Abstract
Rapid diagnosis of infectious diseases and timely initiation of appropriate treatment are critical determinants that promote optimal clinical outcomes and general public health. Conventional in vitro diagnostics for infectious diseases are time-consuming and require centralized laboratories, experienced personnel and bulky equipment. Recent advances in biosensor technologies have potential to deliver point-of-care diagnostics that match or surpass conventional standards in regards to time, accuracy and cost. Broadly classified as either label-free or labeled, modern biosensors exploit micro- and nanofabrication technologies and diverse sensing strategies including optical, electrical and mechanical transducers. Despite clinical need, translation of biosensors from research laboratories to clinical applications has remained limited to a few notable examples, such as the glucose sensor. Challenges to be overcome include sample preparation, matrix effects and system integration. We review the advances of biosensors for infectious disease diagnostics and discuss the critical challenges that need to be overcome in order to implement integrated diagnostic biosensors in real world settings.
Keywords: iosensor, infectious diseases, matrix effects, microfluidics, sample preparation, system integration
Despite significant progress in prevention, diagnosis and treatment in the last century, infectious diseases have remained as significant global health problems [1–3]. Major challenges for management of infectious diseases include injudicious use of antimicrobials, proliferation of multidrug-resistant (MDR) pathogens, emergence of new infectious agents and ease of rapid disease dissemination due to overpopulation and globalization. Timely diagnosis and initiation of targeted antimicrobial treatment are essential for successful clinical management of infectious diseases [4].
Current diagnosis of clinically significant infectious diseases caused by bacterial (e.g., pneumonia, sepsis, genitourinary tract infections), mycobacterial (e.g., tuberculosis), viral (e.g., HIV, hepatitis), fungal (e.g., candidiasis) and parasitic (e.g., malaria) pathogens rely on a variety of laboratory-based tests including microscopy, culture, immunoassays and nucleic-acid amplification (Table 1). While widely used, these in vitro diagnostics have well-recognized shortcomings. Microscopy lack sensitivity in many clinical scenarios and culture has a significant time delay. Immunoassays such as ELISA, while highly sensitive, are labor intensive and challenging to implement multiplex detection. Nucleic-acid amplification tests such as PCR offer molecular specificity but have complex sample preparation and potential for false positives.
Table 1.
Standard in vitro diagnostics for representative infectious diseases.
| Disease | Sites of infection | Types of analytes | Sample | Diagnostic tests | Challenges toward biosensor diagnosis |
Ref. |
|---|---|---|---|---|---|---|
| HIV/AIDS | Immune system | Viruses; antigens; antibodies; host cells; nucleic acids | Blood; saliva; urine | CD4 T-cell counts; dipstick immunoassay; ELISA; western blot; NAAT; viral load | Viral isolation and viral load determination are challenging; multiple biomarkers are required for definitive diagnosis | [114–116] |
| Tuberculosis | Lung | Mycobacteria; antigens; antibodies; nucleic acids | Sputum; urine; blood | Sputum smear microscopy; IFN-γ release assay; tuberculin skin test; culture; NAAT; ELISA | Sample processing of sputum challenging; lack of established biomarkers | [117–119] |
| Malaria | Liver; blood | Parasites; antigens; antibodies; nucleic acids | Blood | Blood film microscopy; dipstick immunoassay; ELISA; NAAT | Requires expertise to perform microscopy (diagnostic standard); lack of molecular targets for non-falciparum infections | [120–122] |
| Herpes simplex virus | Skin | Viruses; antigens; antibodies; cells; nucleic acids | Skin swab; blood | Culture; ELISA; western blot; direct immunofluorescence assay; NAAT | Viral isolation is challenging; sample processing of skin swabs; need type-specific serological assays | [123–125] |
| Viral hepatitis | Liver | Antigens; antibodies; nucleic acids | Blood; stool | ELISA; recombinant immunoblot assay; NAAT | Multiple serological markers are required for different stages of infection and convalescence | [126,127] |
| Dengue fever | Immune system | Viruses; antigens; antibodies; nucleic acids | Blood | Culture; ELISA; NAAT | Viral isolation is challenging; presence of pre-existing antibodies from a prior heterologous or flavivirus infection can affect the performance of many diagnostic assays | [128,129] |
| Urinary tract infections | Bladder; kidney | Bacteria | Urine | Culture; urine dipstick; urine microscopy | Wide variety of potential pathogens; sample concentration may be difficult; lysis of Gram-positive bacteria is still challenging | [130,131] |
| Influenza virus infections | Respiratory tract | Viruses; antigens; antibodies; nucleic acids | Nasal swab; sputum; blood | Hemaggluti nation- inhibition assay; ELISA; immunofluorescence assay; single radial hemolysis; culture; NAAT | Sample processing of sputum and nasal swab processing; strain-specific assays are essential due to the wide diversity of the virus | [132–134] |
| Gastroenteritis | Gl tract | Bacteria; viruses, parasites; leukocytes; toxins; antigens; antibodies; nucleic acids | Stool; blood | Culture; stool Gram’s stain; stool ova and parasites exam; fecal leukocytes; toxin assay; antigen assay; NAAT | Sample processing of stool challenging; wide variety of potential pathogens | [135–137] |
| Sepsis | Bloodstream | Bacteria; fungi; viruses; host immune cells; nucleic acids | Blood | Culture; white blood cell count; immunoassays; FISH; NAAT | Wide panel of pathogens and high individualized nature of the disease can lead to more complicated sensor design; lack of suitable biomarkers for immunoassay | [138–140] |
| Sexually transmitted infections | Genital tract; oropharynx and other mucosal surfaces | Bacteria; fungi; viruses; parasites; protozoa; host immune cells; antibodies; antigens; nucleic acids | Genital swab; urine; blood; skin | NAAT; culture; ELISA; immunochromatographic assay; Papanicolaou test; microscopy; white blood cell count | Multiple biomarkers are required as variant strains can be detected; wide panel of potential pathogens | [141,142] |
| Wound infections | Skin | Bacteria; fungi; viruses; parasites; antigens; antibodies; host immune cells | Wound swab; blood; skin | Blood test; culture; ELISA; NAAT; microscopy; wood’s lamp examination | Wide panel of pathogens can lead to more complicated sensor design | [143,144] |
| Fungal infections | Skin; nails; blood; respiratory tract; urogenital tract; Gl tract | Fungi; antigens; antibodies; nucleic acids | Wound swab; blood; sputum; urine | Culture; microscopy; NAAT; ELISA; blood test; FISH | Combination of tests may be needed to improve invasive fungal infection diagnosis | [145,146] |
NAAT: Nucleic acid amplification test.
Standard process flow for common infectious disease diagnostics includes collection and transport of biological samples (i.e., blood, urine, sputum, cerebrospinal fluid, tissue swabs) from the point of care to a centralized laboratory for sample processing by experienced personnel. After the results become available (usually days), the laboratory notifies the clinicians, who in turn contact the patients and modify the treatment as needed. This inherent inefficiency complicates timely delivery of evidence-based care and has contributed to the injudicious use of antimicrobials. In non-traditional and resource-poor healthcare settings, the shortcomings of standard diagnostics are further highlighted.
A biosensor is an analytical device that converts molecular recognition of a target analyte into a measurable signal via a transducer. The most well-known example in use today is the glucose sensor, which has had a transformative effect on the management of diabetes since its introduction in the current form 30 years ago. Other widely used examples include lateral flow assays such as the home pregnancy test [5,6]. For infectious diseases, biosensors offer the possibility of an easy-to-use, sensitive and inexpensive technology platform that can identify pathogens rapidly and predict effective treatment [7–9]. Advantages include small fluid volume manipulation (less reagent and lower cost), short assay time, low energy consumption, high portability, high-throughput and multiplexing ability [10]. Recent advances in micro- and nanotechnologies have led to development of biosensors capable of performing complex molecular assays required for many of the infectious diseases. In parallel, significant progress has been made toward the understanding of pathogen genomics and proteomics and their interplay with the host [11–13]. Biosensor-based immunoassays may improve the detection sensitivity of pathogen-specific antigens, while multiplex detection of host immune response antibodies (serology) may improve the overall specificity. Further system integration may facilitate assay developments that can integrate both pathogen-specific targets as well as biomarkers representative of host immune responses at different stages of infection [14].
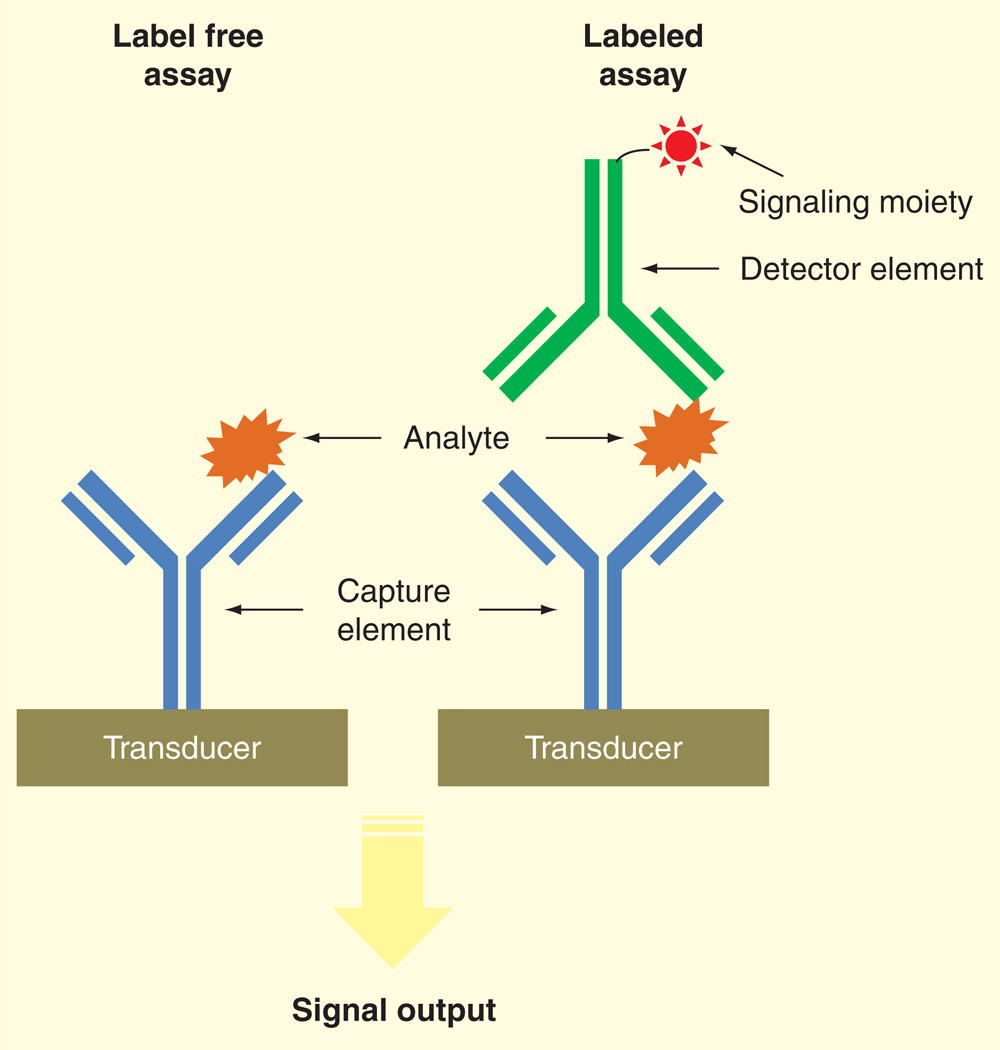
In this review, we focus on advances in biosensor technologies for infectious diseases, with emphasis on distinction among various signal transducer approaches and their potential for clinical translation. Detection strategies are divided into label-free and labeled assays (Figure 1). Label-free assays measure the presence of an analyte directly through biochemical reactions on a transducer surface [15,16]. For labeled assays, the analyte is sandwiched between capture and detector agents, with specific label on the detector agent such as an enzyme, fluorophore, quantum dot or radioisotope, for signal output [17]. Integrated systems based on nucleic-acid amplification tests is another distinct approach for point-of-care diagnosis [18–21], which is not the focus of this review. Finally, the challenges posed by sample preparation, which remains as a ratelimiting factor toward point-of-care diagnostics and clinical translation, will be discussed.
Figure 1.
Schematic representation of label-free and labeled assays to biosensing using antibodies.
Label-free biosensors
Label-free biosensors monitor changes that occur when target analytes bind with molecular capturing elements immobilized on a solid support, or elicit changes in interfacial capacities or resistance [15,16]. Label-free biosensors require only a single recognition element, leading to simplified assay design, decreased assay time and reduction in reagent costs. This recognition mode is especially appropriate for small molecular targets, which can be buried within the binding pocket of the capturing element, leaving little room for interaction with a detector agent that would be required in a labeled assay. Another advantage of label-free method is the ability to perform quantitative measurement of molecular interaction in realtime, allowing continuous data recording. Also, target analytes are detected in their natural form without labeling and chemical modification, thus can be preserved for further analysis. The label-free sensing strategies for various infectious diseases discussed below operate through a binding-event-generated perturbation in optical, electrical or mechanical signals (Table 2).
Table 2.
Examples of label-free detection strategies.
| Label-tree assay |
Technology | Advantages | Disadvantages | Ret. |
|---|---|---|---|---|
| Optical transducer | Surface plasmon resonance | Real-time detection; possibility of high throughput | Sensitive to sample matrix effects; sensor surface functionalization challenging; bulky optical equipment | [23,24] |
| Electrical transducer | Redox electrochemistry (amperometric) | Simple sensor design; detection platform amenable to inexpensive and miniaturization | Redox species required to increase current production; no real-time detection; sensitive to sample matrix effects | [29,147,148] |
| Impedance spectroscopy | Simple electrode design; real-time detection | Sensitive to sample matrix effects; bulky equipment; data analysis may not be trivial (theoretical model may be required) | [30,31,33] | |
| Potentiometry | Real-time detection; consecutive measurements on different samples are possible | Bulky equipment, sensitive to sample matrix; complicated sample preparation steps; careful control of temperature is essential | [34,149] | |
| Field effect transistor | Real-time detection; stable sensor response; detection platform amenable to POC system | Sensitive to sample matrix effects; complicated sensor fabrication; careful control of temperature is essential | [36,37] | |
| Mechanical transducer | Microcantilever | Real-time detection; multiplex and high throughput are possible | Sensitive to sample matrix effects; careful control of temperature is essential; bulky equipment | [38–41] |
| Quartz crystal microbalance | Simple electrode design; real-time detection; detection platform amenable to POC system | Sensitive to sample matrix effects; careful control of temperature and stress is essential | [42,43,150,151] | |
POC: Point-of-care.
Optical transducer
Optical transducers are widely used due to their high sensitivity with several well-established optical phenomena such as surface plasmon changes, scattering and interferometry [22]. Surface plasmon resonance (SPR) is the excitation of an electromagnetic wave propagating along the interface of two media with dielectric constants of opposite signs, such as metal and sample buffer, by a specific angle of incident light beam [23]. The signal is based on total internal reflection that results in a reduced intensity of the reflected light. The angle at which the resonance occurs is sensitive to any change at the interface, such as changes in refractive index or formation of a nanoscale film thickness due to surface molecular interactions. Therefore, these changes can be measured by monitoring the light intensity minimum shift over time. A bioanalyzer based on SPR was employed for the detection of Escherichia coli O157:H7 and methicillin-resistant Staphylococcus aureus (MRSA) using T4 and BP14 bacteriophages, respectively as capturing elements [24]. Without labeling or enrichment, this SPR bioanalyzer could detect as few as 103 cfu/ml in less than 20 min.
Backscattering interferometry (BI) is another optical detection method used for biosensing [25]. BI systems consist of a coherent single wavelength light source (commonly a low power He-Ne or red diode laser) focused onto a microfluidic channel and a detector to analyze the reflected intensity. Upon coherent-laser illumination of the fluid-filled channel, a highly modulated interference pattern is produced due to sub-wavelength structures in the channel. Analysis of changes in the profile of fringe patterns by the detector located in the direct backscatter direction can facilitate measurement of refractive index changes and allow quantification of molecular binding events. BI can detect both free solution or surface immobilized molecular interactions with unprecedented limits in microfluidic devices (picoliter detection volume) and allows real-time determination of binding constants spanning from micro- to picomole. Kussrow et al. have shown the potential of utilizing BI for rapid detection of purified total human IgG from seropositive syphilis patients using a purified recombinant treponenmal antigen r17, demonstrating the prospect of using this approach for serological diagnosis in clinical samples [26].
Most label-free optical biosensors require precise alignment of light coupling to the sensing area, which is a major drawback for point-of-care applications. Therefore, optical sensing can be significantly improved when this approach is used in an integration scheme. Integrated optics allow several passive and active optical components on the same substrate, allowing flexible development of minimized compact sensing devices, with the possibility of fabrication of multiple sensors on one chip. A novel nanoplasmonic biosensor based on light transmission effect in plasmonic nanoholes and group-specific antibodies for highly divergent strains of rapidly evolving viruses has been developed, allowing direct coupling of perpendicularly incident light with the sensing platforms and minimizes the alignment requirements for light coupling. This system was used to demonstrate the recognition of small enveloped RNA viruses (vesicular stomatitis virus and pseudotyped Ebola) as well as large enveloped DNA viruses (vaccinia virus) at clinically relevant concentrations [27].
Electrical transducer
Electrical analytical methods are common sensing approaches due to their innate high sensitivity and simplicity that can be effectively conjugated to miniaturized hardware. Common types of electrical biosensors that have been applied to infectious disease diagnostics include voltammetric, amperometric, impedance and potentiometric sensors [28]. Voltammetric and amperometric sensors involve current measurement of an electrolyte with a DC voltage applied across the electrode as a function of voltage and time, respectively. An immunosensor based on the amperometric approach has been developed for the detection of hepatitis B surface antigen, a major index of hepatitis B viruses [29]. This sensor contains a glassy carbon electrode modified with an assembly of positively charged poly(allyamine)-branched ferrocene and negatively charged gold nanoparticles (Au NPs). The combination of the biocompatible and stable poly(allyamine)-branched ferrocene composite film with redox activity and the conducting Au NPs with larger specific interfacial area were effective in preventing the leakage of both mediator and antibodies and provided sensitive and selective adsorption to hepatitis B surface antigen in human serum. Impedance-based electrical transducers measure the electrical opposition to current flow at an interface by applying a sinusoidal voltage at a particular frequency or at a wide range of frequencies with a constant direct current bias voltage [30]. The impedance is the ratio between applied sinusoidally varying potential and the derived current response across the interface. An impedance biosensor using carbohydrate α-mannoside for recognition was developed for detecting E. coli ORN 178, a surrogate for the pathogenic E. coli O157:H7, with a detection limit of 102 cfu/ml [31]. Another impedance biosensor has been developed for detection of viral infections during acute phase, which is crucial since replication and shedding may occur before detectable antibodies appear [32]. Shafiee et al. have isolated, enriched HIV-1 and its multiple subtypes with magnetic beads conjugated with anti-gp120 antibodies, and detected the viral lysates with impedance analysis at the acute state of infection (106–108 copies/ml) on an electrode with simple geometry [33].
Potentiometry is another simple and widely used technique based on measurement of potential or charge accumulation using a high impedance voltmeter with negligible current flow. An immunosensor developed based on the potentiometric transduction capabilities of single-walled carbon nanotubes (SWCNTs) in combination with the recognition capabilities of protein-specific RNA aptamers was exploited for determining variable surface glycoproteins (VSGs) from African Trypanosomes [34]. Similar to antibodies, apatmers are small synthetic RNA/DNA molecules that can form secondary and tertiary structures capable of specifically binding to various molecular targets [35]. A potential shortcoming with RNA-based aptamers is their short half-life due to susceptibility of the phosphodiester backbone and the 5´ and 3´-termini to ribonucleases and exonucleases, respectively. Nuclease-resistant RNA aptamer sensors were synthesized based on 2´ F-substituted C- and U-nucleotides. The hybrid nanostructured (VSG-specific and nuclease-resistant RNA aptamers hybridized with SWCNTs) potentiometer demonstrated VSG protein detection at attomolar concentrations in blood.
A closely related electrical sensor is the field effect transistor (FET). In this technology, the current-carrying capability of a semiconductor is varied by the application of an electric field due to nearby charged particles. In most cases, the sensor response is interpreted as a result of a shift of the flat-band or threshold voltage of the field-effect structure, which is due to the binding process at a gate electrode or at the current carrying element. A biosensor for detecting the pathogenic yeast Candida albicans was developed based on a FET, in which a network of SWCNTs functionalized with monoclonal anti-Candida antibodies acts as the conductor channel [36]. These specific binding sites for yeast membrane antigens provided a sensitive limit of detection as low as 50 cfu/ml. Another FET-based biosensor involved an In2O3 nanowire functionalized with antibody mimic proteins (AMPs) for detection of nucleocapsid protein, a biomarker of severe acute respiratory syndrome [37]. Similar to aptamers, AMPs are engineered in vitro to target specific analytes. Tailor-made AMPs are stable over a wide range of pH and electrolyte concentrations and can be produced in large quantity at relatively low cost, making them an ideal capturing element for biosensor surface specification. This FET-based platform has been used to demonstrate nucleocapsid protein detection in complex media at sensitivities comparable with ELISA.
Mechanical transducer
Advances in micro- and nanofabrication technologies have facilitated the emergence of micro- and nanoscale mechanical transducers capable of detecting changes in force, motion, mechanical properties and mass that come along with molecular recognition events [38,39]. Among different mechanical biosensors, cantilever and quartz crystal microbalances (QCMs) are the most established techniques. Mechanical bending of a micro- or nanocantilever is monitored as analytes bind, with optical readout typically used to detect the deflection or change in stress/strain profile of the cantilever. In one example, a cantilever array was functionalized with carbohydrate molecules as capture agents for E. coli [40]. In this work, the gold-coated top sides of the cantilever array functionalized with self-assembled layers of distinct mannosides allowed the reproducible real-time detection of different E. coli strains including ORN 208, 178 and 206, with sensitivity range over four orders of magnitude. As the E. coli strains used bind to mannose but not galactose, a structurally similar carbohydrate, an internal reference cantilever with galactose was included to assess non-specific binding and account for non-specific reactions, including small changes in pH, refractive index or reactions occurring on the underside of the cantilever. Liu et al. expanded the applications of the cantilever-based sensor from a cell-screening tool to a real-time cell growth monitor to provide new insights into drug–cell interactions [41]. They demonstrated real-time growth monitoring of Saccharomyces cerevisiae yeast strains, YN94-1 and YN94-19, on the polymer cantilevers. The enhanced sensitivity of the static mode microcantilever-based system differentiated the effects of both withholding essential nutrients (synthetic complete-uracil) and drug (5´-fluoroorotic acid) interactions with yeast cells. Further, compared with silicon nitride cantilevers, polymer microcantilever sensors can be fabricated at lower cost with laser micromachining and offer higher sensitivity due to the rubbery modulus of the polyimide used.
Piezoelectric detection, such as a QCM, measures variations in resonant frequency of an oscillating quartz crystal in response to the changes in surface-adsorbed mass due to a bio-recognition event. The application of an external potential to a piezoelectric material, such as quartz, produces internal mechanical stresses that induce an oscillating electric field, which then initiates an acoustic wave throughout the crystal in a direction perpendicular to the plate surfaces. The resonance frequency shift in a QCM can be influenced by many factors, such as changes in mass, viscosity, dielectric constant of the solution and the ionic status of the crystal interface with the buffer solution. Peduru Hewa et al. [42] developed a QCM-based immunosensor for detection of influenza A and B viruses. By conjugating Au NPs to the anti-influenza A or B monoclonal antibodies, a detection limit of 1 × 103 pfu/ml for laboratory cultured preparations and clinical samples (nasal washes) was achieved. In 67 clinical samples, the QCM-based immunosensor was comparable with standard methods such as shell vial and cell culture and better than ELISA in terms of sensitivity and specificity. Another strategy for enhancing the sensitivity and specificity of QCM-based biosensors involves fabrication of molecular imprinted film on a QCM chip. Molecularly imprinted polymers are a powerful tool for fabrication of synthetic recognition elements. For example, Lu et al. developed a biomimetic sensor based on epitope imprinting for detection of HIV-1 glycoprotein gp41, an important index of disease progression and therapeutic response [43]. The advantages of epitope-mediated imprinting over traditional protein imprinting approaches include higher affinity, less non-specific binding and lower cost. For this sensor, dopamine was used as the functional monomer and polymerized on the surface of a QCM chip in the presence of a synthetic peptide analogous to residues 579–613 of gp41. The sensor allowed direct quantitative detection of gp41 with a detection limit of 2 ng/ml, which is comparable with ELISA. The sensor also showed satisfactory performance of detecting gp41 spiked in human urine samples, demonstrating the potential for point-of-care application. Another example proposed by Tokonami et al. utilized a molecularly imprinted polymer film consisting of overoxidized polypyrrole (OPPy) in combination with QCM for direct bacterial detection at concentrations as low as 103 cfu/ml within 3 min [44]. Furthermore, the bacterial cavities created in the OPPy film had high selectivity and were able to distinguish target bacteria, Pseudomonas aeruginosa, in a mixture of similar shaped bacteria including Acinetobacter calcoaceticus, E. coli and Serratia marcescens.
As label-free schemes generally do not include signal amplification, improvement of specificity and sensitivity of a given device depends largely on the proper selection and combination of capturing elements and transducers. With continuing advances in biochemistry and molecular biology, it is anticipated that the diversity of capturing elements with higher affinity, specificity and stability will continue to expand. A major challenge for clinical application of label-free biosensors remains in translating the technologies from detection in laboratory solutions to real-world clinical samples, such as blood, serum and urine. The complex sample matrices of clinical samples can lead to non-specific binding and aberrant signals. For example, charge-based label-free biosensors are highly sensitive to changes in pH, ionic strength and environmental temperature. Nanowires often require sample desalting prior to detection of analyte, and microcantilevers require sensitive temperature regulators [38,45]. Also, non-specific binding events may contribute a measurable signal indistinguishable from the specific target analyte signal. A number of strategies have been developed to mitigate the sample matrix effect. One of the most common approaches is to exploit hydrophilic ‘antifouling’ surfaces, such as polyethylene glycol and its derivatives [46]. It has been shown that a polyethylene glycolmodified surface was sufficiently robust for biomarkers detection with clinically relevant sensitivity in undiluted blood serum by electrochemical impedance spectroscopy [47]. Zwitterionic polymers, which are highly hydrophilic and electrically neutral in nature, have also received much attention as antifouling interfaces. Several groups have shown that a coating of polycarboxybetaine methacrylate, a zwitterionic-based material, on the sensor surface, prevents non-specific adsorption of proteins from blood serum and enhances the antibody target-binding affinity, making label-free detection in clinical samples a possibility [48,49].
Labeled biosensors
Labeled assays are the most common and robust method of biosensing. Classically, in labeled assays, the analyte is sandwiched between the capture and detector agents [50]. Capture agents are typically immobilized on a solid surface such as electrodes, glass chips, nano- or microparticles, while detector agents are typically conjugated to signaling tags, such as fluorophores, enzymes or NPs [17]. As with label-free assays, optical, electrical or mechanical transducers can be coupled to the signaling tag. Examples of sensor–tag interactions include optical sensors used to detect fluorescent [51], colorimetric [52] or luminescent tags [53], electrochemical sensors used to detect redox reactions from enzyme tags [54] and magnetoresistive sensors used to detect magnetic tags [55]. With these systems, quantitative or semi-quantitative detection of analyte is possible by relating the signal generated to the amount of analyte captured. In general, capture and detector elements have different binding sites, thus the specificity increased and the background reduced. However, the multistep protocol can make the assay more costly and complicated.
ELISA is the standard sandwich immunoassay for infectious disease applications in clinical laboratories [50]. ELISA typically uses a capture antibody and a detector antibody modified with an enzyme tag for catalyzing the conversion of chromogenic substrate into colored molecules. In quantitative ELISA, the optical density of the colored product from the sample is compared with a standard serial dilution of a known concentration of the target molecule. Nucleic acids can also be detected with sandwich assays. For example, the Liao group developed an electrochemical sensor assay to detect urinary pathogens in clinical samples based on immobilized capture oligonucleotide and labeled detector oligonucleotide for detection of bacterial 16S rRNA [54]. Signal is generated by an oxidation-reduction current produced by the enzyme tag conjugated to the detector probe. The best known commercially available sandwich assays are lateral flow immunoassays or immunochromatographic test strips, in which the signal can be measured qualitatively by eye or semi-quantitatively by engineering interfaces such as low-cost laser- and photodiode or amperometric detectors [56]. Most well-known commercially available examples include home pregnancy tests and urinalysis strips. Lateral flow assays have been proposed for saliva- or blood-based HIV tests, blood-based malaria antigen test and serum-based tuberculosis test [6]. Advantages of lateral flow assays include low cost, minimal to no sample preparation and straightforward interpretation of the results [57]. Disadvantages include relatively poor sensitivity for many of the clinically relevant targets and qualitative or semi-quantitative results. To improve the limit of detection, recent efforts have focused on signal amplification. Promising development in the field of nanotechnology over the years has facilitated the functionalization of NPs with different biological molecules, which turns them into ideal labels for various signal amplification processes in the biosensor platforms. Due to their high surface-to-volume ratio, NPs are attractive means of signal amplification to improve sensitivity and versatility of biosensing devices [9,17,58]. Labeled biosensors-based biobarcode, metal NPs and magnetic NPs labeling are reviewed (Table 3).
Table 3.
Examples of labeled detection strategies.
| Labeled assay |
Technology | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Redox electrochemistry (amperometric) | Detection platform amenable to POC system; easy integration with other electric field-driven modules | No real-time detection; multiple steps assay | [14,70,152] | |
| Bio-barcode | Detection platform amenable to POC system; easily interpreted results | No real-time detection; complicated protocol for probe preparations; multiple steps assay | [59,60] | |
| Metal nanoparticles | Detection platform amenable to POC system; easily interpreted results; multiplex | No real-time detection; temperature fluctuations can affect the results; multiple steps assay | [61–66] |
POC: Point-of-care.
Biobarcode
One of the most promising NP-based approaches is the biobarcode amplification (BCA) assay, which is able to detect both proteins and nucleic acids without enzymatic reactions [52,59]. BCA involves a sandwich assay with targets captured with micro- or nanoparticles conjugated with oligonucleotides (barcode DNA) as surrogates for signal amplification. With every target captured, many strands of barcode DNA are released for subsequent detection with other means such as electrochemical or optical. BCA was recently applied to detection of HIV-1 capsid (p24) antigen, a useful marker for predicting CD4+ T-cell decline, disease progression and early detection of HIV-1 infection [60]. The detection scheme used an anti-p24-coated microplate to first capture viral p24, followed by a biotinylated detector antibody. A streptavidin-coated NP-based biobarcode DNAs was next introduced for signal amplification, followed by detection using a chip-based scanometric method. A detection range of 0.1–500 pg/ml was demonstrated, which was 150-fold more sensitive than conventional ELISA. When tested with clinical blood samples, 100% negative and positive predictive values were found in 30 and 45 samples, respectively. Also, it could detect HIV-1 infection 3 days earlier than ELISA in seroconverted samples.
Metal nanoparticles
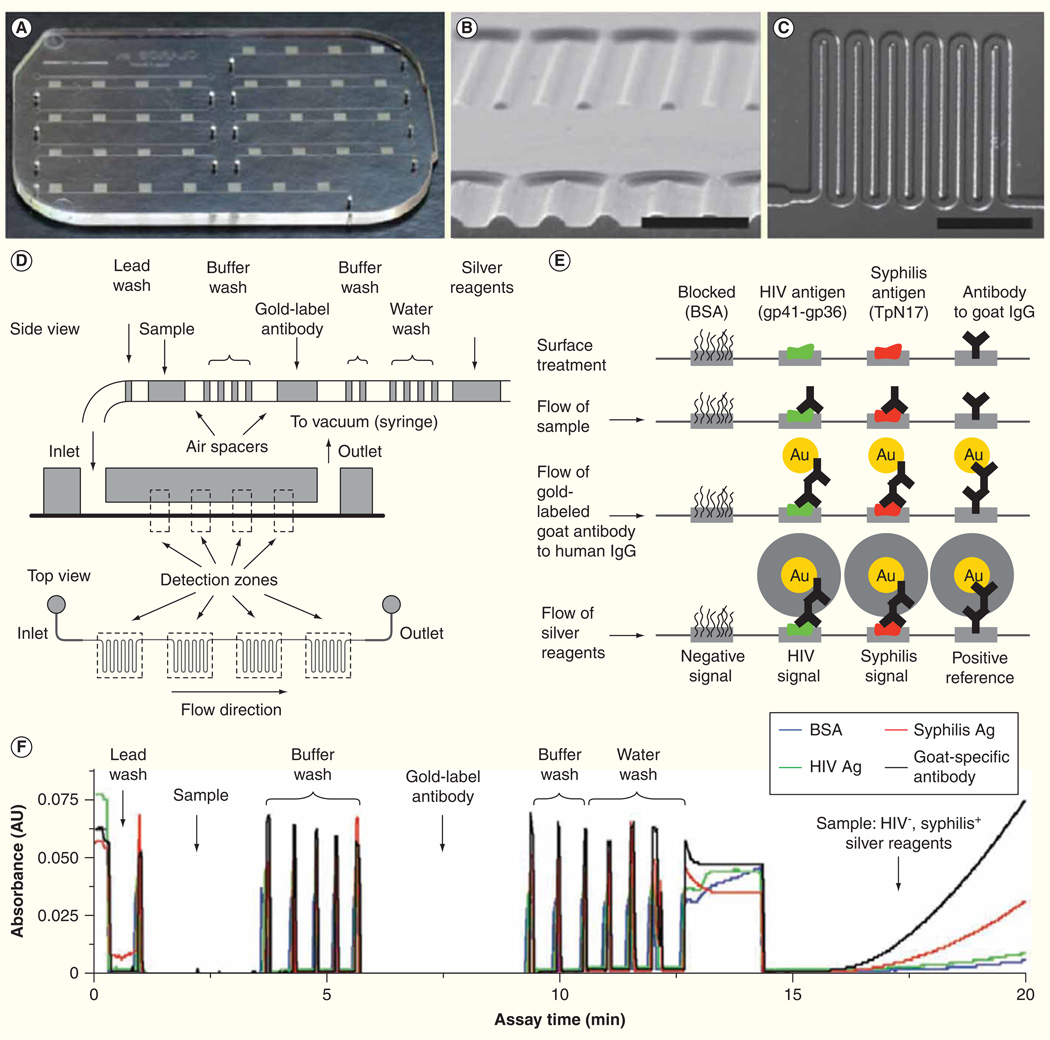
Metal NPs can also serve as signal amplification labels for bio-recognition processes based on their unique optical properties [61,62]. Gold and silver NPs exhibit plasmon absorbance bands in the visible light spectrum that are determined by the size of the respective particles. Therefore, the spectral shifts due to the aggregation of metal NPs have prompted numerous studies to develop optical biosensors with biomaterial-metal NPs hybrid systems as the detection amplifiers. One such sensor is part of an integrated microfluidic chip using gold-labeled antibodies for simultaneous diagnosis of HIV and syphilis from 1 µl of whole blood (Figure 2) [63]. The signal amplification occurs via the reduction of silver ions onto Au NPs inside a millimeter-sized meandering channel design. The optical density of the silver film is detected and can be quantified with the low-cost optics or qualitatively by eye. Initial studies indicate this integrated biosensor is comparable with commercial ELISA kits with near 100% sensitivity and 98–100% specificity for HIV and 82–100% sensitivity and 97–100% specificity for syphilis.
Figure 2. Integrated microfluidic system for multiplexed detection of HIV and syphilis.
(A) Photograph of microfluidic chip. (B) Cross-section of microchannels. Scale bar, 500 µm. (C) The design of channel meanders. Scale bar, 1 mm. (D) Schematic diagram of passive fluid delivery of preloaded reagents over four detection zones based on vacuum generated by a disposable syringe. (E) Illustration of reactions at different detection steps. Signal amplification was achieved by the reduction of silver ions on gold nanoparticle-conjugated antibodies. Signals can be read quantitatively with low-cost optics or qualitatively by eyes. (F) Real-time monitoring of absorbance signals at the detection zones.
Adapted with permission from [63] © Macmillan Publishers Ltd. (2011).
Magnetic nanoparticles
Magnetic NPs-coupled detectors for biosensing can be used for signal amplification with the advantage that they are amenable to use in solution phase sandwich assays such as diagnostic magnetic resonance [55,64]. A major advantage of solution phase assays is significantly faster assay times compared with diffusion-dependent surface structure-based assays. With diagnostic magnetic resonance, both the capture and detection agents are in solution and linked to magnetic particles. When an analyte of interest is present, the magnetic particles cluster as the antibodies bind the analyte. The clusters of magnetic particles are more efficient at dephasing nuclear spins of the adjacent water protons, causing a decrease in the spin-spin relaxation time, resulting in a quantifiable signal. Chung el al. have presented a magneto-DNA platform targeting bacterial 16S rRNAs capable of profiling a panel of 13 bacterial species from clinical samples including urine, pleural fluid, biliary fluid, ascitic fluid and blood [65]. Near single bacterium sensitivity can be achieved by three signal amplification steps including reverse transcription-PCR amplification of the 16S rRNA, polymeric bead capture and enrichment of target DNA and magnetic amplification with magnetically labeled beads conjugated to target DNA (a single magnetic NPs can affect billions of surrounding water molecules) [66]. Two drawbacks of the system are the requirement for manual sample preparation and the PCR experiment is a separate step from the nuclear magnetic resonance-based sensor.
Antimicrobial susceptibility tests
While accurate pathogen identification is the key to diagnosis, assessing pathogen antimicrobial susceptibility is an important parameter in the management of infection. Rapid antimicrobial susceptibility test (AST) can expedite appropriate therapy to impact clinical outcome and may reduce emergence and transmission of MDR pathogens. As the rates of MDR pathogens and new infectious diseases rise, the administration of appropriate treatment in a timely manner becomes more challenging using current tests [67]. Hence, a rapid diagnostic system that combines pathogen identification and AST would meet a significant clinical need [68]. Antimicrobial susceptibility can be determined phenotypically by measuring bacterial growth/growth inhibition in the presence of a drug, or genotypically with PCR-based assays to identify genetic mechanisms that confer resistance [69].
Phenotypic ASTs are the mainstay in the clinical microbiology laboratory. These tests typically require isolation of the pathogen and long incubation time accounting for the lag time of 24–72 h from sample collection to completion of analysis. Recent studies have demonstrated development of biosensor and microfluidic devices for rapid AST. Mach et al. demonstrated rapid AST from clinical urine samples by direct culture of infected urine in the presence of antibiotic followed by electrochemical detection of 16S rRNA levels as a measure of cell growth [70]. The AST assay was completed in 3.5 h with 94% agreement with standard AST. Another rapid AST approach used an electrochemical biosensor for detection of precursor rRNAs (pre-rRNA), an intermediate state in formation of mature rRNA and a marker for cell growth [71]. The specificity of the assay was validated with inhibitors of pre-rRNA synthesis and processing (rifampin/rifampicin and chloramphenicol) and a DNA gyrase inhibitor (ciprofloxacin). A decline in pre-rRNA was detectable within 15 min in drugsusceptible bacteria but not in resistant strains. Another approach used optical detection for a single cell AST where bacteria were cultured in with/without antibiotic in microchannels. Individual uropathogenic E. coli cells were confined to bacterium-width microchannels with dielectrophoresis (DEP), an electrokinetically driven short-range particle trapping force, applied through an integrated microelectrode [72]. Growth was measured with an epifluorescence microscope and AST profile determined within 1 h. Another microfluidic platform for AST was based on stress activation of biosynthetic pathways [73]. In this assay, S. aureus bound to the bottom of a microfluidic channel, was subjected to mechanical shear stress and enzymatic stress with subinhibitory concentrations of a bactericidal agent resulting in cell wall damage. Subsequent treatment with the antibiotic oxacillin interfered with the repair process, resulting in rapid cell death of susceptible S. aureus strains, while resistant bacteria remained viable under the same conditions. Cell viability was monitored using a vital dye and AST results were established based on normalized fluorescence values after 60 min. This approach correctly designated oxacillin susceptibility of 16 clinical relevant S. aureus strains.
In general, microfluidic approaches are promising for the miniaturization and rapid determination of antimicrobial susceptibility [68,74–77]. These approaches can potentially be integrated with multiple functionalities into portable chips, which in turn can facilitate AST at the point of care. Additional work is needed to confirm the accuracy of these devices with respect to current clinical ASTs.
Sample preparation
Advances in biosensor technology and signal amplification have led to highly sensitive detection of pathogen-specific and host immunity biomarkers. However, sample preparation is increasingly recognized as the critical bottleneck in translating biosensors from the laboratory to clinics [78]. Sample preparation involves enrichment of target analyte, removal of matrix inhibitors and sample volume reduction. The strategy of sample preparation depends on the type of biological sample, the sample volume and the target analyte concentration (Table 4). Sample preparation begins with specimen collection: a blood draw to assess serum analytes, a buccal swab to collect somatic cells, a lumbar puncture for cerebrospinal fluid or a collection cup for urine, stool or sputum samples. After collection, samples needed to be loaded on the sensing device for preparation and analysis. Whereas specimen loading can be relatively easy for aqueous samples (i.e., blood, urine, saliva and spinal fluid) [79], additional steps such as digestion and homogenization are necessary for viscous or solid samples (i.e., stool and sputum) [64,80]. On-chip sample preparation becomes essential for direct analysis of raw biological samples on detection platforms. Unique features of microfluidics such as small features size (from nanometers to hundreds of micrometers), the laminar nature of fluid flow, fast thermal relaxation, length scale matching with the electric double layer, low fluid volume handling, short assay time and low power consumption make these techniques ideal for point-of-care sample preparation [10]. A number of microfluidics-based sample preparation platforms based on three major sample preparation steps, separation, concentration and lysis, are reviewed here.
Table 4.
Biological samples and sample preparation considerations.
| Biological matrix |
Sample volume (ml) |
Advantages | Challenges | |
|---|---|---|---|---|
| Blood | 0.1–5 | Well established; baseline concentrations of cellular and extracellular constituents remain largely constant; rapid changes of analyte concentrations during diseased states | Complex matrix; need for separation into different blood components; high background; high viscosity; wide dynamic range of analytes | [47–49,60,63,83–86,98] |
| Urine | 1–100 | Ease of access; non-invasive; abundant; less complex matrix than blood | Wide ranging pH; high conductivity; need to concentrate analytes; biomarkers not well characterized | [54,65,70,87,94,111,152] |
| Saliva | 1–5 | Ease of access; non-invasive | Complex matrix; requires stimulation to obtain samples; variability in getting sufficient sample volume; biomarkers not well characterized | [79,100,153] |
| Sputum | 1–5 | Ease of access; less invasive; localizes to tissue of interest | Very high viscosity; complex matrix; limited sample volume for analysis | [80,154,155] |
| Stool | >1 g | Ease of access; non-invasive | Very challenging matrix; need dilution and separation; high background | [64,156,157] |
| Tissue swab (e.g., buccal, nasal, wound, vagina) | <1 | Ease of access; less invasive; localizes to tissue of interest | Low and variable sample volume; complex matrix; need to dissociate analyte from the swab; low analytes concentration | [102,144,158] |
Separation
Many diagnostic assays are dependent on an initial separation step. Separation is particularly common with blood samples that are commonly fractionated into plasma, white blood cell-rich buffy coat and red blood cells. The conventional means of blood separation in clinical laboratories are centrifugation and filtration. Centrifugation is highly efficient but requires a dedicated instrumentation that is challenging to integrate with other steps of sample preparation. While filtration is a cost-effective alternative to centrifugation, common problems include membrane clogging and hemolysis under high pressure. Microfluidic-based alternatives of separation are under active investigation to facilitate integration with advanced biosensors.
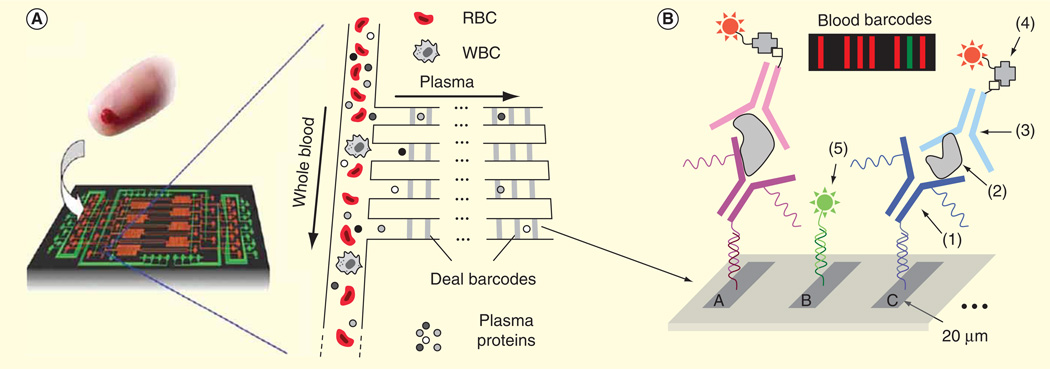
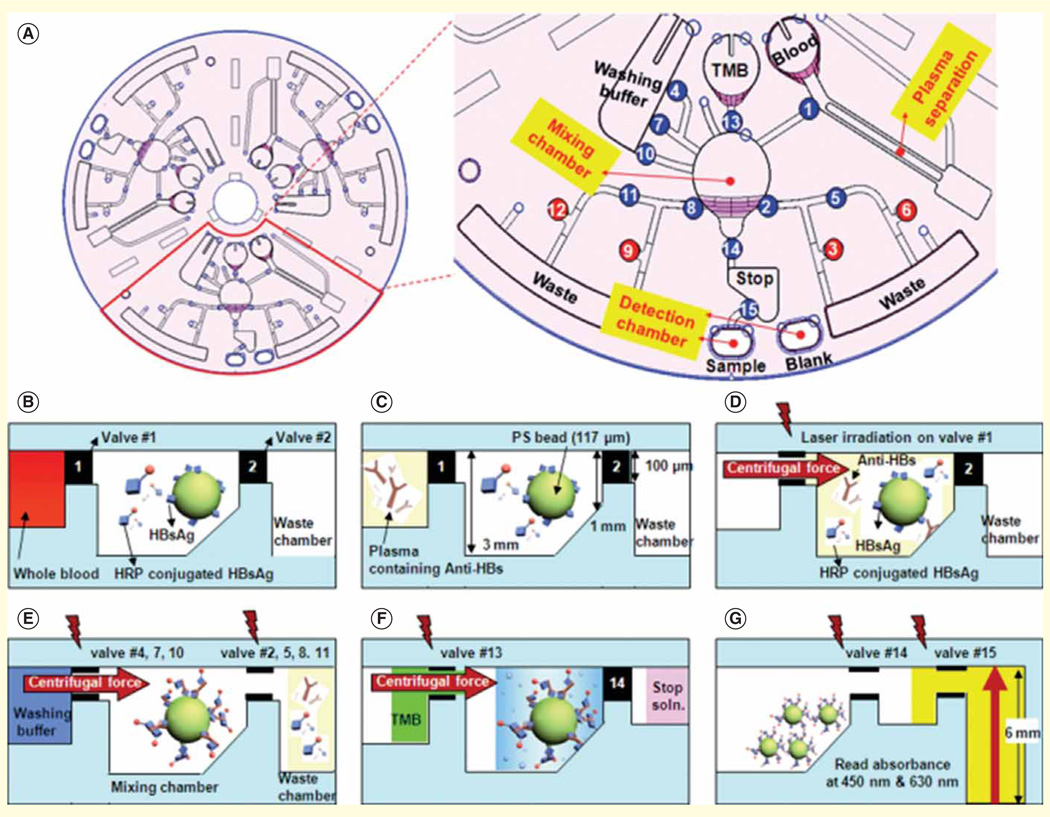
One microfluidic technique for rapid separation of plasma from a finger prick of whole blood is based on the Zweifach–Fung bifurcation effect [81]. This effect relies on the behavior of blood cells at a branch point in a microfluidic channel, where the blood cells will travel into a channel with a higher flow rate and plasma will end up in the lower flow rate channel. One system integrated a Zweifach–Fung-based microfluidic module with a DNA-encoded antibody arrays for rapid onchip blood separation and measurement of a panel of plasma proteins using fluorescence detection [82]. The 10-min assay time from sample collection to detection allows robust detection of proteins that otherwise would rapidly degrade in blood samples (Figure 3). Another low-cost plasma separation approach utilizes red blood cell agglutination in a paper-based microfluidic format [83]. Wax hydrophobic barriers printed on paper were used to filter out agglutinated red blood cells, while the plasma was wicked through the paper substrate onto the test readout zones where a colorimetric assay was used to detect analytes of interest. Lab-on-a-disc is another promising microfluidic technology, which takes advantage of centrifugal forces for various fluid manipulations including plasma separation [84]. With the lab-on-a-disc format, a fully automated immunoassay from whole blood has been demonstrated for detection of hepatitis B virus antigen and antibodies (Figure 4).
Figure 3. Integrated blood barcode chip for multiplexed detection of protein.
(A) Schematic of plasma separation based on Zweifach-Fung effect from a finger prick of blood. Plasma separation channels are integrated with multiple DNA-encoded antibody barcode arrays for protein detection. (B) A, B, C represent different DNA codes. (1) is the DNA-antibody conjugate, (2) is plasma protein, (3) is biotin-labeled detection antibody, (4) is streptavidin-Cy5 fluorescence probe and (5) is complementary DNA-Cy3 reference probe. The inset is a barcode of protein biomarkers with the signal measured by fluorescence detection. The green bar denotes as an alignment marker.
RBC: Red blood cells; WBC: White blood cells.
Adapted with permission from [82] © Macmillan Publishers Ltd. (2008).
Figure 4. Integrated lab-on-a-disc platform for detection of hepatitis B virus.
(A) Schematic of the disc showing the microfluidic layout and function of different compartments. The number indicates the order of operation. (B–G) Illustration of the reactions on the disc.
Adapted with permission from [84] © The Royal Society of Chemistry (2009).
Although various microfluidic-based approaches can effectively separate plasma from whole blood, many of these techniques are limited to small volume (up to about 100 ml) due to the physical restriction of the flow rates. However, clinical samples for bioanalysis are often on the milliliter scale, making large-scale fluid manipulation important especially for low concentration target analytes. Microfluidic chips that can be adapted for higher flow rates and higher volumes for plasma separation are being investigated. One disc-based device is capable of processing 2 ml of whole blood yielding high purity plasma in less than half the time of commercial plasma preparation tubes [85]. Another approach used the temperature effects to generate high flow rates (between 50 and 200 µl/min) with 1 ml of blood sample [86].
Separation is a key step not just for processing whole blood, the sample matrix can impact biosensing from most clinical samples. The matrix effect occurs when the interfering compounds of a sample, such as, abundant proteins, cells, immunoglobulins or debris, alter the final readout of the biosensor by either increasing the background or reducing the signal, ultimately lowering the sensitivity of the assay [87]. Sample dilution with a buffer solution can be sufficient to reduce matrix effects for detection of abundant target analytes. However, in most cases, more complicated sample preparation procedures including concentration and separation are required. Thus, separating and concentrating the targets from a larger volume of samples is a critical sample preparation strategy that can both improve the detection limit by increasing the signal (target concentration) and reducing the noise (matrix effect).
Concentration
With real clinical samples in the milliliter scale and relatively low concentrations of target analyte, both separation and concentration are frequently needed for biosensor detection. Beadbased analyte capture integrated with microfluidic systems have been demonstrated for efficient sample concentration, due to fast diffusion and high surface-to-volume ratio of beads in solution, which provides more binding sites for target analytes or pathogen [88,89]. However, bead manipulation is usually limited by the applied flow condition. To add another degree of freedom for particle manipulation, magnetic beads have been used. Magnetic bead capture relies on mixing and capturing of the targets with the capture agent functionalized beads, followed by application of a magnetic field to capture and wash the bead-target hybrids. Lien et al. used an immunomagnetic bead (IMB)-based system for rapid detection of influenza A virus [90]. Monoclonal antibody-conjugated IMBs were used to target different strains of influenza A virus such as A/H1N1 and A/H3N2 in serum specimens. The limit of detection is about 5 × 10−4 hemagglutin units (HAU), which is three orders of magnitude better than bench top systems using flow cytometry. More importantly, this automated microfluidic assay could be completed within 15 min, which is about 1/10 the time required for the comparable manual assay. Similarly, Soelberg et al. have employed IMBs for surface plasmon detection of staphylococcal enterotoxin B (SEB) from patient stool samples [64]. With this approach, 100 pg/ml SEB in stool samples was easily detected, which is an order of magnitude more sensitive than other commercial assay kits.
Alternative microfluidic devices utilize electrokinetics for sample manipulation. Electrokinetics involve the study of the movement and behavior of particles in suspension when they are under the influence of electric fields. Among different electrokinetics techniques, DEP is one of the most promising approaches for separating and concentrating bacteria and cells as it is a short-range particle force that can directly act on a particle [91]. When the particle is subjected to an electric field, a dipole is induced in a polarizable particle. If the electric field is non-uniform, the particles will experience a net force toward (positive DEP force) or away (negative DEP force) from the electrode surface depending on the conductivity and permittivity of the particles, the surrounding medium and the applied electrical frequency. The magnitude of the DEP force is also proportional to the particle volume, thus allowing efficient separation of different size particles or cells. However, the main challenge of positive DEP trapping is that it is not effective in biological fluids that have high conductivity (≥1 S/m) [92]. To overcome this limitation, Park et al. combined a negative DEP-based separation channel with positive DEP traps that can continuously separate and trap E. coli from either human cerebrospinal fluid or whole blood samples [93]. The proposed platform can take 1 ml volumes of crude biological sample and concentrate target cells into a submicroliter volume with approximately 104-fold of concentration. In another effort, Gao et al. have designed a hybrid electrokinetics device [91] combining short range electrophoresis and DEP particle force, with long range AC electrothermal fluid flow for continuous label-free isolation of bacteria, such as E. coli, Acinetobacter baumannii and Bacillus globigii from biological samples, such as urine and buffy coat with a concentration efficiency of over three orders of magnitude [94].
In addition to bead-based and electrokinetic assays, other simple, low-cost microfluidics target concentration platforms are being developed. Zhang et al. have reported a disposable polymer microfluidic device employing evaporation-induced dragging effect to perform rapid concentration of fluorescently tagged E. coli [95]. The recovery concentration was above 85% or initial bacterial concentrations lower than 1 × 104 cfu/ml. At the lowest initial concentration, 100 cfu/ml, 100 µl of bacteria in solution was concentrated into 500 nl droplets with greater than 90% efficiency in 15 min. However, evaporation-induced concentration will concentrate all components of the sample and may exacerbate the matrix effect. Therefore, this approach may only be feasible for clinical samples in conjunction pre-cleaning steps to remove interfering matrix components of the sample. Another microfluidic approach imitates the functions of centrifuge yet operates without moving parts or external forces [96]. The ‘centrifuge-on-a-chip’ employs fluid vortices to passively trap cells using purely hydrodynamic forces. This approach has been used for high-throughput selective enrichment of cells from 10 ml blood samples into smaller µl volumes at the high flow rate (ml/min scale), followed by an automated fluorescent labeling detection assay on the trapped cells.
Lysis
For many assays, cell lysis to release intracellular components including nucleic acids, proteins and organelle is an essential step in sample preparation. Mechanical, electrical, chemical and thermal lysis methods have been demonstrated in microfluidic platforms [97]. Mechanical lysis involves the generation of shear force through the application of high pressure, rapid agitation or sonication to crush cells. One system for cell lysis and DNA analysis uses phononic lattices to generate surface acoustic wave-induced rotational vortices to mechanically lyse red blood cells and malarial parasitic cells present in a drop of blood [98]. Subsequent real-time PCR analysis also used surface acoustic wave as the heating element and showed that the integrity of the genomic DNA was maintained for efficient analysis. Many systems use friction and collisions between cells and beads in solution for mechanical lysis. One such platform uses a magnetically actuated bead-beating system on a compact disc (CD)-based centrifugal microfluidic platform [99]. This system includes a stationary stand with permanent magnets beneath the CD and magnetic lysis disks inside the CD. As the CD spins over the stationary magnets, the magnetic lysis disks oscillate inside the chambers, resulting in mechanical impact and sample shearing. Biological validation of this platform was tested using Bacillus subtilis spores and clinical nasopharyngeal aspirates for respiratory virus detection. Although mechanical lysis can be adapted to different cell types, it often requires cooling to remove heat produced by the dissipation of the mechanical energy.
Thermal lysis makes use of high temperature to denature cell membrane proteins and damage the cell to promote release of cytoplasmic contents. A short pulse of approximately 100°C is sufficient to break the cell membrane without damaging nucleic acid, yet prolonged heat treatment may cause irreversible denaturation of DNA. This lysis approach is most commonly integrated in microfluidic systems with PCR-based genetic assays as a single embedded resistive heater can provide heat for thermal lysis and PCR [100,101]. This approach has been verified for lysis and detection of influenza viral particles (infA/H1, infA/H3 and infB) in nasopharyngeal swab samples [102]. In this assay, amplification and detection were done sequentially by one-step RT-PCR and an optical detection module with a limit of detection of 100 copies for all influenza viruses. Although thermal lysis requires no chemical reagents, consistent high heat will lead to the denaturation of proteins and may interfere with subsequent assays. Also, large sample volumes require much more energy to heat. Thermal lysis may be improved by increasing the pressure inside the chamber to speed up lysis and use lower temperatures.
Chemical lysis makes use of buffers or other lytic agents to break down cell walls and membranes [103]. The chemical agent used for lysis depends on the target cell type and target molecule, the type of clinical samples and the detection mechanisms. For example, ammonium chloride is effective at lysing erythrocytes but no other mammalian cell types, which can be useful in clinical samples such as urine and blood when specific lysis of erythrocytes is desired. In the electrochemical detection of 16S rRNA of uropathogens, lysozyme followed by sodium hydroxide has been shown to be efficient for lysis of both Gram-positive and Gram-negative bacteria from clinical urine samples [54]. While chemical lysis is simple and effective for wide range of samples, it requires wet chemical storage and mixing, which adds complexity in a microfluidic setting.
Exposure of cells to high-intensity pulsed electric fields can lyse cells due to the dielectric breakdown of the cell membrane. The electric field strength required to reach the threshold to initiate cell lysis depends on cell shape and size, as well as membrane composition. Lam et al. demonstrated bacterial lysis with nanostructured microelectrodes using 100 V, 10 ms DC pulses at a frequency of 1 Hz for 20 s [104]. This lysis approach has been validated with different bacteria, such as, E. coli, Staphylococcus saprophyticus, S. aureus and MRSA using RT-PCR to measure lysis. While electrical lysis is reagentless and quick, the high voltage in high conductivity physiological buffers can lead to chemical electrolysis, undesirable localized heating and denaturation of proteins.
The four main lysis methods described above have their advantages and challenges in terms of time, adaptability to different cell types, heat generation to samples and interference with the subsequent assays. In order to develop an efficient lysis approach, combinations of the aforementioned methods have been designed. For instance, a hybrid chemical and mechanical lysis approach involves directing the bacterial cell through a porous polymer monolith assisted with detergent lytic conditions [105]. With this method, both Gram-negative and Grampositive bacteria were successfully lysed in human blood samples.
System integration
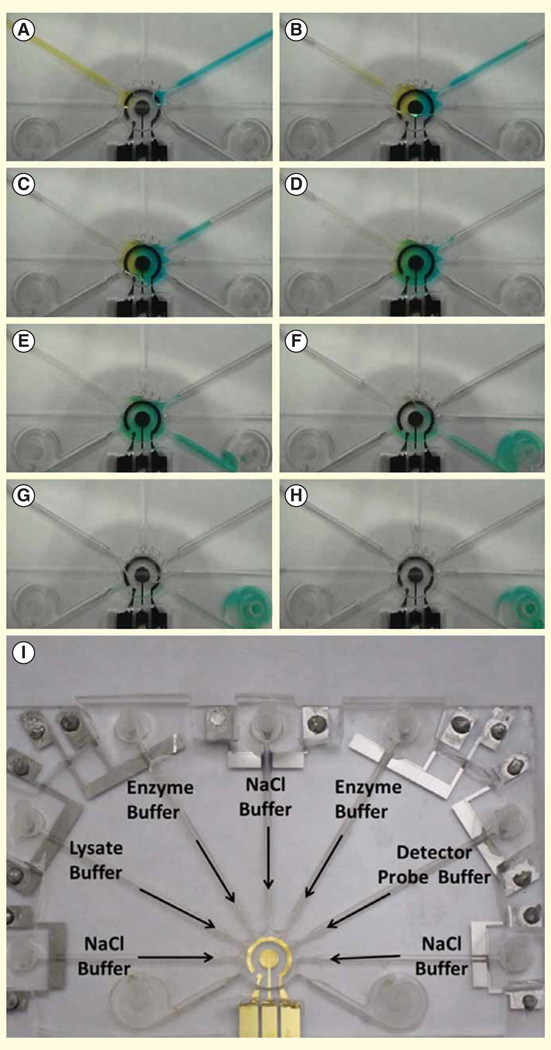
The ideal standalone platform would allow the user to simply add sample, click start then view the results. However, fully integrated systems that bring together the components of sample preparation and analyte detection remain a critical challenge for technology transfer from laboratories to the clinical market [56,106]. Recent system-oriented microfluidic strategies that facilitate system integration include multilayer soft lithography, multiphase microfluidics, electrowetting-on-dielectric, electrokinetics and centrifugal microfluidics [107]. Microfluidic sample preparation steps such as concentration, mixing, pumping and separation can be achieved with these strategies, which make system integration a straightforward task with consistent and monolithic fabrication technologies. Another crucial element for system integration is the detection module. For example, many of the optical detection strategies require a bulky microscope and laser source, which are not practical in clinical settings. Recently, research has focused on portable detection system based on optical, electrical and magnetic sensing. For optical detection, lens-free digital microscopy has been implemented with miniaturized and cost-effective optical components mechanically attached to a camera unit of a cell phone [108]. Images of micro-sized objects, for example, red blood cell and white blood cells can be observed with these portable systems and they will be useful in delivering health information through telecommunication, especially in remote settings. For electrical detection, electrochemistry is a promising candidate for lab-on-a-chip [109]. Not only can the electrical signal be processed by conventional electronics, the miniaturization and integration of the electrochemical transducer into a microfluidic platform is viable. Moreover, as the electrochemical sensor share a similar electrode interface with other electric field-driven microfluidic platforms, such as, electrowetting-on-dielectric [110] and electrokinetics [111], integrating sample preparation components with the detection module is simplified. A multifunctional electrode approach has been demonstrated recently showing the implementation of electrokinetic-induced mixing directly on an electrochemical sensor, resulting in signal enhancement for detecting urinary tract infections (Figure 5) [112]. With the lab-on-a-disc format, a fully automated immunoassay from whole blood has been demonstrated for detection of hepatitis B virus antigen and antibodies (Figure 4) [84]. Compared with over 2 h for conventional ELISA, the lab-on-a-disc assay was complete in less than 30 min with a similar limit of detection. Indeed, as there is no ‘one size fits all’, system integration solutions must be tailored for the intended application with design inputs from all stakeholders.
Figure 5. Demonstration of fluid manipulation with food color dyes in an integrated electrode platform for detection of bacterial 16S rRNA.
(A & B) Electrolytic pumping of two color food dyes into the mixing and sensing chamber in the center. (C & D) Electrokinetic mixing of the color food dyes on top of the electrochemical sensing electrode. (E–H) Electrolytic pumping of washing buffer into the sensing chamber and delivered to the waste reservoirs. (I) Photograph of the universal electrode array for implementing the electrochemical assay for bacterial 16S rRNA.
Reprinted with permission from [112] © IEEE (2013).
Expert commentary
Infectious diseases are ideal applications for the emerging biosensor technologies. For many infectious diseases, rapid diagnosis and timely initiation of effective treatment can be critical for patient outcome and public health. When integrated with advanced microfluidic systems, biosensor can form the foundation of rapid point-of-care devices with the potential to positively impact patient care. As the rate of emergence of MDR pathogens and new infectious diseases continues to increase, an ideal diagnostic system will include pathogen identification, AST and host immune response.
While significant improvements in sensitivity and specificity have been achieved in recent years, the commercialization of biosensors for infectious diseases is still in its infancy [56]. For assay development, both label-free and labeled assays have their advantages and limitations. Label-free assays allow ease of sample preparation and quantitative real-time measurement, but suffer from matrix effects and potential for non-specific bindings. While the multistep protocols for labeled assays make them modestly more complicated, the incorporation of multiple binding events increases specificity and amplification tags improve sensitivity. Taken together, labeled assays appear to have a greater potential for clinical translation, particularly in dealing with real-world samples.
Translating sample preparation techniques imposes a challenging bottleneck for point-of-care device development. The matrix effect of clinical samples presents an important problem for many biosensor devices and needs to be addressed with each device/matrix/analyte scenario. Most biosensors demonstrate excellent performance with the pristine samples such as pure bacterial/viral cultures or purified biomolecules isolated from clinical samples. Promising performance is also commonly observed in spiked samples. Not uncommonly, the matrix becomes more complex in the setting of active infection. For example, healthy urine contains few cells, yet upon infection, the urine can go from a clear salt solution to a cloudy mixture of bacteria, white blood cells, red blood cells and epithelial cells. This change can lead to clogging of microchannels and reduction of the signal. Therefore, even the most promising sensors need to be critically evaluated with the clinical samples spanning the anticipated range of the analyte. A number of recent studies have demonstrated chip-compatible sample preparation strategies that can start with original clinical samples in microfluidic systems [78], yet the complexity in reducing milliliter sample volume down to microliter volume has not been fully addressed.
Finally, system integration remains the most critical challenge for the technology transfer from laboratories into the clinics. Although successful detection mechanisms, microfluidics-based sample preparation strategies and detector modules have been demonstrated separately, hurdles remain in the integration of these modules into a fully automated, standalone platform that is easily operated by a non-technical end user. If these issues can be adequately addressed, it will significantly increase the likelihood of translating research grade biosensors from the research laboratories into the fields and clinics.
Five-year view
Although the potential of microfluidics technology to benefit point-of-care diagnostics has been demonstrated for decades, it is unlikely that integrated lab-on-a-chip system that can directly deal with raw samples will be on market in the next 5 years. The driving force for commercialization fully relies on the cost–effectiveness delivered by the technology, which involves not only the cost, but also the real clinical benefits of the test measured based on disability-adjusted life-years [113]. From the technological point of view, one way to maximize the benefits delivered is to develop a universal integrated system that can efficiently handle a wider range of clinical samples such as urine, blood, saliva for different infectious viruses or bacteria. Another challenge for moving toward the practical goal is the gap between the innovative concepts at the academic level and the clinical validation, which is mainly due to the inaccessibility of the raw samples for most of the researchers in the fields of microfluidics and their limited experiences on the marketable devices. Moving forward, more comprehensive collaboration among academies, healthcare units and industries is the key for the realization of the real lab-on-a-chip devices.
Key issues.
Current in vitro diagnostics require centralized laboratory, experienced personnel and large expensive equipment.
Timely diagnosis and initiation of targeted antimicrobial treatment with portable, sensitive, specific and cost-effective biosensor technologies are keys to clinical management in decentralized and resource-poor settings.
Label-free assays may allow quantitative real-time measurement, yet suffer from a significant degree of non-specific bindings and aberrant signals with analytes in complex matrices.
In labeled assays, incorporation of multiple binding events and amplification tags increase specificity and sensitivity, yet multistep protocols increase the assay complexity.
Matrix effects of clinical samples present a common problem for biosensor technologies and biosensors demonstrating promising performance with original clinical samples are rare.
Developing chip-based sample preparation strategies in microfluidic systems for enrichment of target analytes, removal of matrix inhibitors and sample volume reduction is essential for translating biosensors from laboratory to clinic.
System integration of three major modules of a biosensor, which are detection mechanism, microfluidics-based sample preparation strategies and a transducer, into a fully automated and standalone platform remains the most critical challenge for point-of-care device commercialization.
Acknowledgments
JC Liao and PK Wong were supported by NIH/NIAID grant U01 AI082457.
This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending or royalties.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Franca RF, Da Silva CC, De Paula SO. Recent advances in molecular medicine techniques for the diagnosis, prevention, and control of infectious diseases. Eur J Clin Microbiol Infect Dis. 2013;32(6):723–728. doi: 10.1007/s10096-013-1813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fauci AS, Morens DM. 200 NEJM ANNIVERSARY ARTICLE the perpetual challenge of infectious diseases. N Engl J Med. 2012;366(5):454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- 3. Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–144. doi: 10.1146/annurev.bioeng.10.061807.160524. • Overview of point-of-care diagnostic technologies for global health issues
- 4.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nat Rev Microbiol. 2004;2(3):231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 5.Luong JH, Male KB, Glennon JD. Biosensor technology: technology push versus market pull. Biotechnol Adv. 2008;26(5):492–500. doi: 10.1016/j.biotechadv.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Ngom B, Guo Y, Wang X, Bi D. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: a review. Anal Bioanal Chem. 2010;397(3):1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- 7.Foudeh AM, Fatanat Didar T, Veres T, Tabrizian M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip. 2012;12(18):3249–3266. doi: 10.1039/c2lc40630f. [DOI] [PubMed] [Google Scholar]
- 8.Pejcic B, De Marco R, Parkinson G. The role of biosensors in the detection of emerging infectious diseases. Analyst. 2006;131(10):1079–1090. doi: 10.1039/b603402k. [DOI] [PubMed] [Google Scholar]
- 9.D’orazio P. Biosensors in clinical chemistry - 2011 update. Clin Chim Acta. 2011;412(19–20):1749–1761. doi: 10.1016/j.cca.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 11.Fournier PE, Raoult D. Prospects for the Future Using Genomics and Proteomics in Clinical Microbiology. Annu Rev Microbiol. 2011;65:169–188. doi: 10.1146/annurev-micro-090110-102922. [DOI] [PubMed] [Google Scholar]
- 12.Hodges EN, Connor JH. Translational control by negative-strand RNA viruses: methods for the study of a crucial virus/host interaction. Methods. 2013;59(2):180–187. doi: 10.1016/j.ymeth.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mairiang D, Zhang HM, Sodja A, et al. Identification of new protein interactions between dengue fever virus and its hosts, human and mosquito. PLoS One. 2013;8(1):e53535. doi: 10.1371/journal.pone.0053535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohan R, Mach KE, Bercovici M, et al. Clinical validation of integrated nucleic acid and protein detection on an electrochemical biosensor array for urinary tract infection diagnosis. PLoS One. 2011;6(10):e26846. doi: 10.1371/journal.pone.0026846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt HK, Armani AM. Label-free biological and chemical sensors. Nanoscale. 2010;2(9):1544–1559. doi: 10.1039/c0nr00201a. [DOI] [PubMed] [Google Scholar]
- 16.Rapp BE, Gruhl FJ, Lange K. Biosensors with label-free detection designed for diagnostic applications. Anal Bioanal Chem. 2010;398(6):2403–2412. doi: 10.1007/s00216-010-3906-2. [DOI] [PubMed] [Google Scholar]
- 17.Ju H, Zhang X, Wang J. NanoBiosensing. Springer, NY, USA: 2011. Signal amplification for nanobiosensing; pp. 39–84. [Google Scholar]
- 18.Scanvic A, Courdavault L, Sollet JP, Le Turdu F. Interest of real-time PCR Xpert MRSA/SA on GeneXpert (R) DX System in the investigation of staphylococcal bacteremia. Pathol Biol. 2011;59(2):67–72. doi: 10.1016/j.patbio.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 19.Ioannidis P, Papaventsis D, Karabela S, et al. Cepheid GeneXpert MTB/RIF Assay for Mycobacterium tuberculosis Detection and Rifampin Resistance Identification in Patients with Substantial Clinical Indications of Tuberculosis and Smear-Negative Microscopy Results. J Clin Microbiol. 2011;49(8):3068–3070. doi: 10.1128/JCM.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Zhang Y, Lin S, et al. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv. 2011;29(6):830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad F, Hashsham SA. Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: a review. Anal Chim Acta. 2012;733:1–15. doi: 10.1016/j.aca.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 22.Citartan M, Gopinath SC, Tominaga J, Tang TH. Label-free methods of reporting biomolecular interactions by optical biosensors. Analyst. 2013;138(13):3576–3592. doi: 10.1039/c3an36828a. [DOI] [PubMed] [Google Scholar]
- 23.Guo X. Surface plasmon resonance based biosensor technique: a review. J Biophoton. 2012;5(7):483–501. doi: 10.1002/jbio.201200015. [DOI] [PubMed] [Google Scholar]
- 24.Tawil N, Sacher E, Mandeville R, Meunier M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens Bioelectron. 2012;37(1):24–29. doi: 10.1016/j.bios.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 25.Kussrow A, Baksh MM, Bornhop DJ, Finn MG. Universal sensing by transduction of antibody binding with backscattering interferometry. Chembiochem. 2011;12(3):367–370. doi: 10.1002/cbic.201000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kussrow A, Enders CS, Castro AR, et al. The potential of backscattering interferometry as an in vitro clinical diagnostic tool for the serological diagnosis of infectious disease. Analyst. 2010;135(7):1535–1537. doi: 10.1039/c0an00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanik AA, Huang M, Kamohara O, et al. An optofluidic nanoplasmonic biosensor for direct detection of live viruses from biological media. Nano Lett. 2010;10:4962–4969. doi: 10.1021/nl103025u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo X, Davis JJ. Electrical biosensors and the label free detection of protein disease biomarkers. Chem Soc Rev. 2013;42(13):5944–5962. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- 29.Qiu J-D, Huang H, Liang R-P. Biocompatible and label-free amperometric immunosensor for hepatitis B surface antigen using a sensing film composed of poly(allylamine)-branched ferrocene and gold nanoparticles. Microchim Acta. 2011;174(1–2):97–105. [Google Scholar]
- 30.Daniels JS, Pourmand N. Label-free impedance biosensors: opportunities and challenges. Electroanalysis. 2007;19(12):1239–1257. doi: 10.1002/elan.200603855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo X, Kulkarni A, Doepke A, et al. Carbohydrate-based label-free detection of Escherichia coli ORN 178 using electrochemical impedance spectroscopy. Anal Chem. 2012;84(1):241–246. doi: 10.1021/ac202419u. [DOI] [PubMed] [Google Scholar]
- 32.Patel P, Klausner JD, Bacon OM, et al. Detection of acute HIV infections in high-risk patients in California. J Acquir Immune Defic Syndr. 2006;42(1):75–79. doi: 10.1097/01.qai.0000218363.21088.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafiee H, Jahangir M, Inci F, et al. Acute on-chip HIV detection through label-free electrical sensing of viral nano-lysate. Small. 2013;9(15):2553–2563. doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zelada-Guillen GA, Tweed-Kent A, Niemann M, et al. Ultrasensitive and real-time detection of proteins in blood using a potentiometric carbon-nanotube aptasensor. Biosens Bioelectron. 2013;41:366–371. doi: 10.1016/j.bios.2012.08.055. [DOI] [PubMed] [Google Scholar]
- 35.Song S, Wang L, Li J, et al. Aptamer-based biosensors. TrAC Trends Anal Chem. 2008;27(2):108–117. [Google Scholar]
- 36.Villamizar RA, Maroto A, Rius FX. Improved detection of Candida albicans with carbon nanotube field-effect transistors. Sens Actuators B Chem. 2009;136(2):451–457. [Google Scholar]
- 37.Ishikawa FN, Chang HK, Curreli M, et al. Label-Free, Electrical Detection of the SARS Virus N-Protein with Nanowire Biosensors Utilizing Antibody Mimics as Capture Probes. ACS Nano. 2009;3(5):1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamayo J, Kosaka PM, Ruz JJ, et al. Biosensors based on nanomechanical systems. Chem Soc Rev. 2013;42(3):1287–1311. doi: 10.1039/c2cs35293a. [DOI] [PubMed] [Google Scholar]
- 39.Arlett JL, Myers EB, Roukes ML. Comparative advantages of mechanical biosensors. Nat Nanotechnol. 2011;6(4):203–215. doi: 10.1038/nnano.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mader A, Gruber K, Castelli R, et al. Discrimination of Escherichia coli strains using glycan cantilever array sensors. Nano Lett. 2012;12(1):420–423. doi: 10.1021/nl203736u. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, Schweizer LM, Wang W, et al. Label-free and real-time monitoring of yeast cell growth by the bending of polymer microcantilever biosensors. Sens Actuators B Chem. 2013;178:621–626. [Google Scholar]
- 42.Peduru Hewa TM, Tannock GA, Mainwaring DE, et al. The detection of influenza A and B viruses in clinical specimens using a quartz crystal microbalance. J Virol Methods. 2009;162(1–2):14–21. doi: 10.1016/j.jviromet.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu CH, Zhang Y, Tang SF, et al. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens Bioelectron. 2012;31(1):439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 44.Tokonami S, Nakadoi Y, Takahashi M, et al. Label-free and selective bacteria detection using a film with transferred bacterial configuration. Anal Chem. 2013;85(10):4925–4929. doi: 10.1021/ac3034618. [DOI] [PubMed] [Google Scholar]
- 45.Zhang GJ, Ning Y. Silicon nanowire biosensor and its applications in disease diagnostics: a review. Anal Chim Acta. 2012;749:1–15. doi: 10.1016/j.aca.2012.08.035. [DOI] [PubMed] [Google Scholar]
- 46.Statz AR, Meagher RJ, Barron AE, Messersmith PB. New peptidomimetic polymers for antifouling surfaces. J Am Chem Soc. 2005;127(22):7972–7973. doi: 10.1021/ja0522534. [DOI] [PubMed] [Google Scholar]
- 47.Bryan T, Luo X, Bueno PR, Davis JJ. An optimised electrochemical biosensor for the label-free detection of C-reactive protein in blood. Biosens Bioelectron. 2013;39(1):94–98. doi: 10.1016/j.bios.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 48.Brault ND, Gao C, Xue H, et al. Ultra-low fouling and functionalizable zwitterionic coatings grafted onto SiO2 via a biomimetic adhesive group for sensing and detection in complex media. Biosens Bioelectron. 2010;25(10):2276–2282. doi: 10.1016/j.bios.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Kirk JT, Brault ND, Baehr-Jones T, et al. Zwitterionic polymer-modified silicon microring resonators for label-free biosensing in undiluted human plasma. Biosens Bioelectron. 2013;42:100–105. doi: 10.1016/j.bios.2012.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peruski AH, Peruski LF. Immunological Methods for Detection and Identification of Infectious Disease and Biological Warfare Agents. Clin Vaccine Immunol. 2003;10(4):506–513. doi: 10.1128/CDLI.10.4.506-513.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Yu Q, Duan Y. Fluorescent labels in biosensors for pathogen detection. Crit Rev Biotechnol. 2013 doi: 10.3109/07388551.2013.804487. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 52.Nam JM, Jang KJ, Groves JT. Detection of proteins using a colorimetric bio-barcode assay. Nat protocols. 2007;2(6):1438–1444. doi: 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]
- 53.Sapsford KE, Pons T, Medintz IL, Mattoussi H. Biosensing with luminescent semiconductor quantum dots. Sensors. 2006;6(8):925–953. [Google Scholar]
- 54.Mach KE, Du CB, Phull H, et al. Rapid molecular diagnosis of urinary tract infections. J Urol. 2009;181(4):138–139. [Google Scholar]
- 55.Haun JB, Yoon TJ, Lee H, Weissleder R. Magnetic nanoparticle biosensors. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2010;2(3):291–304. doi: 10.1002/wnan.84. [DOI] [PubMed] [Google Scholar]
- 56. Chin CD, Linder V, Sia SK. Commercialization of microfluidic point-of-care diagnostic devices. Lab Chip. 2012;12(12):2118–2134. doi: 10.1039/c2lc21204h. ••Discussion of challenges on commercialization of microfluidic point-of-care devices
- 57.Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal Chem. 2010;82(1):3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- 58.Wang EC, Wang AZ. Targeted Nanoparticles and their Applications in Biology. Integr Biol. 2014;6:9–26. doi: 10.1039/c3ib40165k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301(5641):1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 60.Tang SX, Zhao JQ, Storhoff JJ, et al. Nanoparticle-based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr. 2007;46(2):231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 61.Cao X, Ye Y, Liu S. Gold nanoparticle-based signal amplification for biosensing. Anal Biochem. 2011;417(1):1–16. doi: 10.1016/j.ab.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 62.Lin M, Pei H, Yang F, et al. Applications of Gold Nanoparticles in the Detection and Identification of Infectious Diseases and Biothreats. Adv Materials. 2013;25(25):3490–3496. doi: 10.1002/adma.201301333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chin CD, Laksanasopin T, Cheung YK, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17(8):1015–1019. doi: 10.1038/nm.2408. •An integrated microfluidic device for multiplex detection of infectious diseases directly from whole blood
- 64.Soelberg SD, Stevens RC, Limaye AP, Furlong CE. Surface Plasmon Resonance Detection Using Antibody-Linked Magnetic Nanoparticles for Analyte Capture, Purification, Concentration, and Signal Amplification. Anal Chem. 2009;81(6):2357–2363. doi: 10.1021/ac900007c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung HJ, Castro CM, Im H, et al. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat Nanotechnol. 2013;8(5):369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14(8):869–874. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore DA, Shah NS. Alternative methods of diagnosing drug resistance–what can they do for me? J Infect Dis. 2011;204(Suppl 4):S1110–S1119. doi: 10.1093/infdis/jir448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pulido MR, Garcia-Quintanilla M, Martin-Pena R, et al. Progress on the development of rapid methods for antimicrobial susceptibility testing. J Antimicrob Chemother. 2013;68(12):2710–2717. doi: 10.1093/jac/dkt253. [DOI] [PubMed] [Google Scholar]
- 69.Bergeron MG, Ouellette M. Preventing antibiotic resistance through rapid genotypic identification of bacteria and of their antibiotic resistance genes in the clinical microbiology laboratory. J Clin Microbiol. 1998;36(8):2169–2172. doi: 10.1128/jcm.36.8.2169-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mach KE, Mohan R, Baron EJ, et al. A biosensor platform for rapid antimicrobial susceptibility testing directly from clinical samples. J Urol. 2011;185(1):148–153. doi: 10.1016/j.juro.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Halford C, Gonzalez R, Campuzano S, et al. Rapid antimicrobial susceptibility testing by sensitive detection of precursor rRNA using a novel electrochemical biosensing platform. Antimicrob Agents Chemother. 2013;57(2):936–943. doi: 10.1128/AAC.00615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Y, Gao J, Zhang DD, et al. Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal Chem. 2013;85(8):3971–3976. doi: 10.1021/ac4004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalashnikov M, Lee JC, Campbell J, et al. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip. 2012;12(21):4523–4532. doi: 10.1039/c2lc40531h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen CH, Lu Y, Sin ML, et al. Antimicrobial susceptibility testing using high surface-to-volume ratio microchannels. Anal Chem. 2010;82(3):1012–1019. doi: 10.1021/ac9022764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim KP, Kim YG, Choi CH, et al. In situ monitoring of antibiotic susceptibility of bacterial biofilms in a microfluidic device. Lab Chip. 2010;10(23):3296–3299. doi: 10.1039/c0lc00154f. [DOI] [PubMed] [Google Scholar]
- 76.Tang Y, Zhen L, Liu J, Wu J. Rapid antibiotic susceptibility testing in a microfluidic pH sensor. Anal Chem. 2013;85(5):2787–2794. doi: 10.1021/ac303282j. [DOI] [PubMed] [Google Scholar]
- 77.Tawil N, Mouawad F, Levesque S, et al. The differential detection of methicillin-resistant, methicillin-susceptible and borderline oxacillin-resistant Staphylococcus aureus by surface plasmon resonance. Biosens Bioelectron. 2013;49:334–340. doi: 10.1016/j.bios.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 78. Ritzi-Lehnert M. Development of chip-compatible sample preparation for diagnosis of infectious diseases. Expert Rev Mol Diagn. 2012;12(2):189–206. doi: 10.1586/erm.11.98. ••Review of most recent development in on-chip sample preparation for microfluidic-based diagnosis of infectious diseases.
- 79.Fu E, Chinowsky T, Nelson K, et al. SPR imaging-based salivary diagnostics system for the detection of small molecule analytes. Ann NY Acad Sci. 2007;1098:335–344. doi: 10.1196/annals.1384.026. [DOI] [PubMed] [Google Scholar]
- 80.Cattamanchi A, Davis JL, Pai M, et al. Does bleach processing increase the accuracy of sputum smear microscopy for diagnosing pulmonary tuberculosis? J Clin Microbiol. 2010;48(7):2433–2439. doi: 10.1128/JCM.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang S, Undar A, Zahn JD. A microfluidic device for continuous, real time blood plasma separation. Lab Chip. 2006;6(7):871–880. doi: 10.1039/b516401j. [DOI] [PubMed] [Google Scholar]
- 82.Fan R, Vermesh O, Srivastava A, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26(12):1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang X, Forouzan O, Brown TP, Shevkoplyas SS. Integrated separation of blood plasma from whole blood for microfluidic paper-based analytical devices. Lab Chip. 2012;12(2):274–280. doi: 10.1039/c1lc20803a. [DOI] [PubMed] [Google Scholar]
- 84. Lee BS, Lee JN, Park JM, et al. A fully automated immunoassay from whole blood on a disc. Lab Chip. 2009;9(11):1548–1555. doi: 10.1039/b820321k. •A fully automated system integrated with sample preparation modules for diagnosis of hepatitis B infection from whole blood.
- 85.Amasia M, Madou M. Large-volume centrifugal microfluidic device for blood plasma separation. Bioanalysis. 2010;2(10):1701–1710. doi: 10.4155/bio.10.140. [DOI] [PubMed] [Google Scholar]
- 86.Rodriguez-Villarreal AI, Arundell M, Carmona M, Samitier J. High flow rate microfluidic device for blood plasma separation using a range of temperatures. Lab Chip. 2010;10(2):211–219. doi: 10.1039/b904531g. [DOI] [PubMed] [Google Scholar]
- 87.Chiu ML, Lawi W, Snyder ST, et al. Matrix effects - a challenge toward automation of molecular analysis. JALA. 2010;15(3):233–242. [Google Scholar]
- 88.Mulvaney SP, Cole CL, Kniller MD, et al. Rapid, femtomolar bioassays in complex matrices combining microfluidics and magnetoelectronics. Biosens Bioelectron. 2007;23(2):191–200. doi: 10.1016/j.bios.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 89.Dimov IK, Garcia-Cordero JL, O’grady J, et al. Integrated microfluidic tmRNA purification and real-time NASBA device for molecular diagnostics. Lab Chip. 2008;8(12):2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 90.Lien KY, Hung LY, Huang TB, et al. Rapid detection of influenza A virus infection utilizing an immunomagnetic bead-based microfluidic system. Biosens Bioelectron. 2011 doi: 10.1016/j.bios.2011.03.006. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao J, Sin ML, Liu T, et al. Hybrid electrokinetic manipulation in high-conductivity media. Lab Chip. 2011;11(10):1770–1775. doi: 10.1039/c1lc20054b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krishnan R, Sullivan BD, Mifflin RL, et al. Alternating current electrokinetic separation and detection of DNA nanoparticles in high-conductance solutions. Electrophoresis. 2008;29(9):1765–1774. doi: 10.1002/elps.200800037. [DOI] [PubMed] [Google Scholar]
- 93.Park S, Zhang Y, Wang TH, Yang S. Continuous dielectrophoretic bacterial separation and concentration from physiological media of high conductivity. Lab Chip. 2011;11(17):2893–2900. doi: 10.1039/c1lc20307j. [DOI] [PubMed] [Google Scholar]
- 94.Gao J, Riahi R, Sin ML, et al. Electrokinetic focusing and separation of mammalian cells in conductive biological fluids. Analyst. 2012;137(22):5215–5221. doi: 10.1039/c2an35707k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang JY, Do J, Premasiri WR, et al. Rapid point-of-care concentration of bacteria in a disposable microfluidic device using meniscus dragging effect. Lab Chip. 2010;10(23):3265–3270. doi: 10.1039/c0lc00051e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mach AJ, Kim JH, Arshi A, et al. Automated cellular sample preparation using a Centrifuge-on-a-Chip. Lab Chip. 2011;11(17):2827–2834. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- 97.Kim J, Johnson M, Hill P, Gale BK. Microfluidic sample preparation: cell lysis and nucleic acid purification. Integr Biol. 2009;1(10):574–586. doi: 10.1039/b905844c. [DOI] [PubMed] [Google Scholar]
- 98.Reboud J, Bourquin Y, Wilson R, et al. Shaping acoustic fields as a toolset for microfluidic manipulations in diagnostic technologies. Proc Natl Acad Sci USA. 2012;109(38):15162–15167. doi: 10.1073/pnas.1206055109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Siegrist J, Gorkin R, Bastien M, et al. Validation of a centrifugal microfluidic sample lysis and homogenization platform for nucleic acid extraction with clinical samples. Lab Chip. 2010;10(3):363–371. doi: 10.1039/b913219h. [DOI] [PubMed] [Google Scholar]
- 100.Oblath EA, Henley WH, Alarie JP, Ramsey JM. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva. Lab Chip. 2013;13(7):1325–1332. doi: 10.1039/c3lc40961a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marshall LA, Wu LL, Babikian S, et al. Integrated printed circuit board device for cell lysis and nucleic acid extraction. Anal Chem. 2012;84(21):9640–9645. doi: 10.1021/ac302622v. [DOI] [PubMed] [Google Scholar]
- 102.Wang C-H, Lien K-Y, Hung L-Y, et al. Integrated microfluidic system for the identification and multiple subtyping of influenza viruses by using a molecular diagnostic approach. Microfluid Nanofluid. 2012;13(1):113–123. [Google Scholar]
- 103.Omiatek DM, Santillo MF, Heien ML, Ewing AG. Hybrid capillary-microfluidic device for the separation, lysis, and electrochemical detection of vesicles. Anal Chem. 2009;81(6):2294–2302. doi: 10.1021/ac802466g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lam B, Fang Z, Sargent EH, Kelley SO. Polymerase chain reaction-free, sample-to-answer bacterial detection in 30 minutes with integrated cell lysis. Anal Chem. 2012;84(1):21–25. doi: 10.1021/ac202599b. [DOI] [PubMed] [Google Scholar]
- 105.Mahalanabis M, Al-Muayad H, Kulinski MD, et al. Cell lysis and DNA extraction of gram-positive and gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip. 2009;9(19):2811–2817. doi: 10.1039/b905065p. [DOI] [PubMed] [Google Scholar]
- 106.Sorger PK. Microfluidics closes in on point-of-care assays. Nat Biotechnol. 2008;26(12):1345–1346. doi: 10.1038/nbt1208-1345. [DOI] [PubMed] [Google Scholar]
- 107. Sin ML, Gao J, Liao JC, Wong PK. System integration - a major step toward lab on a chip. J Biol Eng. 2011;5:6. doi: 10.1186/1754-1611-5-6. •Review of system integration approaches for point-of-care devices
- 108.Gurkan UA, Moon S, Geckil H, et al. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol J. 2011;6(2):138–149. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kimmel DW, Leblanc G, Meschievitz ME, Cliffel DE. Electrochemical sensors and biosensors. Anal Chem. 2012;84(2):685–707. doi: 10.1021/ac202878q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karuwan C, Sukthang K, Wisitsoraat A, et al. Electrochemical detection on electrowetting-on-dielectric digital microfluidic chip. Talanta. 2011;84(5):1384–1389. doi: 10.1016/j.talanta.2011.03.073. [DOI] [PubMed] [Google Scholar]
- 111.Sin ML, Liu T, Pyne JD, et al. In situ electrokinetic enhancement for self-assembled-monolayer-based electrochemical biosensing. Anal Chem. 2012;84(6):2702–2707. doi: 10.1021/ac203245j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sin ML, Gau V, Liao JC, Wong PK. A universal electrode approach for automated electrochemical molecular analyses. J Microelectromech Syst. 2013;22(5):1126–1132. doi: 10.1109/JMEMS.2013.2253545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Laxminarayan R, Mills AJ, Breman JG, et al. Advancement of global health: key messages from the Disease Control Priorities Project. Lancet. 2006;367(9517):1193–1208. doi: 10.1016/S0140-6736(06)68440-7. [DOI] [PubMed] [Google Scholar]
- 114.Clarke JR. Molecular diagnosis of HIV. Expert Rev Mol Diagn. 2002;2(3):233–239. doi: 10.1586/14737159.2.3.233. [DOI] [PubMed] [Google Scholar]
- 115.Bourlet T, Memmi M, Saoudin H, Pozzetto B. Molecular HIV screening. Expert Rev Mol Diagn. 2013;13(7):693–705. doi: 10.1586/14737159.2013.829703. [DOI] [PubMed] [Google Scholar]
- 116.Branson BM. State of the art for diagnosis of HIV infection. Clin Infect Dis. 2007;45:S221–S225. doi: 10.1086/522541. [DOI] [PubMed] [Google Scholar]
- 117.Al-Zamel FA. Detection and diagnosis of Mycobacterium tuberculosis. Expert Rev Anti Infect Ther. 2009;7(9):1099–1108. doi: 10.1586/eri.09.92. [DOI] [PubMed] [Google Scholar]
- 118.Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48(1):229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Van Rie A, Page-Shipp L, Scott L, et al. Xpert (R) MTB/RIF for point-of-care diagnosis of TB in high-HIV burden, resource-limited countries: hype or hope? Expert Rev Mol Diagn. 2010;10(7):937–946. doi: 10.1586/erm.10.67. [DOI] [PubMed] [Google Scholar]
- 120.Hawkes M, Kain KC. Advances in malaria diagnosis. Expert Rev Anti Infect Ther. 2007;5(3):485–495. doi: 10.1586/14787210.5.3.485. [DOI] [PubMed] [Google Scholar]
- 121.Moody A. Rapid Diagnostic Tests for Malaria Parasites. Clin Microbiol Rev. 2002;15(1):66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wongsrichanalai C, Barcus MJ, Muth S, et al. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am J Trop Med Hyg. 2007;77(6):119–127. [PubMed] [Google Scholar]
- 123.Strick L, Wald A. Type-specific testing for herpes simplex virus. Expert Rev Mol Diagn. 2004;4(4):443–453. doi: 10.1586/14737159.4.4.443. [DOI] [PubMed] [Google Scholar]
- 124.Corey L. Laboratory diagnosis of herpes-simplex virus-infections - principles guiding the development of rapid diagnostic-tests. Diagn Micrbiol Infect Dis. 1986;4(3):S111–S119. doi: 10.1016/s0732-8893(86)80049-9. [DOI] [PubMed] [Google Scholar]
- 125.Ryan C, Kinghorn G. Clinical assessment of assays for diagnosis of herpes simplex infection. Expert Rev Mol Diagn. 2006;6(5):767–775. doi: 10.1586/14737159.6.5.767. [DOI] [PubMed] [Google Scholar]
- 126.Bayliss J, Nguyen T, Lesmana CR, et al. Advances in the molecular diagnosis of hepatitis B infection: providing insight into the next generation of disease. Semin Liver Dis. 2013;33(2):113–121. doi: 10.1055/s-0033-1345714. [DOI] [PubMed] [Google Scholar]
- 127.Nainan OV, Xia G, Vaughan G, Margolis HS. Diagnosis of hepatitis a virus infection: a molecular approach. Clin Microbiol Rev. 2006;19(1):63–79. doi: 10.1128/CMR.19.1.63-79.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wiwanitkit V. Dengue fever: diagnosis and treatment. Expert Rev Anti Infect Ther. 2010;8(7):841–845. doi: 10.1586/eri.10.53. [DOI] [PubMed] [Google Scholar]
- 129.Tang KF, Ooi EE. Diagnosis of dengue: an update. Expert Rev Anti Infect Ther. 2012;10(8):895–907. doi: 10.1586/eri.12.76. [DOI] [PubMed] [Google Scholar]
- 130.Hooton TM, Bradley SF, Cardenas DD, et al. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin Infect Dis. 2010;50(5):625–663. doi: 10.1086/650482. [DOI] [PubMed] [Google Scholar]
- 131.Schmiemann G, Kniehl E, Gebhardt K, et al. The diagnosis of urinary tract infection: a systematic review. Deutsch Arztebl Int. 2010;107(21):361–367. doi: 10.3238/arztebl.2010.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Katz JM, Hancock K, Xu XY. Serologic assays for influenza surveillance, diagnosis and vaccine evaluation. Expert Rev Anti Infect Ther. 2011;9(6):669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 133.Dimaio MA, Sahoo MK, Waggoner J, Pinsky BA. Comparison of Xpert Flu rapid nucleic acid testing with rapid antigen testing for the diagnosis of influenza A and B. J Virol Methods. 2012;186(1–2):137–140. doi: 10.1016/j.jviromet.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chauhan N, Narang J, Pundir S, et al. Laboratory diagnosis of swine flu: a review. Artif Cell Nanomed B. 2013;41(3):189–195. doi: 10.3109/10731199.2012.716063. [DOI] [PubMed] [Google Scholar]
- 135.Guerrant RL, Van Gilder T, Steiner TS, et al. Practice guidelines for the management of infectious diarrhea. Clin Infect Dis. 2001;32(3):331–351. doi: 10.1086/318514. [DOI] [PubMed] [Google Scholar]
- 136.O’Ryan M, Lucero Y, O’Ryan-Soriano MA, Ashkenazi S. An update on management of severe acute infectious gastroenteritis in children. Expert Rev Anti Infect Ther. 2010;8(6):671–682. doi: 10.1586/eri.10.40. [DOI] [PubMed] [Google Scholar]
- 137.Feeney SA, Armstrong VJ, Mitchell SJ, et al. Development and clinical validation of multiplex TaqMan(R) assays for rapid diagnosis of viral gastroenteritis. J Med Virol. 2011;83(9):1650–1656. doi: 10.1002/jmv.22162. [DOI] [PubMed] [Google Scholar]
- 138.Bauer M, Reinhart K. Molecular diagnostics of sepsis - where are we today? Int J Med Microbiol. 2010;300(6):411–413. doi: 10.1016/j.ijmm.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 139.Mancini N, Carletti S, Ghidoli N, et al. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin Microbiol Rev. 2010;23(1):235–251. doi: 10.1128/CMR.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kotsaki A, Giamarellos-Bourboulis EJ. Molecular diagnosis of sepsis. Expert Opin Med Diagn. 2012;6(3):209–219. doi: 10.1517/17530059.2012.667799. [DOI] [PubMed] [Google Scholar]
- 141.Graseck AS, Shih SL, Peipert JF. Home versus clinic-based specimen collection for Chlamydia trachomatis and Neisseria gonorrhoeae. Expert Rev Anti Infect Ther. 2011;9(2):183–194. doi: 10.1586/eri.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Munson E, Napierala M, Schell RF. Insights into trichomoniasis as a result of highly sensitive molecular diagnostics screening in a high-prevalence sexually transmitted infection community. Expert Rev Anti Infect Ther. 2013;11(8):845–863. doi: 10.1586/14787210.2013.814429. [DOI] [PubMed] [Google Scholar]
- 143.Stevens DL, Bisno AL, Chambers HF, et al. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin Infect Dis. 2005;41(10):1373–1406. doi: 10.1086/497143. [DOI] [PubMed] [Google Scholar]
- 144.Bonham PA. Swab cultures for diagnosing wound infections: a literature review and clinical guideline. J Wound Ostomy Continence Nurs. 2009;36(4):389–395. doi: 10.1097/WON.0b013e3181aaef7f. [DOI] [PubMed] [Google Scholar]
- 145.Oz Y, Kiraz N. Diagnostic methods for fungal infections in pediatric patients: microbiological, serological and molecular methods. Expert Rev Anti Infect Ther. 2011;9(3):289–298. doi: 10.1586/eri.10.168. [DOI] [PubMed] [Google Scholar]
- 146.Peman J, Zaragoza R. Combined use of nonculture-based lab techniques in the diagnosis and management of critically ill patients with invasive fungal infections. Expert Rev Anti Infect Ther. 2012;10(11):1321–1330. doi: 10.1586/eri.12.128. [DOI] [PubMed] [Google Scholar]
- 147.Liang RP, Fan LX, Huang DM, Qiu JD. A label-free amperometric immunosensor based on redox-active ferrocene-branched chitosan/multiwalled carbon nanotubes conductive composite and gold nanoparticles. Electroanalysis. 2011;23(3):719–727. [Google Scholar]
- 148.Shi WT, Ma ZF. A novel label-free amperometric immunosensor for carcinoembryonic antigen based on redox membrane. Biosens Bioelectron. 2011;26(6):3068–3071. doi: 10.1016/j.bios.2010.11.048. [DOI] [PubMed] [Google Scholar]
- 149.Koncki R. Recent developments in potentiometric biosensors for biomedical analysis. Anal Chim Acta. 2007;599(1):7–15. doi: 10.1016/j.aca.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 150.Hiatt LA, Cliffel DE. Real-time Recognition of Mycobacterium tuberculosis and Lipoarabinomannan using the Quartz Crystal Microbalance. Sens Actuators B Chem. 2012;174:245–252. doi: 10.1016/j.snb.2012.06.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cheng CI, Chang YP, Chu YH. Biomolecular interactions and tools for their recognition: focus on the quartz crystal microbalance and its diverse surface chemistries and applications. Chem Soc Rev. 2012;41(5):1947–1971. doi: 10.1039/c1cs15168a. [DOI] [PubMed] [Google Scholar]
- 152.Pan Y, Sonn GA, Sin MLY, et al. Electrochemical immunosensor detection of urinary lactoferrin in clinical samples for urinary tract infection diagnosis. Biosens Bioelectron. 2010;26(2):649–654. doi: 10.1016/j.bios.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kaufman E, Lamster IB. The diagnostic applications of saliva - a review. Crit Rev Oral Biol Med. 2002;13(2):197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 154.Thanyani ST, Roberts V, Siko DGR, et al. A novel application of affinity biosensor technology to detect antibodies to mycolic acid in tuberculosis patients. J Immunol Methods. 2008;332(1–2):61–72. doi: 10.1016/j.jim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 155.Zuo BL, Li SM, Guo Z, et al. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal Chem. 2004;76(13):3536–3540. doi: 10.1021/ac035367b. [DOI] [PubMed] [Google Scholar]
- 156.Chua AL, Yean CY, Ravichandran M, et al. A rapid DNA biosensor for the molecular diagnosis of infectious disease. Biosens Bioelectron. 2011;26(9):3825–3831. doi: 10.1016/j.bios.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 157.Huang JL, Yang GJ, Meng WJ, et al. An electrochemical impedimetric immunosensor for label-free detection of Campylobacter jejuni in diarrhea patients’ stool based on O-carboxymethylchitosan surface modified Fe3O4 nanoparticles. Biosens Bioelectron. 2010;25(5):1204–1211. doi: 10.1016/j.bios.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 158.Mahilum-Tapay L, Laitila V, Wawrzyniak JJ, et al. New point of care Chlamydia Rapid Test - bridging the gap between diagnosis and treatment: performance evaluation study. BMJ. 2007;335(7631):1190–1194. doi: 10.1136/bmj.39402.463854.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]