Abstract
Over-expression of chemokine receptor 4 (CXCR4) is present in a majority of cancers, has been linked to an aggressive phenotype, and may indicate the metastatic potential of primary tumor. Several CXCR4 targeted therapeutics are in clinical trials and the development of the corresponding imaging agents is an area of active interest. Previously, 64Cu-labeled imaging agents for CXCR4 have provided clear images of CXCR4-bearing tissues in relevant experimental models but demonstrated fast washout from tissues harboring receptor. Addition of stabilizing bridges is known to provide more robust chelator-Cu(II) complexes. In addition, bridged cyclam-based CXCR4 binding agents demonstrated increased receptor residence times relative to existing agents. Based on that knowledge we synthesized several bridged cyclam analogs of AMD3465, a monocyclam-based CXCR4 imaging agent, to increase the retention time of the tracer bound to the receptor to allow for protracted imaging and improved target-to-non-target ratios. Specific accumulation of two radiolabeled, cross-bridged analogs ([64Cu] RAD1–24 and [64Cu]RAD1–52) was observed in U87-stb-CXCR4 tumors in both PET/CT imaging and biodistribution studies. At 90 min post-injection of radiotracer, tumor-to-muscle and tumor-to-blood ratios reached 106.05 ± 17.19 and 28.08 ± 4.78, respectively, for cross-bridged pyrimidine analog [64Cu]RAD1–52. Receptor blockade performed in vivo denoted target binding specificity. The biodistribution and PET/CT imaging studies with the radiolabeled bridged cyclams demonstrated longer tumor retention and comparable uptake to [64Cu]AMD3465, though [64Cu]AMD3465 demonstrated superior overall pharmacokinetics.
Introduction
Chemokine receptor 4 (CXCR4) is a seven transmembrane spanning G-protein-coupled receptor. Its primary endogenous ligand is chemokine ligand 12 (CXCL12). CXCR4 is involved in AIDS as co-receptor for the entry of HIV into T-cells, and known to play major roles in several diseases including rheumatoid arthritis, systemic lupus erythematosis and cancer [1]. CXCR4 is over-expressed in more than 23 tumor types as well as metastases, and this overexpression results in poor outcome. Activation of CXCR4 by CXCL12 leads to G-protein–coupled signaling through extracellular signal-regulated kinases (ERK1/2), Akt effectors, and mitogen-activated protein kinase (MAPK) pathways, promoting cell survival, proliferation, and chemotaxis [2]. The CXCR4-CXCL12 axis mediates resistance to conventional as well as targeted therapies by recruiting myeloid bone marrow-derived cells to facilitate tumor recurrence and metastasis and by promoting angiogenesis [1,3]. Neutralization of CXCR4/CXCL12 chemotaxism using anti-CXCR4 antibodies, peptide antagonists or low-molecular-weight agents significantly reduces metastasis [1,4,5]. Due to its therapeutic importance, several inhibitors of this signaling pathway are now in clinical trials (BKT140 by Biokine Therapeutics Ltd, and BMS-936564 by Bristol-Myers Squibb). CXCR4 is also expressed in several normal tissues [6] suggesting that quantitative knowledge of its presence non-invasively, which can be accomplished by imaging, would be beneficial for therapeutic guidance.
Imaging of CXCR4 expression can be achieved using radiolabeled peptides, antibodies and small molecules, such as cyclams and benzimidazoles [7,8,9,10,11,12,13,14,15]. Conjugates based on T140, a 14-residue peptide, and FC131, a cyclic pentapeptide, have been used to image CXCR4 [7,16]. Bimodal (SPECT and Optical) and optical imaging agents based on peptides, CXCL12 or antibodies have also been described [15]. Amongst all the imaging agents previously reported, the ones based on the pentapeptide CPCR4 labeled with 68Ga and both low molecular weight agents AMD3100 and AMD3465 labeled with 64Cu, demonstrated superior pharmacokinetics and image contrast [17,18,19].
The inherent affinity of the cyclam moiety present in AMD3100 and AMD3465 for metal coordination has been used to synthesize 64Cu-labeled imaging agents [17,18]. It has been shown that both of those agents can be used to image graded levels of CXCR4 expression in a variety of tumor models [9,17,18]. Of those two agents [64Cu]AMD3465 demonstrated the highest target-to-non-target ratios offering a suitable scaffold for further optimization and probe development [17].
Synthesis of side- and cross-bridged analogs of AMD3100 has been performed to improve the affinity of cyclam-based agents to CXCR4 [20]. Complexes of copper with cross-bridged cyclams have been shown to be six to eight times more stable than their non-bridged counterparts [20,21,22]. Based on that knowledge, we synthesized analogs of the high-affinity CXCR4 inhibitor AMD3465 that possess the bridged cyclam motif to improve affinity to CXCR4, to increase stability of the metal complex and to improve the residence time of the analogs once bound to the receptor, allowing for increased tumor retention and protracted imaging. In the present study, we are reporting the synthesis, radiolabeling and biological evaluation of several bridged cyclams as imaging agents for CXCR4. Using subcutaneous brain tumor models stably expressing CXCR4, we demonstrate that cross-bridged analogs of AMD3465 can be used as highly specific positron emission tomography (PET) imaging agents with comparable tumor uptake to that of [64Cu]AMD3465. The high target-to-non-target ratios and longer tumor retention times observed in biodistribution studies support further optimization of the cross-bridged class of agents.
Materials and Methods
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise mentioned. Bridged cyclams 9 and 16 were purchased from Macrocyclics. Reactions were monitored using thin layer chromatography (TLC) on 0.20 mm pre-coated silica gel (EMD, catalog no: HX074938) and visualized by ultraviolet light (254 nm), I2, or 1% ninhydrin in EtOH. Purification was achieved by flash chromatograph using silica gel (Bodman, Aston, PA; MP SiliTech 32–63 D 60 Ǻ), high-performance liquid chromatography (HPLC) or Sep-Pak cartridges (Waters, catalog no: WAT036925). Yields refer to chromatographically and spectroscopically pure compounds. Proton and carbon-13 NMR spectra were obtained on Bruker ultrashield™ 400 MHz. Chemical shifts (δ) for both proton and carbon-13 were reported in ppm with respect to NMR solvent residuals used. Signal patterns are indicated as s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of doublets; b, broad. Low-resolution ESI mass spectra were recorded on Bruker Daltonics Esquire 3000 Plus spectrometer. Higher-resolution mass spectra were obtained by Bruker MicrO-TOF II at the University of Notre Dame Mass Spectrometry and Proteomics Facility (Notre Dame, IN). HPLC purification (radiosynthetic and non-radiosynthetic) was performed by Phenomenex C-18 Luna 5 µm, 10 × 250 mm2 semi-preparative column, using a Varian Prostar System (Palo Alto,CA) equipped with a Varian 325 variable wavelength detector and a Bioscan NaI scintillation detector connected to a Bioscan Flow-Count system. All products were determined to be greater than 95% pure by HPLC. Non-radiolabeled eluents were monitored at 220 nm and 266 nm, and 64Cu-radiolabeled eluents were detected at 266 nm. Syntheses of intermediates 3–8 and 17 are shown in the supplementary section.
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-ylmethyl)benzyl)-2-nitro-N-(pyridin-2-ylmethyl)benzenesulfonamide (10)
A mixture of cross-bridged cyclam (9, 0.02 g, 0.088 mmol), 7 (0.043 g, 0.088 mmol) and K2CO3 (0.012 g, 0.088 mmol) in acetonitrile was stirred at room temperature overnight. The undissolved K2CO3 was removed by filtration, and the solvent was removed under reduced pressure. The residue was purified by Sep-Pak cartridge (5–10% MeOH/H2O with 0.1% TFA) to give the desired product 10 (0.02 g, 35% yield). 1H NMR (400 MHz, MeOD): δ 1.57 (bt, J = 18.1 Hz, 2H), 2.11–2.28 (m, 2H), 2.35–2.55 (m, 4H), 2.68–2.86 (m, 6H), 2.92–3.20 (m, 6H), 3.32–3.3.58 (m, 4H), 4.31 (d, J = 15.2 Hz, 1H), 4.43 (d, J = 15.2 Hz, 1H), 4.55 (s, 2H), 4.63 (s, 2H), 7.12–7.27 (m, 6H), 7.55 (m, 2H), 7.77 (t, J = 4.0 Hz, 1H), 7.95 (d, J = 8.0 Hz, 1H), 8.30 (d, J = 4.1 Hz, 1H); 13C NMR (100 MHz, MeOD): δ 19.73, 20.55, 43.13, 47.36, 49.15, 50.32, 51.02, 52.85, 53.47, 55.34, 56.51, 56.74, 57.79, 59.23, 59.58, 125.43, 125.62, 125.86, 129.13, 130.76, 132.08, 133.33, 133.60, 136.01, 139.56, 142.16, 147.26, 149.52, 155.82; ESI-MS m/z: 622.3 [M+1].
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-ylmethyl)benzyl)-2-nitro-N-(pyrimidin-2-ylmethyl)benzenesulfonamide (11)
A mixture of cross-bridged cyclam (9, 0.02 g, 0.088 mmol), crude 8 (0.043 g, 0.088 mmol) and K2CO3 (0.012 g, 0.088 mmol) in acetonitrile was stirred at room temperature overnight. The undissolved K2CO3 was removed by filtration, and the solvent was removed under reduced pressure. The residue was purified by Sep-Pak cartridge (10% MeOH/H2O with 0.1% TFA) to give the desired product 11 (0.018 g, 32% yield). 1H NMR (400 MHz, MeOD): 1.63–1.72 (m, 2H), 2.33-2.29 (m, 1H), 3.79-2.50 (m, 21H), 4.62 (s, 4H), 4.76 (s, 2H), 7.32–7.44 (m, 4H), 7.61–7.73 (m, 4H), 8.05 (d, J = 7.6 Hz, 1H), 8.48 (s, 2H); 13C NMR (100 MHz, MeOD): δ 19.16, 19.48, 41.69, 48.84, 49.59, 51.30, 51.73, 53.07, 54.06, 55.30, 55.73, 56.55, 57.69, 58.13, 60.15, 119.85, 123.81, 127.78, 128.88, 129.19, 130.68, 131.56, 131.74, 132.99, 133.10, 133.89, 138.71, 147.99157.15, 164.83; ESI-MS m/z: 622.3 [M+1].
N-(4-(1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-ylmethyl)benzyl)-1-(pyridin-2-yl) methanamine (12, RAD1–24)
To a stirred solution of 10 (0.027 g, 0.044 mmol) and anhydrous K2CO3 (0.022 g, 0.154 mmol) in anhydrous DMF (12 mL per mmol of the starting cyclam) under N2 was added thiophenol (0.012 g, 0.11 mmol) drop wise. The reaction mixture was stirred at room temperature for 5 h and concentrated in vacuo. The residue was partitioned between EtOAc and H2O. The organic layer was separated, washed with NaHCO3, and brine, then dried over MgSO4 and concentrated under reduced pressure. The residue was purified by Sep-Pak cartridge (5–10% MeOH/H2O with 0.1% TFA) to give the desired product 12 (0.019 g, 98% yield).
For the synthesis of cold Cu(II) bridged cyclam analog, to 2 mg (4.6 µmoles in 20 µL of deionized water) of bridged cyclam was added 100 µL of 0.1 M sodium acetate (pH 8). One mg of CuCl2 was added to the bridged cyclam solution and the pH was adjusted to 7.5 with 0.1 M ammonium acetate. The solution was then incubated at 95°C for 1 h. HPLC purification was performed by Phenomenex C-18 Luna 5 µm, 10 × 250 mm2 analytical column and was monitored at 266 nm (Supplementary Figures 1–4). The desired peak was at 23.6 min was collected, dried and analyzed via mass spectrometry. 1H NMR (400 MHz, D2O): δ 1.27 (t, J = 7.04 Hz, 1H), 1.81 (d, 16.2 Hz, 1H), 2.36 (bs, 2H), 2.62 (m, 4H), 2.88–3.23 (m, 13 H), 3.52–3.73 (m, 4H), 4.43 (s, 2H), 4.54 (d, J = 13.6 Hz, 1H), 4.58 (d, J = 13.6 Hz, 1H), 4.66 (s, 2H); 13C NMR (100 MHz, D2O): δ 18.20, 18.77, 41.83, 46.28, 47.77, 48.37, 48.78, 49.29, 50.84, 52.28, 53.40, 54.97, 56.24, 57.92, 58.22, 126.69, 128.99, 130.73, 130.84, 131.00, 132.28, 132.41, 133.14, 143.83, 143.90, 145.75, 146.84; ESI-MS m/z: 437 [M+1]; HR MicrO-TOFMS m/z [M+1], exact mass calculated for C26H41N6 437.3314, found 437.3387 (err [ppm] −1.8). MicrO-TOFMS m/z [M+] exact mass calculated for Cu(II)-12 498.25, found 499.26.
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-ylmethyl)benzyl)-1-(pyrimidin-2-yl)methanamine (13, RAD1–52)
To a stirred solution of 11 (0.25 g, 0.44 mmol) and anhydrous K2CO3 (0.22 g, 0.154 mmol) in anhydrous DMF (12 mL per mmol) under N2 was added thiophenol (0.12 g, 0.10 mmol) drop wise. The reaction mixture stirred at room temperature for 5 h and concentrated in vacuo. The residue was partitioned between EtOAc and H2O. The organic layer was separated, washed with NaHCO3, and brine, then dried over MgSO4 and concentrated under reduced pressure. The residue was purified by Sep-Pak cartridge (10% MeOH/H2O with 0.1% TFA) to give the desired product RAD1–52 (0.17. g, 98% yield). Cold Cu(II) analog was synthesized as described previously. 1H NMR (400 MHz, D2O): δ 1.77-1.73 (d, J = 15.7 Hz, 2H), 2.31 (bs, 2H), 2.82-2.65 (m, 4H), 3.37-2.95 (m, 12H), 3.66-3.49 (m, 4H), 4.52-4.46 (m, 6H), 7.64-7.50 (m, 5H), 8.79 (d, J = 4.4 Hz, 2H); 13C NMR (100 MHz, D2O): δ 18.11, 18.68, 41.76, 46.14, 47.69, 48.69, 49.19, 49.94, 50.19, 53.35, 54.50, 54.90, 56.15, 57.87, 58.15, 121.13, 128.84, 130.93, 132.25, 132.64, 157.90, 160.18; ESI-MS m/z: 438.3 [M+1]; HR MicrO-TOFMS m/z [M+1] exact mass calculated for C25H40N7 438.3345, found 438.3340 (err [ppm] 4.5). MicrO-TOFMS m/z [M+] exact mass calculated for Cu(II)-13 499.25, found 500.26.
Synthesis of tert-butyl 2-(11-(4-(((2-nitrophenyl)(pyridin-2-ylmethyl)sulfonamido) methyl) benzyl)-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-yl)acetate (14)
To a solution of 10 (0.055 g, 0.088 mmol) in dry CH3CN (1 mL) were added K2CO3 (0.012 g, 0.088 mmol) and t-butyl bromoacetate (13 µL, 0.088 mmol) in portions. The mixture was constantly stirred in the dark for 48 h under N2. The solvent was then removed, and the residue was dissolved in 20% aqueous NaOH (5 mL) at 0°C. The solution was extracted with cold CHCl3. The combined extracts were dried and the solvent was removed by reduced pressure. The residue was purified by Sep-Pak cartridge (6% MeOH/H2O) to give oil 14 (0.57 g, 96% yield). 1H NMR (400 MHz, MeOD): δ 1.32 (s, 9H), 1.62 (t, J = 22.7 Hz, 2H), 2.10–2.25 (bd, J = 18.3 Hz, 1H), 2.40–2.71 (m, 3H), 2.45–2.65 (m, 5H), 2.66–3.00 (m, 4H), 3.10–3.20 (m, 4H). 3.20–3.45 (m, 4H), 3.45–3.75 (m, 4H), 4.38 (d, J = 15.4 Hz, 1H), 4.51 (d, J = 15.4 Hz, 1H), 4.57 (s, 2H), 4.73 (s, 2H), 7.20 (dd, J1 = 7.3 Hz, J2 = 6.6 Hz, 4H), 7.47 (t, J = 9.5 Hz, 1H), 7.54 (d, J = 8.0 Hz, 1H), 7.66 (m, 1H), 7.71 (s, 2H), 7.97 (m, 2H), 8.39 (d, J = 5.0 Hz, 1H); 13C NMR (100 MHz, MeOD): δ 20.81, 20.83, 28.48, 49.71, 50.00, 50.42, 50.32, 52.00, 53.75, 54.96, 55.45, 56.23, 56.87, 58.23, 58.71, 59.01, 59.56, 86.25, 125.99, 126.14, 126.49, 130.7, 132.27, 133.44, 133.71, 133.97, 134.00, 136.21, 139.26, 144.43, 145.68, 149.79, 155.35, 170.10; ESI-MS m/z: 736.4 [M+1].
Synthesis of 2-(11-(4-(((2-nitrophenyl)(pyridin-2-ylmethyl) sulfonamido) methyl) benzyl) -1,4,8,11-tetraazabicyclo[6.6.2]hexadecane-4-yl)acetic acid (15, RAD1–39)
Protected cyclam 14 (0.057 g, 0.085 mmol) was dissolved in CF3COOH (1mL) and stirred under a nitrogen atmosphere for 24 h at room temperature. The solution was concentrated under reduced pressure and dissolved in CH2Cl2 and the solvent was removed in vacuo. The final step was repeated three more times to completely remove CF3COOH. To a stirred solution of acid deprotected 14 and anhydrous K2CO3 in anhydrous DMF (12 mL per mmol) under N2 was added thiophenol drop wise. The reaction mixture was stirred at room temperature for 5 h and concentrated in vacuo. The residue was partitioned between EtOAc and H2O. The organic layer was separated and washed with NaHCO3, brine and evaporated to obtain oil RAD1–39 (98% yield). 1H NMR (400 MHz, MeOD): δ 1.68 (d, J = 16.5 Hz, 2H), 2.24 (d, J = 18 Hz, 1H), 2.35–2.97 (m, 6H), 3.08 (t, J = 13,3 Hz, 1H), 3.21–3.29 (m, 4H), 3.41–3.57 (m, 4H), 3.71 (d, J = 22.0 Hz, 1H), 4.26 (d, J = 16.5 Hz, 1H), 4.27 (d, J = 16.5 Hz, 1H), 4.49 (d, J = 16.5 Hz, 1H), 4.58 (s, 2H), 4.67 (s, 2H), 7.21 (s, 4H), 7.42 (m, 2H), 7.67 (m, 1H), 7.74 (s, 2H), 7.85 (m, 1H), 8.01 (d, J = 6.0 Hz, 1H), 8.35 (d, J = 4.1 Hz, 1H); 13C NMR (100 MHz, MeOD): δ 21.08, 21.17, 50.01, 50.45, 50.68, 51.31, 52.72, 52.81, 53.51, 55.31, 55.82, 56.05, 57.42, 59.21, 59.32, 59.49, 125.43, 125.67, 125.71, 125.89, 130.70, 132.26, 133.62, 133.69, 136.11, 139.13, 142.14, 147.34, 149.53, 155.97, 156.02, 169.91; ESI-MS m/z: 680.2 [M+1].
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[10.2.2]hexadecane-4-ylmethyl)benzyl)-2-nitro-N-(pyridin-2-ylmethyl)benzenesulfonamide (18)
A mixture of side-bridged cyclam (17, 0.16 g, 0.72 mmol), 7 (0.31 g, 0.72 mmol) and K2CO3 (0.10 g, 0.72 mmol) in acetonitrile was stirred at room temperature overnight. The undissolved K2CO3 was removed by filtration, and the solvent removed under reduced pressure. The residue was used in the next step without purification. ESI-MS m/z: 621.5 [M+].
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[10.2.2]hexadecane-4-ylmethyl)benzyl)-2-nitro-N-(pyrimidin-2-ylmethyl)benzenesulfonamide (19)
A mixture of side-bridged cyclam (17, 0.13 g, 0.58 mmol), 8 (0.25 g, 0.58 mmol) and K2CO3 (0.080 g, 0.58 mmol) in acetonitrile was stirred at room temperature overnight. The undissolved K2CO3 was removed by filtration, and the solvent removed under reduced pressure. The residue was taken immediately to the next step without purification (0.14 g, 38% yield). ESI-MS m/z: 621.7 [M+].
N-(4-(1,4,8,11-tetraazabicyclo[10.2.2]hexadecane-4-ylmethyl)benzyl)-1-(pyridin-2-yl) methanamine (20)
To a stirred solution of 18 (0.19 g, 0.31 mmol) and anhydrous K2CO3 (0.15 g, 1.1 mmol) in anhydrous DMF (12 mL per mmol) under N2 was added thiophenol (0.086 g, 0.78 mmol) drop wise. The reaction mixture stirred at room temperature for 5 h and concentrated in vacuo. The residue was partitioned between EtOAc and H2O. The organic layer was separated, washed with NaHCO3, and brine, then dried over MgSO4 and concentrated under reduced pressure. The residue was purified by Sep-Pak cartridge (5–10% MeOH/H2O with 0.1% TFA) to give the desired product 20 (0.050 g, 35% yield). 1H NMR (400 MHz, D2O): δ 1.93 (m, 2H), 2.06 (m, 2H), 2.87–2.93 (m, 5H), 3.04–3.11 (m, 4H), 3.23–3.41 (m, 7H), 3.57–3.75 (m, 4H), 4.10 (m, 2H), 4.37 (s, 2H), 4.59 (s, 2H), 4.67 (s, 2H), 7.56–7.61 (m, 4H), 7.81 (m, 1H), 7.85 (m, 1H), 8.72 (m, 1H); 13C NMR (100 MHz, D2O): δ 21.67, 33.75, 45.11, 45.61, 45.91, 47.91, 48.31, 48.53, 48.75, 49.00, 51.24, 54.68, 62.20, 115.39, 118.28, 126.96, 127.01, 128.41, 131.22, 131.36, 133.39, 134.17, 134.52, 144.15, 146.45, 147.45; MicrO-TOFMS m/z [M+1] exact mass calculated for C26H41N6, 437.3373, found 437.3387 (err [ppm] 3.2)
Synthesis of N-(4-(1,4,8,11-tetraazabicyclo[10.2.2]hexadecane-4-ylmethyl)benzyl)-1-(pyrimidin-2-yl)methanamine (21)
To a stirred solution of 19 (0.14 g, 0.22 mmol) and anhydrous K2CO3 (0.11 g, 0.77 mmol) in anhydrous DMF (12 mL per mmol) under N2 was added thiophenol (0.061 g, 0.55 mmol) drop wise. The reaction mixture was stirred at room temperature for 5 h and concentrated in vacuo. The residue was partitioned between EtOAc and H2O. The organic layer was separated, washed with NaHCO3, and brine, then dried over MgSO4 and concentrated under reduced pressure. The residue was purified by Sep-Pak cartridge (10% MeOH/H2O with 0.1% TFA) to give the desired product 21 (0.038 g, 40% yield). 1H NMR (400 MHz, D2O): δ 2.02 (m, 2H), 2.26 (m, 2H), 2.83–2.92 (m, 2H), 3.04–3.43 (m, 12H), 3.55–3.88 (m, 7H), 3.92 (m, 1H), 4.44 (s, 2H), 4.50 (s, 2H), 4.60 (s, 1H), 4.72 (s, 1H), 7.47 (m, 1H), 7.56–7.62 (4H), 8.76 (m, 2H); 13C NMR (100 MHz, D2O): δ 22.24, 34.14, 45.80, 48.37, 48.60, 48.80, 49.23, 49.45, 49.66, 51.06, 51.24, 54.72, 55.06, 68.75, 112.97, 115.87, 118.77, 121.67, 122.26, 131.97, 134.49, 134.68, 134.91, 158.99, 161.25; MicrO-TOFMS m/z [M+] exact mass calculated for C25H39N7 437.1939, found 437.1935 (err [ppm] −1.0).
General Procedure for radiolabeling of bridged cyclams with Copper-64
To 100–200 µg of bridged cyclam analog (in 20 µL of deionized water) was added 100 µL of 0.1 M sodium acetate (pH 8) and mixed by pipetting the solution up and down several times. Then 37–74 MBq (1–2 mCi) of [64Cu]CuCl2 in HCl was added to the bridged cyclam solution and the pH was adjusted to 7.5 with 0.1 M ammonium acetate. The mixture was heated at 95°C for 1 h. Radio-HPLC purification was performed using a Phenomenex C-18 Luna 5 µm, 10 × 250 mm2 semi-preparative column using elution conditions described in the supplementary information. The UV absorbance was measured at 266 nm. The desired peaks, 23.6 for 64Cu[RAD1–24] and 23 min for 64Cu[RAD1–52], were collected, concentrated under vacuum and diluted in saline for cell and animal studies.
Cell lines
All cell culture reagents were purchased from Invitrogen (Carlsbad, CA) unless otherwise specified. The human glioblastoma cell line U87 (CXCR4low) and CHO-K1 cell lines were purchased from American Type Culture Collection (ATCC) and cultured in our laboratory in MEM and F-12K medium respectively, both supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin, and 100 mg/mL of streptomycin. A U87 cell line stably transfected with human CD4 and CXCR4 (U87-stb-CXCR4, CXCR4high) was obtained from the NIH AIDS Research Reference Reagent Program and cultured in DMEM supplemented with 15% FBS, 1 µg/mL puromycin, 300 µg/mL G418, 100 units/mL of penicillin, and 100 mg/mL of streptomycin [24]. Cell lines were maintained in a humidified incubator at 37°C with 5% CO2. The CHO-K1 cell line stably expressing SNAP-CXCR4 was generated in our laboratory, and maintained in RPMI supplemented with 10% FBS, 1 µg/mL puromycin, 2 mg/mL G418, 100 units/mL of penicillin, and 100 mg/mL of streptomycin.
Flow cytometry
Surface CXCR4 receptor expression levels were analyzed using a CXCR4 monoclonal antibody (Clone 44716) conjugated to Phycoerythrin (PE) (R&D Systems, Minneapolis, MN) according to procedures described previously [25].
Competitive binding assays
Affinity of the selected analogs for CXCR4 was determined by a FRET-based multi-concentration competitive binding assay using CHO1-SNAP-CXCR4 cells labeled with Lumi4-Tb against fluorescent CXCL12. Cyclam analog stocks were prepared in water and diluted with Tag-lite medium prior to their use in the assay. The assay procedure was adapted from work published by Zwier et al. [26]. CHO1-SNAP-CXCR4 cells were prepared for in vitro assays according to manufacturer’s recommendation (Cisbio Bioassays). Briefly, subconfluent CHO1-SNAP-CXCR4 cells in T-175 cm2 flasks were rinsed with 10 mL of Tag-lite labeling medium and incubated with 100 nM SNAP-Lumi4-Tb in Tag-lite buffer for 1 h at 37 °C. 10,000 cells per well were used to carry out binding assays in 384-well plates. SDF1α-Red (15 nM) was mixed with increasing concentrations of cyclam analog (1×10−4 M to 1×10−13 M) and incubated at room temperature for 2 h. Homogeneous Time-Resolved Fluorescence (HTRF) with excitation at 340 nm and emissions at 620 and 665 nm (delay 50 µs, window time 400 µs, measurement time 1 s) were measured using a Perkin Elmer Victory3 1420 multi-label counter. Binding assays were done in triplicates and three independent experiments were performed. All calculations were done using GraphPad Prism 4 software (GraphPad Software, Inc., San Diego, CA). IC50 and Ki values were calculated by fitting the data to a sigmoidal dose response curve and the Cheng-Prusoff equation (with derived Ki=19 for SDF1-α-Red at the concentration of 15 nM), respectively.
Animal models
All experimental procedures using animals were conducted according to protocols approved by the Johns Hopkins Animal Care and Use Committee. Female NOD/SCID mice, six to eight weeks old, weighing between 25 to 30 g, were purchased from The Johns Hopkins Immune Compromised Animal Core. Mice were implanted subcutaneously (s.c.) with U87 and U87-stb-CXCR4 cells (4 × 106 cells/100 µL) in opposite flanks. Animals were used for biodistribution and PET/CT imaging experiments when the tumor size reached approximately 200 – 400 mm3.
PET/CT imaging and analysis
Whole-body PET and CT images were acquired on an eXplore VISTA small animal PET (GE) and an X-SPECT small SPECT/CT system (Gamma Medica Ideas, Northridge, CA), respectively. For imaging studies, mice were induced with 3% and maintained under 1.5% isoflurane (v/v). Whole body PET images (n = 3, 2 bed positions, 15-min emission/position) were acquired at 90 minutes after injection of ~9.25 MBq (250 µCi) of radiotracer. For binding specificity studies, a separate group of mice (n = 2) were injected with a blocking dose of 25 mg/kg of AMD3465 administered subcutaneously 30 min prior to the injection of [64Cu]bridged cyclam. After each PET scan, a CT scan was acquired in 512 projections for anatomic co-registration. PET data were corrected for decay and dead time, and reconstructed using the three-dimensional ordered subsets-expectation maximization algorithm (3D-OSEM). The percentage of injected dose per cc (%ID/cc) values were calculated based on a calibration factor using a known quantity of radioactivity. Volume-rendered images were generated using Amira 5.3.3 software (Visage Imaging Inc.).
Ex vivo biodistribution
NOD/SCID mice harboring U87 and U87-stb-CXCR4 xenografts were injected intravenously with 740 kBq (20 µCi) of [64Cu]bridged cyclam in 200 µL of saline. Different groups of mice (n=4 per group) were sacrificed 30, 60, 90, 120, 240 and 480 minutes after injection of the tracer. Blood, tumors and selected tissues were harvested, weighed and the radioactivity in the tissues was measured in an automated gamma spectrometer. To demonstrate the in vivo specificity of [64Cu]bridged cyclams [64Cu]RAD1–24 and [64Cu]RAD1–52, mice received blocking doses of 25 mg/kg of AMD3465 subcutaneously 1 h before the injection of the radiotracer, and ex vivo biodistribution studies were performed 90 min after the radiotracer injection. Aliquots of the injected dose were counted as reference standards for the calculation of %ID/g values. All values were normalized and are reported as percent injected dose per gram of tissue (%ID/g).
Data analysis
Statistical analysis was performed using Prism 4 (GraphPad Software). An unpaired two tailed t-test was used and P-values < 0.05 were considered to be significant.
Results
The synthesis of cross-bridged cyclam analogs commenced with the reaction of 2-(aminomethyl)pyridine 1 or 2-pyrimidinemethanamine 2 with 2-nitrobenzenesulfonyl chloride to form sulfonamides 3 and 4, respectively (Scheme 1). Subsequent substitution with 4-bromomethylbenzyl alcohol resulted in the formation of alcohols 5 and 6. Chlorides 7 and 8 were then produced by reaction of mesyl chloride with alcohols 5 and 6. Protected amines 7 and 8 were then reacted with commercially available cross-bridged cyclam 9 to form protected bridged cyclam analogs 10 and 11, respectively. Facile deprotection using thiophenol yielded final cross-bridged analogs RAD1–24 and RAD1–52 (Figure 1A).
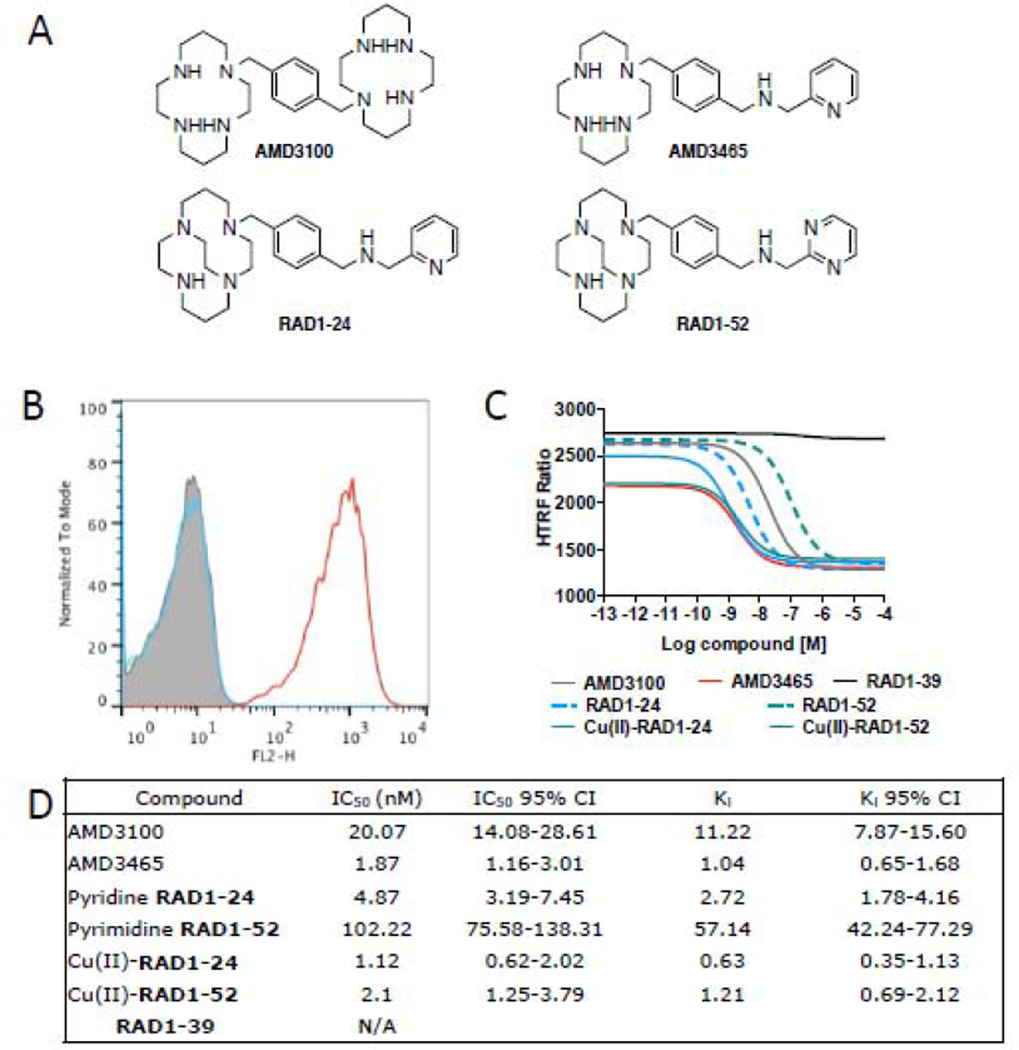
Figure 1.
A) Structures of known and newly synthesized cyclam-based CXCR4 inhibitors; B) Evaluation of surface CXCR4 expression in U87 cancer cell lines by flow cytometry. Grey, isotype; blue, U87, red, U87-stb-CXCR4; C) Inhibition binding assays of SDF1-Red (15 nM) with increasing concentrations of analyzed inhibitors, non-linear curve fit data is shown; D) tabulated IC50 and Ki values for analyzed CXCR4 inhibitors; reported at 95% confidence level.
Acid RAD1–39 was synthesized in three steps from pyridine 10, beginning with the addition of t-butylbromoacetate to give protected acid 14 (Scheme 1). Subsequent deprotection produced the free carboxylic acid, which was taken to the next step without purification. Deprotection of the sulfonamide group yielded RAD1–39.
Side-bridged cyclams 20 and 21 were synthesized starting with the reduction of commercially available glyoxal cyclam 16 (Scheme 2) [23]. Cyclam 16 was refluxed in toluene for ten days to yield side-bridged cyclam 17. Side-bridged cyclam 17 was used immediately in subsequent steps. Side-bridged cyclam 17 was reacted with either protected pyridine 7 or pyrimidine 8 to afford compounds 18 and 19 in moderate yields. Subsequent deprotection yielded final side-bridged cyclams 20 and 21. Side-bridged cyclams, however, decomposed quickly even at −20°C as observed by proton NMR and mass spectroscopy. Hygroscopic nature of other side-bridged cyclam analogs has been previously reported, further confirming our observations [27].
Radiolabeling of cross-bridged cyclams RAD1–24, RAD1–52 and RAD1–39 using [64Cu]CuCl2 required heating to 95°C (Scheme 3). In general, a solution of [64Cu]CuCl2-HCl and bridged cyclam analog was adjusted to pH 7.5 and heated for 45 min to yield radiolabeled analogs. [64Cu]RAD1–24 and [64Cu]RAD1–52 were obtained in 78 and 73%, respectively, while carboxylic acid [64Cu]RAD1–39 was obtained in 45% non-decay corrected radiochemical yield. Side-bridged cyclams 20 and 21 proved to be unstable. For that reason, we chose to evaluate only the cross-bridged cyclams as PET imaging agents.
Copper-64 labeled compounds were purified by HPLC and co-injected with their unlabeled counterparts to assure purity and identity (Supplemental Figures 1–4). Specific activities (± standard error of the mean, SEM) for radiolabeled cyclams [64Cu]RAD1–24 and [64Cu]RAD1–52 were 3.74 ± 1.10 GBq/ µmol (101.33 ± 29.81 mCi/µmol) and 5.60 ± 0.57 GBq/ µmol (151.50 ± 15.50 mCi/ µmol), respectively, while the specific radioactivity of the [64Cu]CuCl2 obtained from the vendor ranged from 33.3–518 GBq/ µmol (0.9–14 Ci/µmol). Radiolabeled cyclams [64Cu]-RAD1–24, RAD1–52 and RAD1–39 were then used for in vivo imaging and ex vivo biodistribution in mice bearing U87-stb-CXCR4 and control U87 tumors.
Receptor expression and inhibition constants
Flow cytometry analysis revealed that nearly 95% of U87-stb-CXCR4 cells and 2 to 5% of U87 control cells have surface CXCR4 expression (Figure 1B). Binding affinities were measured relative to the binding of fluorescent CXCL12 to CXCR4. The bridged-cyclam analogs demonstrated affinities in the low-nanomolar range, similar to those previously reported for AMD3100 and ADM3465 [17,18]. The copper (II) coordinated analogs showed considerably increased affinity to CXCR4 compared to the parent analogs. The Ki values were in the following order: Cu(II)-RAD1–24> AMD3465 ≥ Cu(II)-RAD1–52 > RAD1–24 > RAD1–52>AMD3100. The carboxylic acid analog RAD1–39 did not demonstrate specific binding (Figures 1C and 1D).
PET imaging
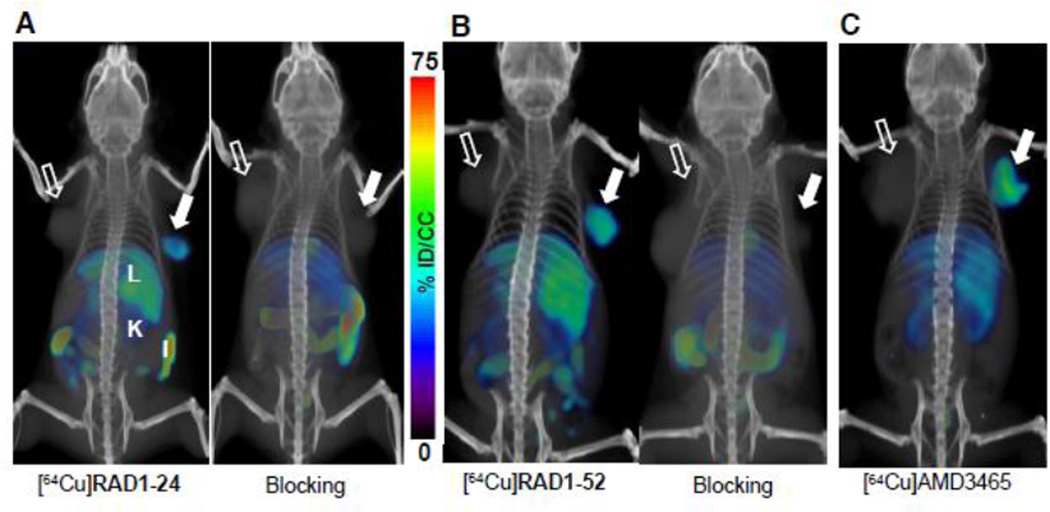
Using U87 and U87-stb-CXCR4 model to characterize the biodistribution and pharmacokinetics of [64Cu]AMD3100 and [64Cu]AMD3465, we observed an optimal contrast at 90 min post-injection [17,18]. To facilitate cross-comparison among different agents, we acquired whole-body PET/CT images of radiolabeled analogs [64Cu]RAD1–24, RAD1–52 and RAD1–39 at 90 min post-injection. We injected ~9.25 MBq (250 µCi) of each tracer to mice bearing U87 and U87-stb-CXCR4 tumors. PET imaging acquired after the injection of pyridine [64Cu]RAD1–24 demonstrated clearer and higher accumulation of radioactivity in the U87-stb-CXCR4 tumor compared to the control U87 tumor (Figure 2A). Highest uptake was observed in the liver and kidneys. Similarly, 90 min post-injection of [64Cu]RAD1–52, the highest accumulation of radioactivity was observed in the U87-stb-CXCR4 tumor and the liver, followed by the kidneys (Figure 2B). The carboxylate analog [64Cu]RAD1–39 (Supplemental Figure 5) did not demonstrate any CXCR4-specific accumulation and its distribution was largely confined to the gallbladder, gastrointestinal system and liver.
Figure 2.
PET/CT imaging of CXCR4 expression in subcutaneous brain tumor xenografts with pyridine [64Cu]RAD1–24 and pyrimidine [64Cu]RAD1–52. NOD/SCID mice bearing U87 and U87-stb-CXCR4 glioblastoma xenografts on the left and right flanks, respectively, were given ~9.25 MBq (250 µCi) of 64Cu-labeled radiotracers via tail vein injection, and PET/CT images were acquired in two bed positions (15 min/bed) under 1.5% isofluorane anesthesia. A) Volume-rendered whole body images of [64Cu]RAD1–24 at 90 minutes post-injection of tracer (left), 25 mg/kg of AMD3465 blocking dose followed by [64Cu]RAD1–24 at 90 minutes post-injection of tracer (right); B) volume-rendered whole body images of [64Cu]RAD1–52 at 90 minutes post-injection of tracer (left), 25 mg/kg of AMD3465 blocking dose followed by [64Cu]RAD1–52 at 90 minutes post-injection of tracer (right); and C) volume-rendered whole body image of [64Cu]AMD3465 at 90 minutes post-injection of tracer. Filled arrow, U87-stb-CXCR4 tumor (right); unfilled arrow, control U87 tumor (left); L - liver, K - kidney, I - intestines.
Biodistribution and blocking studies
To confirm the results obtained with PET, biodistribution studies were conducted at various time points after the injection of the radiotracers (Tables 1 and 2). At 90 min post-injection of [64Cu]RAD1–24, the highest degree of radioactivity was observed in the liver (84.72 ± 9.78 %ID/g), kidneys (36.00 ± 3.46 %ID/g), and in the U87-stb-CXCR4 tumor (18.61 ± 2.29 %ID/g). Uptake in the U87-stb-CXCR4 tumors was retained for several hours and reached the highest value of 19.60 ± 5.55 %ID/g 4 h post-injection. At 90 min post-injection of [64Cu]RAD1–52, the highest uptake was observed in the liver (89.18 ± 4.29 %ID/g), followed by the U87-stb-CXCR4 tumor (36.12 ± 4.85 %ID/g) and the kidneys (35.33 ± 2.15 %ID/g). Tumor-to-muscle ratios for [64Cu]RAD1–24 and [64Cu]RAD1–52 reached 61.10 ± 16.90 and 106.05 ± 17.19 respectively, while tumor-to-blood ratios reached 11.54 ± 2.17 and 28.08 ± 4.78, respectively. Because specific radioactivity may have an effect on tumor uptake and the receptor density is variable due to continuous culture of U87-stb-CXCR4 cells, we compared the biodistribution of these agents with that of [64Cu]AMD3465 synthesized on the same day. These results indicate that tumor uptake from pyrimidine [64Cu]RAD1–52 is very similar to that of [64Cu]AMD3465 (Figure 2). Biodistribution studies were not pursued for [64Cu]RAD1–39 due to lack of demonstration of any site-selective binding in in vitro assays as well as in in vivo imaging.
Table 1. Biodistribution of [64Cu]RAD1–24.
Pyridine [64Cu]RAD1–24 ex vivo biodistribution studies in NOD/SCID mice bearing subcutaneous U87 and U87-stb-CXCR4 brain tumor xenografts; n=3–4 for each time point; reported as percent injected dose per gram tissue (%ID/g).
| Time (minutes) | ||||||
|---|---|---|---|---|---|---|
| Organ/Tissue | 30 | 60 | 90 | 120 | 240 | 480 |
| Blood | 3.90 ± 0.57 | 2.38 ± 0.20 | 1.71 ± 0.17 | 1.51 ± 0.24 | 1.64 ± 0.28 | 1.82 ± 0.26 |
| Heart | 2.46 ± 0.52 | 2.12 ± 0.18 | 1.67 ± 0.25 | 1.56 ± 0.28 | 1.97 ± 0.09 | 1.87 ± 0.22 |
| Lungs | 14.68 ± 3.21 | 10.62 ± 1.72 | 8.43 ± 1.06 | 6.57 ± 2.50 | 8.09 ± 1.59 | 6.10 ± 1.29 |
| Liver | 79.45 ± 20.93 | 91.54 ± 6.03 | 84.72 ± 9.78 | 92.83 ± 20.46 | 101.04 ± 8.94 | 81.98 ± 11.66 |
| Stomach | 2.38 ± 0.51 | 2.13 ± 0.31 | 1.86 ± 0.21 | 1.92 ± 0.44 | 2.32 ± 0.35 | 1.91 ± 0.23 |
| Spleen | 21.15 ± 5.34 | 18.61 ± 1.23 | 16.45 ± 2.76 | 14.87 ± 2.40 | 15.20 ± 1.80 | 12.15 ± 1.95 |
| Kidney | 34.00 ± 5.69 | 39.71 ± 4.03 | 36.00 ± 3.46 | 33.44 ± 3.03 | 35.40 ± 4.75 | 24.76 ± 1.12 |
| Small Intestines | 6.70 ± 2.72 | 4.97 ± 0.96 | 4.63 ± 1.16 | 3.69 ± 1.10 | 4.73 ± 1.19 | 3.62 ± 0.63 |
| Muscle | 0.46 ± 0.08 | 0.36 ± 0.05 | 0.35 ± 0.05 | 0.36 ± 0.07 | 0.34 ± 0.02 | 0.27 ± 0.05 |
| Bladder | 2.55 ± 0.58 | 4.40 ± 2.86 | 4.14 ± 4.78 | 1.81 ± 0.57 | 2.27 ± 0.23 | 1.22 ± 0.41 |
| Pancreas | 1.31 ± 0.18 | 1.38 ± 0.16 | 1.27 ± 0.28 | 1.49 ± 0.05 | 1.70 ± 0.20 | 1.30 ± 0.11 |
| U87 | 5.89 ± 7.01 | 2.54 ± 0.47 | 1.98 ± 0.49 | 2.13 ± 0.72 | 2.72 ± 0.85 | 2.92 ± 0.77 |
| U87-stb-CXCR4 | 13.26 ± 9.83* | 18.95 ± 4.88* | 18.61 ± 2.29* | 18.17 ± 2.03* | 19.60 ± 5.55* | 14.13 ± 7.98* |
P<0.0001 compared to U87 control tumor uptake.
Table 2. Biodistribution of [64Cu]RAD1–52.
Pyrimidine [64Cu]RAD1–52 ex vivo biodistribution studies in NOD/SCID mice bearing subcutaneous U87 and U87-stb-CXCR4 brain tumor xenografts; n=3–4 for each time point; reported as percent injected dose per gram tissue (%ID/g).
| Time (minutes) | ||||||
|---|---|---|---|---|---|---|
| Organ/Tissue | 30 | 60 | 90 | 120 | 240 | 450 |
| Blood | 3.08 ± 0.47 | 1.90 ± 8.81 | 1.42 ± 0.21 | 1.33 ± 0.14 | 1.14 ± 0.29 | 1.02 ± 0.11 |
| Heart | 2.03 ± 0.15 | 1.62 ± 0.15 | 1.24 ± 0.72 | 1.81 ± 0.19 | 1.47 ± 0.15 | 1.45 ± 0.24 |
| Lungs | 10.73 ± 1.06 | 8.12 ± 0.61 | 7.60 ± 1.11 | 7.12 ± 0.89 | 6.94 ± 1.69 | 5.69 ± 0.23 |
| Liver | 79.17 ± 5.73 | 83.46 ± 6.21 | 89.18 ±4.29 | 90.96 ± 16.76 | 79.09 ± 4.85 | 79.81 ± 7.16 |
| Stomach | 3.32 ± 0.27 | 2.85 ± 0.28 | 2.98 ± 0.73 | 3.26 ± 0.64 | 2.41 ± 0.33 | 2.54 ± 0.32 |
| Spleen | 15.10 ± 0.92 | 12.89 ± 1.18 | 15.03 ± 2.15 | 15.85 ± 4.04 | 12.78 ± 1.43 | 11.03 ± 1.06 |
| Kidney | 30.74 ± 1.51 | 31.07 ± 2.62 | 35.33 ± 2.15 | 34.70 ± 9.75 | 28.22 ± 3.07 | 25.45 ± 3.38 |
| Small Intestines | 8.45 ± 5.25 | 4.86 ± 0.52 | 6.49 ± 1.12 | 6.96 ± 0.79 | 4.33 ± 1.11 | 3.98 ± 0.34 |
| Muscle | 0.49 ± 0.03 | 0.40 ± 0.01 | 0.40 ± 0.05 | 0.34 ± 0.03 | 0.30 ± 0.02 | 0.29 ± 0.03 |
| Bladder | 3.47 ± 0.62 | 2.15 ± 0.32 | 3.30 ± 0.95 | 2.05 ± 0.98 | 2.00 ± 0.35 | 1.63 ± 0.96 |
| Pancreas | 1.61 ± 0.08 | 1.61 ± 0.19 | 1.84 ± 0.29 | 1.88 ± 0.32 | 1.60 ± 0.25 | 1.66 ± 0.09 |
| U87 | 4.78 ± 0.21 | 5.67 ± 0.30 | 6.12 ± 0.97 | 6.49 ± 0.60 | 6.87 ± 1.01 | 5.82 ± 1.32 |
| U87-stb-CXCR4 | 26.79 ± 2.98* | 29.26 ± 3.81* | 36.11 ± 4.84* | 26.00 ± 4.53* | 23.07 ± 1.34* | 22.52 ± 4.09* |
P<0.0001 compared to U87 control tumor uptake.
Blocking studies were performed to confirm the specificity of radiotracers [64Cu]RAD1–24 and [64Cu]RAD1–52 for CXCR4 (Figure 2). Radiotracers were administered following an injection of AMD3465 (at a dose of 25 mg/kg), and biodistribution studies were performed 90 min post radiotracer injection (Table 3). Blocking of [64Cu]RAD1–24 with AMD3465 resulted in decreased radioactivity uptake in the U87-stb-CXCR4 tumor by 85%, in the liver by 55% and in kidneys by 23% compared to the non-blocking distribution. Administration of [64Cu]RAD1–52, following a blocking dose of AMD3465 resulted in greater than 90% reduction in radioactivity uptake in the U87-stb-CXCR4 tumor. Radiotracer uptake in liver decreased by 41% while the uptake in the kidneys increased by 42%, perhaps due to enhanced renal clearance from increased free radioligand upon receptor blockade.
Table 3. Comparison of [64Cu]RAD1–24, [64Cu]RAD1–52 and [64Cu]AMD3465.
Blocking biodistribution studies of various analogs performed in NOD/SCID mice bearing subcutaneous U87 and U87-stb-CXCR4 brain tumor xenografts in the same cohort of mice; Mice were sacrificed at 90 minutes post-injection of radiotracer, n=3–4 for each cohort; reported as percent injected dose per gram tissue (%ID/g).
| Compound | |||||
|---|---|---|---|---|---|
| Organ/Tissue | [64Cu]12 | [64Cu]12 (block) | [64Cu]13 | [64Cu]13 (block) | [64Cu]AMD3465 |
| Blood | 1.44 ± 0.10 | 1.23 ± 0.04 | 0.64 ± 0.05 | 0.96 ± 0.19 | 0.83 ± 0.24 |
| Heart | 1.49 ± 0.16 | 1.42 ± 0.04 | 0.81 ± 0.06 | 0.85 ± 0.17 | 1.08 ± 0.38 |
| Lungs | 6.16 ± 0.34 | 2.70 ± 0.24 | 3.15 ± 0.15 | 2.21 ± 0.56 | 3.04 ± 0.17 |
| Liver | 58.03 ± 3.68 | 26.46 ± 3.20 | 61.47 ± 2.75 | 34.85 ± 1.22 | 31.20 ± 1.49 |
| Stomach | 1.82 ± 0.22 | 1.77 ± 0.09 | 1.04 ± 0.12 | 0.91 ± 0.41 | 2.46 ± 0.15 |
| Spleen | 11.58 ± 0.86 | 3.95 ± 0.61 | 7.89 ± 0.47 | 6.48 ± 2.18 | 3.65 ± 0.07 |
| Kidney | 19.07 ± 1.67 | 14.73 ± 1.82 | 22.94 ± 1.76 | 32.55 ± 8.75 | 34.79 ± 0.88 |
| Small Intestines | 3.70 ± 0.25 | 15.05 ± 5.87 | 2.16 ± 0.24 | 31.54 ± 31.00 | 3.32 ± 0.14 |
| Muscle | 0.28 ± 0.04 | 0.32 ± 0.02 | 0.17 ± 0.02 | 0.27 ± 0.12 | 0.29 ± 0.02 |
| Bladder | 1.11 ± 0.07 | 6.92 ± 2.62 | 0.82 ± 0.09 | 7.25 ± 5.29 | 4.36 ± 0.72 |
| Pancreas | 1.35 ± 0.26 | 1.38 ± 0.05 | 0.77 ± 0.14 | 0.84 ± 0.14 | 1.48 ± 0.09 |
| U87 | 3.92 ± 0.54 | 2.29 ± 0.19 | 2.57 ± 0.44 | 1.50 ± 0.16 | 5.35 ± 1.15 |
| U87-stb-CXCR4 | 16.48 ± 1.43 | 2.50 ± 0.41 | 17.71 ± 0.64 | 3.82 ± 1.99 | 30.64 ± 2.47 |
| Ratios | |||||
| U87-stb-CXCR4/U87 | 4.43±1.38 | 7.71±3.36 | 6.44±2.45 | ||
| U87/Muscle | 15.30±7.01 | 15.84±7.01 | 18.43±8.23 | ||
| U87-stb-CXCR4/Muscle | 61.09±16.89 | 106.05±17.18 | 104.17±9.97 | ||
| U87/Blood | 2.81±1.05 | 4.13±1.80 | 4.83±2.26 | ||
| U87-stb-CXCR4/Blood | 11.54±2.16 | 28.08±4.78 | 27.26±2.70 | ||
Discussion
Synthesis of cross-bridged AMD3465 analogs proved facile. Both pyridine and pyrimidine-based agents showed high in vitro specificity to CXCR4. PET imaging studies with radiolabeled analogs in mice bearing tumors showed CXCR4-specific uptake and prolonged tumor retention. Our studies demonstrate that the cross-bridged cyclam motif could be used as a scaffold for further development of CXCR4-targeted imaging agents.
While the synthesis of the cross-bridged analogs was straightforward, the side-bridged cyclam analogs proved difficult to synthesize due to their hygroscopic nature. Mass spectrometry and NMR data indicated that the side-bridged cyclams oxidized to form oxygen-bridged dimeric species, even when care was taken to avoid exposure to oxygen. That instability made the side-bridged analogs unsuitable for further development.
Binding affinity studies showed that the affinity of RAD1–24 and RAD1–52 for CXCR4 increased for Cu(II)-coordinated compounds compared to the parent compounds. Copper(II)-coordinated RAD1–52 displayed a 48-fold higher affinity for CXCR4 compared to the parent pyrimidine cyclam. A similar increase in affinity with metal coordinated cyclam analogs was observed previously by Gerlach et al., with AMD3100 [28]. AMD3100 showed a 7-fold increase in affinity for CXCR4 when bound to Cu(II) due to enhanced interactions between the cyclam and aspartate 262 in the receptor binding pocket [28]. Similarly, configurationally restricted analogs of AMD3100 were shown to adopt a single configuration in solution when bound to Zn(II) or Cu(II), improving in vitro CXCR4 binding properties [20] due to increased interactions between the restricted Zn(II) complex and aspartate residues in the receptor binding pocket [20]. It is likely that the rigidity of Cu(II)-coordinated RAD1–52 enhances the interactions between the complex and aspartate residues in the binding pocket, thereby increasing the binding affinity and target tissue residence time.
Cross-bridged cyclams RAD1–24 and RAD1–52 were radiolabeled in high yield and in both cases the desired product was the only one formed, as shown by HPLC. PET/CT imaging clearly demonstrated a high uptake of [64Cu]RAD1–24 and [64Cu]RAD1–52 in U87-stb-CXCR4 tumors compared to the CXCR4low control U87 tumors. Although RAD1–52 showed lower in vitro affinity compared to RAD1–24, a significant increase in binding affinity of the cold Cu analog relative to RAD1–52 was evident from the in vitro binding studies, which was further confirmed by the high tumor uptake observed in vivo with the radiolabeled analog. Blocking studies using AMD3465 reduced the uptake of [64Cu]RAD1–24 and [64Cu]RAD1–52 in CXCR4 overexpressing tumors by 85–90%, demonstrating their specificity for CXCR4. Those studies further suggested that these agents bound to the same site as that of AMD3465 and AMD3100. In biodistribution studies, [64Cu]RAD1–24 and [64Cu]RAD1–52 demonstrated consistently similar uptake over 8 h in the U87-stb-CXCR4 tumors. From the time of maximum uptake observed (90 min) to 240 min post-injection of the radiotracers, nearly 100% and 70% of radioactivity were retained in the U87-stb-CXCR4 tumors in mice injected with [64Cu]RAD1–24 and [64Cu]RAD1–52, respectively. Using the same tumor model, we have previously shown that less than 50% of [64Cu]AMD3465 uptake was retained within the same time frame [17]. These results suggest that cross-bridged cyclams possess increased residence time with respect to binding to CXCR4. Similar results were also observed with non-radioactive AMD3100 analogs [29].
Sun et al., synthesized radiolabeled cross-bridged cyclams with two pendant carboxylic acid groups. They demonstrated superior in vivo biodistribution characteristics in normal rats, clearing rapidly from the blood, liver and kidneys compared to the cross-bridged cyclams [21]. Based on the stability observed with those agents, we hypothesized that similar carboxylic acid analogs would improve the image contrast of the cross-bridged AMD3465 analogs. The pyridine-based carboxylic acid analog RAD1–39 we synthesized showed no CXCR4 specific in vitro affinity or in vivo uptake in the tumors. Although carboxylic acid-based analogs are known to bind Cu strongly, the negative charge of the carboxylate may reduce the affinity of RAD1–39 to the highly acidic and negatively charged CXCR4 surface [30]. Also, if the cyclam moiety is binding within the pocket in proximity to AspIV:20 (Asp171), in analogy with AMD3465 [31], presence of the carboxyl group in RAD1–39 may provide significant steric hindrance and electrostatic repulsion leading to decrease of its affinity towards CXCR4.
Studies on the Cu chelating properties of cyclams have demonstrated that cross-bridged cyclams are kinetically more stable than their non-bridged counterparts [21,22]. In the case of [64Cu]RAD1–24 and [64Cu]RAD1–52, higher liver uptake was observed compared to [64Cu]AMD3465. Blocking studies revealed a reduction in liver uptake by nearly 50%, suggesting that about half of the liver uptake might be due either to transchelation of Cu, or to other nonspecific uptake, in addition to the receptor-mediated binding. Both AMD3100 and AMD3465 proved to be metabolically stable [17,18] and Cu-chelation stability of cyclams and bridged-cyclams has been well characterized [21]. While the metabolic stability of these complexes was not the primary focus of the current work, possible transchelation and percentage of contribution of the traschelated radioactivity to the liver uptake observed need to be further characterized to pursue the development of cross-bridged cyclams as [64Cu]-labeled imaging agents. The high-affinity, high tumor-to-background ratios and prolonged target tissue residence observed with [64Cu]RAD1–24 and [64Cu]RAD1–52 suggests further development and optimization of the cross-bridged cyclam scaffold for CXCR4 imaging.
Supplementary Material
Acknowledgements
We would like to thank the University of Wisconsin for supplying 64CuCl2 and Gilbert Green for PET/CT imaging assistance and Dr. Marie-France Penet for critical reading of the manuscript. This work was supported by R01CA166131 (SN), The Alexander and Margaret Stewart Trust (SN), DOD W81XWH-12-BCRP-IDEA (SN), and resources provided through P30 CA006973 and P50 CA103175.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting information available
Methods, HPLC conditions, chromatograms for the radioactive analogs and PET/CT image of [64Cu]RAD1–39 are available in supplementary information.
References
- 1.Wong D, Korz W. Translating an Antagonist of Chemokine Receptor CXCR4: from bench to bedside. Clin Cancer Res. 2008;14:7975–7980. doi: 10.1158/1078-0432.CCR-07-4846. [DOI] [PubMed] [Google Scholar]
- 2.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 3.Salvucci O, Bouchard A, Baccarelli A, Deschenes J, Sauter G, Simon R, Bianchi R, Basik M. The role of CXCR4 receptor expression in breast cancer: a large tissue microarray study. Breast Cancer Res Treat. 2006;97:275–283. doi: 10.1007/s10549-005-9121-8. [DOI] [PubMed] [Google Scholar]
- 4.Kang H, Watkins G, Douglas-Jones A, Mansel RE, Jiang WG. The elevated level of CXCR4 is correlated with nodal metastasis of human breast cancer. Breast. 2005;14:360–367. doi: 10.1016/j.breast.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Kitayama J, Kazama S, Nagawa H. Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res. 2003;5:R144–R150. doi: 10.1186/bcr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta SK, Pillarisetti K. Cutting edge: CXCR4-Lo: molecular cloning and functional expression of a novel human CXCR4 splice variant. J Immunol. 1999;163:2368–2372. [PubMed] [Google Scholar]
- 7.Hanaoka H, Mukai T, Tamamura H, Mori T, Ishino S, Ogawa K, Iida Y, Doi R, Fujii N, Saji H. Development of a 111In-labeled peptide derivative targeting a chemokine receptor, CXCR4, for imaging tumors. Nucl Med Biol. 2006;33:489–494. doi: 10.1016/j.nucmedbio.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson O, Weiss ID, Kiesewetter DO, Farber JM, Chen X. PET of tumor CXCR4 expression with 4-18F-T140. J Nucl Med. 2010;51:1796–1804. doi: 10.2967/jnumed.110.079418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobson O, Weiss ID, Szajek L, Farber JM, Kiesewetter DO. 64Cu-AMD3100--a novel imaging agent for targeting chemokine receptor CXCR4. Bioorg Med Chem. 2009;17:1486–1493. doi: 10.1016/j.bmc.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuil J, Buckle T, Yuan H, van den Berg NS, Oishi S, Fujii N, Josephson L, van Leeuwen FW. Synthesis and evaluation of a bimodal CXCR4 antagonistic peptide. Bioconjugate chemistry. 2011;22:859–864. doi: 10.1021/bc2000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Z, Zhan W, Zhu A, Yoon Y, Lin S, Sasaki M, Klapproth JM, Yang H, Grossniklaus HE, Xu J, Rojas M, Voll RJ, Goodman MM, Arrendale RF, Liu J, Yun CC, Snyder JP, Liotta DC, Shim H. Development of a unique small molecule modulator of CXCR4. PLoS ONE. 2012;7:e34038. doi: 10.1371/journal.pone.0034038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimmagadda S, Pullambhatla M, Pomper MG. Immunoimaging of CXCR4 expression in brain tumor xenografts using SPECT/CT. J Nucl Med. 2009;50:1124–1130. doi: 10.2967/jnumed.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiss ID, Jacobson O, Kiesewetter DO, Jacobus JP, Szajek LP, Chen X, Farber JM. Positron Emission Tomography Imaging of Tumors Expressing the Human Chemokine Receptor CXCR4 in Mice with the Use of (64)Cu-AMD3100. Mol Imaging Biol. 2011 doi: 10.1007/s11307-010-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobson O, Weiss ID. CXCR4 chemokine receptor overview: biology, pathology and applications in imaging and therapy. Theranostics. 2013;3:1–2. doi: 10.7150/thno.5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuil J, Buckle T, van Leeuwen FW. Imaging agents for the chemokine receptor 4 (CXCR4) Chemical Society reviews. 2012;41:5239–5261. doi: 10.1039/c2cs35085h. [DOI] [PubMed] [Google Scholar]
- 16.Tamamura H, Tsutsumi H, Masuno H, Fujii N. Development of low molecular weight CXCR4 antagonists by exploratory structural tuning of cyclic tetra- and pentapeptide-scaffolds towards the treatment of HIV infection, cancer metastasis and rheumatoid arthritis. Curr Med Chem. 2007;14:93–102. doi: 10.2174/092986707779313499. [DOI] [PubMed] [Google Scholar]
- 17.De Silva RA, Peyre K, Pullambhatla M, Fox JJ, Pomper MG, Nimmagadda S. Imaging CXCR4 Expression in Human Cancer Xenografts: Evaluation of Monocyclam 64Cu-AMD3465. J Nucl Med. 2011;52:986–993. doi: 10.2967/jnumed.110.085613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer Res. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourni E, Demmer O, Schottelius M, D'Alessandria C, Schulz S, Dijkgraaf I, Schumacher U, Schwaiger M, Kessler H, Wester HJ. PET of CXCR4 expression by a (68)Ga-labeled highly specific targeted contrast agent, Journal of nuclear medicine : official publication. Society of Nuclear Medicine. 2011;52:1803–1810. doi: 10.2967/jnumed.111.098798. [DOI] [PubMed] [Google Scholar]
- 20.Valks GC, McRobbie G, Lewis EA, Hubin TJ, Hunter TM, Sadler PJ, Pannecouque C, De Clercq E, Archibald SJ. Configurationally restricted bismacrocyclic CXCR4 receptor antagonists. J Med Chem. 2006;49:6162–6165. doi: 10.1021/jm0607810. [DOI] [PubMed] [Google Scholar]
- 21.Sun X, Wuest M, Weisman GR, Wong EH, Reed DP, Boswell CA, Motekaitis R, Martell AE, Welch MJ, Anderson CJ. Radiolabeling and in vivo behavior of copper-64-labeled cross-bridged cyclam ligands. J Med Chem. 2002;45:469–477. doi: 10.1021/jm0103817. [DOI] [PubMed] [Google Scholar]
- 22.Hubin TJ, McCormick JM, Collinson SR, Alcock NW, Busch DH. Ultra rigid cross-bridged tetraazamacrocycles as ligands - the challenge and the solution. Chemical communications. 1998:1675–1676. [Google Scholar]
- 23.Yamamoto H, Maruoka K. Regioselective Carbonyl Amination Using Diisobutylaluminum Hydride. Journal of the American Chemical Society. 1981;103:4186–4194. [Google Scholar]
- 24.Bjorndal A, Deng HK, Jansson M, Fiore JR, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman DR, Fenyo EM. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. Journal of Virology. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nimmagadda S, Pullambhatla M, Stone K, Green G, Bhujwalla ZM, Pomper MG. Molecular imaging of CXCR4 receptor expression in human cancer xenografts with [64Cu]AMD3100 positron emission tomography. Cancer research. 2010;70:3935–3944. doi: 10.1158/0008-5472.CAN-09-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zwier JM, Roux T, Cottet M, Durroux T, Douzon S, Bdioui S, Gregor N, Bourrier E, Oueslati N, Nicolas L, Tinel N, Boisseau C, Yverneau P, Charrier-Savournin F, Fink M, Trinquet E. A Fluorescent Ligand-Binding Alternative Using Tag-lite (R) Technology. Journal of biomolecular screening. 2010;15:1248–1259. doi: 10.1177/1087057110384611. [DOI] [PubMed] [Google Scholar]
- 27.Jones-Wilson TM, Deal KA, Anderson CJ, McCarthy DW, Kovacs Z, Motekaitis RJ, Sherry AD, Martell AE, Welch MJ. The in vivo behavior of copper-64-labeled azamacrocyclic complexes. Nucl Med Biol. 1998;25:523–530. doi: 10.1016/s0969-8051(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 28.Gerlach LO, Jakobsen JS, Jensen KP, Rosenkilde MR, Skerlj RT, Ryde U, Bridger GJ, Schwartz TW. Metal ion enhanced binding of AMD3100 to Asp262 in the CXCR4 receptor. Biochemistry. 2003;42:710–717. doi: 10.1021/bi0264770. [DOI] [PubMed] [Google Scholar]
- 29.Khan A, Nicholson G, Greenman J, Madden L, McRobbie G, Pannecouque C, De Clercq E, Ullom R, Maples DL, Maples RD, Silversides JD, Hubin TJ, Archibald SJ. Binding optimization through coordination chemistry: CXCR4 chemokine receptor antagonists from ultrarigid metal complexes. J Am Chem Soc. 2009;131:3416–3417. doi: 10.1021/ja807921k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenkilde MM, Gerlach LO, Hatse S, Skerlj RT, Schols D, Bridger GJ, Schwartz TW. Molecular mechanism of action of monocyclam versus bicyclam non-peptide antagonists in the CXCR4 chemokine receptor. J Biol Chem. 2007;282:27354–27365. doi: 10.1074/jbc.M704739200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.