Abstract
The question of where retroviral DNA becomes integrated in chromosomes is important for understanding (i) the mechanisms of viral growth, (ii) devising new anti-retroviral therapy, (iii) understanding how genomes evolve, and (iv) developing safer methods for gene therapy. With the completion of genome sequences for many organisms, it has become possible to study integration targeting by cloning and sequencing large numbers of host–virus DNA junctions, then mapping the host DNA segments back onto the genomic sequence. This allows statistical analysis of the distribution of integration sites relative to the myriad types of genomic features that are also being mapped onto the sequence scaffold. Here we present methods for recovering and analyzing integration site sequences.
Keywords: Integration sites, HIV, Retroviral DNA, Genomic
1. Introduction
Following binding of a retrovirus to a sensitive cell, the viral and cellular membranes fuse and the viral core is released into the cytoplasm. The viral genomic RNA is then reverse transcribed, yielding a double stranded cDNA copy of the viral RNA genome. The complex of viral DNA and proteins (the “preintegration complex” or “PIC”) next carries out the covalent attachment of the viral cDNA to host DNA. The integration step completes the formation of a provirus, which contains all the information necessary for the synthesis of the viral RNAs and proteins and formation of new virions (for reviews see [1,2]).
The DNA breaking and joining reactions that mediate retroviral cDNA integration are well understood (reviewed in [1–4]). A linear form of the viral cDNA serves as the immediate precursor of the integrated provirus. Prior to integration, integrase removes in most cases two nucleotides from the 3′ end of each LTR (long terminal repeat), exposing recessed 3′ hydroxyl groups (a minority of viruses use a slight variation of this theme—for example human immunodeficiency virus [HIV] type 2 clips a trinucleotide from its downstream LTR). This terminal cleavage step may serve to remove heterogeneous extra nucleotides occasionally added to the cDNA ends by reverse transcriptase [5,6] and promote the formation of a stable complex [7,8]. Integrase then catalyzes attack by the recessed 3′ hydroxyl groups on phosphodiester bonds on each target DNA strand so as to join each viral DNA 3′ end to protruding 5′ ends in the target. The points of joining on each strand of the target DNA are separated by 4–6 base pairs depending on the retrovirus involved. Unfolding of this integration intermediate yields gaps at each junction between viral and host DNA, and a 5′ two-base flap derived from the viral DNA. Gap repair to connect the remaining DNA strands is probably carried out by host DNA repair systems. The terminal cleavage and first strand transfer reactions can be modeled in vitro with purified integrase [9–15] and the gap repair step modeled with purified DNA repair enzymes [16,17].
The target for HIV DNA integration is the human genome. The roughly 3.4 billion bases of the euchromatic portion of the human genome have now been sequenced [18,19], though the centromere sequences remain inaccessible with available technology. There are believed to be ~30,000 protein coding genes, with the exons comprising ~1.5% of the genome and the transcription units ~33%. However, these numbers remain surprisingly soft: non-protein-coding genes are hard to detect and may be quite abundant, and several reports suggest that low level transcription may take place in what were thought to be intergenic regions [20,21]. It could still turn out that almost all euchromatic human DNA is transcribed at some level, with the recognized transcription units simply transcribed more frequently.
With the completion of the draft human genome sequence in 2001 [18,19], it became possible to study integration target site selection by retroviruses using high-throughput DNA sequencing. In our first genome-wide study of retroviral DNA integration [22], we mapped and analyzed 524 sites of HIV cDNA integration in human SupT1 cells. To isolate integration site clones, cells were infected with HIV or an HIV-based vector, and then genomic DNA was harvested 2–3 days after infection. The period of cell growth after infection was minimized to reduce the chances of selection of particular integration sites during cell proliferation. DNA was harvested, cleaved with a restriction enzyme, and linkers were then ligated onto the cleaved DNA ends. Integration site sequences were amplified using primers complementary to the linker and the viral DNA end. PCR products were then cloned into a plasmid, transformed into bacteria, and sequenced in a 96-well plate format. The first survey of HIV integration sites in the SupT1 T-cell line revealed that transcription units were strongly favored as integration targets [22]. Global analysis of cellular transcription using Affymetrix gene expression microarrays indicated that active genes were preferential integration targets, particularly genes that were activated in cells after infection by HIV-1. These data documented unexpectedly strong biases in integration site selection and suggested that HIV had evolved to maximize transcription after integration by targeting active genes.
Next, it was of interest to ask whether similar results would be seen in other human cell types, particularly primary cells. Integration site surveys were thus carried out in many primary cell types and cell lines [22–29]. In all of these data sets, HIV DNA integration was found to be favored in transcription units as well. Integration site selection by retroviruses has been the topic of several recent reviews [4,30–32].
Murine leukemia virus (MLV) integration targeting was initially characterized in a publication from Shawn Burgess’s laboratory [33]. They reported the striking discovery that MLV favored integration near gene 5′ ends and CpG islands, and only showed a weak preference for transcription units.
Avian sarcoma-leukosis virus (ASLV) was next characterized and showed the most random distribution of integration sites of the three retroviruses, only weakly favoring transcription units, and not strongly favoring gene 5′ ends or CpG islands [23,34]. Thus, to our surprise, each of the three retroviruses studied showed a unique pattern of integration targeting. Summarizing over many recent studies, integration targeting preferences appear to be consistent within the different retroviral genera but generally different between genera.
A variety of subsequent studies have focused on testing models for possible mechanisms of integration targeting. Three models (which are not mutually exclusive) are as follows:
1.1. Tethering
One simple possibility is that integration complexes bind to cellular proteins that are bound at specific locations on chromosomes. Such tethering interactions are well-documented for the retrovirus- related retrotransposons of yeast [35–37], where binding of the element-encoded integrase proteins to cellular DNA-binding proteins has been shown convincingly to mediate integration targeting. According to this idea, the different favored integration sites for the different retroviruses would be a consequence of binding to different cellular tethering proteins. Additional support for this possibility comes from studies of artificial fusions of integrase proteins to sequence-specific DNA binding domains—in these studies, the fusion integrases have been shown to direct favored integration near DNA recognition sites for the added binding domain [38–44]. In principle, any component of the PIC, viral or cellular, could serve as a docking point for a cellular tethering factor that targets retroviral integration. Among the candidates on the viral side are MA, Vpr, and IN. Cellular factors proposed to be associated with PICs include BAF [45], HMGA1 [46], EED [47], p300 [48], Ini-1 [49,50], and LEDGF/p75 [26,51–59].
Considerable data, presented elsewhere in this volume, indicates that LEDGF/p75 has good credentials as a tethering factor for HIV integration targeting [26,58–60].
1.2. Open chromatin
This idea holds that much of the DNA in human cells is inaccessible due to tight wrapping in chromosomal proteins, so that only those regions that are particularly exposed can serve as integration targets. This is probably the first idea proposed for a mechanism of integration targeting in vivo, based on the apparent proximity of sequenced sites of MLV integration to DNase I hypersensitive sites [61–63]. Considerable data indicates that retroviral integration takes place on nucleosome-bound DNA [64–69]. Contemporary studies confirm that retroviral DNA integration is often favored near DNase I cleavage sites and epigenetic marks positively associated with transcription [29,69,70].
Another type of analysis has also provided tentative support for this idea. HIV integration is significantly less frequent in alphoid repeats than expected by chance [22,71]. Alphoid repeats are mostly found in centromeres and are wrapped in centromeric heterochromatin. This observation suggests that centromeric heterochromatin is disfavored for integration, though the mechanistic basis is not known.
1.3. Cell cycle
Retroviruses differ in the cell cycle timing of integration, which could potentially influence integration targeting. HIV can infect cells arrested at several phases of the cell cycle. MLV, in contrast, requires that cells pass through mitosis to allow integration [72–74]. It is likely that the state of chromatin changes during progression of the cell cycle—thus the point in the cell cycle when integration takes place might influence the selection of integration target sites. Two HIV integration site data sets were generated after infection of growth-arrested cells [27,28]. These showed only slightly different patterns of integration compared to dividing cells, and both showed favored integration in active transcription units. Thus, for the moment, there is not strong data to support differences in targeting due to infection at different cell cycle stages.
Studies of integration targeting are also critical for gene therapy. Gene therapy has successfully restored function in children with inherited immunodeficiencies [75,76]—however, adverse events have also taken place in which the vector containing the therapeutic trans gene integrated near a proto-oncogene 5′ end and activated transcription, thereby contributing to leukemogenesis [77–81]. Thus there is intense interest in integration targeting in the gene therapy field, focusing on devising safer gene therapy [30,81,82].
Given these intriguing questions, many groups have become interested in profiling integration site distributions. Some methods are summarized below.
2. Overview of the method
In a typical experiment, cells are transduced with the integrating vector of interest and incubated for sufficient time to allow integration of the new DNA sequences. For infection of cultured cells with a retroviral vector, two days is typically sufficient time for integration to take place. When longer incubations are allowed, selection for integration sites that promote growth of the host cells may become a factor. In some cases this may be the main question under study, for example in analyzing possible adverse events during gene therapy.
To allow efficient isolation of integration site junction fragments, it is helpful if possible to maximize the number of integrated copies per cell. Of course, for some protocols this will not be feasible, for example in analyzing samples from patients treated by therapeutic gene transfer.
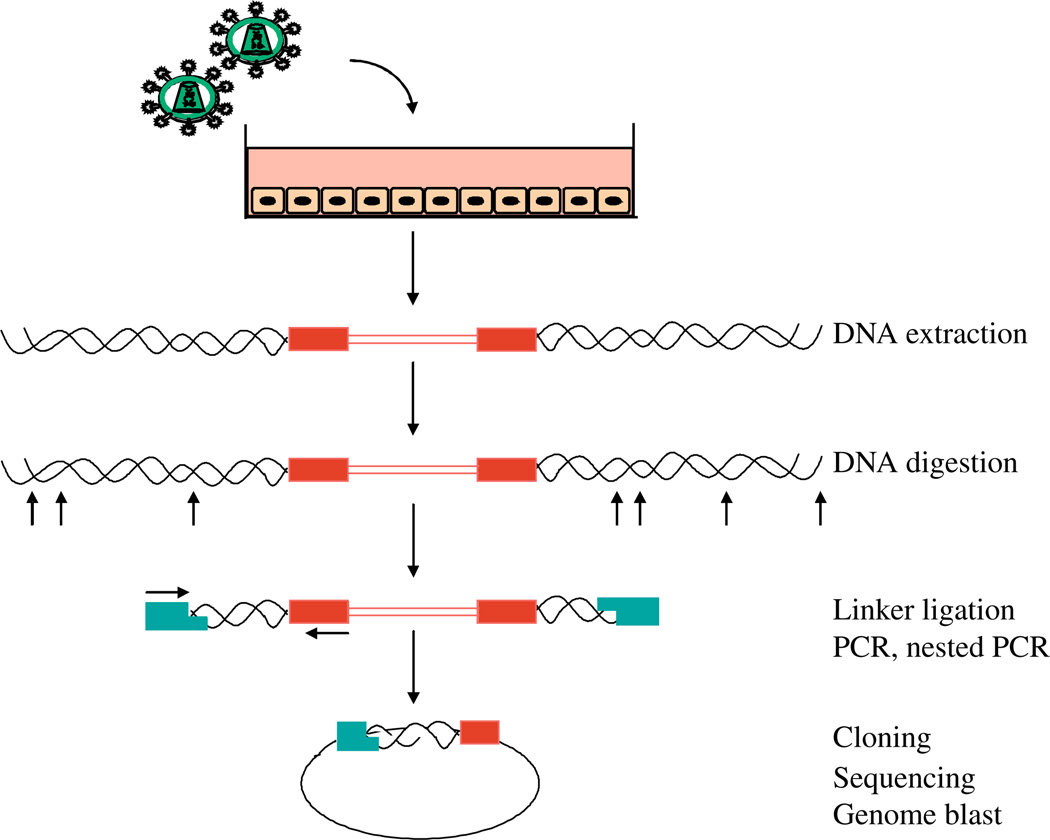
The initial steps of the integration site isolation method are adapted from the Genome Walker Kit (catalog number 1803-1, Clontech, Takara Bio USA, Madison, WI). DNA is extracted from transduced cells, and then cleaved with one or more restriction enzymes (Fig. 1). DNA linkers are then ligated to the cleaved ends. The linkers are specially designed to require PCR amplification to originate within the integrated element, and not at the linker. This is accomplished by attaching a blocking group to one 3′ end within the linker (amino-modifier AmC7-Q), which prevents extension. The 5′ end of the linker contains a single stranded extension. The first amplification primer is identical to the 5′ overhang of the linker, so that the sequence must first be copied by a polymerase primed in the vector. This suppresses linker-to-linker amplification of genomic DNA segments lacking vector sequences.
Fig. 1.
Diagram of the integration site recovery method. Red boxes, viral LTRs; blue rectangles, linker used during ligation.
Considerable care needs to be exercised in the choice of restriction enzyme used for cleaving genomic DNA. For retroviruses or retroviral vectors, the LTR sequence is duplicated. For this reason, primers binding to the LTRs can bind to both copies and allow extension with a polymerase. Special steps are needed to avoid amplification of an “internal fragment” from within the viral vector. This can be achieved by cleaving genomic DNA with a restriction enzyme that does not cleave within the vector, or by using a second enzyme cleavage step after linker ligation to clear out the internal fragment. In addition, restriction cleavage should not be carried out with enzymes that do not yield ends suitable for ligation, nor with enzymes that have the CpG dinucleotide in the recognition sequence (CpG is rare in vertebrate genomes and unevenly distributed).
Two rounds of PCR (nested PCR) are used to recover DNAs containing host–virus DNA junctions. The vector primers need to be designed to lie reasonably close to the host–virus DNA junction, so that sequence reads yield a useful segment of flanking host cell DNA. Amplified vector–host junctions are then cloned and sequenced. Reads are then trimmed to remove plasmid, primer, and viral sequences, aligned with the target genome, and analyzed statistically.
3. Step-by-step protocol
3.1. Objective
A common type of study begins with infecting cells with an HIV-based vector containing a green fluorescent protein (GFP) marker gene, which is pseudotyped with the pantropic envelop glycoprotein G from vesicular stomatitis virus (VSV-G) to provide for a relatively high multiplicity of infection (MOI). The resulting integration sites are then cloned from the infected cells (that is, a segment of host cellular DNA at junctions with HIV) for downstream analyses. To accomplish this, treat viral stocks derived from DNA transfections with RNase-free DNaseI (about 10 U/100 µl viral supernatant) (catalog number 04716728001, Roche Applied Science, Mannheim, Germany) for 1 h at 37 °C (this will decrease plasmid carryover in the subsequent steps), and infect cells with the HIV-derived vector or wild type HIV at high MOI (usually 10; where the MOI was determined on 293T cells, in order to obtain ideally 30–70% of infected cells). Let the cells grow for 48 h to allow integration—this time allows for efficient provirus formation but minimizes selection of cells during growth after integration. Choose a seeding density of cells that will allow you to have a nearly confluent plate after 48 h. Perform fluorescence-activated cell sorting (FACS) with an aliquot of cells to determine the infection rate (proportion of GFP positive cells). Infection rates of 10% or higher are preferred, though it is possible to recover integration sites from samples infected at lower efficiencies.
In the protocol below, besides working up samples of genomic DNA, it is recommended to work up a sample with a model integration site cloned in bacteria, to allow optimization of PCR conditions and genomic DNA cloning steps. In the example below, plasmid SINcPPTeGFP is used for this purpose. We describe a protocol where a cocktail of AvrII, NheI, and SpeI restriction enzymes is used to cleave the genomic DNA.
3.2. Procedure
3.2.1. Genomic DNA extraction
1. Harvest cells (control or HIV-infected), pellet them, and proceed to genomic DNA extraction, following instructions for the DNeasy tissue kit (catalog number 69506, Qiagen, Valencia, CA). Measure the concentration of the purified DNA by spectrophotometry (OD260).
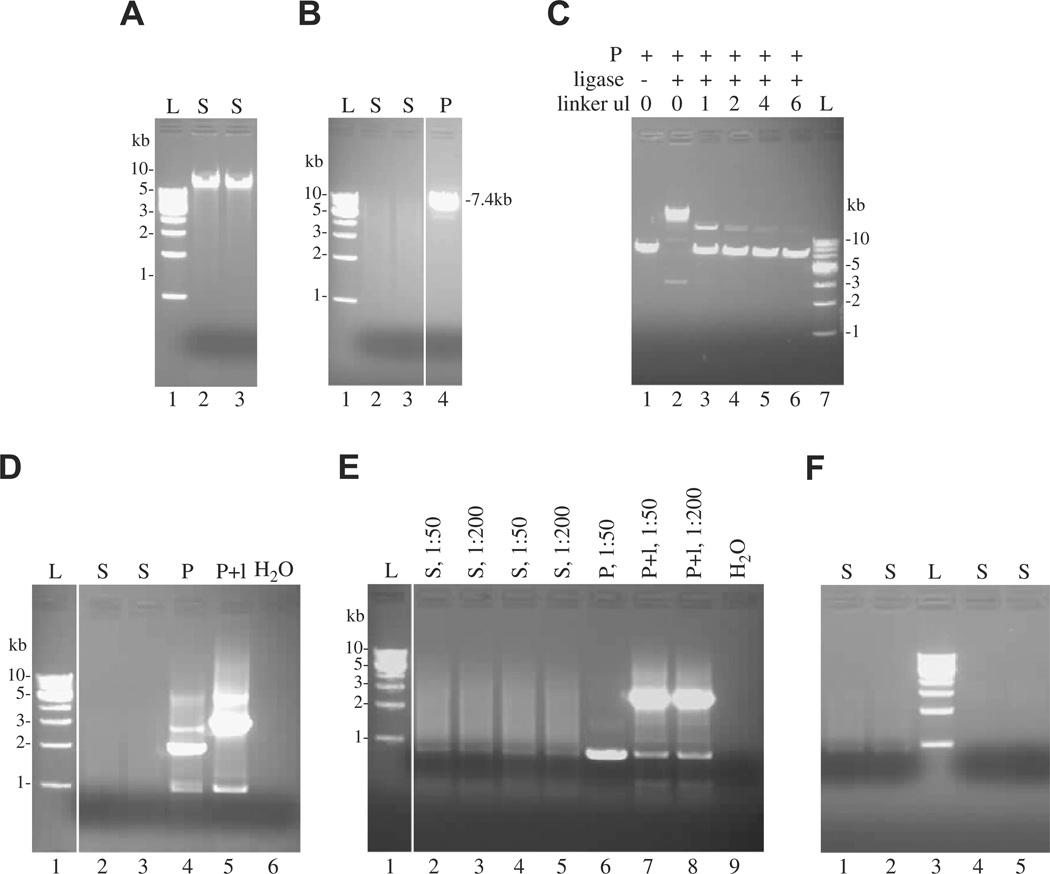
Check 5 µl on an agarose gel (Fig. 2A).
Fig. 2.
Analysis of DNA products by native agarose gel electrophoresis and ethidium bromide staining. (A) Verification of the presence of high molecular weight genomic DNA upon extraction of HIV-infected cells. (B) Efficiency of enzymatic digestion with AvrII+NheI+SpeI, with linearization of the plasmid control (at 7.4 kb). (C) Titration of linker (in µl) in the ligation procedure with the plasmid as control, in presence and in absence of the ligase. Lanes 1 and 4 are used as controls for subsequent PCRs. (D) First PCR with one primer annealing in the LTR and one primer annealing in the linker. The plasmid control should give a band at 3.3 kb. (E) Nested PCR with 1:50 and 1:200 dilutions of the first PCR. (F) Purification of the samples to be subsequently cloned into TOPO TA vector (lanes 1 and 2) or TOPO XL vector (lanes 4 and 5). L, 1 kb DNA ladder; S, sample; P, plasmid SINcPPTeGFP as control; l, linker.
3.2.2. DNA digestions
2. Genomic DNA digestion (enzymes, reaction buffer, and bovine serum albumin (BSA) from New England Biolabs, Ipswich, MA).
1.5 µg genomic DNA
30 µl NEB2 10× reaction buffer
12 µl AvrII (4 U/µl)
6 µl SpeI (10 U/µl)
6 µl NheI (10 U/µl)
3 µl BSA (100× stock)
→ top up to 300 µl with H2O
(digest both infected and uninfected DNA)
3. Plasmid DNA digestion
Digest 20 µg of plasmid pSINcPPTeGFP [83,84] as a control/optimization template.
7 µl NEB2 10× reaction buffer
7 µl AvrII (4 U/µl)
→ add H2O to 70 µl
Digest genomic and plasmid DNAs at 37 °C for 3 h.
Check 5 µl on an agarose gel to ensure proper digestion occurred (Fig. 2B).
3.2.3. DNA purification
Remove proteins via extraction with organic solvents, and recover the digested DNAs by ethanol precipitation.
4. Add 230 µl of H2O to the control plasmid digestion in order to obtain 300 µl final. Add an equal volume (300 µl) of a 1:1 mixture of phenol/chloroform to both digests. Vortex vigorously and centrifuge for 5 min at 13,000 rpm at room temperature in an Eppendorf centrifuge. Transfer the aqueous phase into a new Eppendorf tube. Add 300 µl of chloroform, vortex vigorously, and repeat centrifugation. Transfer the aqueous phase into a new Eppendorf tube.
Add 1/10th volume (30 µl) of 3 M sodium acetate, pH 5.2, and mix (quick vortex). Add 2.5 volumes (825 µl) of ice cold 100% ethanol. Mix by inversion, and incubate on dry ice for 5 min. Centrifuge at 13,000 rpm for 1 h at 4 °C to pellet DNA. Aspirate the supernatant and invert the Eppendorf tube on a kimwipe. Let air dry for 10–30 min. Resuspend the pellet in 10 µl H2O, and let sit a moment to help dissolve the DNA.
5. Alternatively, the DNA can be purified using commercially-available spin columns; we prefer the Strataprep PCR purification kit (catalog number 400771; Stratagene, La Jolla, CA). For this, double the genomic DNA digestion to 3 µg DNA in 600 µl final volume. Purify the DNA using the manufacturer’s recommendations; elute in 50 µl 10 mM Tris pH 8.0. Split the plasmid control digestion onto two columns, and elute each column with 50 µl 10 mM Tris pH 8.0.
3.2.4. Linker preparation
Prepare the linker by annealing two adapter oligonucleotides.
6. The mNAS and HincII adapters are added to the final concentrations of 10 µM each to buffer containing 10 mM Tris pH 8.0, 0.1 mM EDTA. Incubate for 5 min at 90 °C in a PCR machine, then cool 1 °C every 3 min until the temperature reaches ⩽20 °C. Alternatively, switch off the PCR machine, and let the tubes cool down to room temperature over 2–4 h. Store the annealed oligonucleotides at 4 °C until use (but 1 week maximum).
| mNAS adapter | 5′-[Phosp]CTAGGCAGCCCG[AmC7-Q] |
| HincII adapter | 5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGC |
Phosp, 5′ phosphate modification; AmC7-Q, 3′ amino-modifier; underlined, HincII site.
3.2.5. Linker ligation
Ligase and reaction buffer are from New England Biolabs.
7. For genomic DNA, mix:
2 µl 10× ligase buffer
1 µl T4 DNA ligase (400 U/µl)
7 µl adapter mix
10 µl digested genomic DNA in H2O
(20 µl total)
If the DNA was purified using a spin column, modify this step as follows:
4 µl 10× ligase buffer
2 µl T4 DNA ligase (400 U/µl)
1 µl H2O
7 µl adapter mix
26 µl digested genomic DNA
(40 µl total)
8. For plasmid DNA, prepare:
2 µl 10× ligase buffer
1 µl T4 DNA ligase (400 U/µl)
0 or 2 µl adapter mix
1 µl control plasmid linearized with AvrII (2 µg)
→ top up to 20 µl with H2O
For plasmid digests purified by spin column:
2 µl 10× ligase buffer
1 µl T4 DNA ligase (400 U/µl)
0 or 2 µl adapter mix
10 µl control plasmid linearized with AvrII (2 µg)
→ top up to 20 µl with H2O
Incubate the ligation mixtures overnight at room temperature. Add 80 µl of low TE buffer (10 mM Tris pH 8.0–0.1 mM EDTA) to the genomic DNA mixture (60 µl if using the alternate purification) to attain a final DNA concentration of 10–15 ng/µl, and 47 µl low TE buffer for the control plasmid ligation (30 ng/µl final DNA concentration). Check 10 µl on an agarose gel to ensure proper ligation occurred (Fig. 2C).
3.2.6. Nested PCR amplification of linker ligation products
Use two PCR rounds to amplify host–HIV sequences from the ligation reaction mixtures.
9. The first round, PCRI, utilizes HIV LTR-specific primer SB-76 (5′-GAGGGATCTCTAGTTACCAGAGTCACA) and linker-specific primer ASB-9 (5′-GACTCACTATAGGGCACGCGT); use the Advantage 2 PCR Polymerase Mix from Clontech (catalog number 639201). In separate reactions, analyze DNA prepared from: control (mock-infected) cells, HIV-infected cells, and control plasmid digest (processed with versus without the linker in the previous ligation step). It is also important to include a no-DNA (H2O-only) control reaction. Mix:
0.5 µl primer ASB-9 (15 µM) → 300 nM final
0.5 µl primer SB-76 (15 µM) → 300 nM final
2.5 µl 10× 2PCR buffer → 1× final
0.5 µl 50× dNTP stock (10 mM each) → 0.2 mM each final
0.5 µl Advantage 2PCR Polymerase mix
5 µl DNA sample (or H2O)
→ top up to 25 µl with H2O
PCR cycling conditions :
1× 94 °C for 2 s
7× 94 °C, 2 s; 72 °C, 3 min
37× 94 °C, 2 s; 67 °C, 3 min
1× 67 °C, 4 min
Hold 4 °C
Check 5 µl on an agarose gel (Fig. 2D).
10. PCRI samples are diluted at the following levels for analysis in the nested PCRII step: mock-infected cells, 1:50 (2 µl PCRI product + 98 µl H2O) and 1:200 (2 µl PCRI product + 398 µl H2O); HIV-infected cells, 1:50 and 1:200; digested control plasmid (processed without the linker), 1:50; digested control plasmid (processed with the linker), 1:50 and 1:200. Primer ASB-1 (5′-AGCCAGAGAGCTCCCAGGCTCAGATC) is specific for the HIV LTR, whereas ASB-16 (5′-GTCGACGGCCCGGGCTGCCTA) anneals to the linker. Mix:
0.5 µl primer ASB-1 (15 µM) → 300 nM final
0.5 µl primer ASB-16 (15 µM) → 300 nM final
2.5 µl 10× 2PCR buffer → 1× final
0.5 µl 50× dNTP stock (10 mM each) → 0.2 mM each final
0.5 µl Advantage 2PCR Polymerase mix
1 µl DNA sample (dilution of PCRI product, or H2O)
→ top up to 25 µl with H2O
PCR cycling conditions:
1× 94 °C for 2 s
7× 94 °C, 2 s; 72 °C, 3 min
37× 94 °C, 2 s; 67 °C, 3 min
1× 67 °C, 20 min
Hold 4 °C
Check 5 µl on an agarose gel (Fig. 2D).
3.2.7. Gel purification of PCR products
PCRII products are purified following agarose gel electrophoresis using the S.N.A.P. UV-Free Gel Purification Kit (Invitrogen catalog number K2000).
11. Prepare a 0.8% agarose gel in TAE (40 mM Tris base, 20 mM acetate, 1 mM EDTA) buffer (50 ml minigel). Add 40 µl Crystal Violet Solution (2 mg/ml). Add 4 µl 6× Crystal Violet loading dye to 20 µl of PCRII product. Analyze a mixture of 1 µl 6× Crystal Violet loading dye, 2 µl 6× gel loading dye (Promega catalog number G190A, Madison, WI), and 3 µl H2O as a no-DNA marker to allow the visualization of bromophenol blue and xylene cyanol dye migration positions. Load the gel (leave one or two empty lanes between the “marker” and the sample). Use different gels for samples originating from different cell lines to minimize possible cross-contamination between samples. Run about 10 min at 90 V; the crystal violet will migrate towards the negative pole. Isolate a gel piece extending from the bromophenol blue dye to the loading well of the gel, and cut it in two halves: The upper half, containing relatively large 3–6 kb fragments, will be used for cloning with the TOPO XL cloning kit, and the lower half, containing smaller 0.5–3 kb fragments, will be used for cloning with the TOPO TA cloning kit. Weigh the gel pieces, and purify the DNA using the S.N.A.P. kit according to the manufacturer’s instructions. Elute the DNA in 40 µl low TE buffer. Check 10 µl of the purified products on a 1% agarose gel (Fig. 2F).
3.2.8. TOPO cloning and bacterial transformation
We use the TOPO TA cloning kit for sequencing (catalog number K4575) and TOPO XL PCR Cloning kit with OneShot TOP10 electro-competent Escherichia coli (catalog number K4700) from Invitrogen.
12. XL cloning. Mix 4 µl of the purified PCRII XL (3–6 kb DNA fragments) product with 1 µl TOPO XL vector DNA. Incubate at room temperature for 5 min. Add 1 µl of 6× TOPO Stop solution. Quick spin, and place the tube on ice.
13. TA cloning. Mix 4 µl of the purified PCRII TA (0.5–3 kb DNA fragments) product, 1 µl of a 1:4 dilution of Salt Solution, and 1 µl TOPO TA vector. Incubate for 30 min at room temperature. Quick spin, and place the tube on ice.
14. TOP10 electrocompetent cell transformation. Pipet 2 µl of each TOPO reaction into one 50 µl vial of TOP10 bacteria. Do not mix by pipetting, but flick the tube gently. Place the tube back on ice for 5 min. Transfer the mixture into prechilled 2 mm electroporation cuvettes, and place back on ice. Tap the cuvette on the bench to ensure the bacteria are on the bottom of the cuvette, and to eliminate bubbles. Dry the cuvette walls with a kimwipe (this will prevent arcing problems). Electroporate at 2.5 kV (preset bacterial program number 2 on BioRad GenePulser System). Very rapidly add 450 µl (for XL) or 250 µl (for TA) SOC medium, mix by pipetting up and down, and transfer into a polypropylene round bottom tube (catalog number 2059 Falcon, Becton–Dickinson, NJ). Incubate the bacteria at 37 °C for 1 h at 225 rpm. Distribute the XL reaction onto four LBagar-50 µg/ml kanamycin plates: two plates each with 50 µl, and the other two with 200 µl. Also distribute the TA reaction onto 4 LBagar-50 µg/ml kanamycin plates: two plates each with 30 µl, and the remaining two with 120 µl. Incubate the plates overnight (~17 h) at 37 °C.
3.2.9. Colony picking and sequencing
15. Transfer colonies to individual wells of a 96-well plate containing 100 µl LB-50 µg/ml kanamycin, in duplicate (the second plate is prepared as a backup). Incubate the plates overnight at 37 °C. To the backup plate, add 34 µl LB-60% glycerol to each well, and freeze the plate at −80 °C. Submit the other plate to a core facility or company for plasmid extraction and sequencing using M13-reverse (5′-CAGGAAACAGCTATGAC) and M13-forward (5′-TGTAAAACGACGGCCAGT) primers.
4. Processing sequences
Integration site sequences are received from a sequencing center and housed in a MySql database. Sequences are initially trimmed to remove the viral LTR sequence and linker sequence if present. Each sequence is queried against the host cell genome using a local BLAT server. Integration site coordinates (chromosome, base position, and orientation) are calculated based on the best alignment hit for each sequence and entered into the database. Local annotation is then downloaded and added to the integration site record.
The collection of sites is sorted to recover high quality matches. Typically, sequences are required to have (1) a perfect match to the vector terminus, (2) a 98% match to genomic DNA, (3) a unique best hit to the host genome, and (4) a match to host cell DNA beginning within 3 bp of the end of the vector DNA. Trimmed sequences passing these quality controls are stored in separate databases.
A fraction of sequences are found to have high quality matches to multiple sequences in the genome under study. Such multiple hits can be ignored if small in number. In the statistical analysis, the multiple hits can be counted as 1/n of an integration site at each of the n equally high scoring genomic locations.
5. Comments on the statistical analysis
For many statistical comparisons of integration sites, it is natural to compare integration frequency near genomic features to the expectation from random integration. However, there is a danger in using random sites—the experimental integration sites were isolated after cleavage of genomic DNA with restriction enzymes, which was recently shown to introduce a recovery bias [85].
To account for restriction site bias in recovery, the random controls are “matched” to have the same potential bias. Random sites are matched to each experimental integration site so that they are randomly distributed, but constrained to lie the same distance from a restriction enzyme recognition site as the experimental site [23,70]. Comparison of experimental sites to the “matched random controls (MRCs)” washes out any potential restriction bias.
Note that restriction bias can have another consequence, which is that specific sites may be difficult to isolate if the distribution of nearby restriction cleavage sites is unfavorable. This is a major issue in essentially all the gene therapy literature on integration sites. However, this is generally not critical in surveys aimed at characterizing genomic features positively or negatively associated with integration frequency.
A question that often arises is “How many integration sites do I need to sequence?” To call a trend in the data, one needs to have a significant difference between the experimental integration sites and the matched random controls. That is, one needs a p value <0.05, which corresponds to making an erroneous claim for an effect when there is no effect (Type I error) less than one time in 20.
Use of p-values, however conflates the strength of the effect with the sample size. To obtain a significant difference with a small sample, a quite strong difference is required. To obtain significance with a large sample, quite modest differences may often be sufficient.
For example, imagine that you are characterizing integration frequency in transcription units for a newly discovered retrovirus. For the retrovirus, 40% of integration sites are in transcription units, while for the matched random control, only 30% are in transcription units. Whether or not this achieves significance depends on the sample size. If you have 100 integration sites and 100 matched random controls, analysis by Fisher’s exact test (two tailed) yields p = 0.1819, a difference that does not achieve significance. However, if you sequence 1000 integration sites, and prepare 1000 matched random controls, then Fisher’s exact test yields p < 0.0001 for the same difference in proportions. In practice, if possible, it can be helpful to carry out a pilot experiment to obtain an initial indication of the strength of trend of interest, and then scale the full study appropriately.
The section below provides an overview of some of the more advanced aspects of statistical analysis of integration sites, written for readers with statistical training. A comprehensive description can be found in [70].
The expected number of integration events, Y, given genomic location, L, (i.e. chromosome, position, strand) is represented by a log-linear model
where xil is the value of genomic feature ‘i’ at location ‘l’ and βi is an unknown constant governing the effect of that feature. Features include 0/1 indicators for whether ‘l’ is in a gene, exon, or other kind of region of interest, quantitative measures such as position weight matrix scores for the local DNA sequence or counts of genes in neighborhoods of ‘l’, and terms for interactions among such features. Model fitting and inference is based on the conditional logit model as implemented by the clogit function of the R survival library [86] using a ‘nested case-control’ approach (as reviewed by [87]) when control genomic locations are matched to integration sites based on distances to restriction sites for the enzyme(s) used to recover the integration event. When random controls are employed, logistic regression as implemented in the glm R function [88] is used. Logistic regression is also used when two different types of integration events (from integration complexes ‘a’ and ‘b’, say) are compared in a common framework; in terms of the earlier model for expected integration events, the coefficients in the logistic regression, logit(Pr(IC = a|x1, x2, ⋯)) = α + ∑ixijθi, turn out to be θi = βia − βib, allowing their interpretation as the difference in effect of xi on integration of complex ‘a’ versus ‘b’.
In screening quantitative variables, transformation of regressors to a uniform distribution on [−1,1] diminishes the leverage of extreme data points due to highly skewed variables, which makes for easier comparisons of strength of effect of different variables. Also, interpretation of the fitted coefficient is convenient; it gives the effect of increasing the variable by 50 percentile points. Parametric spline functions accommodate non-linear effects; biological considerations (e.g. where transcription starts) sometimes dictates placement of knots.
Some features like gene density and GC density must be tallied in a window (or windows) of specified width(s). The best choice for this (these) window(s) is unknown a priori. We approach this problem by fitting a model that includes basis vectors for a variety of choices and use shrinkage methods to avoid overfitting. This method was especially helpful in revealing that HIV integration sites prefer 50 kb wide GC poor ‘valleys’ surrounded by GC rich regions several megabases in width in the Wang et al., 2007 study [69].
Generally, control of overfitting is achieved by use of penalized likelihood methods [89] or by Bayesian model averaging as implemented in bic.surv [90]. K-fold crossvalidation [91] provides honest estimates of prediction error and is also used for tuning ridge parameters of penalized likelihoods.
The method used to control Type I error due to multiple testing depends on the particular hypothesis/hypotheses and setup. They include Holm’s [92] p-value adjustment when closely related hypotheses are studied (e.g. a dozen different measures of gene density are screened to see if gene density affects integration), omnibus general linear hypothesis tests to compare the profiles of effects of two different integration complexes (which implicitly control Type I error rates by constructing a single overarching test), and estimation of false discovery rates [93] when screening many candidates for useful leads. (It often happens that statistical significance is a secondary concern, because very high levels of significance may be achieved for many features, whose influence ranges from biologically trivial to important.)
These model based methods are supplemented by machine learning techniques [94], especially the randomForest algorithm, which has great flexibility in terms of detecting effects due to complicated combinations of features, resists being driven by a few observations of high leverage, measures the importance of each feature, and provides honest estimates of prediction error automatically.
6. Concluding remarks
The study of integration target site selection has taken off in recent years with the availability of complete gene sequences and new high-throughput methods. In all likelihood ongoing methods development will provide a wealth of new research opportunities, including, for example, the new deep sequencing techniques [95,96].
Acknowledgments
We are grateful to members of the Bushman laboratory for help and suggestions. This work was supported by NIH Grants AI52845 and AI66290 to F.D.B., the University of Pennsylvania Center for AIDS Research, and the Penn Genomic Frontiers Institute. A.C. was supported in part by a fellowship from the Swiss National Science Foundation.
References
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruse. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 2.Bushman FD. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 3.Asante-Appiah E, Skalka AM. Adv. Virus Res. 1999;52:351–369. doi: 10.1016/s0065-3527(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 4.Grandgenett DP. Proc. Natl. Acad. Sci. USA. 2005;102:5903–5904. doi: 10.1073/pnas.0502045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MD, Farnet CM, Bushman FD. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel PH, Preston BD. Proc. Natl. Acad. Sci. USA. 1994;91:549–553. doi: 10.1073/pnas.91.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellison V, Brown PO. Proc. Natl. Acad. Sci. USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bushman FD, Fujiwara T, Craigie R. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 10.Bushman FD, Craigie R. J. Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craigie R, Fujiwara T, Bushman F. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 12.Sherman PA, Fyfe JA. Proc. Natl. Acad. Sci. USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bushman FD, Craigie R. Proc. Nat. Acad. Sci. USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 15.Katzman M, Katz RA, Skalka AM, Leis J. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoder K, Bushman FD. J. Virol. 2000;74:11191–11200. doi: 10.1128/jvi.74.23.11191-11200.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brin E, Yi J, Skalka AM, Leis J. J. Biol. Chem. 2000;275:39287–39295. doi: 10.1074/jbc.M006929200. [DOI] [PubMed] [Google Scholar]
- 18.Lander E. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Venter JC. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 20.Katayama S. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 21.Carninci P. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 22.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewinski M, Bisgrove D, Shinn P, Chen H, Verdin E, Berry CC, Ecker JR, Bushman FD. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. J. Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 27.Ciuffi A, Mitchell RS, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman FD. Mol. Ther. 2006;13:366–373. doi: 10.1016/j.ymthe.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. Mol. Ther. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 29.Lewinski MK, Yamashita M, Emerman M, Ciuffi A, Marshall H, Crawford G, Collins F, Shinn P, Leipzig J, Hannenhalli S, Berry CC, Ecker JR, Bushman FD. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman A. Proc. Natl. Acad. Sci. USA. 2005;102:1275–1276. doi: 10.1073/pnas.0409587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 32.Ciuffi A, Bushman FD. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 2006;22:388–395. doi: 10.1016/j.tig.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Li Y, Crise B, Burgess SM. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 34.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandmeyer S. Proc. Natl. Acad. Sci. USA. 2003;100:5586–5588. doi: 10.1073/pnas.1031802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu Y, Dai J, Fuerst PG, Voytas DF. Proc. Natl. Acad. Sci. USA. 2003;100:5891–5895. doi: 10.1073/pnas.1036705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boeke JD, Devine SE. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 38.Bushman FD. Science. 1995;267:1443–1444. doi: 10.1126/science.7878462. [DOI] [PubMed] [Google Scholar]
- 39.Goulaouic H, Chow SA. J. Virol. 1996;70:37–46. doi: 10.1128/jvi.70.1.37-46.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katz RA, Merkel G, Skalka AM. Virology. 1996;217:178–190. doi: 10.1006/viro.1996.0105. [DOI] [PubMed] [Google Scholar]
- 41.Bushman F, Miller MD. J. Virol. 1997;71:458–464. doi: 10.1128/jvi.71.1.458-464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushman FD. Mol. Ther. 2002;6:570–571. [PubMed] [Google Scholar]
- 43.Tan W, Dong Z, Wilkinson TA, Barbas CF, Chow SA. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushman FD. Proc. Natl. Acad. Sci. USA. 1994;91:9233–9237. doi: 10.1073/pnas.91.20.9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee MS, Craigie R. Proc. Natl. Acad. Sci. USA. 1994;91:9823–9827. doi: 10.1073/pnas.91.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farnet C, Bushman FD. Cell. 1997;88:1–20. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 47.Violot S, Hong SS, Rakotobe D, Petit C, Gay B, Moreau K, Billaud G, Priet S, Sire J, Schwartz O, Mouscadet JF, Boulanger P. J. Virol. 2003;77:12507–12522. doi: 10.1128/JVI.77.23.12507-12522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. EMBO J. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 50.Yung E, Sorin M, Pal A, Craig E, Morozov A, Delattre O, Kappes JC, Ott D, Kalpana G. Nat. Med. 2001;7:920–926. doi: 10.1038/90959. [DOI] [PubMed] [Google Scholar]
- 51.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 52.Turlure F, Devroe E, Silver PA, Engelman A. Front. Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- 53.Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. J. Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Llano M, Delgado S, Vanegas M, Poeschla EM. J. Biol. Chem. 2004;279:55570–55577. doi: 10.1074/jbc.M408508200. [DOI] [PubMed] [Google Scholar]
- 55.Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla EM. J. Cell Sci. 2005;118:1733–1743. doi: 10.1242/jcs.02299. [DOI] [PubMed] [Google Scholar]
- 56.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 57.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 58.Marshall H, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman F. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciuffi A, Diamond T, Hwang Y, Marshall H, Bushman FD. Hum. Gene Ther. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- 60.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panet A, Cedar H. Cell. 1977;11:933–940. doi: 10.1016/0092-8674(77)90304-x. [DOI] [PubMed] [Google Scholar]
- 62.Rohdewohld H, Weiher H, Reik W, Jaenisch R, Breindl M. J. Virol. 1987;61:336. doi: 10.1128/jvi.61.2.336-343.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vijaya S, Steffan DL, Robinson HL. J. Virol. 1986;60:683–692. doi: 10.1128/jvi.60.2.683-692.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pryciak PM, Varmus HE. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 65.Pryciak PM, Sil A, Varmus HE. EMBO J. 1992;11:291–303. doi: 10.1002/j.1460-2075.1992.tb05052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pryciak P, Muller H, Varmus HE. Proc. Natl. Acad. Sci. USA. 1992;89:9237–9241. doi: 10.1073/pnas.89.19.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pruss D, Reeves R, Bushman FD, Wolffe AP. J. Biol. Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 68.Pruss D, Bushman FD, Wolffe AP. Proc. Natl. Acad. Sci. USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berry C, Hannenhalli S, Leipzig J, Bushman FD. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carteau S, Hoffmann C, Bushman FD. J. Virol. 1998;72:4005–4014. doi: 10.1128/jvi.72.5.4005-4014.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roe T, Reynolds TC, Yu G, Brown PO. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamashita M, Emerman M. J. Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamashita M, Emerman M. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, Selz F, Hue C, Certain S, Casanova JL, Bousso P, Deist FL, Fischer A. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 76.Hacein-Bey-Abina S, Le Deist F, Carlier F, Bouneaud C, Hue C, De Villartay JP, Thrasher AJ, Wulffraat N, Sorensen R, Dupuis-Girod S, Fischer A, Davies EG, Kuis W, Leiva L, Cavazzana-Calvo M. N. Engl. J. Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 77.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 78.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, Fischer A. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 79.Thrasher AJ, Gaspar HB, Baum C, Modlich U, Schambach A, Candotti F, Otsu M, Sorrentino B, Scobie L, Cameron E, Blyth K, Neil J, Abina SH, Cavazzana-Calvo M, Fischer A. Nature. 2006;443:E5–E6. doi: 10.1038/nature05219. (discussion E6–7). [DOI] [PubMed] [Google Scholar]
- 80.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, MacIntyre E, Dal Cortivo L, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bushman FD. J. Clin. Invest. 2007;117:2083–2086. doi: 10.1172/JCI32949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Palma M, Montini E, Santoni de Sio FR, Benedicenti F, Gentile A, Medico E, Naldini L. Blood. 2005;105:2307–2315. doi: 10.1182/blood-2004-03-0798. [DOI] [PubMed] [Google Scholar]
- 83.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 84.Zufferey R, Dull T, Mandel R, Bukovsky A, Quiroz D, Naldini L, Trono D. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang GP, Garrigue A, Ciuffi A, Ronen K, Leipzig J, Berry C, Lagresle-Peyrou C, Benjelloun F, Hacein-Bey-Abina S, Fischer A, Cavazzana-Calvo M, Bushman FD. Nucleic Acids Res. 2008;36:e49. doi: 10.1093/nar/gkn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Therneau T, Lumley T. Survival: survival analysis, including penalised likelihood. R package version 2.03. 2006. [Google Scholar]
- 87.Breslow NE. J. Am. Stat. Soc. 1996;91:14–28. doi: 10.1080/01621459.1996.10476660. [DOI] [PubMed] [Google Scholar]
- 88.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2006. [Google Scholar]
- 89.Gray R. JASA. 1992;87:942–951. [Google Scholar]
- 90.Raftery A, Hoeting J, Volinsky C, Painter I. BMA: Bayesian Model Averaging. R package version 3.03. 2006. [Google Scholar]
- 91.Stone M. J. R. Stat. Soc., Ser. B—Methodol. 1974;36:111–147. [Google Scholar]
- 92.Holm S. SJS. 1979;6:65–70. [Google Scholar]
- 93.Efron B, Tibshirani R. Genet. Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 94.Liaw A, Wiener M. R News. 2002;2:18–22. [Google Scholar]
- 95.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bushman FD, Hoffmann C, Ronen K, Malani N, Minkah N, Rose HM, Tebas P, Wang GP. AIDS. 2008;22:1411–1415. doi: 10.1097/QAD.0b013e3282fc972e. [DOI] [PMC free article] [PubMed] [Google Scholar]