Abstract
Chaperones are abundant cellular proteins that promote the folding and function of their substrate proteins (clients). In vivo, chaperones also associate with a large and diverse set of co-factors (co-chaperones) that regulate their specificity and function. However, how these co-chaperones regulate protein folding and whether they have chaperone-independent biological functions is largely unknown. We have combined mass spectrometry and quantitative high-throughput LUMIER assays to systematically characterize the chaperone/co-chaperone/client interaction network in human cells. We uncover hundreds of novel chaperone clients, delineate their participation in specific co-chaperone complexes, and establish a surprisingly distinct network of protein/protein interactions for co-chaperones. As a salient example of the power of such analysis, we establish that NUDC family co-chaperones specifically associate with structurally related but evolutionarily distinct β-propeller folds. We provide a framework for deciphering the proteostasis network, its regulation in development and disease, and expand the use of chaperones as sensors for drug/target engagement.
INTRODUCTION
Perturbation of the proteostasis network has been implicated in many diseases ranging from neurodegeneration, to cancer, to Mendelian disorders (Powers et al., 2009). At the same time, preclinical models and clinical results with drugs that target central modules of the network, such as proteasome or Hsp90 inhibitors, have shown that targeting the network has high therapeutic potential (Trepel et al., 2010). It is clear, however, that we need a more detailed understanding of the proteostasis network to decipher how exactly it is perturbed in disease and to develop more effective and specific therapeutics.
Chaperones are the most prominent class of proteins that shape this network. They transiently bind thousands of substrate proteins (clients) and promote their folding, trafficking, and degradation (Saibil, 2013). Systematic proteomic approaches have started to uncover the client protein ensembles of chaperones (Calloni et al., 2012; Yam et al., 2008; Zhao et al., 2005). However, previous studies have employed widely varying methods and model organisms, making it a challenge to quantitatively compare results and integrate them into a coherent model. Perhaps more importantly, however, chaperones do not function in isolation. Rather, they dynamically associate with a diverse set of cofactors, or co-chaperones. Co-chaperones provide a host of auxiliary functions to chaperones, ranging from regulating the rate of client release to recruiting specific clients to the core chaperone (Echtenkamp and Freeman, 2012). A growing body of evidence suggests co-chaperones play much more than a supportive role. Some “co-chaperones” possess intrinsic chaperone activity themselves (Freeman et al., 1996), and others independently regulate cellular processes that are distinct from those of the canonical chaperones (Echtenkamp et al., 2011). Yet, both the client-protein specificity and possible chaperone-independent functions of most co-chaperones remain enigmatic.
Here, we have taken a systematic and integrative approach, surveying the physical interaction landscape of all known Hsp90 co-chaperones and several known Hsp70 cochaperones. We combine mass spectrometry and quantitative LUMIER assays to characterize the client specificity of co-chaperones and begin to decipher the proteostasis network as a whole.
RESULTS
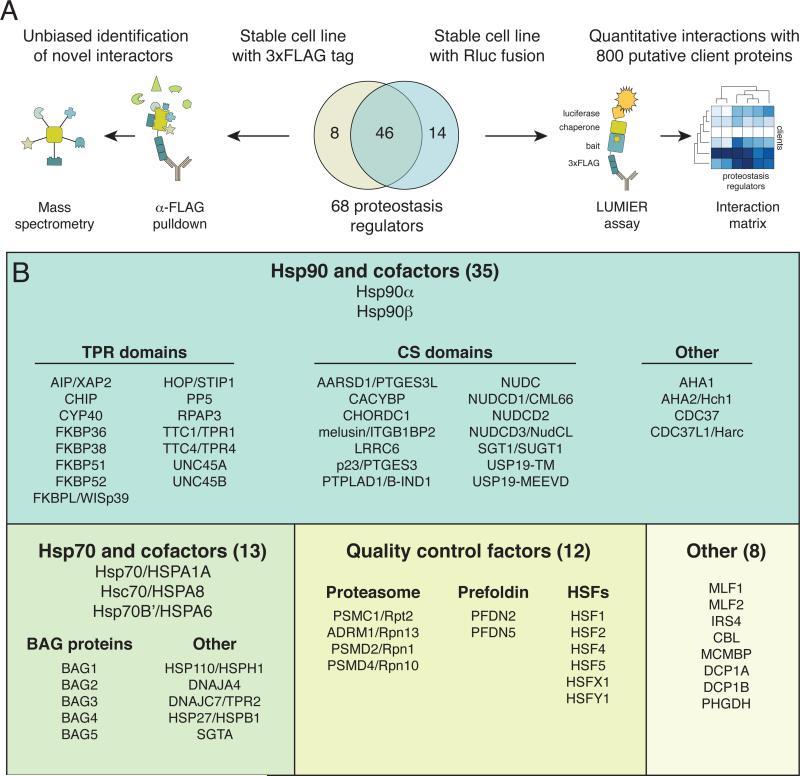
To systematically characterize the cytoplasmic proteostasis network in human cells, we generated two sets of stable 293T cell lines. One set consisted of 54 proteins tagged with a 3xFLAG-V5 epitope for affinity purification coupled to mass spectrometry (AP-MS), the other of 60 proteins fused to Renilla luciferase for a quantitative LUMIER assay (Taipale et al., 2012; Figure 1A). Importantly, almost all surveyed proteins were tagged in the C terminus. This ensured that the interactions represent steady-state post-translational chaperone/client interactions rather than transient co-translational interactions (as nascent chains cannot be captured with a C-terminal tag).
Figure 1. Two-pronged approach for characterizing the proteostasis network in human cells.
A) 67 chaperones, co-chaperones or quality control factors tagged with either 3xFLAG tag or Renilla luciferase were stably expressed in 293T cells. Interactors were identified either by affinity purification followed by mass spectrometry (APMS; 3xFLAG tagged proteins) or by LUMIER assay (Renilla-tagged proteins).
B) Chaperones, co-chaperones, protein quality control factors and other proteins characterized in this study. See also Figure S1.
Proteins were selected for our analysis based on multiple criteria (Figure 1B). First, we cloned all previously identified and characterized Hsp90 co-chaperones and Hsp70 nucleotide-exchange factors. Second, we cloned a group of proteins with either a tetratricopeptide repeat (TPR) domain or a CS (CHORD and Sgt1) domain, as most Hsp90 co-chaperones contain one or more of these domains (Taipale et al., 2010). Third, we selected several other proteostasis regulators, including four subunits of the 19S proteasome regulatory particle, two subunits of the prefoldin complex, and six HSF family transcription factors. Finally, during the course of the project we identified eight prominent co-chaperone interactors, which were cloned for systematic analysis (Figure 1B).
Unbiased discovery of chaperone/cochaperone/client interactions with AP-MS
Co-chaperones were affinity-purified from cell lysates with anti-FLAG beads and interacting proteins identified with mass spectrometry. To distinguish significant interactors from background noise, we used the SAINT algorithm (Teo et al., 2013). To validate the AP-MS interactions with an orthogonal method, we used the LUMIER assay. In this assay, a prey protein fused to Renilla luciferase is stably expressed in 293T cells. Putative interactors (baits) are tagged with a 3xFLAG epitope and transfected into the reporter cell line. Cells lysates are then incubated on 384-well plates coated with an anti-FLAG antibody, leading to capture of the bait protein. Interaction of the bait protein with the tested chaperone can be then quantified as luminescence (Barrios-Rodiles et al., 2005). As a cutoff for high-confidence interactions, we used LUMIER score ≥7 (with an estimated upper bound for FDR 4.4%).
To characterize the validation rate of AP-MS interactions, we cloned 423 interacting proteins that scored SAINT AvgP ≥0.5 in any one of the AP-MS experiments. These proteins were tested for interaction with all 54 baits with LUMIER. Using a stringent cutoff for SAINT (AvgP ≥0.85; estimated FDR 1.8%), 28% of interactions identified in AP-MS were validated by LUMIER, and 81% of them were novel (Figure S1A, S1B and Table S1). Conversely, 35% of interactions that scored positive in LUMIER had an AvgP score ≥0.85 (Figure S1C). These validation rates are consistent with the observation that any one protein/protein interaction assay can detect about one-third of all interactions without compromising specificity (Braun et al., 2009).
Both the validation rate and the overlap with published interactions decreased with lower scores, supporting our selection of a stringent cutoff (Figure S1A, S1B, S1C). We also validated that SAINT was the best computational algorithm for identifying high-confidence interactors in our dataset (Figure S1E). Finally, we investigated if the location of the epitope tag affected the interactions identified by assaying several proteins again with a tag in the other terminus. The results were highly similar (Figure S2A, S2B).
Global features of the AP-MS interaction network: Chaperone complexes
We identified 486 high-confidence interactions for the 54 tagged baits by AP-MS. The number of identified interactions did not correlate with bait protein expression level, suggesting that there were no systematic biases introduced by exogenous expression (Figure S1E). Notably, our chaperone-focused AP-MS network was much larger and more interconnected than chaperone interaction networks that could be recovered from previous large-scale studies (Figure S1F, S2D, and S2E).
The AP-MS network revealed two tiers of organization. The first tier connected all but six bait proteins into a central network with multiple edges between chaperones and their clients (Figure 2). Two sub-networks emerged within this central network, corresponding to known Hsp90 and Hsp70 chaperone complexes (Figure 2, blue and orange squares, respectively). These two sub-networks were bridged by a unique set of co-chaperones (Figure 2, tan squares). Among these were the well-known bridging factors HOP/STIP1, TPR2/DNAJC7 and CHIP/STUB1, validating our approach (Brychzy et al., 2003; Schmid et al., 2012; Xu et al., 2002). Other bridging factors in this first tier of organization included members of the Hsp40 chaperone family (DNAJB1, DNAJB6), the HSP70-binding protein 1 (HSPBP1), the TPR domain protein EDRF1, and the E3 ligase NRDP1/RNF41.
Figure 2. The proteostasis network in human cells characterized by mass spectrometry.
Protein-protein interactions were identified by affinity-purification followed by mass spectrometry (AP-MS), and filtered using the SAINT algorithm with cutoff AvgP ≥ 0.85. Proteins are shown as rectangles, and lines represent interactions between the proteins. Bait proteins are indicated by dark edges. The width of the edges corresponds to the number of spectral counts identified for each interaction. Examples of biologically coherent interactions are indicated in colors. See also Figure S1 and S2.
Local features of the AP-MS interaction network: unique chaperone/client interactions
The second tier of organization consisted of co-chaperone/client interactions. For example, we identified several protein kinases co-purifying with CDC37, a known kinase-specific Hsp90 co-chaperone (Taipale et al., 2012). CDC37L1/Harc, a protein that is 62% similar to CDC37 (Figure S3A; Scholz et al., 2001), similarly interacted very strongly with Hsp90 and several of its co-chaperones (Figure 2, 3B). Otherwise, however, the interactions of these co-chaperones were unique. For example, CDC37L1 interacted with the bridging factor HOP whereas CDC37 co-purified with AHA1 (Figure 3B), but even more strikingly, CDC37L1 did not associate with any kinases in AP-MS (Figure 2, 3B). CDC37L1 lacks the very N-terminus of CDC37, which is required for kinase interaction and the cellular function of CDC37 (Shao et al., 2003), and able to mediate strong interaction with ARAF when fused to CDC37L1 (Figure S3B). Thus, our findings establish that the CDC37L1 has evolved a unique position in the Hsp90 chaperone machinery and that due to its divergent N-terminus CDC37L1 does not associate with kinase clients.
Figure 3. Unique associations of FKBP and BAG family co-chaperones.
A) The FKBP (FK506-binding protein) family of Hsp90 co-chaperones are characterized by one or more TPR domains that can interact with Hsp90 and the FKBP domainchk.
B) The interaction network of FKBP family co-chaperones. Selected unique interaction partners and protein classes are indicated.
C) The natural compound OSW-1 disrupts the interaction between OSBP and FKBP36. 3xFLAG-tagged OSBP was transfected into 293T cells stably expressing FKBP36-Renilla luciferase fusion. One hour before cell lysis, cells were treated with a dilution series of the indicated compounds. The interaction between OSBP and FKBP36 was then measured with LUMIER. Error bars indicate standard deviation.
D) BAG proteins are a family of homologous Hsp70 co-factors that interact with Hsp70 through their BAG domain.
E) The interaction network of BAG family co-chaperones. Selected unique interaction partners are indicated.
F) BAG4 co-localizes with the P body component DCP1A and regulates P body assembly. Top panel: YFP-BAG4 (green) was transfected into HeLa cells, which were then fixed and stained for endogenous DCP1A (red), a component of P bodies. DNA was stained with Hoechst 33342 (cyan). Bottom panel: in cells where YFP-BAG4 is expressed at high levels, endogenous DCP1A appears diffuse. See also Figure S3
Unique associations of FKBP family co-chaperones
Many Hsp90 co-chaperones are members of multi-protein families and share significant homology with each other. One of the most prominent of these co-chaperone groups is the FK506-binding protein family, or FKBPs. These proteins share an FK506-binding domain and one or more TPR domains, which confer interaction with Hsp90 (Figure 3A). Our AP-MS dataset robustly detected shared interactions between these co-chaperones and several members of the Hsp90 chaperone machinery (Figure 3B), but it also revealed distinct associations for each.
FKBP51 (aka FKBP5), but none of the other FKBPs, associated with four distinct protein families, pointing to an unexpectedly diverse repertoire of clients. First, FKBP51 interacted with a subset of the kinases that interacted with CDC37 (Figure 3B). Second, FKBP51 interacted with the Argonaute proteins AGO1 and AGO2, known Hsp90 clients involved in small RNA biogenesis (Iwasaki et al., 2010). Third, FKBP51 associated with three transcription factors (EGLN1, PDCD2, ANKMY2), all of which contain a MYND zinc finger domain, suggesting that this domain represents a novel Hsp90-interacting protein fold.
Perhaps most surprisingly, we found that FKBP51 interacted with MCM4 and MCMBP (Figure 3B), two subunits of the MCM complex involved in DNA replication initiation and fork progression. FKBP51 purification recovered the most peptides for one particular subunit of the complex, MCMBP (Jagannathan et al., 2012). We validated this interaction by affinity purification of 3xFLAG-tagged MCMBP. Indeed, MCMBP interacted with all members of the MCM complex and with FKBP51 (Figure S3C). Hsp90, however, was not enriched in the MCMBP purifications and we did not detect an interaction between Hsp90 and MCMBP by LUMIER assay or by co-immunoprecipitation (Figure S3D, S3E), suggesting that FKBP51 associates with MCMBP independently of Hsp90. Thus, our results reveal an unexpected Hsp90-independent link between FKBP51 and genome maintenance. Corroborating our results, a recent systematic siRNA screen identified FKBP51 as a factor modulating the cellular response to DNA damage (Cotta-Ramusino et al., 2011).
Although their interactomes were significantly more compact than FKBP51, all of the other FKBPs also exhibited unique interactions with other proteins (Figure 3B). For example, the FKBP38 (aka FKBP8) interactome suggested a link to G-protein signaling through interaction with PDCL, which acts as a chaperone for G protein gamma subunits (Lukov et al., 2006; Figure 3B). In contrast, the highly homologous co-chaperone FKBP36 (aka FKBP6) did not interact with PDCL, but instead associated with the oxysterol-binding protein OSBP (Burgett et al., 2011; Figure 3B).
FKBP36 as a sensor for drug/target interactions
We recently reported that Hsp90 and Hsp70 can be used as sensors for drug/target interactions in living cells (Taipale et al., 2013), as they are exquisitely sensitive to the conformational status of the client protein. Binding of a small molecule conformationally stabilizes the protein it targets, decreasing the association of the target with chaperones. Recently, a class of natural products (ORPphilins) with potent activity against cancer cell lines was shown to target OSBP (Burgett et al., 2011). Discovering the OSBP/FKBP36 interaction by AP-MS provided an opportunity to investigate the general applicability of the chaperone assay for new types of drug/target interactions.
We therefore asked if the ORPphilin OSW-1 would disrupt the interaction between FKBP36 and OSBP, as measured by LUMIER. Indeed, OSW-1 treatment led to the dissociation of OSBP from FKBP36 (Figure 3C). The potency of OSW-1 in disrupting the interaction was 60 nM (EC50), very close to the Ki of the compound (26 nM; Burgett et al., 2011). Next, we tested four structural analogs of OSW-1. Two of them (compounds 6 and 7) are active in cellular assays, whereas compounds 9 and 10 are inactive (Figure S3F; Burgett et al., 2011). Consistent with their different cellular effects, compounds 9 and 10 did not disrupt the OSBP/FKBP36 interaction, whereas compounds 6 and 7 did (Figure 3C). Thus, the biological activity of the OSW-1 family compounds is reflected in their ability to disrupt the interaction between OSBP and FKBP36. Disruption of the interaction could be either caused by overlapping binding sites for FKBP36 and OSW-1 or by thermodynamic stabilization of the OSBP fold. In any case, the use of chaperones and co-chaperones as sensors of drug/target interactions is likely to be very broadly applicable.
Unique associations of BAG family co-chaperones
BAG proteins comprise a family of homologous co-chaperones for Hsp70. All regulate Hsp70's ATPase activity, interacting with Hsp70 through a conserved BAG domain in their C-termini (Figure 3D; Kampinga and Craig, 2010). Little is known, however, about their biological functions and whether they contribute to Hsp70 client specificity. Again, our AP-MS results pointed to unique biological connections for at least four of the five family members (Figure 3E).
BAG1 interacted with the E3 ubiquitin ligase Listerin (LTN1), involved in ribosomal quality control (RQC) of stalled polypeptides (Bengtson and Joazeiro, 2010). As BAG1 is known to regulate the degradation of at least some Hsp70 and Hsp90 clients (Tsukahara and Maru, 2010), our results suggest it may be also involved in the degradation of proteins targeted by the RQC. As reported before, BAG3 associated with the small heat shock proteins Hsp22/HSPB8 and Hsp27/HSPB1 (Fuchs et al., 2010). In addition, we detected a robust interaction with HSF1, the master regulator of the heat-shock response (Figure 3E).
BAG5 and BAG4 interacted with protein complexes that have not previously been associated with chaperones. BAG5 purification revealed the spindle checkpoint components Mad1/MAD1L1 and Mad2/MAD2L1 as the most prominent interactors (Schuyler et al., 2012). BAG4, in contrast, interacted with three central components of the mRNA decapping complex, DCP1A, EDC3 and DDX6 (Figure 3E). These three proteins localize to cytoplasmic structures known as processing bodies, or P bodies, which are involved in mRNA decapping, degradation, and translational silencing (Eulalio et al., 2007).
To test whether BAG4 also localizes to P bodies, we transfected HeLa cells with EYFPBAG4. When EYFP-BAG4 was expressed at low levels, it co-localized with endogenous DCP1A in cytoplasmic foci (Figure 3F). Because overexpression of P body components often disrupts P body formation (Eulalio et al., 2007), we also examined cells where BAG4 was expressed at high levels. In such cells, EYFP-BAG4 was localized more diffusely in the cytoplasm and DCP1A localization also became diffuse (Figure 3F, lower panel). Thus, BAG4 overexpression disrupted P body organization. We then investigated if this effect was dependent on the association of BAG4 with Hsp70. We introduced a point mutation in the BAG domain of BAG4 (D424A) that disrupts its association with Hsp70 (Briknarová et al., 2002 and Figure S3G). However, mutant BAG4 still localized to P bodies and disrupted P body organization at high expression levels (Figure S3H). Our data thus establish BAG4 as a novel component of P bodies and suggest that this function is independent of Hsp70.
Quantitative profiling of the human chaperone/client landscape
The network we uncovered by AP-MS was surprisingly compact and likely represented only the most abundant chaperone/client interactions. To further expand the network and to investigate more quantitatively how client proteins are integrated within it, we systematically surveyed pairwise interactions between clients and co-chaperones with LUMIER. To this end, we constructed a panel of 800 query proteins (Table S1). This set included known Hsp90 clients (www.picard.ch), a subset of kinases, E3 ligases and transcription factors that we previously identified as Hsp90 clients (Taipale et al., 2012), and the 423 proteins that we used for characterizing the overlap between AP-MS interactors and LUMIER assay (see above). After filtering out proteins that were not expressed at detectable levels, our dataset comprised 40,604 pairwise assays, each performed in duplicate.
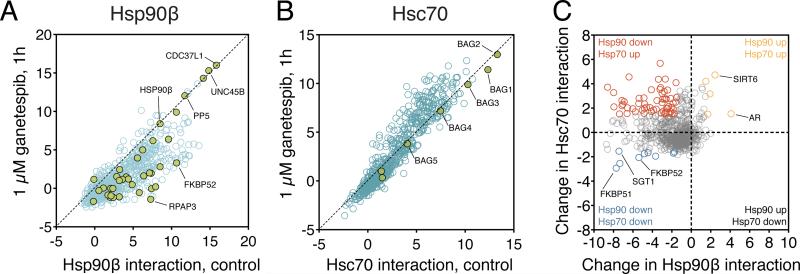
We first examined the effects of the Hsp90 inhibitor ganetespib on Hsp90 and Hsp70 interactions (Ying et al., 2012). This served two purposes. First, Hsp90 inhibition leads to dissociation of most known clients from Hsp90 (Taipale et al., 2012). Although dissociation does not directly prove that the interactor is a client, it provides supportive evidence. Second, this allowed us to test the generality of client handoff from Hsp70 to Hsp90. Individual, earlier studies have shown that Hsp90 inhibition is accompanied by accumulation of clients with the upstream chaperone Hsp70 (Xu et al., 2002). Our platform enabled us systematically test this model. To this end, cells expressing Renilla tagged Hsp90β or Hsc70 were transfected with the bait protein collection and treated with 1 μM ganetespib for 1 hour before LUMIER assay.
Ganetespib treatment had a strong effect on most Hsp90/client interactions. Of the 630 unique proteins that we detected in LUMIER assay, 46% significantly decreased their interaction with Hsp90β (change in LUMIER score >1.5, adjusted p-value <0.05; Figure 4A). Using a binary cutoff for Hsp90β interactions (LUMIER score ≥7), 81% of high-confidence interactors (84/104) decreased their interaction with the chaperone (Table S1). Notably, one third (7/20) of those that still interacted with Hsp90β were known cochaperones (Figure 4A, Table S1). Yet, even some co-chaperones lost their interaction with Hsp90. This is consistent with the observation that Hsp90 inhibitors stabilize a specific conformation of the chaperone, leading to differential co-chaperone interactions (Gano and Simon, 2010). The effect of ganetespib on Hsc70 interactions was more subtle but still clearly detectable. 16% of tested proteins increased their interaction with Hsc70 upon ganetespib treatment. This was particularly noticeable for proteins that interacted strongly with Hsc70 (Figure 4B). None of the tested Hsp70 co-chaperones were affected by inhibitor treatment (Figure 4B).
Figure 4. Effect of transient Hsp90 inhibition on chaperone interactions.
A) Hsp90 inhibition leads to dissociation of most clients from Hsp90β. Hsp90β was surveyed for interaction with 800 proteins with LUMIER assay. Cells were treated for 1 hour with 1 µM ganetespib or left untreated before the assay. Hsp90 co-chaperones are shown as green circles. Interaction strength was quantitated as LUMIER scores. B) Hsp90 inhibition leads to stronger association of some proteins with Hsc70. Hsc70 was assayed for interaction with 800 proteins as in (A). Hsp70 cochaperones are shown as green circles.
C) Comparison of the effects of ganetespib on Hsp90β and Hsc70 interactions. The plot shows the change in interaction of 800 tested proteins with Hsp90β and Hsc70 (change is defined as LUMIER score drug – LUMIER score control). Proteins that show differential association with both chaperones are indicated in orange (both increase), blue (both decrease) or red (decrease in Hsp90β interaction, increase in Hsc70 interaction). Selected proteins are labeled.
We then compared the effects of ganetespib on Hsp90β and Hsc70 interactions (Figure 4C). Most proteins that decreased their association with Hsp90β did not significantly change their interaction with Hsc70. Those that did, however, generally associated more strongly with Hsc70 (red circles, Figure 4C). This group of proteins was enriched in kinases (p < 0.0001, Fisher's exact test; Table S1). A few, mostly co-chaperones, decreased their interaction with both Hsp90β and Hsc70 (blue circles, Figure 4C). Interestingly, five proteins increased their interaction with both Hsp90 and Hsp70 after drug treatment (orange circles, Figure 4C). They might represent clients chaperoned in a distinct manner.
Taken together, these experiments demonstrate that Hsp90 inhibition leads to an almost global loss of Hsp90/client interactions. This is accompanied by a more subtle increase in Hsp70 interaction for many clients, in particular kinases. The results also suggest that the vast majority of these interactions are true chaperone/client interactions.
Unbiased clustering of proteostasis regulators by protein interaction profiles
The application of quantitative analyses has often revealed unexpected associations between genes in gene expression and genetic interaction profiles (Eisen et al., 1998; Tong et al., 2004). The quantitative readout of our assay allowed us to exploit this approach for the analysis of protein-protein interactions. That is, rather than employing statistical cutoffs to determine significant interactions, we used the entire dataset and treated interaction scores as quantitative variables. Although LUMIER scores do not directly correspond to biophysical parameters such as affinity or stoichiometry, correlations in the interaction profiles can reveal novel relationships between proteins.
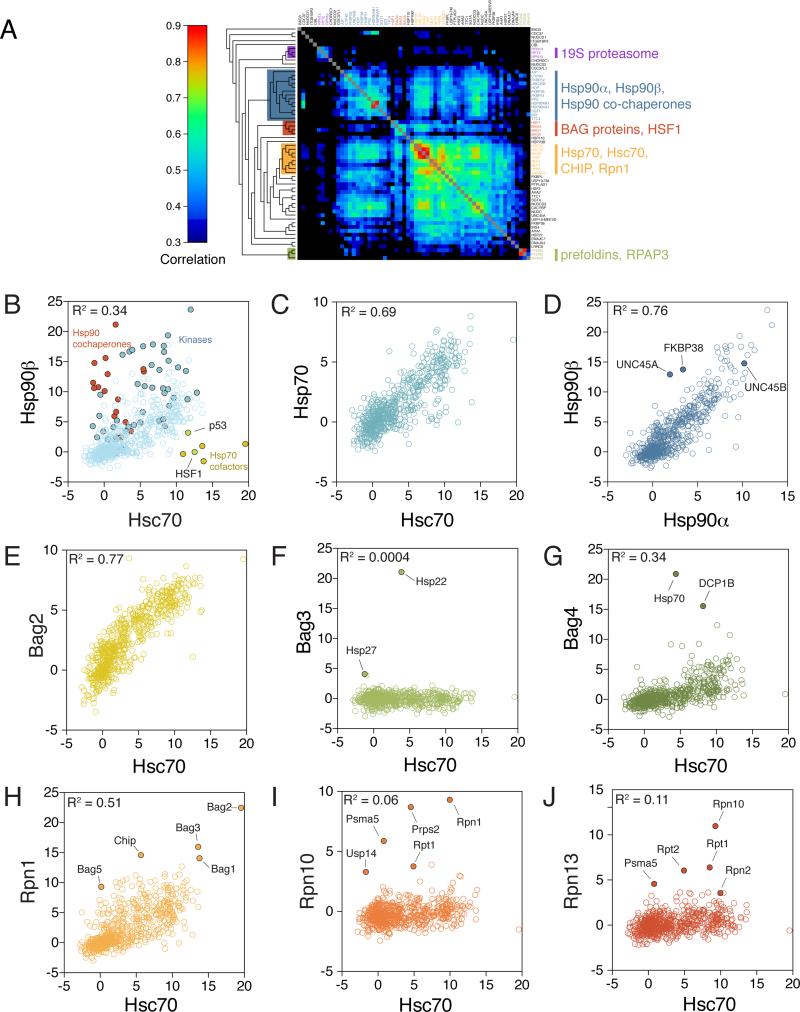
We first clustered chaperones and co-chaperones based on their similarities in client-interaction profiles. This recovered well-known biological complexes (Figure 5A). For example, Hsp70 and Hsp90 machineries formed distinct clusters: Hsp70 clustered together with Hsc70, their nucleotide-exchange factor BAG2, and the E3 ligase CHIP (Figure 5A, orange cluster), whereas Hsp90 and many of its co-chaperones formed a separate group (Figure 5A, blue cluster). Similarly, three components of the proteasome regulatory particle (PSMD4, PSMC1 and ADRM1) clustered together, as did the two subunits of the prefoldin complex, PFDN2 and PFDN5 (Figure 5A). Notably, some of the clusters formed by LUMIER interactions were different from those recovered in APMS. RPAP3 interacted strongly with the Hsp90 machinery in AP-MS, but its interaction profile in LUMIER connected it more tightly with prefoldin subunits. The most likely reason is that the chaperone complexes in AP-MS were mainly connected by interactions between complex members, whereas LUMIER clusters tend to be determined by shared interactions of cluster members. The recovery of well-established, biologically coherent chaperone clusters interacting with a diversity of known substrate proteins validates our approach.
Figure 5. Clustering of chaperones, co-chaperones and quality control based on similarities in client interaction profiles.
A) Chaperones, co-chaperones and protein quality control factors were clustered based on their interaction profiles with 800 query proteins. Clustering corresponds well to known biological complexes. Three components of the 19S proteasome (purple) cluster together, as do two prefoldin subunits (green). Hsp70 and its cofactors (yellow) and BAG proteins (red) cluster separately from Hsp90 and its cochaperones (blue).
B) Significant correlation between Hsc70 and Hsp90 client interaction profiles. 800 proteins (light blue) were assayed for interaction with Hsc70 or Hsp90 by LUMIER. Hsp90 co-chaperones (red) interact exclusively with Hsp90 and Hsp70 co-chaperones (ochre) with Hsc70. Kinases (filled blue circles) interact generally more strongly with Hsp90 than with Hsc70, whereas p53 and HSF1 (green) prefer Hsc70.
C) The client interaction profiles of Hsp70 isoforms Hsc70 and Hsp70 are highly similar.
D) The client interaction profiles of Hsp90 isoforms Hsp90α and Hsp90β are highly similar. Most clients (blue) interact with Hsp90α and Hsp90β to a similar degree. The co-chaperones UNC45A and FKBP38 (filled blue circles) interact more strongly with Hsp90β. In contrast, UNC45A paralog UNC45B interacts with both isoforms to a similar degree.
E) BAG2 interaction profile is almost identical to that of Hsc70, suggesting that it is a general co-factor for Hsp70 chaperones. In contrast, BAG3 (F) and BAG4 (G) are more specific, interacting primarily with small heat shock proteins Hsp22 and Hsp27 (BAG3) and the mRNA decapping factor DCP1B (BAG4)
H) Rpn1 is a component of the proteasome regulatory particle and its interaction profile correlates significantly with that of Hsc70. In contrast, Rpn10 (I) and Rpn13 (J), also components of the core particle, mainly interact with other subunits of the proteasome. See also Figure S4
Paralogous chaperones have very similar client and co-chaperone interactions
We first examined the interaction profiles of the Hsp90 and Hsp70 family chaperones. As expected, co-chaperones showed distinct specificities for Hsp90 (Figure 5B, red circles) or Hsp70 (Figure 5B, ochre circles). However, most clients interacted with both Hsp90β and Hsc70 to a similar degree (Figure 5B, light blue circles). Consequently, the interaction profiles of the chaperones were themselves correlated (R2=0.34). Yet, some clients clearly preferred one chaperone over the other. Hsp90β interacted particularly strongly with kinases (Figure 5B, filled blue circles), whereas the transcription factors p53 and HSF1 were among the most Hsp70-biased interactors (Figure 5B, green circles).
We then compared the interaction profiles of different Hsp70 and Hsp90 isoforms. Virtually all eukaryotic genomes encode multiple isoforms of both of these chaperones, even within the same cellular compartment (Powers and Balch, 2013). However, it is not known if they have distinct client-protein preferences. The three cytoplasmic Hsp70 isoforms we profiled (Hsp70, Hsc70, and Hsp70B’) had very similar interaction profiles across the 800 proteins we tested (Figure 5C, S4B). Similarly, the interaction profiles for Hsp90α and Hsp90β correlated strongly (R2=0.76; Figure 5D). A few co-chaperones had strong isoform preferences. BAG proteins interacted more strongly with Hsp70B’ than with Hsp70 or Hsc70 (Figure S4B and data not shown). The Hsp90 co-chaperone UNC45A, but not its homolog UNC45B, had been previously described as an Hsp90β-specific co-chaperone (Chadli et al., 2008) and this held true in our assay. We detected a similar preference for FKBP38 (Figure 5D). We further validated the isoform preference of UNC45A and FKBP38 by co-immunoprecipitation with the endogenous Hsp90 isoforms (Figure S4A). These outliers provide an avenue for investigating distinctions in the functions of specific chaperone isoforms. By and large, however, Hsp70 and Hsp90 isoforms interacted with the same clients and co-chaperones, and did so with very similar affinities.
BAG2 and RPN1 are tightly connected to the Hsp70 machinery
Our AP-MS interaction network suggested unique roles for each of the BAG proteins except for BAG2 (Figure 3E). Illustrating the power of combining unbiased AP-MS analysis with the quantitative nature of the LUMIER assay, we uncovered a striking interaction pattern for BAG2. Across the 800 queried proteins, BAG2 interactions were remarkably similar to those of Hsc70 (R2=0.77; Figure 5E). In contrast, the other BAG proteins had highly specific interactions that complemented our results from AP-MS. BAG3 showed strong association only with the small heat-shock protein Hsp27 (Figure 5F), and BAG4 with the mRNA decapping complex member DCP1B (Figure 5G), while BAG1 interacted with several proteasome subunits (Figure S4C). Thus, BAG2 appears to be a general co-factor for Hsp70 with little client protein preference, whereas other BAG proteins have unique clients.
The interaction profile of RPN1/PSMD2, a subunit of the proteasome regulatory particle, was also highly correlated with that of Hsc70 (R2=0.51; Figure 5H). RPN1 interacted strongly with four BAG proteins and with the ubiquitin ligase CHIP. This was in contrast to the three other subunits (RPN10, RPN13, RPT2) that primarily interacted with other proteasome subunits (Figure 5I, 5J, S4E). Although all four subunits are part of the base of the proteasome regulatory particle (Lander et al., 2012), RPN1 clustered together with Hsp70 rather than with the other proteasome subunits (Figure 5A). RPN1 is thought to act as a scaffold protein that binds and recruits diverse proteasome-associated factors to the proteasome (Finley, 2009). The correlation between RPN1 and Hsc70 interaction profiles suggests that RPN1 could also serve as a bridge between protein folding by the Hsp70 machinery and protein degradation by the proteasome.
Another factor that correlated well with Hsc70 was the ubiquitin ligase CHIP (Figure S4E). CHIP binds Hsp90 and Hsp70 with similar affinities and can regulate the degradation of chaperone clients (Kundrat and Regan, 2010). However, LUMIER revealed an interaction profile that correlated with Hsc70 (R2=0.74) even more strongly than with Hsp90β (R2=0.38; Figure S4F, S4G). Indeed, hierarchical clustering placed it together with Hsp70 proteins rather than with Hsp90 (Figure 5A). This finding suggests that CHIP is most tightly coupled to the Hsp70 chaperone machinery.
Quantitative client profiling reveals unique co-chaperone specificities
We next focused on the client specificity of chaperones and co-chaperones, and clustered the 800 tested clients based on their LUMIER interaction patterns. This analysis revealed several client groups that shared specific chaperones or co-chaperones (Figure 6). For example, seven members of the cytoplasmic RNA polymerase assembly complex, R2TP, interacted with the prefoldin subunits PFDN2 and PFDN5. However, the R2TP subunits formed two distinct clusters (Figure 6B). The four proteins in the first cluster (PFDN2 itself, VBP1, UXT, and PDRG1) are all prefoldin-like proteins and interacted primarily with PFDN2 and PFDN5. The second cluster (URI1, POLR3A, and RPAP3) interacted also with RPAP3 and Hsp90 in addition to the prefoldins (Figure 6B). Yet another distinct prefoldin interaction module consisted of G protein beta subunits, linking prefoldins to GPCR signaling (Figure 6J). While prefoldins have generally been thought to participate primarily in the folding of actin and tubulin (Lundin et al., 2010), our results expand the specificity of this little-characterized chaperone system.
Figure 6. Quantitative view of the human protein folding landscape.
800 query proteins (arranged in columns) were assayed for interaction with 60 different chaperones, co-chaperones and quality control factors (rows) with a quantitative LUMIER assay. Query proteins were clustered based on their interaction profiles. Some of the biologically coherent clusters are highlighted in more detail. Proteins that share the same fold or are part of the same biological complex in each cluster are indicated in color. (A) LRR proteins (red) and Argonaute proteins (orange) form distinct clusters. LRR proteins interact strongly with SGT1, while Argonaute proteins associate with PP5. (B) The R2TP complex members (purple) form two separate clusters. (C) Hsp90 co-chaperone cluster. (D) Kinases (orange) cluster together and interact specifically with CDC37 but not with CDC37L1. (E) NUDCD1 associates with DEAH/DEAD box helicases (green). (F) BAG proteins that cluster together interact strongly with Hsp70 proteins, Rpn1, Hsf1 and Hsf2. (G) Kelch domain protein cluster (brown) with NUDCD3. (H) Proteasome cluster. (I) RCC1 repeat protein FBXO24 (purple) interacts with NUDCD2. (J) G protein [.gamma] subunits (green) interact with prefoldins.
Hierarchical clustering of the LUMIER data revealed several additional co-chaperone modules. Proteins with kinase domains clustered together, as was expected from their known preference for CDC37 (Figure 6D). Leucine-rich repeat (LRR) proteins and Argonaute proteins also formed distinct clusters. LRR proteins interacted particularly strongly with the SGT1 co-chaperone, whereas Argonaute proteins bound the protein phosphatase PP5 and p23, both well-characterized Hsp90 co-chaperones (Figure 6A).
NUDC family co-chaperones associate with distinct β-propeller folds
Perhaps the most striking client specificity we uncovered by LUMIER involved the NUDC family of co-chaperones. The human genome encodes four evolutionary related proteins in this family (Figure S5A). NUDC proteins have been found to associate with the Hsp90 complex, but the biological roles of these co-chaperones are largely unknown (Zheng et al., 2011).
We noticed that three of the four NUDC proteins associated with a group of proteins that had no functional relationship to each other, yet contained structurally related folds. NUDC interacted strongly with WD40 repeat proteins (Table S1). NUDCD3, in contrast, interacted with proteins with Kelch domains (Figure 6G and Table S1). For NUDCD2, the most significant interacting protein was FBXO24, an RCC1 repeat protein (Figure 6I). Since WD40, Kelch, and RCC1 domains all have β-propeller folds, we reasoned that NUDC proteins might represent a novel family of β-propeller specific co-chaperones.
To more rigorously test the specificity of NUDC proteins, we cloned into our LUMIER vector 275 genes that contained a predicted β-propeller domain. In addition, we cloned 156 genes with leucine-rich repeats (LRR), because several LRR proteins clustered together as SGT1-interacting clients (Figure 6A). Finally, we included as controls 80 kinases that were strong Hsp90 and Cdc37 clients (Taipale et al., 2012). We quantitatively assayed the interaction of these 511 proteins with SGT1, CDC37, and all four NUDC family members (Figure 7A). Except for NUDCD1, each co-chaperone showed a striking client preference. As expected, CDC37 interacted virtually exclusively with kinases (p<0.0001, Mann-Whitney test; Figure 7A). Similarly, although SGT1 interacted with some non-LRR proteins, its interactions with LRR proteins were significantly stronger than with other domains (p<0.0001). NUDC selectively associated with WD40 repeats, NUDCD2 with RCC1 repeats, and NUDCD3 with Kelch domains (p<0.0001 for each; Figure 7A). Although there was additional weak crosstalk between some of these co-chaperones and their clients, these specific associations stood out. We further validated the specificity of NUDC and NUDCD3 with AP-MS. Endogenous NUDC but not NUDCD3 co-purified with 3xFLAG tagged WD40 protein FBXW2, whereas endogenous NUDCD3 (but not NUDC) co-purified with five different 3xFLAG-tagged Kelch domain proteins in 293T cells (Figure S5B).
Figure 7. NUDC family proteins are specific co-chaperones for β-propeller domains.
A) NUDC family co-chaperones, SGT1, and CDC37 were assayed with a quantitative LUMIER assay for interaction with 80 kinases, 156 LRR domain proteins, and 275 proteins with a β-propeller domain. Bait proteins are organized by domain (annotated below) and rank-sorted based on their interaction with a specific co-chaperone (kinases with CDC37, LRRs with SGT1, WD40 with NUDC, RCC1 with NUDCD2, Kelch with NUDCD3).
B) Co-chaperones recognize specific domains in their clients. Indicated full-length proteins or truncated constructs were tagged with a 3xFLAG epitope and transfected into 293T cells. Their interaction with endogenous Hsp90 (top panel) or with endogenous, specific co-chaperone (middle panel) was assayed by co-immunoprecipitation. For NUDCD2, a 3xHA tagged construct was co-transfected with FBXO24 and the blot probed with an anti-HA antibody.
C) Evolution of the NUDC protein family and their client specificity. NUDC, NUDCD2 and NUDCD3 each recognize distinct β-propeller folds. NUDCD1, in contrast, associates with proteins with an unrelated fold (RNA helicases and the COPI complex), but it itself contains a β-propeller domain. The COPI complex image used with permission from Science Magazine.
D) Genomes that encode the Kelch-domain specific co-chaperone NUDCD3 contain significantly more proteins with Kelch domains than genomes without NUDCD3. The number of Kelch domains was analyzed in each of 147 fully sequenced eukaryotic proteomes. Normalized Kelch domain abundance (as a fraction of total number of proteins) was compared between species that have NUDCD3 ortholog and those that do not. Histograms display the t-statistic distribution for 10,000 random eukaryotic proteins that were assayed for similar evolutionary cooccurrence with Kelch domains. Orange line shows the t-statistic for NUDCD3 and black lines show the t-statistic for three other co-chaperones that are not Kelch-specific.
E) Evolutionary analysis of LRR domain evolution with LRR-specific co-chaperone SGT1, performed as in D. See also Figure S5.
We then tested if the co-chaperone specifically bound the kinase, β-propeller, or LRR domain by co-immunoprecipitation (Figure 7B and S5A). As reported before, CDC37 interacted with the kinase domain of ARAF and SGT1 interacted with the LRR domain of FBXL2 (Figure 7B). NUDC interacted with the WD40 domain of FBXW2; NUDCD2 with the RCC1 domain of FBXO24; and NUDCD3 with the Kelch domain of KLHL38 (Figure 7B). In each case, the interaction with these isolated domains was as strong as with the full-length protein. These results establish that the evolutionarily related NUDC co-chaperones recognize a specific β-propeller fold in their clients (Figure 7C).
NUDCD1 was the only member of the NUDC family that did not interact with β-propeller domain proteins in our query set. However, AP-MS and LUMIER revealed that it did interact strongly with multiple DEAH/DEAD box RNA helicases and several subunits of the COPI complex, which regulates retrograde signaling between Golgi compartments (Figure 2 and 6E). Although two members of the COPI complex (COP-[.alpha] and COP-[.gamma]2) contain β-propeller domains, we could not detect an interaction between these subunits and NUDCD1 (data not shown). Interestingly, however, in contrast to other NUDC proteins, NUDCD1 itself contains a β-propeller domain (Figure 7C and S5A), connecting also this co-chaperone to β-propeller domains.
Co-chaperones may facilitate the evolutionary diversification of protein folds
We next asked whether the emergence of fold-specific co-chaperones might have enabled the diversification of their client protein folds during evolution. Chaperones with broad client specificities can promote evolution by providing a buffering mechanism against destabilizing mutations (Jarosz et al., 2010; Tokuriki and Tawfik, 2009). Domain-specific co-chaperones might facilitate evolution in a similar, but domain-specific manner. We therefore analyzed the genomes of 147 fully sequenced organisms. We calculated the number of proteins with co-chaperone-specific protein folds (LRR, WD40, Kelch, and RCC1) and asked if the number of such proteins was larger in genomes that contain the specific co-chaperone compared to those without the co-chaperone (after controlling for nonspecific expansion of the proteome). Genomes that contained the NUDCD3 or SGT1 co-chaperones showed a striking, and highly significant, enrichment for their client folds (Kelch and LRR, respectively; Figure 7D and 7E). The associations of NUDC and NUDCD2 with their cognate client folds were not statistically significant (data not shown). These results suggest that the evolution and diversification of LRR domains and Kelch repeats may have been promoted by the emergence of co-chaperones specific to these folds.
DISCUSSION
We have systematically and quantitatively characterized the human chaperone/co-chaperone/client interactome in human cells. The broad and quantitative nature of our approach allowed the analysis of protein-protein interaction data by hierarchical clustering, illuminating unexpected and highly specific connections between chaperones and particular biological processes. We validated many novel interactions by orthogonal interaction assays and by functional assays, but as in any endeavor of this size, a large fraction of the network remains unexplored. We highlight here only the most salient insights our analysis uncovered, to encourage others to explore this resource in their own investigations.
Co-chaperones and protein complex assembly
Systematic studies of the components of the proteostasis networks have almost exclusively focused on chaperones rather than their co-factors. Our data suggest a surprisingly diverse role for co-chaperones in particular cellular processes, including spindle assembly (BAG5 and MAD proteins), DNA replication (FKBP51 and the MCM complex), mRNA decapping (BAG4 and P bodies), and GPCR signaling (prefoldins and G protein [.beta] subunits). These cellular processes are completely unrelated, yet conceptually they share key features. The co-chaperone interactors are individual components of much larger multiprotein complexes, and in each case, these complexes must be assembled in a specific location at a specific time. Further, their assembly is often regulated by dramatic conformational changes in the associated proteins.
Our results suggest that co-chaperones are broadly involved in the assembly of multiprotein complexes. On the one hand, co-chaperones provide a means to recruit the Hsp90 or the Hsp70 chaperone system to very specific biological processes. In contrast to Hsp70, which generally recognizes unfolded proteins with exposed hydrophobic stretches, co-chaperones associate with proteins that have specific domains. Presumably, they associate with domains that are at least transiently recognizable yet retain a level of conformational flexibility that guides them to the chaperone machinery. Recruitment of Hsp70 by these co-chaperones would thus create a local pool of the chaperone to facilitate transitions between conformational states. On the other hand, certain co-chaperone interactions are independent of the core chaperones (e.g. FKBP51 and the MCM complex) and likely serve specific roles that do not require extensive structural rearrangements driven by chaperones. It will be of great interest to determine whether those functions evolved from the initial chaperone interaction or vice versa.
Domain-specific co-chaperones, client protein recognition and protein fold evolution
Our analysis revealed that like the well-known kinase specificity of CDC37, other cochaperones also have distinct specificities. SGT1 interacts particularly strongly with clients with leucine-rich repeats. (This had been suggested previously (Kadota et al., 2010), but awaited systematic testing.) We also uncovered previously unsuspected specificities for the poorly characterized NUDC family co-chaperones. These evolutionarily related co-chaperones recognized distinct but structurally homologous β-propeller domains (Figure 7C). The interaction patterns of all the domain-specific cochaperones were analogous to that of CDC37. That is, although the co-chaperones clearly preferred a specific protein fold, neither they nor Hsp90 associated with all members of that family. Furthermore, the clients did not phylogenetically cluster, but were scattered throughout the evolutionary tree (Figure S5C). For those with which they did interact, the strength of interactions varied over a broad continuum (Figure S5C). Thus, they point to a dynamic process of evolutionary diversification still at work.
In addition to their specific co-chaperones, many β-propeller proteins interacted with Hsp90, Hsp70, and prefoldins. WD40 domain proteins have also been shown to associate with the TRiC/CCT chaperonin (Yam et al., 2008). Why might β-propeller domains require so many chaperones and a dedicated system of co-chaperones? The canonical β-propeller structure provides a clue. β-propellers are composed of repeating units of four antiparallel beta-sheets arranged around a ring. β-sheets from the last repeat are often circularly permuted to the first repeat, closing the ring. We suggest that these proteins have a particularly high requirement for chaperone proteins to keep the β-propeller soluble before ring closure.
β-propellers from different families share very little sequence homology, and therefore it has been difficult to resolve whether they evolved from a common ancestral fold or if the fold is an example of convergent evolution (Chaudhuri et al., 2008; Hudson and Cooley, 2008). Our results suggest common ancestry is the more parsimonious scenario. That is, the evolution of β-propeller folds by duplication and diversification was facilitated by the evolutionary expansion of the NUDC family. This seems more plausible than the alternative scenario, where unrelated repeats happened to converge on the same fold while requiring highly related NUDC chaperones.
Our evolutionary analysis revealed that Kelch and LRR domains co-evolve with their cochaperones NUDCD3 and SGT1, respectively. Eukaryotic proteomes with these cochaperones contain a larger fraction of respective client protein folds than proteomes without them. Although it is possible that these co-chaperones evolved as a response to an expanded repertoire of client protein folds, we consider it more likely that the emergence of NUDCD3 and SGT1 promoted the divergence of the clients. That chaperones can buffer genetic variation in several model organisms and in experimental evolution supports this interpretation (Jarosz et al., 2010; Tokuriki and Tawfik, 2009).
Concluding remarks
The architecture of the chaperone/co-chaperone/client interaction network reported here has broad implications for human biology and medicine. A large fraction of human diseases, from cystic fibrosis to cancer and to neurodegeneration, is now known to ultimately stem from problems in the folding of specific proteins. But these proteins do not misfold in isolation. Rather, their misfolding ramifies at a system-wide level to impinge on critical cellular functions (Powers et al., 2009), derailing the homeostatic control of protein folding, trafficking, and degradation in a tissue-specific manner. The interaction network we have uncovered provides a robust framework for systematically dissecting the effects of such perturbations and for characterizing the unique features of the network in different tissues and cellular states.
In addition, these interactions can be exploited to study drug/target interactions in living cells (Taipale et al., 2013). We further expanded this approach here to another co-chaperone, FKBP36. It is very likely that many chaperone/client interactions will prove amenable to such an analysis. Furthermore, given that the approach relies on fundamental biophysical principles rather than a specific functional readout, it should be generally applicable to proteins that have been traditionally difficult to assay for small molecule binding. Thus, our results provide not only a springboard for deciphering how the protein homeostasis network is dynamically rewired in various disease states, but also a platform for evaluating the therapeutic potential of small molecules that could ameliorate these perturbations.
EXPERIMENTAL PROCEDURES
Clones and cell lines
All clones originated from the human ORFeome collection 7.1 or they were cloned from cDNA with PCR. Clones were transferred into a mammalian expression vector with a 3xFLAG-V5 epitope tag or a pLenti6-based lentiviral vector containing a 3xFLAG-V5 epitope tag or Renilla luciferase protein. Stable polyclonal 293T cell lines were established by lentiviral infection and expression of each protein was verified by western blotting or by luciferase assay. Mutant constructs were created with site-directed mutagenesis. All clones used in the study were validated by sequencing or by restriction digestion.
LUMIER assay
LUMIER assay was performed as previously described (Taipale et al., 2012) with one modification. Instead of calculating the prey/bait (luminescence/ELISA) ratio, we used normalized luminescence Z-scores as a quantitative interaction measure. ELISA values were used to remove bait proteins that were not detectably expressed the assay (see Extended Experimental Procedures for details).
AP-MS
AP-MS was performed as previously described (Kean et al., 2012) except that sodium molybdate was included in the lysis buffer to help preserve chaperone client interactions. Samples were analyzed on AB SCIEX 5600 TripleTOF by data dependent acquisition (DDA). See Extended Experimental Procedures for details and for AP-MS data analysis.
Hierarchical clustering
Co-chaperones and client proteins were organized by hierarchical clustering with average linkage and centered Pearson correlation. Only bait proteins that were detectably expressed (as measured by ELISA) in at least 50 of the 60 experiments were included in the analysis.
Datasets
All datasets can be accessed online at http://prohits-web.lunenfeld.ca. All interactions have also been submitted to BioGrid and the IMEx consortium (imexconsortium.org) through IntAct (Orchard et al., 2014) and assigned the identifier IM-22301.
Supplementary Material
HIGHLIGHTS.
Client interactions for >60 chaperones and co-chaperones mapped by AP-MS and LUMIER
Characterization of cellular roles of co-chaperones and their client specificities
NUDC family co-chaperones associate with β-propeller domains (Kelch, WD40 and RCC1)
Co-chaperones may promote the evolutionary diversification of client folds
ACKNOWLEDGEMENTS
We thank Y. Freyzon and M. Fischer for technical help, M. Shair for OSW-1, and A. Nesvizhskii and members of the Lindquist and Gingras labs for comments and suggestions. We are grateful to M. Vidal and D. Hill for providing the human ORFeome. The website prohits-web.lunenfeld.ca is designed and maintained by J. Zhang and F. Liu. This work was supported by the Canadian Institutes of Health Research (MOP-84314 to ACG) and US National Institutes of Health (5R01GM94231 to ACG and GM081871 to BB). MT was supported by HFSP and Margaret and Herman Sokol Postdoctoral Award. ACG is the Canada Chair in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics. SL is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
MT, SL, and ACG conceived the project and designed experiments. MT and IK established stable cell lines and performed all LUMIER assays. ZYL performed AP-MS experiments. BL provided mass spectrometry guidance to ZYL and helped develop the AP-MS method. GT and JP, supervised by BB, developed the LUMIER scoring algorithm and JP performed co-evolutionary analyses. HC performed statistical analyses of AP-MS data. MT and SL wrote the paper with significant contributions from ACG, GT and JP. GT and JP contributed equally to this work.
REFERENCES
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, De Smet A-S, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Meth. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briknarová K, Takayama S, Homma S, Baker K, Cabezas E, Hoyt DW, Li Z, Satterthwait AC, Ely KR. BAG4/SODD protein contains a short BAG domain. J Biol Chem. 2002;277:31172–31178. doi: 10.1074/jbc.M202792200. [DOI] [PubMed] [Google Scholar]
- Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WMJ. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. Embo J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgett AWG, Poulsen TB, Wangkanont K, Anderson DR, Kikuchi C, Shimada K, Okubo S, Fortner KC, Mimaki Y, Kuroda M, et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nature Chemical Biology. 2011;7:639–647. doi: 10.1038/nchembio.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang H-C, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Chadli A, Felts SJ, Toft DO. GCUNC45 Is the First Hsp90 Co-chaperone to Show {alpha}/{beta} Isoform Specificity. J Biol Chem. 2008;283:9509–9512. doi: 10.1074/jbc.C800017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri I, Söding J, Lupas AN. Evolution of the beta-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- Cotta-Ramusino C, McDonald ER, Hurov K, Sowa ME, Harper JW, Elledge SJ. A DNA damage response screen identifies RHINO, a 9-1-1 and TopBP1 interacting protein required for ATR signaling. Science. 2011;332:1313–1317. doi: 10.1126/science.1203430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echtenkamp FJ, Freeman BC. Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochim Biophys Acta. 2012;1823:668–673. doi: 10.1016/j.bbamcr.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Echtenkamp FJ, Zelin E, Oxelmark E, Woo JI, Andrews BJ, Garabedian M, Freeman BC. Global Functional Map of the p23 Molecular Chaperone Reveals an Extensive Cellular Network. Mol Cell. 2011;43:229–241. doi: 10.1016/j.molcel.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI. Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Fuchs M, Poirier DJ, Seguin SJ, Lambert H, Carra S, Charette SJ, Landry J. Identification of the key structural motifs involved in HspB8/HspB6-Bag3 interaction. Biochem J. 2010;425:245–255. doi: 10.1042/BJ20090907. [DOI] [PubMed] [Google Scholar]
- Gano JJ, Simon JA. A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol Cell Proteomics. 2010;9:255–270. doi: 10.1074/mcp.M900261-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. Phylogenetic, structural and functional relationships between WD- and Kelch-repeat proteins. Subcell. Biochem. 2008;48:6–19. doi: 10.1007/978-0-387-09595-0_2. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 Chaperone Machinery Mediates ATP-Dependent RISC Loading of Small RNA Duplexes. Mol Cell. 2010:1–8. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Jagannathan M, Sakwe AM, Nguyen T, Frappier L. The MCM-associated protein MCM-BP is important for human nuclear morphology. J Cell Sci. 2012;125:133–143. doi: 10.1242/jcs.089938. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu Rev Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci. 2010;35:199–207. doi: 10.1016/j.tibs.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean MJ, Couzens AL, Gingras A-C. Mass spectrometry approaches to study mammalian kinase and phosphatase associated proteins. Methods. 2012;57:400–408. doi: 10.1016/j.ymeth.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry. 2010;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukov GL, Baker CM, Ludtke PJ, Hu T, Carter MD, Hackett RA, Thulin CD, Willardson BM. Mechanism of assembly of G protein betagamma subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J Biol Chem. 2006;281:22261–22274. doi: 10.1074/jbc.M601590200. [DOI] [PubMed] [Google Scholar]
- Lundin VF, Leroux MR, Stirling PC. Quality control of cytoskeletal proteins and human disease. Trends Biochem Sci. 2010;35:288–297. doi: 10.1016/j.tibs.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers E, Morimoto R, Dillin A, Kelly J, Balch W. Biological and Chemical Approaches to Diseases of Proteostasis Deficiency. Annu Rev Biochem. 2009 doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Powers ET, Balch WE. Diversity in the origins of proteostasis networks - a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Mol Cell Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid AB, Lagleder S, Gräwert MA, Röhl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M, et al. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. Embo J. 2012;31:1506–1517. doi: 10.1038/emboj.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz GM, Cartledge K, Hall NE. Identification and characterization of Harc, a novel Hsp90-associating relative of Cdc37. J Biol Chem. 2001;276:30971–30979. doi: 10.1074/jbc.M103889200. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Wu Y-F, Kuan VJ-W. The Mad1-Mad2 balancing act - a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci. 2012;125:4197–4206. doi: 10.1242/jcs.107037. [DOI] [PubMed] [Google Scholar]
- Shao J, Irwin A, Hartson SD, Matts RL. Functional dissection of cdc37: characterization of domain structure and amino acid residues critical for protein kinase binding. Biochemistry. 2003;42:12577–12588. doi: 10.1021/bi035138j. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of hsp90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Whitesell L, Santagata S, Zhang J, Liu Q, Gray NS, Lindquist S. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotech. 2013;31:630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras A-C, Choi H. SAINTexpress: Improvements and additional features in Significance Analysis of Interactome software. J Proteomics. 2013;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tsukahara F, Maru Y. Bag1 directly routes immature BCR-ABL for proteasomal degradation. Blood. 2010;116:3582–3592. doi: 10.1182/blood-2009-10-249623. [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam AY, Xia Y, Lin H-TJ, Burlingame A, Gerstein M, Frydman J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Du Z, Sun L, Foley KP, Proia DA, Blackman RK, Zhou D, Inoue T, Tatsuta N, Sang J, et al. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol Cancer Ther. 2012;11:475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu Y-C, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zheng M, Cierpicki T, Burdette AJ, Utepbergenov D, Janczyk PŁ, Derewenda U, Stukenberg PT, Caldwell KA, Derewenda ZS. Structural features and chaperone activity of the NudC protein family. J Mol Biol. 2011;409:722–741. doi: 10.1016/j.jmb.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.