SUMMARY
In bacteria, most secretory proteins are translocated across the plasma membrane by the interplay of the SecA ATPase and the SecY channel. How SecA moves a broad range of polypeptide substrates is only poorly understood. Here, we show that SecA moves polypeptides through the SecY channel by a “push and slide” mechanism. In its ATP-bound state, SecA interacts through a two-helix finger with a subset of amino acids in a substrate, pushing them into the channel. A polypeptide can also passively slide back and forth when SecA is in the predominant ADP-bound state, or when SecA encounters a poorly interacting amino acid in its ATP-bound state. SecA performs multiple rounds of ATP hydrolysis before dissociating from SecY. The proposed “push and slide” mechanism is supported by a mathematical model and explains how SecA allows translocation of a wide range of polypeptides. This mechanism may also apply to hexameric polypeptide-translocating ATPases.
INTRODUCTION
Many bacterial proteins, including most secretory proteins, are transported across the plasma membrane by a process that is similar to protein translocation across the endoplasmic reticulum membrane in eukaryotes (for review, see Park and Rapoport, 2012). Translocation occurs through a channel that is formed from a conserved heterotrimeric membrane protein complex, called the SecY complex in bacteria and archaea, and the Sec61 complex in eukaryotes. The complex consists of a large α-subunit (SecY or Sec61p) that spans the membrane ten times, and two smaller β- and γ-subunits (called SecG and SecE in bacteria) (Van den Berg et al., 2004). In bacteria, the SecY channel can either associate with the ribosome to translocate proteins during their synthesis (co-translational translocation), or it can cooperate with the cytosolic ATPase SecA to transport polypeptides after completion of their synthesis (post-translational translocation).
The SecA ATPase uses the energy of ATP hydrolysis to transport polypeptides through the SecY channel. SecA is a multi-domain protein (Figure S1; available online) that contains two nucleotide-binding domains (NBD1 and NBD2), which bind the nucleotide at their interface and move relative to one another during the ATP hydrolysis cycle (Hunt et al., 2002). SecA also contains a polypeptide-crosslinking domain (PPXD), a helical wing domain (HWD), and a helical scaffold domain (HSD). The latter consists of a long helix and two shorter ones that form a two-helix finger (Figure S1). When SecA binds to the SecY channel, the two-helix finger inserts into the cytoplasmic opening of the channel (Zimmer et al., 2008). A translocating polypeptide chain passes by the loop between the two helices before entering the SecY channel (Bauer and Rapoport, 2009). This fingertip loop contains a conserved Tyr residue that is required for efficient translocation (Erlandson et al., 2008a).
SecA is related to polypeptide-translocating hexameric ATPases. These ATPases include the regulatory subunit of the proteasome, p97/VCP/Cdc48p, and the Clp proteins. Each subunit in a hexameric ring contributes a loop that protrudes into the central pore and contains a conserved aromatic residue (DeLaBarre et al., 2006; Glynn et al., 2009; Hinnerwisch et al., 2005; Martin et al., 2008; Wang et al., 2001). During the ATPase cycle, the loop is thought to move inside the central pore and push the polypeptide chain through the hexameric ring. It has been proposed that a similar mechanism applies to SecA, with the two-helix finger functioning analogously to the central loops of the hexameric ATPases (Erlandson et al., 2008a).
How SecA and the hexameric ATPases move polypeptides chains is only poorly understood. All these ATPases translocate a large number of substrates, and it is unclear how they interact with a broad range of different amino acid sequences. It is also unknown whether the ATPases have a defined step size, i.e. translocate a fixed number of amino acids of the polypeptide substrate during each ATP hydrolysis cycle, analogously to how helicases move along nucleic acid strands (Pyle, 2008). However, the tracks of helicases consist of regularly spaced phosphate groups. In contrast, polypeptide substrates would present different residues during each translocation step, and it is unclear whether amino side chains or the polypeptide backbone are recognized. In the case of SecA, it is not even clear whether SecA is processive, i.e. whether it remains bound to the SecY channel until the entire polypeptide chain is translocated. It is also conceivable that SecA molecules continuously bind and dissociate from SecY, with each binding event resulting in the translocation of a polypeptide segment. How the ATP hydrolysis cycle is utilized to move a polypeptide chain is also only poorly understood. Specifically, it is unknown whether SecA interacts with polypeptide substrates in their ATP- or ADP states, or even in both states.
Here, we have developed new methodology that allows us to study the dynamic interaction of SecA with SecY during translocation. We show that polypeptide movement through the SecA-SecY complex is driven by a combination of passive sliding and occasional “pushing” by SecA’s two-helix finger. This “push and slide” model explains how SecA can translocate polypeptides containing a wide range of sequences.
RESULTS
SecA shows moderate processivity during translocation
Our strategy to study SecA function is to use translocation intermediates, thus avoiding the complicated and non-synchronous assembly of substrate, SecA, and SecY channel during initiation of translocation. The prior assembly of a translocation intermediate makes it possible to study SecA during the actual transport process, with all substrate molecules starting translocation at the same time.
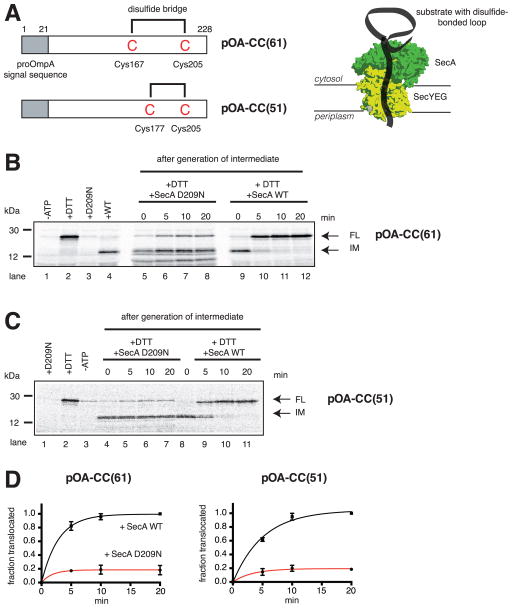
To generate a translocation intermediate, we used derivatives of proOmpA (pOA), a protein that is post-translationally secreted into the periplasm of E. coli. pOA-CC(51) and pOA-CC(61) contain disulfide bonded cysteines separated by 28 and 38 amino acids, respectively, followed by a segment of 23 residues (Erlandson et al., 2008b) (Figure 1A). These proteins were synthesized in vitro in the presence of 35S-methionine, denatured in urea, and incubated with E. coli SecA and proteoliposomes containing E. coli SecY complex. For both substrates, subsequent protease treatment resulted in the generation of a fragment (Figures 1B and 1C, lane 4), which is significantly smaller than the size of the full-length protein (lane 2). The size difference originates from the fact that the disulfide- bridged loop in the substrates is too large to move through the SecY channel, leaving the C-terminus accessible to the protease. When dithiothreitol (DTT) was added to reduce the disulfide bridge, no intermediate was formed; the C-terminus was completely translocated, resulting in protection of the full-length protein (Figures 1B and 1C, lane 2).
Figure 1. SecA is moderately processive.
(A) Generation of translocation intermediates. The substrates pOA-CC(61) and pOA-CC(51) contain a signal sequence (grey box) and cysteines (C) at the indicated positions, which form a disulfide-bonded loop that prevents complete transport through the complex of SecA and SecYEG (shown in green and yellow, respectively).
(B) A translocation intermediate was generated with pOA-CC(61), wild type (WT) SecA, ATP, and proteoliposomes containing SecYEG. Then, a 64fold molar excess of either the dominant-negative mutant SecA D209N or WT SecA was added together with DTT. Samples were taken at the indicated time points, treated with proteinase K, and analyzed by SDS-PAGE and autoradiography. In lanes 1–4, the indicated components were added or omitted before generation of the intermediate. IM and FL indicate protease-protected fragments, corresponding to the intermediate and fully translocated pOA-CC(61), respectively. Molecular weight markers are indicated (in kDa).
(C) As in (B), but with pOA-CC(51).
(D) Quantification of the experiments in (B) and (C). Plotted is the amount of FL relative to the final levels of FL seen with WT SecA (means and standard deviations of three experiments).
We used the translocation intermediates to investigate whether SecA is processive. In one extreme, SecA would remain bound to the SecY channel throughout translocation of a polypeptide chain, in the other extreme, it would dissociate and rebind during each ATP hydrolysis cycle. To test SecA dissociation during translocation, we employed a dominant-negative SecA mutant (SecA D209N) (Economou et al., 1995), which can replace wild type SecA from the SecY channel (Figure S2A). The SecA mutation is in the Walker B motif and affects primarily ATP-hydrolysis (Economou et al., 1995).
When DTT and additional wild type SecA were added, the translocation intermediates gradually disappeared, and full-length substrates appeared instead (Figures 1B, lanes 9–12 and Figure 1C, lanes 8–11). In the presence of a 64-fold excess of SecA D209N, only about 20% of all substrate molecules completed translocation (Figure 1B lanes 5–8 and Figure 1C, lanes 4–7; quantification in Figure 1D). Importantly, SecA D209N addition at the beginning of the translocation reaction prevented the generation of the intermediate (Figure 1B, lane 3; Figure 1C, lane 1), demonstrating that it is indeed a dominant inhibitor. Similar results were obtained with proOmpA derivatives in which the segment following the second cysteine was extended (pOA-CC(120) and pOA-CC(160); Figures S2B,C; quantification in Figures S2D,E). These results show that some molecules of SecA never dissociate from the SecY channel during the translocation of the C-terminal tail of the substrates, while others dissociate and rebind. Thus, SecA exhibits a moderate degree of processivity.
Rebinding of SecA to the SecY channel facilitated by lipid interaction
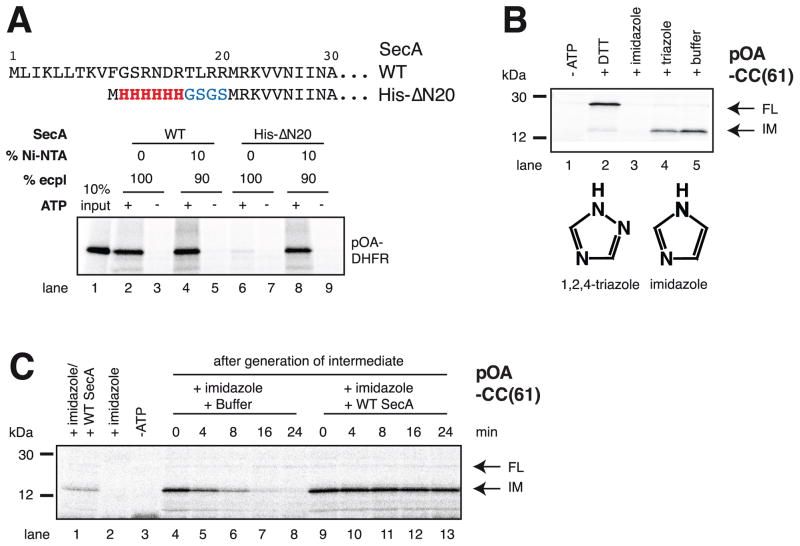
Next we asked how dissociated SecA rebinds to the translocating SecY channel. SecA can bind directly to the SecY complex in detergent, but it can also interact with negatively charged lipids (Hendrick and Wickner, 1991). Based on the crystal structure of the SecA-SecY complex (Zimmer et al., 2008), lipid binding is probably mediated by the N-terminus of SecA, as this is the only domain that could come close to the lipid surface (Figure S1A). We therefore removed the first 20 amino acids of SecA and substituted them with a 6-His tag and short linker (SecA His-ΔN20). In contrast to wild type SecA, SecA His-ΔN20 did not support translocation of a 35S-methionine-labeled substrate into proteoliposomes containing SecY complex and E. coli polar lipids (Figure 2A, lane 6). However, translocation activity was restored to wild type levels when the proteoliposomes also contained 10% Ni-NTA lipids, allowing artificial tethering of SecA His-ΔN20 to the membrane surface (Figure 2A, lanes 8 versus 4). Similarly, a translocation intermediate could be formed with SecA His-ΔN20 and Ni-NTA lipid-containing proteoliposomes, but not if the incubation was performed in the presence of imidazole (Figure 2B, lanes 5 versus 3). An inactive imidazole analog (1,2,4-triazole) did not inhibit the formation of the intermediate (Figure 2B, lane 4). The processivity of artificially tethered SecA was about the same as that of wild type SecA (Figures S3A,B). Finally, we found that SecA ΔN20 did not float with liposomes containing acidic lipids, in contrast to wild type SecA (Figures S3C,D). Taken together, these experiments show that the first 20 amino acids are required for lipid interaction, but can be replaced by an artificial membrane tether.
Figure 2. SecA rebinds to the translocating SecY channel through prior interaction with lipids.
(A) N-terminal sequences of wild type (WT) SecA and of the His-ΔN20 mutant. These proteins were incubated with radiolabeled substrate (a fusion of proOmpA and DHFR; pOA-DHFR), ATP, and proteoliposomes containing SecYEG and either 100% E. coli polar lipids (ecpl) or 90% ecpl and 10% Ni-NTA lipids. Translocation of pOA-DHFR was assessed by proteinase K treatment followed by SDS-PAGE and autoradiography. 10% of the input material was loaded in lane 1.
(B) SecA His-ΔN20 was incubated with radiolabeled pOA-CC(61), ATP, and Ni-NTA proteoliposomes containing SecYEG. Hexokinase/glucose (-ATP), DTT, imidazole, or triazole were added, as indicated, before the start of the translocation reaction. The samples were analyzed as in (A). IM and FL indicate protease-protected fragments, corresponding to the intermediate and fully translocated pOA-CC(61), respectively. Molecular weight markers are indicated (in kDa). The lower panel shows the structures of imidazole and triazole.
(C) As in (B), but imidazole was added after generation of the translocation intermediate, together with buffer or WT SecA. In lanes 1–3, the indicated components were added before generation of the intermediate.
To test whether SecA requires lipid interaction for rebinding to the SecY channel, we generated a translocation intermediate with pOA-CC(61), SecA His-ΔN20, and proteoliposomes containing Ni-NTA lipids, and then added imidazole; if SecA His-ΔN20 dissociates from SecY, it would not be able to rebind through lipids. The dissociation of SecA from translocating SecY channels can be determined in backsliding experiments, in which the C-terminal disulfide-bonded loop in the substrate is maintained (by omitting DTT) and protease is added at different time points (Erlandson et al., 2008b). When SecA is lost from SecY, the polypeptide chain is no longer pushed into the channel, the disulfide loop can move backwards, and the protease can cleave at many different sites, generating a heterogeneous population of protected chains that can no longer be visualized as a defined band. The experiment with SecA His-ΔN20 showed that the fragment corresponding to the translocation intermediate indeed gradually disappeared (Figure 2C, lanes 4–8). In contrast, no backsliding was observed when wild type SecA was added together with imidazole (Figure 2C, lanes 9–13). These data confirm that SecA can dissociate from the translocating SecY channel and show that its interaction with lipids is important for rebinding.
SecA-SecY interaction during translocation
To directly measure the interaction between SecA and translocating SecY, we developed a fluorescence resonance energy transfer (FRET) assay. SecA with a unique cysteine was labeled with the FRET acceptor fluorophore Cy5. The SecY complex was modified with the donor fluorophore tetramethyl-rhodamine at a unique cysteine introduced into the cytoplasmic loop of SecG. The labeled residues have an expected distance of ~30Å (Zimmer et al., 2008). SecA and SecY complex were quantitatively labeled and retained their translocation activity (Figure S4A).
For the FRET experiments, we used the translocation substrate pOA-DHFR, a fusion of the first 175 amino acids of proOmpA and E. coli dehydrofolate reductase (DHFR) (Bauer and Rapoport, 2009). In the presence of methotrexate, the C-terminal DHFR moiety forms a tightly folded domain. When pOA-DHFR is translocated into proteoliposomes, an intermediate is generated, in which the large DHFR domain remains outside the vesicles (Figure 3A). Essentially all SecY channels could be saturated with the pOA-DHFR substrate (Figure S4B). When this translocation intermediate was generated with labeled SecA and SecY, a FRET signal was observed, as indicated by a decrease of the donor, and an increase of the acceptor fluorescence (Figure S4C). To follow the dissociation of SecA from SecY, an excess of unlabeled SecA was added to prevent the rebinding of labeled SecA. The FRET signal indeed decreased with time (Figure 3B), whereas it remained constant without SecA addition (Figure S4D), confirming that SecA is continuously dissociating from and re-associating with the SecY complex. In the absence of substrate, SecA dissociated from the SecY channel significantly faster (Figure 3B; half-life shorter than 6 sec versus 34 sec). Fast SecA dissociation was also observed with pOA-DHFR lacking a signal sequence (Figure S4E), which prevents translocation of the substrate, as well as with a SecY mutant (SecY R357E) (Alami et al., 2007), in which the interaction with SecA is compromised (Figure S4F). In addition, SecA dissociated quickly from SecY when the pOA-DHFR substrate was added in the absence of methotrexate (Figure 3C), conditions that allow complete translocation of the substrate into the vesicles. Taken together, these results show that the presence of a translocating substrate in the SecY channel leads to a significant stabilization of the SecA-SecY interaction.
Figure 3. Dynamic interaction between SecA and SecY channel.
(A) A translocation intermediate was generated by fusing proOmpA with dehydrofolate reductase (DHFR) (pOA-DHFR). The DHFR domain folds in the presence of methotrexate (MTX), and stalls translocation. A FRET assay was used to follow the interaction of SecA (in green), labeled with the FRET-acceptor Cy5 (in blue), with SecYEG (in yellow), labeled with the FRET-donor tetramethyl rhodamine (TMR; in red).
(B) SecA-Cy5 was mixed with ATP, MTX, and proteoliposomes containing SecYEG-TMR in the presence or absence of pOA-DHFR. The translocation intermediate was then incubated with a 25fold molar excess of unlabeled SecA, and the resulting decay of the FRET signal measured over time. The curves represent exponential fits to the data points.
(C) As in (B), but in the presence or absence of MTX.
(D) As in (B), but with ATP, ATPγS, or hexokinase/glucose (HK/GL) added to the reactions.
(E) As in (D), but without substrate.
To test whether the SecA-SecY interaction is affected by the nucleotide state of SecA, we first generated a translocation intermediate with the pOA-DHFR substrate, using fluorescently labeled SecA and SecY complex in the presence of methotrexate. ATPγS was then added to lock SecA in the ATP-bound state. The addition of unlabeled SecA showed that the dissociation rate of SecA was drastically reduced compared with the situation in which ATP is continuously hydrolyzed (Figure 3D). To lock SecA in the ADP-bound state, ATP was converted into ADP by the addition of hexokinase and glucose, a reaction that is completed within 60 sec (Figure S4G). In this case, the FRET signal decayed as fast as with ongoing ATP hydrolysis (Figure 3D). The small difference is due to the presence of hexokinase; when the enzyme was added to the ATP control in the absence of glucose, the dissociation curves were completely superimposable (Figure S4H). These experiments show that SecA cycles between high- and low-affinity states, but, at steady-state with ongoing ATP-hydrolysis, it spends most of its time in the low-affinity, ADP-bound state. In the absence of substrate, SecA dissociated more rapidly from SecY, regardless of the nucleotide state, although a small stabilizing effect was observed in the presence of ATPγS (Figure 3E). Thus, even in the weakly binding ADP-state, SecA retains some contact with the polypeptide substrate. From ATP hydrolysis rates in the literature (7.6 ATP molecules per sec and SecA molecule (Robson et al., 2009)) and the measured dissociation rates in Figure 3 (koff ~0.020/sec), we estimate that, on average, a SecA molecule dissociates from translocating SecY after approximately 260 cycles of ATP hydrolysis (see Extended Experimental Procedures).
Polypeptide interactions with the SecA-SecY complex
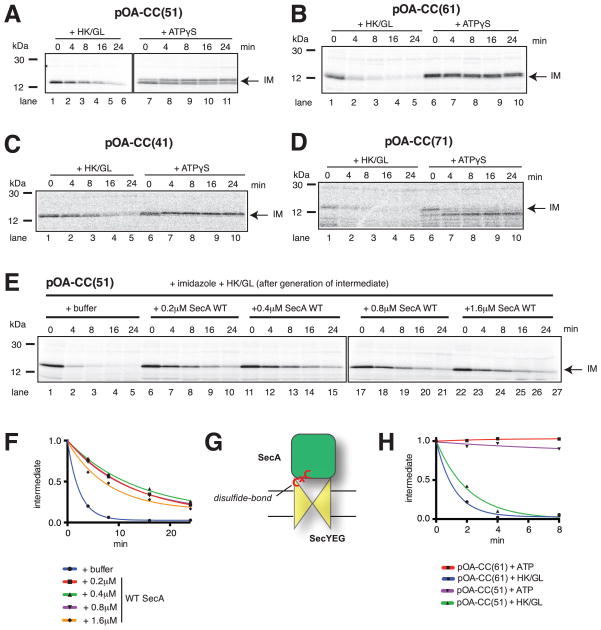
To address how the nucleotide state affects SecA’s interaction with the translocation substrate, we again used a backsliding assay. A translocation intermediate was formed with the pOA-CC(51) substrate, and its backsliding was determined after addition of ATPγS or hexokinase/glucose (Erlandson et al., 2008b). In the presence of ATPγS, no or only little movement of the polypeptide chain was observed (Figure 4A, lanes 7–11). The appearance of slightly larger bands is consistent with SecA-ATP allowing the chain to slide by a few amino acids into the channel before polypeptide movement is arrested. In contrast, backsliding was seen with SecA in its ADP-bound state (Figure 4A, lanes 1–6).
Figure 4. Nucleotide-dependent interaction of SecA with a translocation substrate.
(A) Radiolabeled pOA-CC(51) was mixed with ATP, wild type (WT) SecA, and proteoliposomes containing SecYEG. After generation of the intermediate, DTT was added together with hexokinase/glucose (HK/GL) or ATPγS. Samples were taken at different times, treated with proteinase K, and subjected to SDS-PAGE followed by autoradiography. IM indicates the position of the translocation intermediate. Molecular weight markers are indicated (in kDa).
(B) As in (A), but with pOA-CC(61).
(C) As in (A), but with pOA-CC(41).
(D) As in (A), but with pOA-CC(71).
(E) Radiolabeled pOA-CC(51) was mixed with ATP, SecA His-ΔN20, and Ni-NTA proteoliposomes containing SecYEG. After generation of a translocation intermediate, imidazole was added together with HK/GL and either buffer or WT SecA at various concentrations. The samples were analyzed as in (A).
(F) Quantification of the experiments shown in (E). The amount of IM, relative to the initial level, was plotted over time.
(G) Scheme showing a translocation intermediate generated with SecA disulfide-crosslinked to the SecY complex.
(H) A translocation intermediate was generated with a covalent SecA-SecY complex (see (G)) and either pOA-CC(61) or pOA-CC(51). The intermediates were incubated with either HK/GL or ATP, and backsliding determined as in (A).
Similar results were obtained with other substrates, in which the first cysteine of the disulfide-bonded loop was moved towards the N- or C-terminus, so that different amino acid sequences are located inside the SecA-SecY complex in the translocation intermediate (Figures 4B–D; see schemes in Figure S5A). In all cases, a stable proteolytically protected fragment was generated in ATPγS. With pOA-CC(71), the protected band was slightly smaller (Figure 4D, lanes 7–10), indicating that the polypeptide slid back by a few amino acids before being arrested. Thus, SecA in its ATP-bound state interacts strongly with many positions in a polypeptide chain, but it allows passive diffusion of the substrate at other positions until it encounters a strongly interacting residue in the flanking regions. In the presence of ADP, backsliding was observed with all constructs (Figures 4B–D), indicating that SecA-ADP interacts only weakly with amino acids in the substrate. Nevertheless, different sequences inside the SecA-SecY complex resulted in different backsliding rates (Figure S5B), suggesting that some interaction is maintained.
Backsliding in ADP may occur either after dissociation of SecA-ADP from the SecY channel (Figure 3D), or while SecA-ADP is still bound to the channel. To distinguish between these possibilities, we generated a translocation intermediate with pOA-CC(51), SecA His-ΔN20, and proteoliposomes containing Ni-NTA lipids. Backsliding in ADP was then determined in the presence of imidazole, which prevents the rebinding of SecA His-ΔN20 to the SecY channel. Backsliding was extremely fast (Figure 4E, lanes 1–5; quantification in Figure 4F), indicating that the SecY channel alone allows rapid diffusion of the polypeptide. The backsliding rate was significantly reduced when wild type SecA was added (Figure 4E, lanes 6–27; Figure 4F). Since the rate was independent of the SecA concentration, most backsliding occurred with SecY channels occupied by SecA-ADP. The reduced diffusion of the substrate compared with the channel alone confirms that SecA maintains some interaction with the polypeptide even in its ADP-bound state.
We further demonstrated that passive backsliding can occur with SecA-ADP bound to the SecY channel by using a covalently linked complex. SecA was crosslinked to SecY through a disulfide bond between a cysteine in SecA and a cysteine in SecY (Whitehouse et al., 2012) (Figure 4G; the purity of the complex is shown in Figure S5C). With both pOA-CC(51) and pOA-CC(61), the translocation intermediate remained stable in the presence of ATP, but rapidly disappeared in ADP (Figure 4H), confirming backsliding with SecA-ADP bound to the SecY channel.
Although the measured backsliding rates may not only depend on the amino acid sequence encountered by the SecA-SecY complex, but also on the exact position of proteinase K cleavage sites in the substrate, it is remarkable that they can be in the same range as actual translocation rates (for pOA-CC(61) the half-life of the translocation intermediate is ~2.5 min after addition of hexokinase/glucose, and ~1 min in the presence of ATP and DTT; Figure S5D). Thus, it seems that passive polypeptide movements contribute significantly to the kinetics of translocation.
SecA interacts in a sequence-specific manner with the substrate
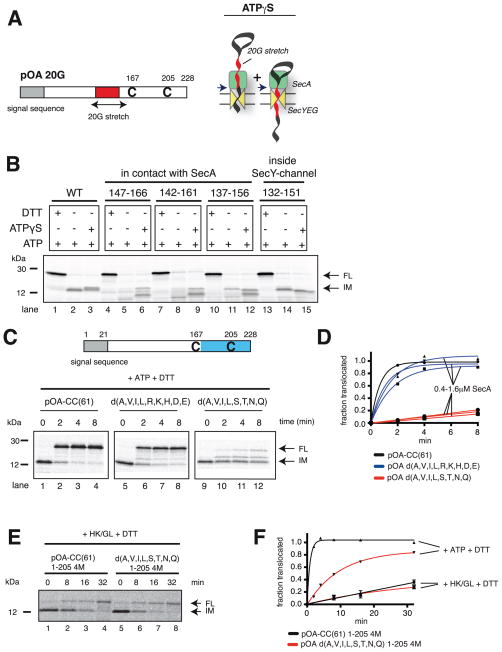
Because SecA interacts with a translocating polypeptide much stronger in its ATP- than in its ADP-bound state (Figure 4A–D), we next tested whether SecA-ATP binds to the backbone of the polypeptide or to amino acid side chains. To this end, we replaced segments of the substrate with stretches of glycines (Figure 5A). When 20 glycines were placed directly in front of the first cysteine of pOA-CC(61), no defined band corresponding to the translocation intermediate was detected (Figure 5B, lane 5 versus 2). Thus, SecA does not seem to be able to push the glycine stretch all the way into the SecY channel, resulting in a heterogeneous population of polypeptide chains, which do not give rise to a defined proteolytic fragment. When the glycine stretch was gradually moved towards the SecY channel, the intermediate started to re-appear (Figure 5B, lanes 8,11,14). It reached the intensity seen with the wild type protein with a distance of 15 residues from the first cysteine (lane 14), a position that places the glycine stretch in the SecY channel, where it can no longer interact with SecA. In the presence of ATP and DTT, all constructs were translocated into the proteoliposomes (Figure 5B, lanes 1,4,7,10,13). Thus, despite the fact that SecA does not interact well with the glycine stretches, it can still translocate these segments.
Figure 5. SecA interacts with amino acid side chains of the polypeptide substrate.
(A) Stretches of 20 consecutive glycines (G) were inserted into the translocation substrate pOA-CC(61) at various distances in front of Cys167. Backsliding in the presence of ATPγS arrests polypeptide movements at interacting amino acids in the flanking regions, giving rise to two defined proteolytically protected fragments (arrows in right scheme).
(B) pOA-CC(61) and derivatives with glycine stretches inserted at the indicated positions were mixed with ATP, wild type SecA, and proteoliposomes containing SecYEG. After generation of a translocation intermediate, ATP or ATPγS were added. The samples were analyzed at different times by treatment with proteinase K, followed by SDS-PAGE and autoradiography. Where indicated, DTT was added at the beginning of the translocation reaction. IM and FL indicate protease-protected fragments, corresponding to the intermediate and fully translocated products, respectively. Molecular weight markers are indicated (in kDa).
(C) Translocation of the C-terminal tail (in blue) was tested with pOA-CC(61) or derivatives in which several amino acids in the segment following the first cysteine were replaced with glycines (the substituted amino acids are given in one-letter code). The radiolabeled substrate was mixed with ATP, SecA His-Δ N20, and Ni-NTA proteoliposomes containing SecYEG. The generated translocation intermediate was incubated with imidazole and wild type (WT) SecA, before addition of DTT. The samples were analyzed as in (B).
(D) Quantification of the experiments in (C). The experiments were performed at different concentrations of SecA. Plotted is the amount of FL relative to the initial level of IM, divided by the ratio determined for wild type substrate at the last time point. A correction was made for the different numbers of methionines in IM and FL (8 versus 9).
(E) A comparison of translocation and passive forward sliding was performed with substrates in which the segment in pOA-CC(61) following the second cysteine was replaced with four methionines (pOA-CC(61) 1-205 4M). The segment in the disulfide-bonded loop was either the wild type sequence or lacked hydrophobic and polar amino acids (d(A,V,I,L,S,T,N,Q)). Translocation intermediates were incubated with hexokinase/glucose (HK/GL), followed by addition of DTT. The appearance of FL was followed by protease protection.
(F) Quantification of the experiment in (E), done as in (D). A correction was made for the different numbers of methionines in IM and FL (8 versus 13). Shown are the means and standard deviation of three experiments. Also shown is translocation in the presence of ATP, determined in parallel.
As observed before (Figure 4B), when a translocation intermediate of wild type pOA-CC(61) was incubated with ATPγS, additional, closely spaced proteolytic fragments appeared (Figure 5B, lane 3 versus 2; see also Figure 4B, lanes 6–10). Similarly, when the glycine stretch was located in the SecY channel, the protected bands in ATPγS were close to that seen in ATP (Figure 5B, lane 15 versus 14). In contrast, with all constructs in which a glycine stretch contacted SecA, two well-separated major bands were seen (Figure 5B, lanes 6, 9,12). The larger and smaller fragments were similar in size to pOA fragments truncated after and before the glycine stretch, respectively (Figure S6A). Thus, it seems that SecA in its ATP-locked state allows a glycine stretch to slide back and forth, but traps the polypeptide chain at residues preceding or succeeding the glycine stretch (see schemes in Figure 5A and S6A). Taken together, these results suggest that SecA in its ATP-bound state interacts with some amino acid side chains, rather than with the backbone of the polypeptide chain.
To test whether SecA’s translocation efficiency depends on the sequence of the substrate, we replaced the 61 amino acids following the first cysteine in pOA-CC(61) with segments, in which different types of amino acids were substituted with glycines, keeping the total number of glycines constant (40 glycines). In one construct, all hydrophobic (A, V, I, L) and charged (R, K, H, D, E) amino acids were replaced, and in another all hydrophobic and non-charged polar (S, T, N, Q) residues (the sequences are shown in Figure S6B). After addition of DTT to the translocation intermediates, the different C-terminal tails were translocated with strikingly different kinetics (Figure 5C,D). The differences are not due to a change in SecA-SecY interaction, as the translocation rates were independent of the SecA concentration (Figure 5D). The slowest translocation rate was seen with a substrate that does not contain long continuous glycine stretches (Figure 5C, lanes 9–12; the longest stretch in the slowest substrate pOA d(A,V,I,L,S,T,N,Q) is four amino acids long), suggesting that other amino acids also do not interact well with SecA in its ATP-bound state.
Next we tested passive sliding with substrates of different sequences. We determined forward sliding rates with SecA in its ADP-bound state, using pOA-CC(61) derivatives, in which the C-terminal tail after the second cysteine was replaced with four methionines (to shorten the translocated segment and increase the sensitivity of detection after labeling with 35S-methionine). Upon depletion of ATP with hexokinase/glucose and addition of DTT, some of the translocation intermediate was converted into fully translocated material (Figures 5E,F). The rate of passive forward sliding was the same whether the translocated segment contained the wild type sequence or a sequence (d(A,V,I,L,S,T,N,Q)) that was only poorly translocated in ATP. The rate did not change when the SecA concentration was increased (Figure S6C,D), confirming that sliding occurs with SecA-ADP bound to the SecY channel. Thus, most of the sequence-dependent interaction with a substrate occurs with SecA in its ATP-bound state. The almost identical passive sliding rates observed with the different substrates also indicates that folding properties are not responsible for their different translocation rates in the presence of ATP.
Translocation requires substrate interaction of SecA’s two-helix finger
Crosslinking and structural data (Erlandson et al., 2008a; Zimmer et al., 2008) suggest that the tip of SecA’s two-helix finger is responsible for the interaction with amino acids in a substrate. To test this hypothesis, we introduced mutations into the fingertip (Figure 6A) and determined the rate at which these SecA mutants translocate different C-terminal sequences of pOA-CC(61). The SecA mutants were normally folded, as shown by gel filtration experiments (Figure S7A). We first generated a translocation intermediate with SecA His-ΔN20, and then added imidazole together with SecA fingertip mutants, thereby replacing SecA His-ΔN20. The rate at which translocation intermediates are converted to fully translocated products was determined by protease protection. Substitution of the SecA loop residue Tyr794 with Gly did not affect the translocation of the wild type sequence (Figure 6B, lanes 5–8; quantification in Figure 6D), whereas the substitution of the two basic residues Arg792 and Lys797 had a moderate effect (Figure 6B, lanes 9–12; Figure 6D). Much stronger effects were seen when the SecA mutants were tested with a substrate in which all hydrophobic amino acids were replaced with glycines d(A,I,L,V) (Figure 6C, lanes 5–12, quantification in Figure 6E; the sequence is shown in Figure S6B), although this substrate was translocated with normal kinetics by wild type SecA (Figure 6C, lanes 1–4). The slower translocation with the SecA mutants was not caused by their reduced binding to SecY, as the same, or even somewhat reduced rates were observed at higher SecA concentrations (Figure S7B,C). Both SecA fingertip mutants also translocated a substrate sequence lacking all polar residues d(S,T,N,Q,R,K,H,D,E) as fast as the wild type sequence (Figure S7D; the sequence is shown in Figure S6B). Taken together, these results show that the fingertip of SecA interacts in a sequence-specific manner with the polypeptide substrate. Several residues in the fingertip loop, including Tyr794, and Arg792/Lys797, seem to be involved in substrate interaction. In fact, Arg792 is similarly conserved as Tyr794 among SecAs from different bacterial species (Figure S7E).
Figure 6. Substrate interaction of SecA’s two-helix finger.
(A) The two-helix finger (THF) of SecA has a loop with the indicated sequence between helices 1 and 2. Residues R792/K797 and Y794 (in red) were replaced by glycines.
(B) Radiolabeled pOA-CC(61) was mixed with ATP, SecA His-ΔN20, and Ni-NTA proteoliposomes containing SecYEG. After generation of a translocation intermediate, imidazole was added together with either wild type (WT) SecA or fingertip SecA mutants. After 20 min, DTT was added, and the conversion of the intermediate (IM) to fully translocated product (FL) was determined by protease protection.
(C) As in (B), but with pOA-CC(61) in which the indicated amino acids following the first cysteine were replaced by glycines.
(D) Quantification of the experiments in (B). Plotted is the amount of FL relative to the final levels of FL seen with WT SecA (means and standard deviations).
(E) As in (D), but for the experiments in (C).
(F) As in (B) with SecA Y794G added at the indicated concentrations, but without DTT and with either buffer (BF), ATP, ATPγS, or hexokinase/glucose (HK/GL) added. The amount of IM, relative to the initial level, was plotted over time.
To further test whether SecA in its ATP-bound state uses Tyr794 at the fingertip to push a polypeptide chain, we generated a translocation intermediate with pOA-CC(61), SecA His-ΔN20, and Ni-NTA lipid-containing proteoliposomes. Imidazole was then added together with SecA Y794G and different nucleotides, and backsliding was determined by protease protection. In the presence of ATPγS, significant backsliding was observed (Figure 6F), in contrast to the situation with wild type SecA, where polypeptide movements were totally arrested (Figure 4A). These data confirm that Tyr794 at the fingertip contributes to the interaction with the polypeptide. With ongoing ATP hydrolysis, the backsliding rate was much slower than in ATPγS (Figure 6F), indicating that movements of the defective two-helix finger can still propel the polypeptide forward, albeit with reduced efficiency. As expected, the backsliding rate with SecA Y794G was very fast in ADP (Figure 6F), even faster than with wild type SecA (Figure 4B), indicating that the two-helix finger maintains some low affinity for the polypeptide chain even in the ADP-bound state. The previously observed sequence-independent forward sliding (Figure 5E,F) may be explained by assuming that weak contacts at several different positions average out over a longer polypeptide stretch.
A translocation model combining power strokes and passive diffusion
Our data indicate that SecA moves a polypeptide into the SecY channel by occasional power strokes, but also permits passive sliding in either direction. To further analyze this “push and slide” mechanism, we developed a mathematical model, which is an extension of previous models of translocation (Liebermeister et al., 2001; Peskin et al., 1993). We assumed that a polypeptide chain moves through the SecY channel one amino acid at a time, either by SecA’s power stroke or by passive diffusion (assuming that a power stroke moves more than one amino acid does not change the qualitative behavior of the system; see Supplement). We also assume that SecA is either in the ATP- or ADP-bound state (Figure 7A). When SecA binds ATP (with a rate constant Kbind), the two-helix finger can interact with some amino acids in the polypeptide chain; the finger would move towards the channel and drag the interacting amino acids with it (power stroke). If the fingertip in SecA-ATP does not interact with the amino acid encountered, the polypeptide chain cannot be pushed, but it can diffuse back and forth (with a diffusion constant DATP). Hydrolysis of ATP (with a rate constant Khyd) would reset the two-helix finger without polypeptide movement. In the resulting ADP-bound state, the polypeptide chain can diffuse in either direction, regardless of the amino acid encountered (with a diffusion constant DADP).
Figure 7. A mathematical model for the “push and slide” mechanism of SecA.
(A) Push and slide model. Amino acids of the translocating polypeptide are shown as circles. Upon ATP binding by SecA (rate constant Kbind), the two-helix finger (in blue) undergoes a power stroke (curved arrow). If it interacts with an amino acid (green), it pushes it forward (green arrow). Otherwise, the amino acid (in red) can diffuse forward or backward (diffusion constant DATP). SecA-ATP is converted into SecA-ADP with the rate constant Khyd, resulting in resetting of the two-helix finger (purple; curved arrow). In the ADP-state, SecA allows passive sliding of any amino acid (diffusion constant DADP). The translocation direction is indicated with a blue arrow.
(B) Modeling of the experiment in Figure 5D. The translocated last 61 residues of the substrates correspond to either wild type (black curve) or mutant sequences (blue and red). The best fit was obtained with the percentages of interacting amino acids indicated in the inset, distributed evenly throughout the segment. Parameters: DATP=DADP=0.04 nm2/sec; Khydr=107/sec; Kbind=11/sec.
(C) Translocation of a 40-residue polypeptide containing interacting (green) and non-interacting (red) amino acids. SecA was assumed to be 90% of the time in its ADP-bound state. The fraction of polypeptide chains translocated to different positions is given at different time points (blue columns; time in sec). Position 40 corresponds to complete translocation. The parameters were as in (B).
(D) As in (C), but with SecA being 10% of its time in the ADP-bound state.
(E) The translocation time (the time its takes to fully translocate 95% of all chains) was plotted over the ATP consumption rate (the average of ATP molecules hydrolyzed per sec during translocation of the entire chain). SecA was assumed to spend different fractions of time in the ADP-bound state (inset). The substrate (50 amino acids) contained 90% or 10% of evenly spaced interacting amino acids.
(F) As in (E), but with translocation rates (inverse of translocation time) expressed as percentage relative to the maximal rate at infinite ATP consumption rate. The calculations were performed for different percentages of evenly spaced interacting amino acids (inset), assuming SecA to be 90% in its ADP-bound state. The broken line indicates the experimentally determined ATP consumption rate (7.6/sec). This corresponds to an optimal situation, as translocation would be slow towards the left, and ATP consumption excessive towards the right.
We adjusted the parameter values of the model by using the reported average ATP-hydrolysis rate (Robson et al., 2009) and our data on passive forward sliding in Figure 5F. The computed time-courses of translocation agree reasonably well with the experimental data in Figure 5D, if the percentage of interacting amino acids is ~50% in wild type proOmpA and significantly lower in the mutant substrates (Figure 7B). Differences to the data, in particular at early time points, may be due to experimental limitations of the translocation assay, or to the simplifying assumptions of the model, i.e. that DADP is the same for all positions, that DADP is equal to DATP, and that interacting and non-interacting amino acids are distributed at equal distances throughout the polypeptide chain (in reality, neither DADP nor DATP are entirely sequence-independent, amino acids may have intermediate affinities for SecA, and strongly or weakly interacting amino acids could be clustered). Nevertheless, the calculations provide additional evidence for idle power strokes of SecA at non-interacting amino acids and for passive sliding of the polypeptide.
The model shows that when SecA-ATP can interact with each amino acid of a substrate, translocation proceeds efficiently, as power strokes are possible at each position (not shown). However, when SecA encounters an imperfect substrate, passive sliding becomes important. With SecA mostly in its ADP-bound state, which we suggest is the physiological situation (Figure 3D), a polypeptide segment containing non-interacting amino acids can rapidly slide through the SecA-SecY complex (Figure 7C). In contrast, if SecA were mostly in the ATP-bound state, polypeptide movement would be dramatically slowed at the beginning of the non-interacting segment (Figure 7D).
The calculations demonstrate that the translocation time for a polypeptide chain decreases as SecA’s ATP consumption rate increases, until a plateau is reached (Figure 7E). With a low frequency of power strokes, translocation is slow, as it primarily relies on passive sliding. On the other hand, too frequent power strokes result in wasteful ATP hydrolysis without accelerating translocation. For a given ATP-consumption rate, translocation is faster if the polypeptide contains a higher percentage of interacting amino acids and if SecA spends more time in its ADP-bound state, during which passive sliding can occur between power strokes (Figure 7E).
Interestingly, SecA appears to operate close to an optimum at which translocation is sufficiently fast, but ATP-consumption not excessive (Figure 7F); regardless of the amino acid composition of a substrate, translocation rates are estimated to be 60–90% of the maximum rates achievable (if the ATP-consumption rate was infinite). Taken together, our model not only explains why SecA spends most of its hydrolysis cycle in ADP, but also how it can translocate polypeptide chains of diverse sequences.
DISCUSSION
We have used translocation intermediates to study the mechanism of SecA function during the actual translocation process. Our results lead to a “push and slide” model for SecA-mediated protein translocation through the SecY channel. In the model, SecA moves the polypeptide chain by both ATP-driven power strokes and by allowing passive sliding. When SecA binds ATP, it interacts through the tip of its two-helix finger with a subset of amino acids of the substrate, the finger moves towards the channel and drags the polypeptide with it. The two-helix finger does not interact strongly with all amino acids encountered, so that power strokes not always result in active pushing. Instead, when the finger faces a non-interacting amino acid, the polypeptide can passively move in either direction. Following ATP-hydrolysis, the two-helix finger would retract from the polypeptide chain, allowing passive sliding regardless of the amino acid encountered. Since SecA spends most of its time during the ATP hydrolysis cycle in the ADP state, passive movements contribute greatly to translocation. However, the pushing phase is required to provide directionality and to increase the efficiency of translocation. The proposed mechanism allows SecA to translocate amino acid sequences with which it interacts only weakly. In vivo the efficiency of translocation may be increased by SecDF pulling on the polypeptide chain on the extracellular side of the membrane (Tsukazaki et al., 2011). In addition, polypeptide chain folding or the binding of periplasmic chaperones could bias the direction of polypeptide sliding.
In contrast to our previous suggestions (Erlandson et al., 2008a), SecA does not translocate a fixed number of amino acids with each power stroke of the two-helix finger, and it does not interact strongly with the translocating polypeptide chain in its ADP-bound state when the two-helix finger resets. The previous model cannot explain how extended segments of non-interacting amino acids, such as glycine stretches, can be translocated, as SecA would undergo ATPase cycles without moving the polypeptide chain forward. However, with the push and slide mechanism, a glycine stretch would passively move either forward into the SecY channel, or backward into the cytosol. If it moves forward, SecA would eventually encounter interacting side chains in the amino acid segment following the glycine stretch and could continue to push. If it moves backward, SecA would eventually interact with amino acid side chains preceding the glycine stretch, and could start over again to push the polypeptide into the channel. The overall result is net movement in the forward direction. Glycine stretches are rare, but other non-interacting amino acids seem to occur frequently in wild type proteins, as shown by passive movements of proOmpA polypeptides when SecA is locked in the ATP-bound state (Figure 4A–D), and by our modeling, which indicates that ~50% of all residues are not actively pushed.
The ATPase cycle of SecA seems to be optimally coupled with the mechanical cycle. SecA probably starts out in the cytosol mostly in its ADP-bound state (~98%, as estimated on the basis of published rate constants (Robson et al., 2009)). When SecA binds to the translocating SecY channel, the rate-limiting step of ADP-release is drastically stimulated (Robson et al., 2009), resulting in rapid exchange of ADP for ATP. SecA still spends most of its time in the ADP-bound state (Figure 3D), but the overall hydrolysis cycle is greatly accelerated, resulting in rapid power strokes. As shown by our modeling, it would be disadvantageous for SecA to spend too much time in the ATP- bound state, as polypeptide movement would be arrested at certain positions. The ATPase cycle appears to represent a compromise that allows rapid translocation of a wide range of sequences, while keeping ATP-consumption low.
Our results suggest that several amino acids in the tip of SecA’s two-helix finger interact with the translocating polypeptide chain. None of the amino acids in the fingertip loop, including Tyr794 (Erlandson et al., 2008a), is absolutely essential. However, in the absence of Tyr794 or two basic residues in the loop (Arg792 and K797) some sequences are only poorly translocated. Several loop residues may therefore interact with amino acids of a substrate, consistent with disulfide crosslinking results (Erlandson et al., 2008a). Perhaps, multiple interactions of the two-helix finger loop increase the range of amino acids that can be pushed, but it remains unclear what exactly the fingertip recognizes; it may be a combination of several properties, such as size and hydrophobicity of amino acids. For hexameric ATPases, it is also unclear what features are recognized by the central loops (Barkow et al., 2009; Too et al., 2013).
Our data suggest that movements of the two-helix finger are required to push a polypeptide into the SecY channel. A model in which the two-helix finger of SecA-ATP binds the polypeptide and moves towards the SecY channel is consistent with a SecA-SecY structure, in which SecA is close to its ATP-bound state and the two-helix finger is inserted deeply into the channel (Zimmer et al., 2008). However, crosslinking of the two-helix finger to SecY does not abolish translocation (Whitehouse et al., 2012), perhaps because the crosslinked complex is conformationally flexible. Single-molecule experiments are required to directly test whether the two-helix finger moves in an ATP-dependent manner.
We can exclude a model in which SecA would bind to and dissociate from the SecY channel during each ATP hydrolysis cycle. Instead, a SecA molecule remains bound for ~260 ATP hydrolysis cycles. Nevertheless, the rate of SecA-dissociation and -rebinding is appreciable. Dissociation occurs with SecA in its ADP-bound state, both because the affinity for SecY is weaker than in ATP, and because SecA spends most of its time in the ADP state. SecA retains some low affinity for substrate in its ADP state, likely through some residual contact of the two-helix finger. In addition, another domain of SecA (the “clamp”) interacts with a short backbone segment of the substrate (Zimmer and Rapoport, 2009). In the absence of a translocating polypeptide, SecA dissociates rapidly from the SecY channel (Figure 3B,C), providing a simple mechanism by which the machinery is reset after termination of translocation.
We show that efficient rebinding of SecA to a translocating SecY channel requires that SecA first interacts through its N-terminus with lipids in the membrane. The initial binding to the membrane surface probably concentrates SecA and restricts diffusion to the plane of the membrane, thus increasing the rate of association with the SecY channel. The indirect binding of SecA to translocation sites may explain, at least in part, the long-known affinity of SecA for lipids (Hendrick and Wickner, 1991).
The proposed push and slide mechanism may also apply to hexameric ATPases that move polypeptide chains. For both ClpX and other hexameric ATPases, it is clear that they interact only weakly with some amino acids, particularly glycines (Tian et al., 2005; Too et al., 2013). Passive backsliding of a polypeptide has also been observed (for example, Too et al., 2013; Kraut et al., 2013). A push and slide model can also explain the puzzling observation that a stretch of 10 amino acids in a polypeptide substrate can be replaced by a large number of segments, including those that lack NH groups or contain ten consecutive methylenes (Barkow et al., 2009). These results may be explained if a non-interacting segment can slide back and forth, but is ultimately moved by interactions of the ClpX loops with amino acids in the flanking regions. Unlike SecA, which deals with unfolded or weakly folded substrates, hexameric ATPases can unfold proteins, and may therefore have reduced periods of passive sliding. Sliding would not be required for the related ATPases that function as helicases, as they face substrates with regularly repeating interaction sites.
EXPERIMENTAL PROCEDURES
Plasmids used in this study are listed in the Supplemental Table and details of the experiments are described in Extended Experimental Procedures.
Protein purifications and translocation assays
SecA (amino acids 1-831, with all endogenous cysteines replaced with serines, referred to as wild type), SecYEG with all endogeneous cysteines replaced with serines, and pOA-DHFR were purified essentially as described before (Bauer and Rapoport, 2009). Reconstitutions were essentially done as described (Kusters et al., 2010). All substrates were synthesized in vitro in rabbit reticulocyte lysate in the presence of 35S-methionine, precipitated with ammonium sulfate, and resuspended in 6 M urea. Translocation reactions were performed essentially as described (Erlandson et al., 2008a), in the presence of 0.2 μM SecYEG in proteoliposomes, 0.2–1.6 μM SecA, and in vitro synthesized substrate diluted 1:50. Translocation intermediates were formed in the presence of 400 μM sodium tetrathionate. After translocation at 37°C, samples were treated with proteinase K, precipitated with trichloroacetic acid, and analyzed by SDS-PAGE and autoradiography. SecA His-ΔN20 was dissociated from 1,2-dioleoyl-sn-glycero-3- [(N-(5-amino-1-carboxypentyl) iminoacetic acid) succinyl] (Ni-NTA-DGS) containing proteoliposomes by addition of 250 mM imidazole. The disulfide bond in substrates was reduced with 50 mM DTT.
FRET assays
Single cysteines in SecA (position 21) and SecYEG (position 46 in SecG) were labeled with the maleimide-containing derivatives of Cy5 and tetramethyl rhodamine, respectively. 1.6 μM recombinant pOA-DHFR was mixed with ATP, proteoliposomes containing 0.4 μM TMR-labeled SecYEG, and 0.4 μM Cy5-labeled SecA in the presence or absence of 50 μM methotrexate. After 20 min at 37°C, the fluorescence was measured at 25°C. The FRET donor was excited at 541 nm, donor emission was measured at 577 nm, and acceptor emission at 670 nm. SecA dissociation was followed after addition of 10 μM unlabeled SecA. Where indicated, 5 mM ATPγS or 0.4 U/μl hexokinase/20 mM glucose were added 1 min earlier.
Mathematical modeling
A substrate was modeled as a string of n amino acids, with i amino acids translocated at time t, and associated with SecA-ATP or SecA-ADP. Changes in the states were described by terms for pushing and sliding, as well as ATP-binding and -hydrolysis. Together, these terms give the probability of having i amino acids translocated over time (see Extended Experimental Procedures). The resulting set of differential equations was integrated numerically in Matlab.
Supplementary Material
Highlights.
SecA mechanism is analyzed with translocation intermediates
SecA moves polypeptides through the SecY channel by a push and slide mechanism
Sliding and pushing allows translocation of a wide range of substrates
The model may also apply to polypeptide-translocating hexameric ATPases
Acknowledgments
We thank Karl Erlandson and Stephanie Miller for providing preliminary data, the ICCB for providing access to the plate reader, Alexander Stein for helpful suggestions, and Thomas Güttler, Alexander Stein, and Long Li for critical reading of the manuscript. B. W. B. is the recipient of a fellowship from the Boehringer Ingelheim Fonds. T. A. R. is supported by NIH grant GM05586 and is a Howard Hughes Medical Institute Investigator.
Footnotes
Supplemental information includes seven figures, Extended Experimental Procedures, and one table.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide Translocation by the AAA+ ClpXP Protease Machine. Chem Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer BW, Rapoport TA. Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci USA. 2009;106:20800–20805. doi: 10.1073/pnas.0910550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central Pore Residues Mediate the p97/VCP Activity Required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- Erlandson KJ, Miller SBM, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008a;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlandson KJ, Or E, Osborne AR, Rapoport TA. Analysis of polypeptide movement in the SecY channel during SecA-mediated protein translocation. J Biol Chem. 2008b;283:15709–15715. doi: 10.1074/jbc.M710356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of Asymmetric ClpX Hexamers Reveal Nucleotide-Dependent Motions in a AAA+ Protein-Unfolding Machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Wickner W. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the Central Channel of ClpA Chaperone Mediate Protein Binding, Unfolding, and Translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Hunt JF. Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- Kraut DA. Slippery substrates impair ATP-dependent protease function by slowing unfolding. J Biol Chem. 2013;288:34729–34731. doi: 10.1074/jbc.M113.512533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters I, Bogaart G, Wit J, Krasnikov V, Poolman B, Driessen A. Purification and functional reconstitution of the bacterial protein translocation pore, the SecYEG complex. Methods Mol Biol. 2010;619:131–143. doi: 10.1007/978-1-60327-412-8_8. [DOI] [PubMed] [Google Scholar]
- Liebermeister W, Rapoport TA, Heinrich R. Ratcheting in post-translational protein translocation: a mathematical model. J Mol Bio. 2001;305:643–656. doi: 10.1006/jmbi.2000.4302. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse Pore Loops of the AAA+ ClpX Machine Mediate Unassisted and Adaptor-Dependent Recognition of ssrA-Tagged Substrates. Mol Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-Mediated Protein Translocation Across Membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. Translocation and Unwinding Mechanisms of RNA and DNA Helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Robson A, Gold VAM, Hodson S, Clarke AR, Collinson I. Energy transduction in protein transport and the ATP hydrolytic cycle of SecA. Proc Natl Acad Sci USA. 2009;106:5111–5116. doi: 10.1073/pnas.0809592106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Holmgren RA, Matouschek A. A conserved processing mechanism regulates the activity of transcription factors Cubitus interruptus and NF-κB. Nat Struct Mol Biol. 2005;12:1045–1053. doi: 10.1038/nsmb1018. [DOI] [PubMed] [Google Scholar]
- Too PHM, Erales J, Simen JD, Marjanovic A, Coffino P. Slippery Substrates Impair Function of a Bacterial Protease ATPase by Unbalancing Translocation versus Exit. J Biol Chem. 2013;288:13243–13257. doi: 10.1074/jbc.M113.452524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T, Perederina A, Vassylyev DG, Kohno T, Maturana AD, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474:235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- Whitehouse S, Gold VA, Robson A, Allen WJ, Sessions RB, Collinson I. Mobility of the SecA 2-helix-finger is not essential for polypeptide translocation via the SecYEG complex. J Cell Biol. 2012;199:919–929. doi: 10.1083/jcb.201205191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Rapoport TA. Conformational flexibility and peptide interaction of the translocation ATPase SecA. J Mol Biol. 2009;394:606–612. doi: 10.1016/j.jmb.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.