SUMMARY
The small nuclear RNA (snRNA) genes have been widely used as a model system for understanding transcriptional regulation due to the unique aspects of their promoter structure, selectivity for either RNA Polymerase (Pol) II or III, and because of their unique mechanism of termination that is tightly linked with the promoter. Recently, we identified the Little Elongation Complex (LEC) in Drosophila that is required for the expression of Pol II-transcribed snRNA genes. Here, using Drosophila and mammalian systems, we provide genetic and molecular evidence that LEC functions in at least two phases of snRNA transcription: an initiation step requiring the ICE1 subunit, and an elongation step requiring ELL.
INTRODUCTION
The promoter structure of the human snRNA genes can be distinguished from the promoters of protein-coding genes and other non-coding RNAs transcribed by Pol II by a proximal sequence element (PSE) and an enhancer-like distal sequence element (DSE) (Hernandez, 2001). The PSE is required for basal transcription and recruits the multi-subunit snRNA activator protein complex (SNAPc). The DSE enhances the transcription of snRNA genes and typically consists of an octamer (POU2F1/Oct-1) binding site and a ZNF143 (hStaf) binding site (Jawdekar and Henry, 2008; Murphy et al., 1992; Yoon et al., 1995). snRNA genes can be transcribed by either Pol II or Pol III. Slight differences in snRNA promoter elements may put SNAPc into different conformations that can be differentially recognized by Pol II or Pol III (Kim et al., 2010).
Unlike pre-mRNAs, snRNAs are not spliced or polyadenylated. The snRNA 3’ end formation is dependent on a conserved 3’ box, which directs the processing of the snRNA through the Integrator complex (Baillat et al., 2005; Cuello et al., 1999; Kunkel and Pederson, 1985). The function of the 3’ box is promoter-specific as it is only recognized when transcribed from an snRNA promoter (Hernandez and Weiner, 1986). Many of the factors that have been found to affect snRNA expression are found to primarily affect 3’-end processing (Baillat et al., 2005; Egloff et al., 2007; Ezzeddine et al., 2011; Medlin et al., 2005).
In mammals, the carboxyl-terminal repeat domain (CTD) of the largest subunit of RNA Pol II is comprised of 52 tandem repeats of the heptapeptide Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, which can undergo reversible phosphorylation at Serine 2, Serine 5, and Serine 7 in vivo. Serine 5 phosphorylation is catalyzed by the CDK7 subunit of TFIIH at the initiation stage, which allows the CTD to interact with and activate factors that cap the 5’ end of the nascent RNA (Ho and Shuman, 1999; Trigon et al., 1998). Subsequent phosphorylation of Serine 2 by the positive transcription elongation factor b (P-TEFb), comprising CDK9 and Cyclin T1/T2, is associated with productive transcriptional elongation and 3’ end processing for mRNA genes, but is only required for the 3’ end processing of snRNA genes (Buratowski, 2009; Jacobs et al., 2004; Medlin et al., 2003). CTD phosphorylation at Serine 7 is required for recruitment of the Integrator complex to the snRNA genes and is, therefore, required for 3’ end formation of snRNAs in vivo (Egloff et al., 2007; Egloff et al., 2010).
ELL, a common translocation partner of the mixed-lineage leukemia (MLL) gene in acute myeloid leukemia, was biochemically identified as a single polypeptide from rat liver extracts that could stimulate transcription elongation in vitro and was later shown to associate with transcriptionally active loci in vivo (Eissenberg et al., 2002; Gerber et al., 2001; Shilatifard et al., 1996; Thirman et al., 1994). Recently, we identified two distinct ELL-containing complexes, the Super Elongation Complex (SEC) and the Little Elongation Complex (LEC) (Smith and Shilatifard 2013). SEC is comprised of the ELL family member ELL2, the MLL translocation partners AFF1 and AFF4, either AF9 or ENL, and the CTD kinase P-TEFb (Lin et al., 2010; Luo et al., 2012). SEC, as one of the most active forms of P-TEFb (Luo et al., 2012), is required for several normal cellular functions such as the induction of HSP70 by heat shock and the response of developmental genes to environmental signals, as well as for the misregulation of HOX genes in MLL-AFF1 transformed cells (Lin et al., 2010), and for the activation of HIV transcription (He et al., 2010; Sobhian et al., 2010).
ELL in Drosophila is also found as part of LEC, containing ICE1, ICE2, and EAF. This complex has been shown to specifically regulate the transcription of Pol II-dependent snRNA genes in Drosophila. In mammalian cells, LEC colocalizes with coilin in subnuclear bodies (Polak et al., 2003; Smith et al, 2011). Coilin is found in both Cajal bodies and histone locus bodies. Cajal bodies contain small nuclear ribonucleoprotein particles (snRNPs) and small nucleolar ribonucleoprotein particles (snoRNPs) and are thought to be the sites of maturation of these particles (Kiss, 2004). Histone locus bodies contain the U7 snRNP that participates in histone mRNA 3’ end processing (Nizami et al., 2010).
Here, employing genetic and biochemical methods we characterize the molecular properties of LEC in human cells, and provide evidence that the LEC has dual functions in regulating snRNA gene transcription: ICE1 is required for promoting Pol II occupancy at snRNA genes, while the ELL subunit facilitates transcriptional elongation control of snRNA genes.
RESULTS
Functional Conservation of LEC subunits from Drosophila to Human
We previously reported that an ELL-containing complex in Drosophila, termed LEC, is required for snRNA gene transcription by Pol II. Furthermore, biochemical purifications of human ELL and the knockdown of ELL in mouse ES cells suggested that LEC is conserved in mammals. In order to define LEC's molecular composition and functional significance in mammalian cells, we purified LEC subunits ICE1 and ICE2 from human cells and determined their interactors using mass spectrometric methods (Figure S1A-C). In addition to finding that ELL, ICE1, and ICE2 form a complex similar to what we found in Drosophila, we also found ZC3H8 to be a novel interactor of LEC (Figure S1A-C). ZC3H8 appears to be a bona fide component of LEC in humans as evidenced by colocalization with LEC subunits in subnuclear bodies, and by coimmunoprecipitation studies (Figure S1D-F).
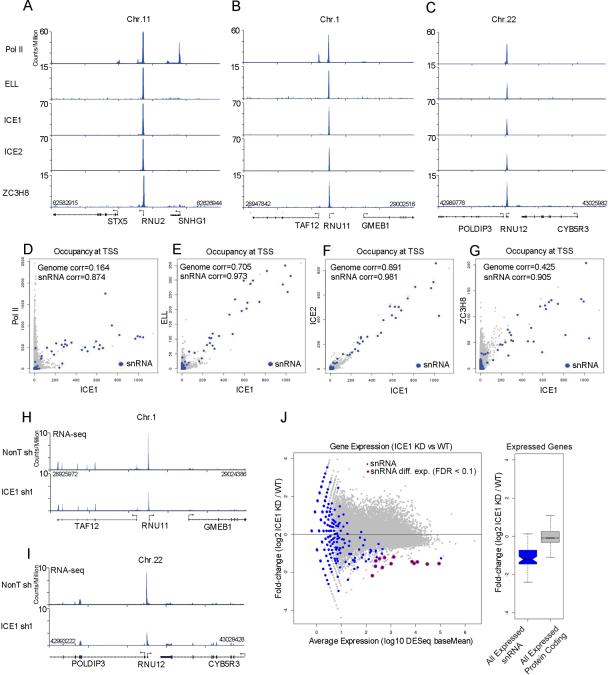
To formally demonstrate that snRNA genes are direct targets of human LEC, we employed ChIP-seq to map the genome-wide distribution of LEC subunits and RNA Pol II in HCT116 cells. We find that all subunits of LEC occupy Pol II-transcribed snRNA genes (Figure 1A-C). The specificity of LEC for Pol II-transcribed snRNA genes is also demonstrated by the high correlation among LEC components at these genes (Figure 1D-G). We further defined high-confidence enriched peak regions for LEC subunits using MACS (FDR < 0.05 and a fivefold enrichment over input) to determine putative binding sites for the complex subunits genome-wide. There were 63 regions in the genome where all subunit peak regions overlapped, corresponding to the promoter regions (±50 bp) of 28 snRNA genes (Table S1). This high fraction of snRNA-bound genes further demonstrates the genome-wide specificity of LEC for snRNAs (p < 2.2 e-16) as tested against finding a similar fraction of snRNA genes from the random sampling (n=100,000) of 63 regions of the genome. As previously suggested by our mouse ES studies (Smith et al., 2011), LEC-bound genes are not strongly bound by SEC components such as AFF4 (Figure S1G).
Figure 1. Functional conservation of the LEC complex in human cells.
(A-C) The human LEC complex is enriched at the promoters of the snRNA genes. Genome browser track examples for the occupancy of human LEC subunits ICE1, ICE2, ELL, and ZC3H8 as well as Pol II in HCT116 cells at the U2 (RNU2), U11 (RNU11), and U12 (RNU12) snRNA genes. Nearby protein-coding genes such as STX5, SNHG1 and TAF12 have high levels of Pol II, but no LEC. (D) Scatter plot depicting the genome-wide occupancy levels of Pol II vs. ICE1, showing that the highest co-bound genes are snRNA genes (blue color). Pearson correlations are shown. (E-G) Scatter plots depicting the genome-wide occupancy levels of ELL (E), ICE2 (F), and ZC3H8 (G) versus ICE1. Occupancy levels were measured as the sum of reads per million (RPM) aligned within 10 bp of all ENSEMBL 69-annotated transcription start sites. (H-J) RNA-seq analysis shows that ICE1 regulates snRNA gene expression. (H-I) Genome browser tracks show examples of RNA-seq data surrounding the RNU11 and RNU12 genes. The expression of RNU11 and RNU12, but not nearby protein-coding genes, is decreased in ICE1 shRNA-treated samples. (J) The left panel shows a MA plot of the genome-wide expression changes as determined by RNA-seq for ICE1 shRNA. The x-axis shows the log10 normalized read abundance (baseMean) of knockdown and non-targeting samples as reported by DESeq (Anders and Huber, 2010). The y-axis shows the log2 fold changes of normalized read abundance of ICE1 knockdown (KD) divided by the wild-type (WT) condition. Annotated snRNA genes are shown in blue. Differentially expressed snRNA genes (FDR<0.1) as reported by DESeq are circled in red. The right panel shows a boxplot of the log2 fold change ICE1 KD divided by WT for expressed snRNA genes (blue box) and all expressed protein coding genes (gray box). A baseMean of 10 was chosen as the cutoff for determining whether a gene is expressed. Box plot colored portions indicate the middle quartiles; the whiskers indicate a maximum of 1.5 of the interquartile range; middle notches indicate a 95% confidence interval of the median. See Figure S1 for related information.
In order to determine whether human LEC subunits contribute to snRNA transcription, we carried out the shRNA-mediated knockdown of ICE1, ICE2, and ZC3H8 in HCT116 cells and the RNA-seq of total RNA depleted for rRNA. Knockdown of ICE1 led to a general reduction of snRNA transcription (Figure 1H-J and Figure S1H) similar to what we observed previously for ELL RNAi in mouse ES cells (Smith et al., 2011). Although ICE1 and ELL are generally required for snRNA transcription by Pol II, we have been unable to find significant global defects in snRNA gene expression after depletion of ICE2 and ZC3H8 (data not shown), suggesting that these subunits function either in context-specific situations or in other steps of snRNA biogenesis. Therefore, we have focused our mechanistic studies of LEC function on defining the role of the ICE1 and ELL subunits in snRNA transcription.
ICE1 is a scaffold for LEC formation and targeting to subnuclear bodies containing coilin
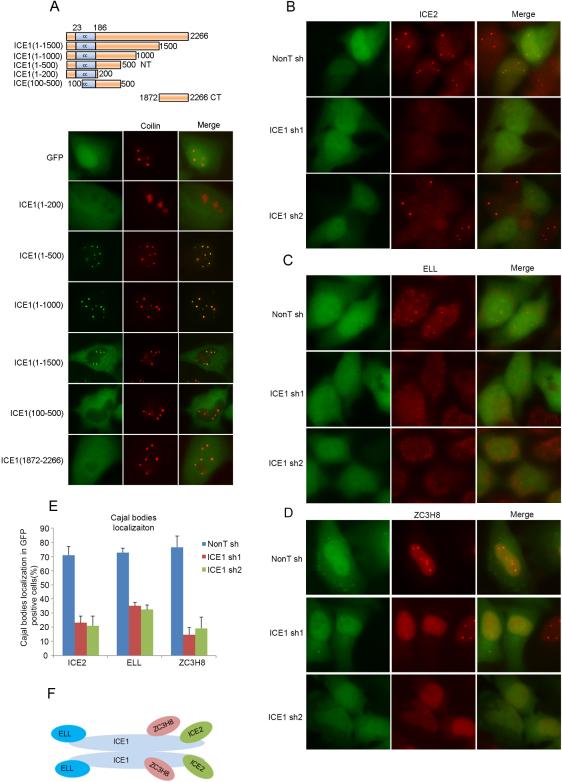
The LEC was previously shown to co-localize with coilin in the subnuclear bodies of HeLa cells (Polak et al., 2003; Smith et al., 2011), while the SEC components such as AFF4 do not (Smith et al., 2011). To define the role of ICE1 in LEC targeting, we used transient transfection with a series of EGFP-tagged ICE1 deletion constructs and assayed protein localization in HeLa cells. Antibodies to coilin were used to mark Cajal and histone locus bodies. We found that the N-terminal 500 amino acids of ICE1 were necessary and sufficient for GFP accumulation in these bodies (Figure 2A). The only predicted domain in this region is a coiled-coil structure spanning between amino acids 23 to 186. The coiled coil is a common structural motif mediating self-interaction and/or interaction with other proteins (Wolf et al., 1997).
Figure 2. ICE1 is a scaffolding protein for LEC and is required for the targeting of LEC to subnuclear bodies.
(A) The recruitment of ICE1 to coilin-positive subnuclear bodies requires its N-Terminal 500 amino acids, which includes a 163 amino acid coiled-coil domain. Different ICE1 truncations were expressed with an N-terminal GFP tag in HeLa cells and the localization of these ICE1 truncations was visualized by fluorescence microscopy. (B-E) Images (B-D) and their quantitation (E) show that HeLa cells depleted of ICE1 by two different shRNA are defective in the localization of ICE2 (B), ELL (C), and ZC3H8 (D) to subnuclear bodies. For (E), more than 100 GFP-positive cells were scored for whether bright nuclear dots of ICE2, ELL, or ZC3H8 were present, indicative of coilin body localization. Error bars represent the standard deviations. (F) Proposed molecular architecture of the LEC complex in human cells. The LEC complex might dimerize through the N-terminus of ICE1 (data not shown). ICE2 and ZC3H8 associate with ICE1 through its C-terminal portion while ELL binds to the N-terminal region (see Figure S2 for related information).
In order to determine if ICE1 is required for colocalization of the LEC subunits with coilin, we depleted HeLa cells of ICE1 with two different shRNAs, each co-expressed in a bicistronic transcript with GFP to allow for marking shRNA expressing cells (Figure 2B-2E and Figure S2A and S2C). Reduced ICE1 staining in the nucleoplasm and subnuclear bodies, as well as reduced mRNA levels were observed, demonstrating the efficiency of these two shRNAs (Figure S2A and S2B). Coilin localization remained normal (Figure S2C). Knockdown of ICE1 led to loss of ICE2, ELL, and ZC3H8 from coilin-containing subnuclear bodies (Figure 2B-E) without affecting the protein levels of these subunits (Figure S2D). ICE1 appears to be central for formation of LEC, as all subunits can be isolated with either N-terminal (ELL) or C-terminal (ZC3H8 and ICE2) fragments of ICE1 (Figure S1B). Furthermore, the association of ICE2 with ELL and ZC3H8 was reduced after ICE1 depletion in HCT116 cells (Figure S2E). This scaffolding function of ICE1 may involve a dimerization of ICE1 through the amino-terminal coiled coil motif, as the Flag-tagged ICE1 N-terminal domain was able to associate with full-length ICE1 (Data not shown). Together, these findings demonstrate that ICE1 is a key protein for LEC complex formation and for targeting LEC's localization with coilin in subnuclear bodies in cells (Figure 2F).
ICE1 mediates LEC recruitment and Pol II occupancy at snRNA genes
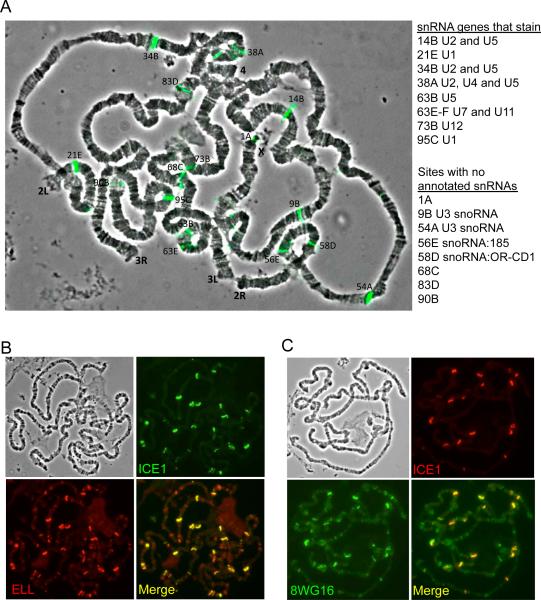
In order to understand ICE1's role in targeting LEC to chromatin in Drosophila, we turned to the giant polytene chromosomes of third instar larval salivary glands. ICE1 is expressed at a very low, or undetectable, level in this tissue, while ELL is concentrated with the Serine 2 phosphorylated form of Pol II at numerous sites including many puff sites indicative of high levels of transcription (Figure S3A-C) (Gerber et al., 2001). We expressed ICE1 using the Gal4-UAS system and a salivary gland-specific driver. In the presence of Gal4, ICE1 protein approached the levels normally found in ovaries (Figure S3A). Polytene squash preparations show that ICE1 and ELL co-accumulate at sites of snRNA genes when ICE1 is overexpressed (Figure 3A-B). However, these sites are not enriched for the Serine 2 phosphorylated form of Pol II (Figure S3D). Remarkably, new sites of strong Pol II staining, which correspond to the sites of strong ICE1 and ELL staining, can be observed in the ICE1 overexpressing glands when using the 8WG16 monoclonal antibody that recognizes the unphosphorylated CTD of Pol II (Figure 3C and S3E).
Figure 3. Overexpression of ICE1 in larval salivary glands re-localizes ELL and Pol II on polytene chromosomes.
(A) Overlay of ICE1 (green) immunostaining and the phase contrast image in ICE1 overexpressing salivary glands. Many of the sites of ICE1 staining correspond to snRNA genes as annotated in Flybase (right panel). A few of the mapped sites correspond to genes annotated as snoRNAs. These particular snoRNAs are LEC targets, as they were previously reported to have high levels of LEC in S2 cells and their levels are reduced after RNAi of ICE1 and ELL in S2 cells (Smith et al., 2011). (B) In ICE1 overexpressing glands, some of the strongest sites of ELL co-localize with the ectopically expressed ICE1. (C) ICE1 overexpression leads to strong occupancy of Pol II at the ectopic sites of ICE1 when visualized with the 8WG16 monoclonal recognizing unmodified Pol II. Phase contrast images for B-C are shown in the upper left panels. See Figure S3 for related information.
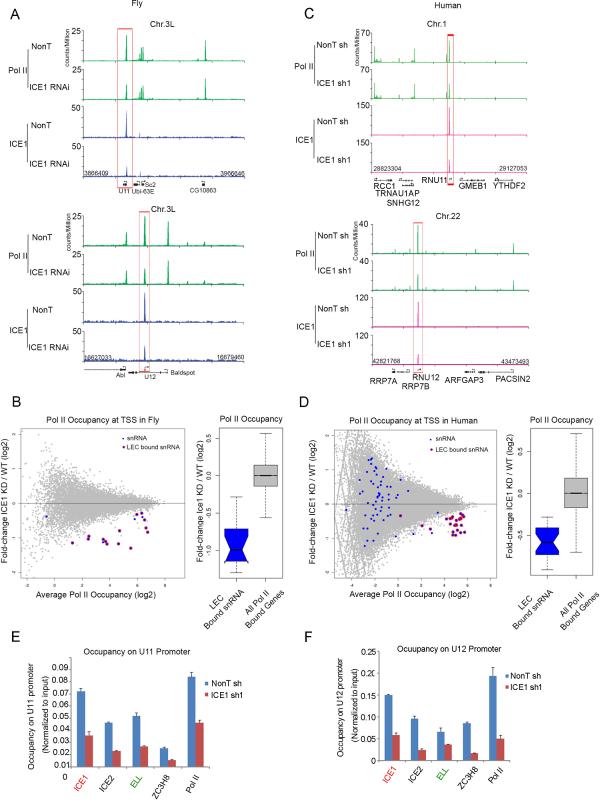
To further explore a possible role of ICE1 in influencing Pol II occupancy at a higher resolution, we performed RNAi knockdown of ICE1 in Drosophila S2 cells followed by ChIP-seq with antibodies recognizing total Pol II (Figures 4A and 4B). We find Pol II occupancy significantly reduced at LEC-bound snRNA genes in S2 cells, such as U11, without affecting Pol II occupancy at nearby protein-coding genes (Figure 4A and S4A). We also found that the LEC-bound snRNA genes were significantly reduced in Pol II occupancy genome-wide as compared to all Pol II-bound protein-coding genes (Wilcox; one-tailed; p = 7.603e-10; Figure 4B). To test the evolutionarily conserved role for ICE1 in the recruitment of Pol II to snRNA genes from Drosophila to human, we also performed Pol II ChIP-seq in HCT116 cells after depletion of ICE1 with shRNA. Pol II occupancy is significantly reduced at LEC-bound snRNA genes in human cells in the absence of ICE1 (Wilcox; one-tailed; p= 1.091e-12; Figure 4C and 4D). However, depletion of ICE2 or ZC3H8 has no major effect on Pol II residency on snRNA genes (Figure S4B). Depletion of ICE1 leads to reduced occupancy of the ICE2, ELL and ZC3H8 subunits of LEC at snRNA genes (Figure 4E and 4F).
Figure 4. ICE1 specifically regulates the occupancy of Pol II and LEC subunits at the snRNA genes.
(A) Genome browser track examples of Pol II occupancy in non-targeting (Non T) and ICE1-depleted Drosophila S2 cells. Pol II occupancy is specifically down-regulated at U11 and U12 snRNA genes, but does not change at their neighboring protein-coding genes, such as Baldspot, Sc2, and Abl. (B) The left panel shows a MA plot of the genome-wide Pol II occupancy after ICE1 depletion in Drosophila S2 cells. The x-axis shows the log2 geometric average of Pol II occupancy as measured as the maximum read sums per million sequenced (RPM) within the TSS and 100 bp downstream of the TSS for all annotated transcription start sites. Only Pol II-bound transcripts are shown that had enriched Pol II regions (MACS p < 0.05) in the wild-type sample within 100 bp of an annotated start site. The y-axis shows the log2 fold change of the Pol II occupancy levels measured by taking the occupancy levels in the knockdown divided by the non-targeting control. Pol II-bound snRNA genes are shown in blue. Some transcriptionally silent snRNA genes are called bound by Pol II using our peak finding criteria due to their proximity to other transcribed genes, so we defined a high confidence LEC target set for start sites that showed a two-fold or greater loss of ICE1 occupancy with 10 bp of the transcription start site in the ICE1 RNAi condition compared to wild-type ICE1 ChIP-seq levels. These genes are highlighted with a red circle. The right panel is a box plot analysis of the log2 fold change in Pol II occupancy after ICE1 RNAi. (C) Genome browser track examples of Pol II occupancy in non-targeting and ICE1-depleted human HCT116 cells. Pol II occupancy is specifically reduced at RNU11 and RNU12 snRNA genes, but does not change at their neighboring protein-coding genes. (D) The left panel shows a MA plot of the genome-wide Pol II occupancy after ICE1 depletion in HCT116 cells, as plotted in Panel B. LEC-bound snRNAs are highlighted with red circles. The snRNA genes with LEC bound in the wild-type condition show a general loss of Pol II after RNAi. The right panel is a box plot representation of the log2 fold change in Pol II after ICE1 knockdown. (E-F) ICE1 is required for recruitment of LEC subunits ICE2, ELL, and ZC3H8 to the RNU11 and RNU12 snRNA genes. ChIP-qPCR was performed with ICE1, ICE2, ELL, ZC3H8, ELL, and Pol II antibodies in control and ICE1-depleted HCT116 cells. Error bars represent the standard deviations. See Figure S4 for related information.
Evidence that ELL enhances transcriptional elongation of snRNA genes by Pol II
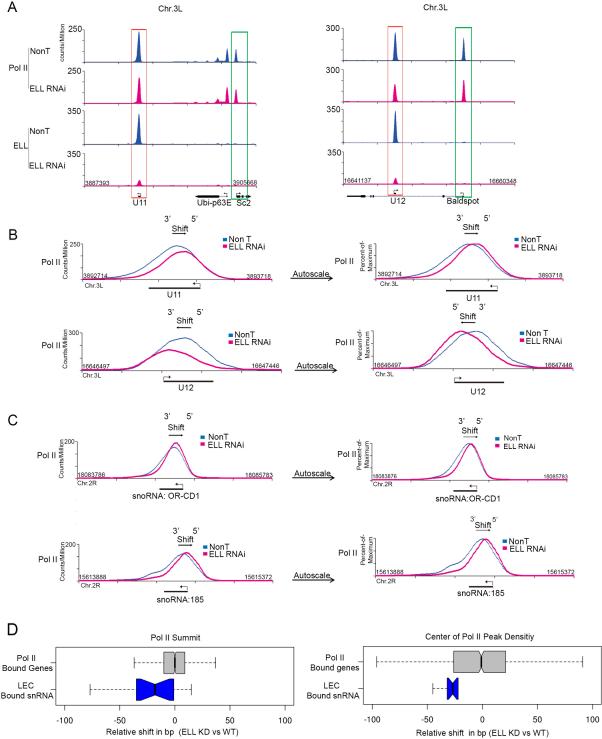
The ELL gene was initially identified as one of the translocation partners of MLL found in hematological malignancies (Rowley, 1998; Thirman et al., 1994). Human and Drosophila ELL were demonstrated to function as Pol II elongation factors capable of increasing the catalytic rate of transcription elongation by reducing transient pausing in vitro (Gerber et al., 2001; Shilatifard, 1998; Shilatifard et al., 1996), but no direct in vivo evidence exists that ELL specifically affects the elongation step of transcription by Pol II. Although snRNA genes are relatively short, sustaining their high rates of transcription may require the elongation activity of ELL. In order to better understand ELL's in vivo role on Pol II's activity, we performed Pol II ChIP-seq after ELL RNAi knockdown in Drosophila S2 cells. In contrast to what is seen with ICE1 knockdown, no significant global change in Pol II occupancy is observed at snRNA genes after ELL RNAi (Wilcox; two-tailed; p=0.4004; Figure S5A). However, occasionally a modest reduction in the Pol II ChIP-seq levels at some snRNA genes after ELL RNAi knockdown can be observed (Figure 5A, red boxes).
Figure 5. ELL knockdown affects the Pol II distribution across snRNA genes.
(A) Genome browser track examples of Pol II occupancy at snRNA and neighboring genes in non-targeting (Non T) and ELL-depleted Drosophila S2 cells. Pol II occupancy at snRNA genes is less affected after ELL knockdown compared to ICE1 depletion (compare to Figure 5A and Figures 4A and S4A), even though ELL itself is greatly reduced at the snRNA genes. (B) Overlay and expanded view of the NonT and ELL knockdown Pol II occupancies at U11 (top panels) and U12 (bottom panels). The peak Pol II signal in the ELL knockdown is shifted in the direction of the transcription start site (TSS) for each gene. Left panels compare the Pol II signals in NonT and knockdown using the same fixed scales. The right panels are scaled to the maximum peak signal of each condition to allow better visualization of the shift in the overall distribution of Pol II after ELL knockdown. (C) Track examples showing a shift in Pol II towards the TSS of two snoRNA genes that are regulated by LEC (Smith et al., 2011). Fixed and percent-of-maximum versions are plotted as in (B). (D) Quantitation of the degree of shift as seen by two distinct measurements. The left panel shows a box plot representation of the distance, in base pairs, from the Pol II peak summit in the wild-type condition to the Pol II peak summit in the ELL RNAi knockdown. The box plot in the right panel instead uses the distance from the center of read density of the Pol II enriched region at each gene. All genes are oriented 5’ to 3’ such that negative values indicate a shift toward the 5’ end of the gene. The set of LEC-bound snRNAs and all Pol II-bound genes are the same as in Figure 4B. See Supplemental Figure S5 for related information.
Although Pol II levels appeared relatively unaffected by loss of ELL, we noticed that when the Pol II signal at snRNA genes is scaled to the maximum of each condition, a shift in the Pol II distribution towards the 5’ end of the gene in the absence of ELL could be observed (Figure 5B, right panels). The Pol II shift is also seen at the small nucleolar RNA (snoRNA) genes that are transcribed by Pol II (Figure 5C), without any obvious reduction in Pol II levels at these genes (Figure S5B). Pol II transcribed genes that are not targets of LEC, such as Sc2 and Baldspot (Figure 5A, green boxes), do not show a shift in the Pol II distribution after ELL RNAi knockdown (Figure S5C). We made two measurements to quantify the degree and specificity of the Pol II shift for snRNA genes after ELL knockdown. For both methods, we annotated the nearest Pol II enriched region for the wild-type and ELL knockdown, if an enriched peak region was found within 100 bp of the annotated transcription start site in the wild-type. We then computed the positions of the summit and the center of density for each peak region. In order to determine a shift, we oriented each transcript start site 5’ to 3’ and computed the shift in base pairs between both the summits and centers of density, respectively. We sought to compare the shift in Pol II peak behavior between the LEC-bound snRNAs and 1,000 random Pol II-bound genes (Figure 5D). We found significant changes in the shift distribution for the LEC-bound genes compared to the random Pol II-bound genes (Wilcox test; one-tailed; summit p = 0.002412; center p = 0.008095).
LEC can regulate genes that are co-transcribed by Pol II and Pol III
Recently, Hernandez and colleagues performed ChIP-seq in IMR90Tert cells for the subunits of Pol II and Pol III, and for the subunits of SNAPc that binds the core promoters of both Pol II and Pol III-transcribed snRNA genes (James Faresse et al., 2012). They were able to identify seven unannotated Pol II transcribed regions that were bound by SNAPc, suggesting that they were controlled similarly to snRNA genes. Six of these Unknown #1-7 genes are also enriched for Pol II in HCT116 cells (Table S2). We measured Pol II occupancy in the wild-type and ICE1 RNAi knockdown HCT116 cells and saw that all six lose Pol II occupancy in the knockdown cells (Table S2), similar to what we found with annotated snRNAs.
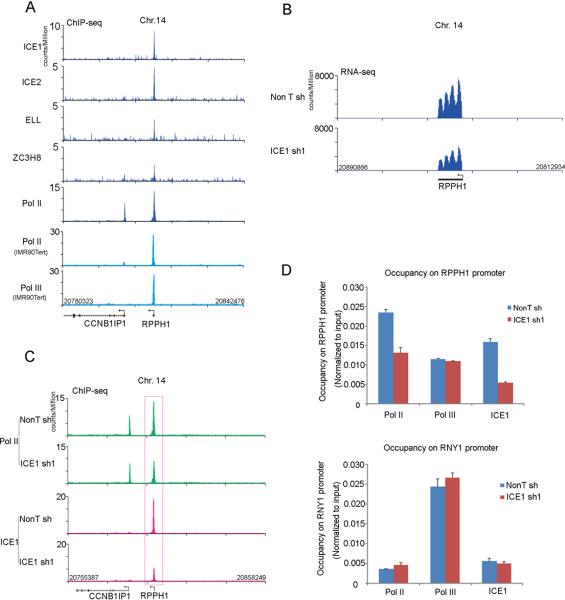
The James Faresse et al. study also reported that one gene, RPPH1, is transcribed by both Pol II and Pol III in IMR90Tert cells. The RPPH1 gene expresses the H1 RNA, the 340 nucleotide catalytic component of the RNase P involved in tRNA maturation (Bartkiewicz et al., 1989). We find that LEC subunits and Pol II are enriched at the RPPH1 gene in HCT116 cells (Figure 6A). Furthermore, H1 RNA levels are reduced when ICE1 is depleted (Figure 6B). Pol II occupancy on RPPH1 is also reduced by ICE1 depletion (Figure 6C). However, Pol III occupancy at RPPH1, or at a gene exclusively transcribed by Pol III, RNY1, is not affected by ICE1 knockdown (Figure 6D). Together these data support the conclusion of James Faresse et al., which showed that Pol II and Pol III are non-redundant for the transcription of the RPPH1 gene.
Figure 6. Human LEC regulates RPPH1 gene expression.
(A) Human LEC subunits are present with Pol II at the RPPH1 gene, a gene that is transcribed by both Pol II and Pol III (James Faresse et al., 2012). Genome browser tracks show the occupancy of human LEC subunits ICE1, ICE2, ELL, and ZC3H8 as well as Pol II at the RPPH1 gene in HCT116 cells. Pol II and Pol III ChIP-seq tracks, shown in light blue, are from (James Faresse et al., 2012). (B) ICE1 regulates RPPH1 gene expression. Genome browser track of RPPH1 gene expression determined by RNA-seq. (C) ICE1 is required for Pol II occupancy at the RPPH1 gene. Genome browser profiles of Pol II occupancy at the RPPH1 gene in non-targeting and ICE1-depleted HCT116 cells are shown. (D) ICE1 is not required for the recruitment of Pol III to the RPPH1 genes. Pol II, but not Pol III, is reduced at the RPPH1 promoter after ICE1 RNAi. Another Pol III transcribed non-coding RNA gene, RNY1, with little or no associated Pol II, is shown for comparison. Error bars represent the standard deviations. See Figure S6 for related information.
We were also surprised that a U6 gene, U6-26, was identified in our list of genes that are co-occupied by all LEC subunits (Figure S6A and Table S1). The U6-26 gene was also found to have the second highest enrichment of Pol II among the list of Pol III-class snRNA genes that were identified by James Faresse and colleagues (Figure S6A and Table S2). Importantly, both ICE1 and Pol II levels are reduced at U6-26 after ICE1 knockdown (Figure S6B and Table S2), further demonstrating that the presence of SNAPc with Pol II is sufficient to identify a LEC-regulated gene.
DISCUSSION
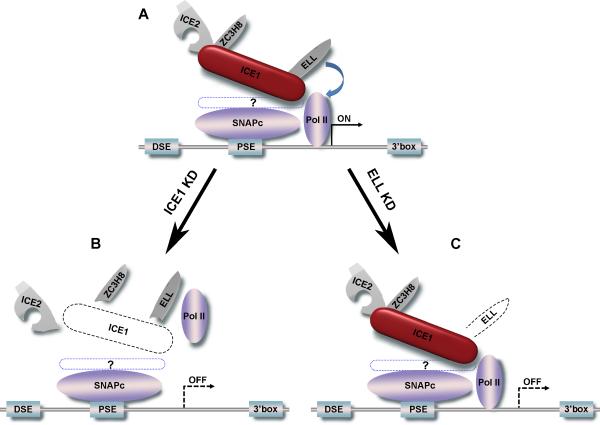
In the present study, we have demonstrated the functional conservation of the Little Elongation Complex (LEC) between flies and humans and have provided evidence that it has roles in both the recruitment and elongation stages of Pol II-dependent snRNA gene transcription. ICE1 is required for complex assembly and for Pol II occupancy at snRNA genes. ELL is required for Pol II elongation as indicated by a 5’ shift in the Pol II distribution at snRNA genes. The function(s) of the other subunits are currently unknown, suggesting that LEC could function as a multi-functional tool (Figure 7) with some subunits functioning at distinct steps or in a context-dependent manner.
Figure 7.
LEC has roles in establishing Pol II occupancy and productive transcription of Pol II at snRNA genes. (A) A typical snRNA gene is composed of a distal sequence element (DSE), a core promoter element (PSE), the transcription unit, and a 3’ box which signals for 3’ RNA processing. The PSE is bound by the small nuclear RNA-activating protein complex (SNAPc), which is recruited to snRNA promoters in early G1 of the cell cycle before Pol II is recruited (James Faresse et al., 2012). LEC, represented as a multi-tool pocketknife, is composed of the scaffolding protein ICE1, the transcription elongation factor ELL, and proteins of unknown function, ICE2 and ZC3H8. How LEC specifically recognizes Pol II transcribed snRNA genes is currently unknown (represented by a dotted blue line encircling a question mark). (B) Knockdown of ICE1 (dashed line) leads not only to the release of LEC components from snRNA genes, but also to the loss of Pol II occupancy, leading to reduced snRNA transcription. (C) Knockdown of ELL (dashed line) leaves LEC largely intact and has a minimal effect on Pol II levels. Instead, knockdown of ELL results in an overall Pol II redistribution towards the 5’ end of the snRNA gene, suggestive of a defect in transcription elongation. Although the role of ICE2 and ZC3H8 are currently unknown, their presence with other LEC components in coilin-stained subnuclear bodies, suggests that LEC could regulate other post-transcriptional steps, such as snRNP maturation or recycling.
In ICE1 knockdown cells from flies or mammals, Pol II occupancy is specifically reduced at snRNA genes (Figure 4) and expressing ICE1 in Drosophila salivary glands leads to new Pol II recruitment to snRNA genes (Figure 3). In the case of ELL knockdown, we did not observe a global reduction in Pol II occupancy at snRNA genes (Figure S5A). Instead, we noticed a shift in Pol II to the 5’ end of the snRNA genes after ELL RNAi (Figures 5B and 5C). Due to the small size of snRNA genes, and since Pol II peaks well into the snRNA gene body, we were not able to use traditional calculations of traveling ratios to describe a change in polymerase distribution along the transcription unit (Wade and Struhl, 2008). Therefore, we developed two metrics to quantify our observed shift, one based on the position of the peak summit of Pol II, and another, which determined the base position at the center of the density of the total Pol II signal around the snRNA gene. Comparing these measurements in wild-type and RNAi conditions allowed us to obtain statistical support for our hypothesis that ELL could play a role in either Pol II release from the start site or in facilitating Pol II processivity in gene bodies (Figure 5D).
The function of ICE2 and ZC3H8 within LEC are currently unknown. ICE2 is conserved in both human and fly LEC, while ZC3H8 has no obvious ortholog in Drosophila. ZC3H8 is comprised of a charged domain and three putative C(3)H-type zinc fingers and belongs to a family of proteins implicated in RNA binding and 3’ end processing. We previously noted that ICE2 is related to proteins that bind the 5’ ends of RNA (Margelevicius et al., 2010). Another protein implicated in LEC function is USPL1. A divergent relative of USPL1, CG8229, was found associated with Drosophila LEC; and a proteomics study found that overexpressed USPL1 co-purified with ICE1 and ELL in human cells (Smith et al., 2011; Sowa et al., 2009). Recently, USPL1 was demonstrated to be a sumo isopeptidase with non-catalytic, but essential functions in cell proliferation and coilin body formation, despite being expressed at very low levels (Schulz et al., 2012). All LEC components localize to coilin-stained bodies. Coilin is found in both Cajal bodies, sites of snRNP maturation and recycling (Kiss, 2004), and histone locus bodies, which are sites of U7 snRNP-dependent histone mRNA 3’ end processing (Nizami et al., 2010). Interestingly, both knockdown of USPL1 (Sowa et al., 2009) and overexpression of the first 200 amino acids of ICE1 (Figure 2A) resulted in the mislocalization of coilin to nucleoli, further suggesting interdependent functions between USPL1, LEC, and coilin-positive subnuclear bodies. Since knockdown of ICE2 and ZC3H8 did not appear to have a major effect on snRNA expression levels, and since these proteins colocalize with ICE1 and ELL (both at snRNA genes and with coilin), we speculate that LEC could regulate snRNP biogenesis from transcription through snRNP maturation and recycling, as described below.
Pol II-dependent spliceosomal snRNAs are modified to carry a 5’-terminal monomethylguanosine (m7G) cap structure and short non-polyadenylated 3’ end sequences (Kiss, 2004). These precursor snRNAs are exported to the cytoplasm and assembled with Sm proteins into snRNPs through the Survival of the Motor Neuron (SMN) protein complex (Kiss, 2004). During snRNP assembly, trimming of ca. 10 nt of the 3’ end of the snRNA can occur (Kiss, 2004). Hypermethylation of the 5’-cap (2,2,7-trimethylguanosine, m3G) also occurs in the cytoplasm and acts as a signal for nuclear import (Kiss, 2004). The snRNPs are then transported into Cajal bodies where the snRNAs can be further modified (i.e. site-specific pseudouridylation and 2’-O-methylation directed by the small Cajal body-specific RNAs (scaRNAs))(Kiss, 2004). The LEC, with putative 5’ RNA and 3’ RNA-binding proteins, and ICE1-dependent colocalization with the Cajal body component coilin, could be regulating or monitoring some of these processes. Evidence that snRNP biogenesis is under feedback regulation comes from a zebrafish study where mutation of SART3, a snRNP4/6 recycling factor, led to an increased expression of a variety of proteins involved in snRNP biogenesis, including ICE1 and USPL1 (Trede et al., 2007). Coilin was recently found to mark not only Cajal bodies, but also histone locus bodies that function in histone 3’ end processing (Nizami et al., 2010). This processing is dependent on the U7 snRNP (Mowry and Steitz, 1987), whose RNA component is one of the direct targets of LEC in Drosophila and humans. LEC might travel with snRNAs from their site of synthesis to their site of final maturation or function in Cajal and histone locus bodies. We postulate that the LEC subunits with unknown functions may participate in some of these processes.
ELL was originally described as an in vitro Pol II elongation processivity factor, but recent studies have implicated the related factor ELL2 as regulating the release of promoter- proximal paused Pol II as part of the active P-TEFb complex SEC (Lin et al., 2010). However, the snRNA genes appear to be relatively independent of the Serine 2 phosphorylation of the CTD (Medlin et al., 2005). Therefore, at snRNA genes, ELL's ability to suppress transient pausing and backtracking by Pol II, as demonstrated in vitro, could be employed in vivo to maintain the high levels of transcription characteristic of this class of genes.
The past several years have seen a growth in the study of the elongation stage of transcription, and the diversity of elongation factors and their function is just beginning to be appreciated (Smith and Shilatifard, 2013). The recent growth in the study of transcription elongation has been brought about by genome-wide approaches, which have the ability to demonstrate the generality of transcription elongation factor function beyond a few model genes, while also revealing the specificity of elongation factors for specific classes of genes within the whole genome. Recently developed nascent RNA sequencing strategies should facilitate detailed molecular studies of the mechanistic properties of diverse transcription elongation factors and how they function in vivo.
EXPERIMENTAL PROCEDURES
Antibodies
Human Pol II (N-20) monoclonal antibody and Pol III antibody (SC-21574) were purchased from Santa Cruz; 8WG16 anti-CTD (MMS-126R) and H5 Ser2P-CTD (MMS-129R) monoclonals were purchased from Covance. Anti-coilin antibody was purchased from Abcam (ab11822). M2 Flag antibody was purchased from Sigma. Beta-tubulin monoclonal (E-7) was purchased from the Developmental Studies Hybridoma Bank. Full-length recombinant ZC3H8 protein, ICE1-CT (aa 1872-2266) and ICE2 NT (aa 1-261) were bacterially expressed as His–tag fusion proteins in pET-16b. Antibodies against ELL1, AFF4, dICE1, dELL, actin, and dRpb1 have been described previously (Lin et al., 2011; Lin et al., 2010; Smith et al., 2011).
ChIP-qPCR and ChIP-seq
For mammalian cells, 5x107 cells (ChIP-seq) or 1x107 (ChIP qPCR) were used for each ChIP assay according to previously described protocols (Lee et al., 2006). Briefly, HCT116 cells were crosslinked with 1% formaldehyde for 10 min at room temperature with rotation, and then crosslinking was quenched by the addition of glycine. Fixed chromatin was sonicated on a Misonix 3000 and used for immunoprecipitation with the indicated antibody. Isolated DNA was analyzed by qPCR using SYBR green on a MyIQ thermal cycler (BioRad). ChIP-seq in Drosophila S2 cells was performed similarly, except that 3x108 S2 cells were used per sample. ChIP-sequencing libraries were prepared with Illunina's Tru-seq DNA sample prep kit. Sequencing data were retrieved through the default Illumina pipeline using Casava V1.8. Reads were aligned to the human genome (UCSC hg19) or fly genome (UCSC dm3) using the Bowtie aligner V0.12.7 allowing uniquely mapping reads only with up to three mismatches to be aligned to the genome (Langmead et al., 2009). Gene annotations are from ENSEMBL 67 for fly and ENSEMBL 69 for human.
Flag purification and MudPIT analyses
Dignam nuclear extracts were prepared from HEK293FRT cells expressing various Flag-tagged proteins (Dignam et al., 1983). Flag-tagged and their associated proteins were purified on anti-Flag (M2) agarose beads in the presence of Benzonase (Sigma). Trichloroacetic acid-precipitated protein mixtures from the Flag purifications were digested with endoproteinase Lys-C and trypsin (Roche) and analyzed by MudPIT as previously described (Wu et al., 2008).
Immunoprecipitation
HCT116 cells were washed with cold phosphate-buffered saline (PBS) twice and lysed in RIPA buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP40, 1% sodium deoxycholate, 0.1% SDS, 1 mM dithiothreitol [DTT]) containing proteinase inhibitors (Sigma) for 30 min at 4°C. After centrifugation at 13,000 rpm for 30 min, the supernatant was incubated with the indicated antibodies and protein A/G PLUS agarose (Santa Cruz) at 4 °C overnight with gentle rotation. The beads were spun down and washed three times with wash buffer (10 mM HEPES [pH7.4], 1 mM MgCl2, 300mM NaCl, 10mM KCl, 0.2%TritonX-100) before boiling in sodium dodecyl sulfate (SDS) loading buffer.
RNA interference (RNAi), RT-qPCR, and RNA-seq
HCT116 cells were infected with lentivirus harboring either Non-targeting shRNA or ICE1 shRNA in the presence of 8 μg/ml of polybrene (Sigma) for 24 hours (Target sequence for ICE1 sh1: 5’-ATAAGAGTCGTTTGCGAAATA-3’, Target sequence for ICE1 sh2: 5’-GCCTAATCAAGTATCAGTTAT-3’). The infected cells were selected with 2 μg/ml puromycin for an extra 48 h before harvesting the cells for RNA extraction. Total RNA was extracted with Trizol (Invitrogen), treated with DNase I (NEB) for 20 min at room temperature, and precipitated with sodium acetate/ethanol in the presence of 1 μg/ml glycogen and dissolved again in RNase-free water. The expression levels were quantitated with a One-step RT-PCR kit (QIAGEN) on a My IQ Thermocycler (Bio-Rad). Relative expression to housekeeping gene GAPDH was determined by using the comparative cycle threshold method. Drosophila ICE1 and ELL RNAi were performed as previously described (LEC paper) except that S2 cells were grown in SFX serum-free media from HyClone. For RNA-seq, 2 μg of total RNA was depleted of ribosomal RNA with the RiboZero kit (Epicentre) and libraries were made with the TruSeq RNA sample Prep Kit (Illumina).
Immunofluorescence
HeLa cells were transfected with pGIPZ control and ICE1-specific shRNA (Open Biosystem, V2LHS_16118 for sh1 and V3LH3_302918 for sh2), which GFP and shRNA are part of a bicistronic transcript allowing the visual marking of shRNA expressing cells. After 48 h, cells were fixed by 1% paraformaldehyde for 10 min, permeabilized with 0.25% Triton X-100 for 10 min, and blocked with 1% bovine serum albumin before antibody incubations. Drosophila polytene chromosome immunofluorescence was performed as previously described (Eissenberg, 2006). For the ICE1 overexpression experiments, a misexpression line, EP-G2779, was obtained from Bloomington (BL27010) and crossed to the Sgs3-Gal4 driver (BL6870) at 27° C.
Supplementary Material
HIGHLIGHTS.
The Little Elongation Complex (LEC) mediates snRNA transcription in flies and humans.
ICE1 is the central scaffold for the assembly and subcellular localization of LEC.
Pol II's recruitment to snRNA genes requires ICE1.
ELL facilitates proper transcription elongation by Pol II at snRNA genes.
ACKNOWLEDGMENTS
We thank Rhonda Egidy, Allison Peak, Anoja Perera, and the rest of the Stowers Molecular Biology core for help with Illumina sequencing. We are grateful to the Stowers Tissue Culture core for assistance with the generation and maintenance of cell lines. We thank Laura Shilatifard for editorial assistance. These studies were supported in part by the National Cancer Institute grants R01CA150265 and R01CA089455 to A.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers. ChIP-seq and RNA-seq data are available from GEO with accession number GSE47938.
SUPPLEMENTAL INFORMATION. Supplemental Information includes six figures and two supplemental tables.
REFERENCES
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello P, Boyd DC, Dye MJ, Proudfoot NJ, Murphy S. Transcription of the human U2 snRNA genes continues beyond the 3′ box in vivo. EMBO J. 1999;18:2867–2877. doi: 10.1093/emboj/18.10.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods in enzymology. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem. 2010;285:20564–20569. doi: 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC. Functional genomics of histone modification and non-histone chromosomal proteins using the polytene chromosomes of Drosophila. Methods. 2006;40:360–364. doi: 10.1016/j.ymeth.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chen J, Waltenspiel B, Burch B, Albrecht T, Zhuo M, Warren WD, Marzluff WF, Wagner EJ. A subset of Drosophila integrator proteins is essential for efficient U7 snRNA and spliceosomal snRNA 3′-end formation. Mol Cell Biol. 2011;31:328–341. doi: 10.1128/MCB.00943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Ma J, Dean K, Eissenberg JC, Shilatifard A. Drosophila ELL is associated with actively elongating RNA polymerase II on transcriptionally active sites in vivo. EMBO J. 2001;20:6104–6114. doi: 10.1093/emboj/20.21.6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Jacobs EY, Ogiwara I, Weiner AM. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol Cell Biol. 2004;24:846–855. doi: 10.1128/MCB.24.2.846-855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet. 2012;8:e1003028. doi: 10.1371/journal.pgen.1003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawdekar GW, Henry RW. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta. 2008;1779:295–305. doi: 10.1016/j.bbagrm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Kang YS, Lai HT, Barakat NH, Magante D, Stumph WE. Identification of SNAPc subunit domains that interact with specific nucleotide positions in the U1 and U6 gene promoters. Mol Cell Biol. 2010;30:2411–2423. doi: 10.1128/MCB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Pederson T. Transcription boundaries of U1 small nuclear RNA. Mol Cell Biol. 1985;5:2332–2340. doi: 10.1128/mcb.5.9.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol. 2012;32:2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margelevicius M, Laganeckas M, Venclovas C. COMA server for protein distant homology search. Bioinformatics. 2010;26:1905–1906. doi: 10.1093/bioinformatics/btq306. [DOI] [PubMed] [Google Scholar]
- Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J. 2005;24:4154–4165. doi: 10.1038/sj.emboj.7600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–934. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247–3261. doi: 10.1128/mcb.12.7.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb Symp Quant Biol. 2010;75:313–320. doi: 10.1101/sqb.2010.75.005. [DOI] [PubMed] [Google Scholar]
- Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ. ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia. Mol Biol Cell. 2003;14:1517–1528. doi: 10.1091/mbc.E02-07-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic-Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa H, Wittbrodt J, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012;13:930–938. doi: 10.1038/embor.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev. 2013;27:1079–1088. doi: 10.1101/gad.215137.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al. The little elongation complex regulates small nuclear RNA transcription. Mol Cell. 2011;44:954–965. doi: 10.1016/j.molcel.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, Benkirane M. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Mol Cell. 2010;38:439–451. doi: 10.1016/j.molcel.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trede NS, Medenbach J, Damianov A, Hung LH, Weber GJ, Paw BH, Zhou Y, Hersey C, Zapata A, Keefe M, et al. Network of coregulated spliceosome components revealed by zebrafish mutant in recycling factor p110. Proc Natl Acad Sci U S A. 2007;104:6608–6613. doi: 10.1073/pnas.0701919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JB, Murphy S, Bai L, Wang Z, Roeder RG. Proximal sequence element-binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.