 10.1016/j.dci.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 132.Bayne CJ. Origins and evolutionary relationships between the innate and adaptive arms of immune systems. Integr Comp Biol. 2003;43:293–299. doi: 10.1093/icb/43.2.293. [DOI] [PubMed] [Google Scholar]
- 133.Soderhall K. Special issue: invertebrate immunity. Dev Comp Immunol. 1999;23:263–266. doi: 10.1016/s0145-305x(99)00009-9. [DOI] [PubMed] [Google Scholar]
- 134.Wu JL, Muroga K. Apoptosis does not play an important role in the resistance of ‘immune’ Penaeus japonicus against white spot syndrome virus. J Fish Dis. 2004;27:15–21. doi: 10.1046/j.1365-2761.2003.00491.x. [DOI] [PubMed] [Google Scholar]
- 135.Bangrak P, Graidist P, Chotigeat W, Phongdara A. Molecular cloning and expression of a mammalian homologue of a translationally controlled tumor protein (TCTP) gene from Penaeus monodon shrimp. J Biotechnol. 2004;108:219–226. doi: 10.1016/j.jbiotec.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 136.Li F, Zhang D, Fujise K. Characterization of fortilin, a novel antiapoptotic protein. J Biol Chem. 2001;276:47542–47549. doi: 10.1074/jbc.M108954200. [DOI] [PubMed] [Google Scholar]
- 137.Zhang D, Li F, Weidner D, Mnjoyan ZH, Fujise K. Physical and functional interaction between myeloid cell leukemia 1 protein (MCL1) and Fortilin. The potential role of MCL1 as a fortilin chaperone. J Biol Chem. 2002;277:37430–37438. doi: 10.1074/jbc.M207413200. [DOI] [PubMed] [Google Scholar]
- 138.Molthathong S, Senapin S, Klinbunga S, Puanglarp N, Rojtinnakorn J, Flegel TW. Down-regulation of defender against apoptotic death (DAD1) after yellow head virus (YHV) challenge in black tiger shrimp Penaeus monodon. Fish Shellfish Immun. 2008;24:173–179. doi: 10.1016/j.fsi.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 139.Lindholm P, Kuittinen T, Sorri O, et al. Glycosylation of phytepsin and expression of dad1, dad2 and ost1 during onset of cell death in germinating barley scutella. Mech Develop. 2000;93:169–173. doi: 10.1016/s0925-4773(00)00254-9. [DOI] [PubMed] [Google Scholar]
- 140.Molthathong S, Buaklin A, Senapin S, Klinbunga S, Rojtinnakorn J, Flegel TW. Up-regulation of Ribophorin l after yellow head virus (YHV) challenge in black tiger shrimp Penaeus monodon. Fish Shellfish Immun. 2008;25:40–46. doi: 10.1016/j.fsi.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 141.Spierings D, McStay G, Saleh M, et al. Connected to death: the (unexpurgated) mitochondrial pathway of apoptosis. Science. 2005;310:66–67. doi: 10.1126/science.1117105. [DOI] [PubMed] [Google Scholar]
- 142.Chang CW, Li HC, Hsu CF, Chang CY, Lo SY. Increased ATP generation in the host cell is required for efficient vaccinia virus production. J Biomed Sci. 2009;16:80. doi: 10.1186/1423-0127-16-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Harzsch S, Benton J, Dawirs RR, Beltz B. A new look at embryonic development of the visual system in decapod crustaceans: neuropil formation, neurogenesis, and apoptotic cell death. J Neurobiol. 1999;39:294–306. [PubMed] [Google Scholar]
- 144.Harzsch S, Miller J, Benton J, Beltz B. From embryo to adult: persistent neurogenesis and apoptotic cell death shape the lobster deutocerebrum. J Neurosci. 1999;19:3472–3485. doi: 10.1523/JNEUROSCI.19-09-03472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buchanan SS, Menze MA, Hand SC, Pyatt DW, Carpenter JF. Cryopreservation of human hematopoetic stem and progenitor cells loaded with trehalose: transient permeabilization via the adenosine triphosphate-dependent P2Z receptor channel. Cell Preserv Tech. 2005;3:212–222. [Google Scholar]
- 146.Crowe JH, Crowe LM, Tablin F. Stabilization of dry mammalian cells: lessons from nature. Integr Comp Biol. 2005;44:542. doi: 10.1093/icb/45.5.810. [DOI] [PubMed] [Google Scholar]
- 147.Menze MA, Clavenna MJ, Hand SC. Depression of cell metabolism and proliferation by membrane-permeable and -impermeable modulators: role for AMP-to-ATP ratio. Am J Physiol Regul Integr Comp Physiol. 2005;288:R501–R510. doi: 10.1152/ajpregu.00490.2004. [DOI] [PubMed] [Google Scholar]
- 148.Hand SC, Hagedorn M. New approaches for cell and animal preservation: lessons from aquatic organisms. In: Walsh PJ, Smith LE, Fleming LE, Solo-Gabriele H, Gerwick WH, editors. Oceans and human health: risks and remedies from the seas. Academic Press; London: 2008. pp. 611–629. [Google Scholar]
- 149.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]