Abstract
The evolution of a variety of important chromophore-dependent biological processes, including microbial light sensing and mammalian color vision, relies on protein modifications that alter the spectral characteristics of a bound chromophore. Three different color opsins share the same chromophore, but have three distinct absorptions that together cover the entire visible spectrum, giving rise to trichromatic vision. The influence of opsins on the absorbance of the chromophore has been studied through methods such as model compounds, opsin mutagenesis, and computational modeling. The recent development of rhodopsin mimic that uses small soluble proteins to recapitulate the binding and wavelength tuning of the native opsins provides a new platform for studying protein-regulated spectral tuning. The ability to achieve far-red shifted absorption in the rhodopsin mimic system was attributed to a combination of the lack of a counteranion proximal to the iminium, and a uniformly neutral electrostatic environment surrounding the chromophore.
Keywords: hCRBPII, red shift, retinal, rhodopsin, rhodopsin mimic, wavelength regulation
Introduction
In order to improve food recognition and increase energy gain, living organisms have adapted to process light for two functions: sensory perception and energy production [1–3]. Conversion of light to chemical energy is performed by the chlorophyll-based photosynthetic machinery of plants and photosynthetic bacteria [4], and proton pumps (e.g. bacteriorhodopsin in archaea) [5]. Light-induced sensory perception systems include the plant photoreceptors (phytochromes, cryptochromes, and phototropin) [6], prokaryotic photo-receptors such as the sensory rhodopsins [7] (e.g. bacteriophytochromes [8]) and the visual opsins responsible for vision in animals [9]. The retinylidene-based opsins are the only class of chromophoric proteins that perform both sensory perception and energy production [10].
The most advanced form of biological light sensor is the eye, which is capable of discerning shapes and movement of objects [11]. These have reached various degrees of sophistication, ranging from monochromatic light detection, through trichromatic in the case of primates [2], to the remarkable example of 16 wavelength-specific opsins in the compound eyes of mantis shrimps [12–14]. Understanding the mechanism of this adaptation is the subject of this review.
This adaptation accompanied the evolution of primate visual opsin into four distinct forms, short wavelength (S), medium wavelength (M), and long wavelength (L)-opsins in the retinal cone cells and rhodopsin in the retinal rod cells [9, 15]. Each of the four visual rhodopsins binds the same chromophore, 11-cis-retinal, as a protonated Schiff base. However, the absorption maximum of this chromophore is uniquely shifted by the protein environment and its coordinated water molecules [16, 17].
What are visual opsins and how do they lead to color vision?
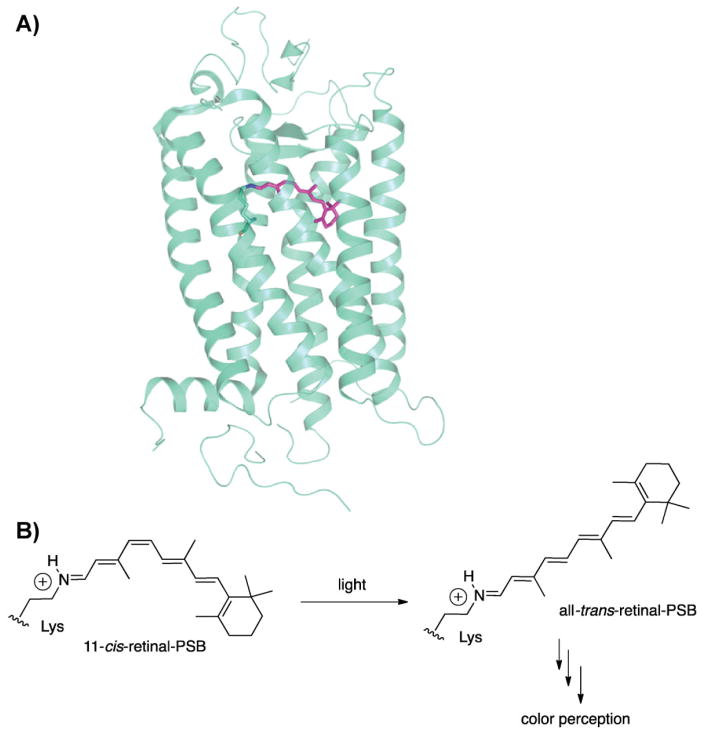
Opsin is a seven-α-helix-transmembrane protein, and is a member of the G-protein coupled receptor (GPCR) family. The 11-cis isomer of vitamin A aldehyde (also called retinal) is covalently linked to a lysine residue inside the binding pocket of the opsin as a protonated Schiff base (PSB)1 (Fig. 1) [18]. Opsins differ from most other members of the GPCR family in that they are activated not by binding of their ligand, but by the absorption of light by their chromophoric ligand [19].
Figure 1.
A: Rhodopsin crystal structure with 11-cis-retinal bound to a lysine residue in the binding pocket as protonated Schiff base. PDB code: 1F88. B: Isomerization of 11-cis-retinal to all-trans-retinal upon light absorption.
Humans have three distinct opsins in the retinal cone cells: short (blue), medium (green), and long wavelength (red), absorbing maximally at 420, 530, and 560 nm, respectively [20]. Besides these cone opsins, humans also have the substantially more abundant rod opsin (rhodopsin) in the retina rod cells. It has an absorption maximum of 500 nm and is mainly responsible for dim light vision. Though all of these opsins bind the same 11-cis-retinal chromophore, their absorption properties are uniquely modulated by the protein environment and its coordinated water molecules of each of the opsins to produce the set of pigments that enable color vision [16, 17].
Rhodopsin is highly sensitive to light, with a quantum efficiency of 67% [21], meaning that for every three photons absorbed by rhodopsin, two of them will result in isomerization of retinal from the 11-cis to the all-trans conformation (Fig. 1). The energy of the remaining one is dissipated in thermal motion. The change in the shape of the chromophore from bent (11-cis) to straight (all-trans) induces a conformational change in the protein that initiates a typical G-protein signaling cascade. Fifty rhodopsins or 203 cone opsins have to be activated to fire up the neurons and lead to vision [22, 23]. The entire process takes only a few milliseconds [24]. Subsequently, all trans-retinal dissociates and converts to 11-cis-retinal in the retinal pigment epithelium cells, which are adjacent to the photoreceptors [25]. Newly generated 11-cis-retinal binds to apoopsins and reconstitute the 11-cis-retinal bound opsins.
Microbial rhodopsins enable microorganisms to sense light
The function of microbial rhodopsins can be generally divided into three categories: (i) proton pumps, such as bacteriorhodopsin, that transport protons against a membrane-separated gradient that is used for energy production [26]; (ii) ion transporters that transport protons or ions across the membrane to maintain homeostasis, such as halorhodopsin [27]; (iii) sensory rhodopsins that can direct the micro-organisms to move toward or away from a light source [28]. A single microorganism can have multiple rhodopsins that absorb distinctly different wavelengths of light, similar to the visual rhodopsins. In an extreme case, six different rhodopsins were found in one archaeon, with absorption maxima ranging from 483 to 578 nm [29].
Although microbial rhodopsins and human visual opsins have low sequence identity, they share a similar protein structure and all are covalently bound to a retinal molecule via a protonated Schiff base. In contrast to visual opsins, microbial rhodopsins have all-trans-retinal bound in the dark state. Light triggers the isomerization of all-trans-retinal, usually to 13-cis-retinal, which causes the migration of an ion through the protein and across the membrane in ion pumps [30]. But contrary to visual opsins, retinal does not dissociate from the microbial rhodopsins after isomerization. Instead, 13-cis-retinal spontaneously converts back to its dark state, all-trans-retinal [31].
Bacteriorhodpsin and halorhodopsin translocate protons and halides against a gradient across the cellular membrane, directly, via the isomerization process [30, 32]. This energy requiring process is not catalytic, meaning that one photon translocates only one proton or one halide. The proton gradient produced by bacteriorhodopsin is subsequently used for the production of chemical energy via an ATP synthase [33]. Sensory rhodopsins have a transducin partner that becomes activated by the conformational change in rhodopsin caused by isomerization [34].
A direct result of isomerization is the drastically lowered pKa of the retinal-PSB, which leads to its deprotonation. This is because the counteranion is moved further away from the PSB and is no longer able to stabilize the positive charge. This is evident in the crystal structure of meta II (all-trans-retinal-bound) bovine rhodopsin, the active state of visual rhodopsin [35]. Zhu et al. recently found that the pKa of the protonated Schiff base is further a function of β-ionone ring orientation [36].
How do opsins regulate the wavelength of their chromophore: Retinal-PSB
Though great progress has been made in understanding many aspects of the opsins, the mechanism by which the protein environment modulates the absorption maxima of a single chromophore, retinal-PSB, over the range from 420 to 600 nm is still debated [37–40]. This represents the most regulated chromophore observed in nature, and provides a good platform to study how protein-chromophore interactions affect a chromophore’s photophysics.
Retinal absorbs at 380 nm in ethanol and it blue shifts to 360 nm when it forms a Schiff base (an imine) with n-butylamine. A large red shift to 440 nm results when the Schiff base is protonated. Retinal bound to opsins as a PSB (an iminium) absorb at different wavelengths. The difference between the protein-bound PSB absorbance and 440 nm is referred to as the “opsin shift”, and represents the protein-induced change in absorption spectrum of the pigment as compared with that of the native protonated retinylidene Schiff base [41].
Wavelength is inversely proportional to the excitation energy of an electron promoted from the ground state to the excited state of a chromophore. Therefore, altering the relative energies of the ground and excited states will lead to alterations in the wavelength of absorption. Lowering the ground state while raising the excited state will result in blue shifted absorption, while raising the ground state and lowering the excited state will lead to red shifted absorption.
Two general approaches to wavelength tuning can be envisioned; a conformational approach (Fig. 2A) and an electronic approach (Fig. 2B). The conformational approach involves controlling the planarity of the polyene by steric interaction with the protein. A more planar polyene results in a higher degree of conjugation and π delocalization, thus a red shifted absorption. Twisting along the single bond results in lower degree of conjugation and a corresponding blue shifted absorption spectrum [42]. The electronic approach takes advantage of the fact that the positive charge localized at the Schiff base becomes more delocalized across the entire polyene upon photo-excitation from the ground state to the excited state [43]. This causes a large difference in dipole moment between the retinal-PSB ground and excited states. Therefore, the polarity of the surrounding environment should exert a strong impact on the energy difference between the ground and excited state of the bound chromophore. More negative polarity close to the Schiff base discourages positive charge delocalization because it would stabilize the positively charged iminium.
Figure 2.
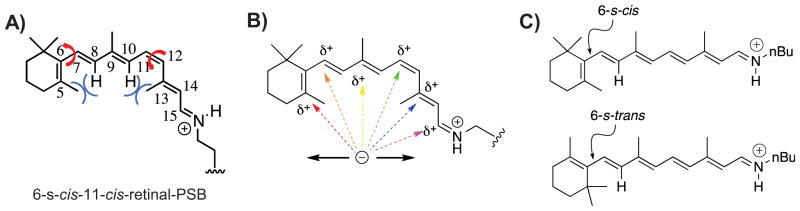
Wavelength regulation mechanisms. A: Rotation along the single bond C6–C7 or twisting along double bond C11–C12 could alter the degree of conjugation for 11-cis-retinal. The whole retinal could be divided into three planes: one composed of the first double bond C5=C6 in the ionone ring; one composed of C7=C8–C9=C10–C11; one composed of C12–C13=C14–C15=N. The three planes are not planar due to steric hindrance of C5–CH3 and C8–H, and C10–H and C13–CH3. B: Resonance of the positive charge along the polyene provides the opportunity for interactions at different locations to stabilize and/or extend the delocalization of the positive charge. It has been hypothesized that different opsins could alter the wavelength of absorption through electrostatic interactions with the polyene at different locations. C: The two different C6/C7 rotomeric configurations for all-trans-retinal PSB.
The protein environment can have a great impact on the retinal-PSB, either through protein-chromophore packing to change the conformation of retinal, or by tuning its binding pocket’s polarity to affect the ground state and excited state energy and the delocalization of the positive charge. However, accurately accounting for the effects of protein environment on the retinal-PSB still remains a big challenge, because of the dynamics of the protein environment and the dependency on the conformation of retinal. Furthermore, the electrostatics of the protein-binding pocket could be changed drastically by the rotation of side chains, and the presence of ordered water molecules within the binding cavity.
Early model compound studies and rhodopsin mutagenesis studies
Visual opsins provide a wide palette of colors for studying the opsin shift. However, opsin is not a trivial protein to work with. As an integral membrane protein, opsin is hard to express, purify and even more so to crystallize [44]. So far, none of the color opsins has been crystallized, only bovine rhodopsin’s structure and squid rhodopsin have been determined [18, 45]. Sequence homology of rhodopsin with the S-, M-, and L-opsins is ca. 40%, although structural data and analysis indicate a high structural homology [46, 47]. M and L-opsin are 95% identical in sequence (in the case of the human proteins) [48]. Mutagenesis studies have suggested that seven amino acids are responsible for the 30 nm difference between these two opsins [48, 49]. Chan et al. introduced three hydroxyl containing amino acids to bovine rhodopsin that resulted in ~20 nm red shift [50]. The bovine rhodopsin crystal structure shows that all three residues are in the ionone ring region, and indicates introduction of polar residues in the ionone region might lead to the red shift.
Structures of sensory rhodopsin II (500 nn), bacteriorhodopsin (560 nm), and halorhodopsin (570 nm) have been determined [51, 26, 27]. Kloppmann et al. performed electrostatic potential calculation on these three crystal structures [40]. The maps of electrostatic potential projected on the van der Waals surface of the retinal for the three rhodopsins suggests that the slightly more negative polarity in the ionone ring region contributes to the red shift observed in bacteriorhodopsin and halorhodopsin versus sensory rhodopsin. However, attempts to shift the wavelength of sensory rhodopsin to that of bacteriorhodopsin by changing the amino acids in the first shell of the binding pocket resulted only in a 24 nm red shift, versus the 60 nm difference observed between the two protein complexes [52, 53]. Based on the sensory rhodopsin II (SRII) crystal structure, Birge and coworkers performed MNDO-PSDCI molecular orbital theory to calculate the spectroscopic properties of bacteriorhodopsin (BR) and SRII [54]. They found that the remaining portion of opsin shift of BR over SRII not accounted by the mutations made by Kamo and coworkers [52, 53] could be explained by the altered position of Arg72 in SrII (Arg82 in BR). In SRII, this side chain has moved away from the chromophore Schiff base nitrogen and closer to the beta-ionylidene ring. This shift in position transfers this positively charged residue from a region of chromophore destabilization in BR to a region of chromophore stabilization in NpSRII, and is considered responsible for the rest of the blue shift in SRII.
Protein-bound water molecules can affect the dielectric environment of the binding pocket, but their role in spectral tuning of rhodopsin is not well known [42]. Recently, Katayama and collegues applied low-temperature Fourier transform infrared (FTIR) spectroscopy to monkey L and M cone opsins to study water vibrations inside the opsin binding pocket [42]. Consistently, the longer the absorbance wavelength of the opsin, the lower the averaged frequencies of water. This might suggest the important role of Protein-bound water molecules in affecting the chromophore’s absorbance. More studies are needed in order to understand the role of water molecules in spectral tuning. High-resolution crystal structure that can clearly show the position of the water molecules would greatly facilitate the studies.
Model analogs of retinal have also suggested aspects of the fundamental mechanism of wavelength regulation. Retinal analogs with ring-locked structures were used to study the contribution of conformational change to the absorption maximum of the retinal-PSB [55]. Molecules that are more planar lead to red shifted absorption spectra. To study the electronic effects on the chromophores’ absorption maxima, Sheves’ group used retinal-PSB analogs with a point charge positioned at different locations along the polyene [56]. Not surprisingly, a positive charge that can promote charge delocalization along the polyene gives the most red shifted species, while a positive charge along the polyene that inhibits delocalization of the iminium along the polyene results in absorption spectra (Fig. 3). Similarly, the counteranion placed at a further distance to the protonated Schiff base led to a red shift (Fig. 3) [57]. This leads to reduced stabilization of the positive charge on the iminium, which promotes positive charge delocalization along the polyene.
Figure 3.
Absorption maxima of retinal analogs in ethanol (top four molecules) and dichloromethane (bottom two molecules), demonstrating wavelength modulations as a result of charge interactions.
Prediction of the upper limit of red shift and computational studies of the retinal-PSB
It is generally believed that isolation of retinal-PSB from its counteranion results in red shifted spectra [58]. The counteranion is critical in stabilizing the positively charged retinal-PSB in a hydrophobic protein pocket through charge compensation. The distance between the counteranion and the iminium plays a crucial role in the absorbance of retinal-PSB [57], as a counteranion perturbs the delocalization of the positive charge along the polyene.
It was predicted from computational studies that counteranion removal will result in maximal positive charge delocalization and cause the retinal-PSB to red shift dramatically to ~600 nm [59]. Anderson et al. developed an electrostatic ion storage ring in Aarhus technique to record the electronic absorption spectrum of all-trans-retinal PSB in the gas phase [60]. It was found that retinal-PSB in the gas phase and thus isolated from a counteranion, has an absorption maximum at ~610 nm. More recently Rajput et al. improved the latter technique and were able to detect an absorption peak with a plateau extending from 530 to 610 nm [38]. The observation above was attributed to the chromophore adopting both 6-s-cis and 6-s-trans rotameric configurations in the gas phase (Fig. 2C). This was further verified by gas phase spectra of retinal analogs that would resemble the structure of the 6-s-cis and 6-s-trans retinal-PSB. Since the 6-s-cis rotamer is highly twisted because of steric repulsion between the gem-dimethyl group and C8–H, the first double bond is significantly less conjugated with the polyene. This results in a large blue shift (absorbing at 530 nm) compared to the 6-s-trans rotomer, which absorbs at 610 nm. These gas-phase studies provide a new perspective for wavelength regulation observed in rhodopsins, suggesting the possibility that the most red shifted rhodopsin pigments are due to better masking of the counteranion by the protein binding pocket.
The development of better computational tools, especially quantum mechanical/molecular mechanics hybrid platforms (QM/MM), have made the rhodopsin system amenable to such studies [61]. Crystal structure and mutagenesis studies on microbial rhodopsin provide a platform to test these computational models. High level theoretical methods are necessary to obtain more accurate calculation of the ground state and excited state energy, in order to obtain the absorption spectra [62, 63]. These calculations have shown that both electrostatic interactions and dispersive interactions due to polarizable aromatic residues play a crucial role in the red shift [64, 65, 53].
The central importance of the retinal-PSB for wavelength tuning suggested by the gas phase studies was tested computationally in the bovine rhodopsin. Indeed, introduction of the counteranion contributed the most blue shift from 610 nm in the gas phase to 486 nm in the protein environment, and other protein interactions counterbalance the counteranion effect and lead to the opsin shift [39].
Rhodopsin mimic engineering: Initial efforts
To avoid the pitfalls of working with either the natural integral membrane rhodopsin proteins, or the isolated chromophores devoid of the protein/chromophore interactions that must be the root of the phenomenon, Wang et al. studied spectral tuning using a novel strategy that is orthogonal to previous efforts [66]. They have developed small cellular proteins to be surrogates of the rhodopsins to study a protein’s effect on retinal-PSB’s absorption. The small cellular proteins they have used, cellular retinoic acid binding protein II and cellular retinol binding protein II, naturally bind ligands similar to the retinal PSB (retinoic acid and retinol or retinal, respectively) and have many significant advantages over the natural systems. They are expressed and purified with a high yield, unusually receptive to mutation, and readily produce crystals that diffract to high resolution (between 1.1 and 1.7 Å typically) [67, 68]. These characteristics allow for exhaustive, systematic analysis both spectroscopically and structurally.
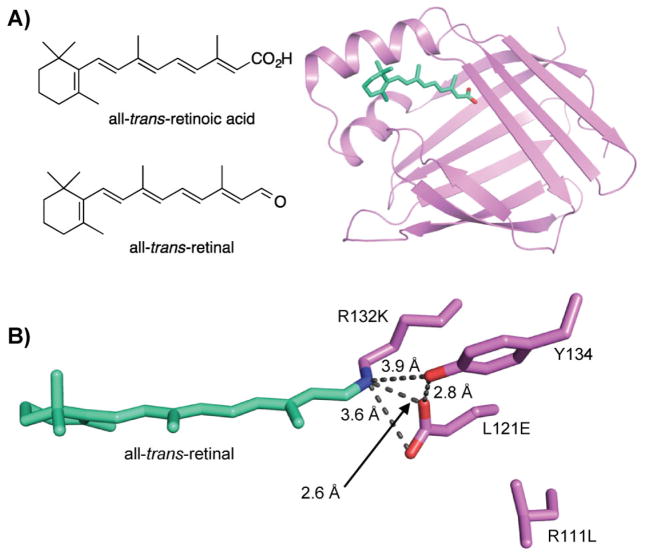
They first started with cellular retinoic acid binding protein II (CRABPII), which naturally binds all-trans-retinoic acid (Fig. 4A). To mimic rhodopsin, the small cellular protein must be re-engineered to bind and form a PSB with retinal. Modeling of the binding pocket led to the introduction of a lysine residue to form a PSB with retinal [69]. In the initial design, hydrophobic residues were introduced to lower the pKa of the newly introduced lysine residue such that the lysine would be more prone for nucleophilic attack to form a Schiff base with retinal. It was later realized that an optimal Bürgi–Dunitz trajectory is more critical for Schiff base formation than the hydrophobicity of the pocket [70, 71]. A triple mutant, R132K:R111L:L121E, was designed to be optimal for formation of a retinal-PSB with high binding affinity to retinal and reasonable pKa of 8.7 in the presence of a counteranion, L121E (Fig. 4B).
Figure 4.
Rhodopsin mimic based on CRABPII. A: Cartoon of the crystal structure of WT CRABPII bound with all-trans-retinoic acid. PDB code: 1CBS. B: Distinct residues in the binding pocket and retinal-PSB in the crystal structure of engineered CRABPII mutant, R132K:R111L:L121E. PDB code: 2G7B.
Full encapsulation of chromophore is crucial
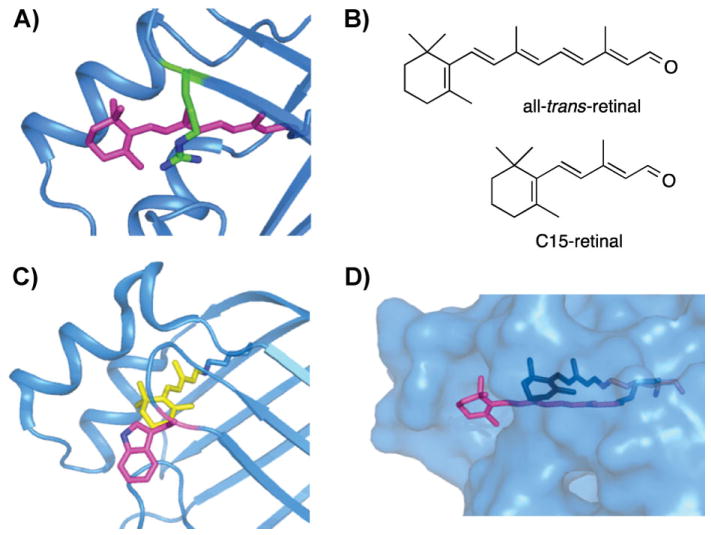
Mutations of the latter triple mutant with the aim of altering electrostatic interactions along the polyene did not result in wavelength regulation. In search of an explanation for the protein’s relative inertness to spectral tuning, it was noticed that the ionone ring of CRABPII was not fully buried in the binding cavity, leaving it exposed to bulk solvent. The aqueous solvent has a much higher dielectric constant (D =78) than the hydrophobic binding pocket of a protein (estimated to be between 2 and 4) [72]; exposure to this higher dielectric constant would buffer the relatively small changes in electrostatics caused by mutations of the binding pocket, resulting in a system that is insensitive to electrostatic changes in the binding cavity. This idea was first tested by using a shorter retinal analog (C15-retinal) by removing two double bonds. Compared to retinal, C15-retinal was fully embedded within the binding pocket (Fig. 5). As a result, introduction of negative polarity near the ionone ring region resulted in a red shift for C15-retinal, while it had no effect on retinal [73].
Figure 5.
Comparison of CRABPII bound with retinal and C15-retinal. A: Crystal structure of CRABPII bound with retinal, with retinal highlighted in magenta. PDB code: 2G7B. B: Chemical structures of all-trans-retinal and all-trans-C15-retinal. C: Crystal structure of CRABPII bound with C15-retinal, with C15-retinal highlighted in yellow, PDB code: 3F8A. D: Overlaid structure of CRABPII bound with retinal and C15-retinal, showing that C15-retinal is fully embedded within the protein binding pocket, while retinal (magenta) is exposed.
The second generation rhodopsin mimic surpassed the theoretical limit
Encouraged by the latter result, the search was on for another protein in the family that would bind retinal more deeply in the binding pocket, while maintaining the attractive features of CRABPII. This led to human cellular retinol binding protein II (hCRBPII). hCRBPII binds retinal fully within the binding pocket and more completely isolates the ligand from the bulk solvent (Fig. 6A) [66]. In contrast to both rhodopsin and the CRABPII-based rhodopsin mimic, a counteranion proximal to the imine was not required for PSB formation. As there is no need to counter balance the counteranion effect, which serves to localize the positive charge at the imine nitrogen, the positive charge on the iminium is more prone to be delocalized along the polyene.
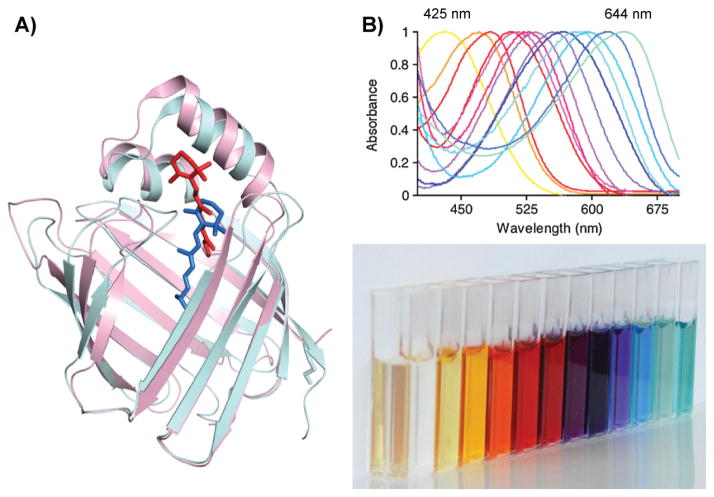
Figure 6.
Rhodopsin mimic based on hCRBPII. A: Overlaid crystal structures of R132K:R111L: L121E-CRABPII (pink, 2G7B) bound with all-trans-retinal (red) and WT hCRBPII (cyan, 2RCT) bound with all-trans-retinol (blue). B: UV spectra of hCRBPII mutants bound to retinal, spanning from 425 to 644 nm and a spectrum of hCRBPII mutant protein solution incubated with retinal in cuvettes.
The hCRBPII rhodopsin mimic proved to be spectacularly successful (Fig. 6B). It was shown that the wavelength of the retinal PSB could be modulated from 425 nm, when negatively charged residues close to the PSB iminium were introduced, to 644 nm, representing a red shift almost 30 nm further than was thought to be theoretically possible [38]. A number of conclusions were drawn from this study of over 300 hCRBPII rhodopsin mimic mutants. First it was clear that electrostatics, particularly in the vicinity of the PSB, played an important role in the mechanism of wavelength tuning. Changes that enhanced localization of positive charge at the iminium invariably lead to blue shifted absorption spectra, while mutations that discouraged positive charge localized at the PSB lead to red shifted spectra. Nonetheless, changes in electrostatics (mutation of a Thr for a Val as in the T51V mutation) were found to be important. On the other hand, the most important changes at the ionone ring end of the chromophore involved better encapsulation of the chromophore, not large electrostatic perturbations. For example, introduction of negatively charged residues in this region did not lead to the expected red shift, presumably because it would lead to localization of the cationic charge and defeat the goal of maximally distributing the cation along the polyene. Mutation of Arg 58 and Ala 33 to Trp led to better sequestration of the binding pocket from bulk solvent, and not only red shifted the spectrum, but also enhanced the electrostatic perturbations caused by other mutations, leading to a much larger redshift overall.
The overall take-home messages of the study were that while blue shifted absorption spectra were often the result of localizing positive charge on the iminium by directing negative electrostatic potential in this region, maximally red shifted absorption spectra are the result of first the removal of a counteranion in the vicinity of the PSB, and second the creation of a uniformly neutral electrostatic environment across the entire polyene. This uniformity necessarily requires the chromophore to be as isolated from the high dielectric constant of the bulk solvent as possible. Only when this is accomplished can other, more subtle variations in electrostatic potential realize their full effect on the spectroscopy of the chromophore. This allows for maximal delocalization of the positive charge, which invariably leads to the most red shifted absorption spectra observed for a retinylidene complex. This is in contrast to the idea that a charge reversal, where negative electrostatics at the ionone ring combined with more positive electrostatic potential at the PSB would lead to the most red shifted absorption spectra [74].
Rhodopsin mimics provide a simpler platform for theoreticians
The rhodopsin mimics derived from hCRBPII cover the entire visible spectrum. The structures of representatives that span the spectrum have been determined, many to high resolution (1.1 Å in some cases) [66] providing a powerful platform for theoreticians to develop methodology, especially with the exact positions of the protein bound water molecules defined. This will allow accurate examination of the effect of the bound water molecules in spectral tuning of retinal-PSB, which is currently unknown. Although the localization of structural waters could have different locations compared to opsins and therefore different influences on spectral tuning, the underlying principle for spectral tuning of the structural water molecules should be similar in both systems. Wavelength tuning in rhodopsin mimic system is not equivalent to opsins in that: all-trans-retinal is used as the chromophore, same as that in microbial rhodopsins, but different from that in visual opsins, where 11-cis-retinal is bound as the chromophore; besides, there is no direct counteranion interaction or water-mediated counteranion interaction in the bathochromically shifted rhodopsin mimic. However, this is a simpler problem than rhodopsin, as the protein is much smaller, more rigid and has only one shell of residues in the binding pocket. Besides, calculations can be directly compared among the different variants with only nine total mutations. The development of robust theoretical models of protein-chromophore interactions will eventually yield theoretical calculations that will closely predict the experimental data. This in turn may result in the engineering of natural rhodopsins for a wide range of applications.
Possibility of spectral tuning of opsins?
The idea of a balanced distribution of the iminium positive charge by fine tuning the electrostatic environment of the binding pocket may also apply to the opsin systems [75]. Assuming that removal of the counteranion would not abolish the function of the opsin, one could start by replacing the counteranion with a neutral polar residue or aromatic residue, similar to that which stabilizes the PSB in the hCRBPII system. Once a stable PSB in the absence of a counteranion is achieved, the iminium positive charge would be delocalized by tuning the electrostatics in the binding pocket.
Optogenetics is a newly developed technique that can spatially and temporally apply light to genetically modified animals to control the activity of neurons in live animals [76]. Channelrhodopsin has been engineered to induce cation influx upon blue light illumination, causing depolarization and subsequent firing of the neuron [77]. Halorhodopsin was used to uptake halide ions across the membrane in neurons to end depolarization, because halorhodopsin absorbs at 570 nm maximally and has little overlap with the absorption of channelrhodopsin [78]. These two microbial rhodopsins could be introduced together to turn on or shut off neuronal activity. Although optical fibers have been introduced to deliver light deep in the tissue to solve the problem of low tissue penetration of short-wavelength light, rhodopsins with longer absorption wavelength are favored for better tissue penetration and orthogonal usage of multiple rhodopsins to control neuronal activities. With the channelrhodopsin crystal structure now available [79], it will be easier to engineer channelrhodopsin for wavelength tuning. One could apply the principles learned from the rhodopsin mimic system by creating a uniform electrostatic environment surrounding the chromophore.
Conclusions and outlook
The mechanisms that allow a protein to modulate the absorption properties of an embedded chromophore has been a topic of interest for a number of years; the retinal-bound rhodopsin system representing a system of particular interest. Both model compound studies, where variants of the chromophore are studied in isolation, and studies of the retinylidene-bound rhodopsin system itself have been pursued. The recently developed soluble rhodopsin mimic based on hCRBPII, using a rational protein engineering strategy, represents a new tool for the study of this phenomenon. Using this system, Wang et al. have demonstrated wavelength tuning over the entire visible range (from 425 to 644 nm) by modifying the protein in only nine positions [66]. They were able to make, purify and study more than 300 mutants and obtain high resolution structures for many of these variants. Drastic wavelength tuning can be obtained by a combination of complete sequestration of the chromophore from solvent, electrostatic tuning, particularly in the vicinity of the PSB, and a uniformly neutral electrostatic potential across the polyene for maximal red shift. The application of these principles in the natural systems may lead to physiologically relevant variants with altered and enhanced spectroscopic properties. For example, super red shifted visual opsins that are sensitive to near IR light might be engineered; more red shifted microbial rhodopsins engineering could lead to near IR sensitive phototaxis and ion channels.
With regards to the application of these rhodopsin mimics, the next reasonable step is to engineer functions into these rhodopsin mimics. Is it possible to induce light dependent isomerization of the retinal-PSB that could result in a protein conformational change of the rhodopsin mimic? This way, a light induced protein-protein interaction could be achieved by evolving a peptide or protein to bind specifically with the rhodopsin mimic after isomerization. It might also be feasible to engineer these widely regulated rhodopsin mimics into chromophoric tags or fluorescent tags if appropriate retinal analogs are used.
Acknowledgments
We thank the Editor and two reviewers for their helpful comments that considerably improved the paper.
Abbreviations
- CRABPII
cellular retinoic acid binding protein II
- GPCR
G-protein coupled receptor
- hCRBPII
human cellular retinol binding protein II
- PSB
protonated Schiff base
Footnotes
Schiff base: an organic C=N derivative with three R-groups, two bound to the C and one bound to the N, the same as imine. Protonated Schiff base is the protonated form of Schiff base, C=NH+, with a proton bound to N, forming a positive charge in N, the same as iminium.
The authors have declared no conflict of interest.
References
- 1.Jacobs GH, Nathans J. The evolution of primate color vision. Sci Am. 2009;300:56–63. doi: 10.1038/scientificamerican0409-56. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs GH. Evolution of colour vision in mammals. Philos T R Soc B. 2009;364:2957–67. doi: 10.1098/rstb.2009.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunt DM, Carvalho LS, Cowing JA, Davies WL. Evolution and spectral tuning of visual pigments in birds and mammals. Philos T R Soc B. 2009;364:2941–55. doi: 10.1098/rstb.2009.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nevo R, Charuvi D, Tsabari O, Reich Z. Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 2012;70:157–76. doi: 10.1111/j.1365-313X.2011.04876.x. [DOI] [PubMed] [Google Scholar]
- 5.Braun-Sand S, Sharma PK, Chu ZT, Pisliakov AV, et al. The energetics of the primary proton transfer in bacteriorhodopsin revisited: It is a sequential light-induced charge separation after all. Biochim Biophys Acta. 2008;1777:441–52. doi: 10.1016/j.bbabio.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losi A, Gaertner W. The evolution of flavin-binding photo-receptors: an ancient chromophore serving trendy blue-light sensors. In: Merchant SS, editor. Annual Review of Plant Biology. Vol. 63. Palo Alto: Annual Reviews; 2012. pp. 49–72. [DOI] [PubMed] [Google Scholar]
- 7.Jekely G. Evolution of phototaxis. Philos T R Soc B. 2009;364:2795–808. doi: 10.1098/rstb.2009.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toh KC, Stojkovic EA, van Stokkum IHM, Moffat K, et al. Fluorescence quantum yield and photochemistry of bacteriophytochrome constructs. Phys Chem Chem Phys. 2011;13:11985–97. doi: 10.1039/c1cp00050k. [DOI] [PubMed] [Google Scholar]
- 9.Palczewski K. Chemistry and biology of vision. J Biol Chem. 2012;287:1612–9. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spudich JL, Yang CS, Jung KH, Spudich EN. Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol. 2000;16:365–92. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 11.Sung C-H, Chuang J-Z. The cell biology of vision. J Cell Biol. 2010;190:953–63. doi: 10.1083/jcb.201006020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osorio D, Marshall NJ, Cronin TW. Stomatopod photoreceptor spectral tuning as an adaptation for colour constancy in water. Vision Res. 1997;37:3299–309. doi: 10.1016/s0042-6989(97)00136-3. [DOI] [PubMed] [Google Scholar]
- 13.Marshall J, Oberwinkler J. The colourful world of the mantis shrimp. Nature. 1999;401:873–4. doi: 10.1038/44751. [DOI] [PubMed] [Google Scholar]
- 14.Cronin TW, Caldwell RL, Marshall J. Sensory adaptation –tunable colour vision in a mantis shrimp. Nature. 2001;411:547–8. doi: 10.1038/35079184. [DOI] [PubMed] [Google Scholar]
- 15.Conway BR, Chatterjee S, Field GD, Horwitz GD, et al. Advances in color science: from retina to behavior. J Neurosci. 2010;30:14955–63. doi: 10.1523/JNEUROSCI.4348-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. How color visual pigments are tuned. Trends Biochem Sci. 1999;24:300–5. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 17.Katayama K, Furutani Y, Imai H, Kandori H. Protein-bound water molecules in primate red- and green-sensitive visual pigments. Biochemistry. 2012;51:1126–33. doi: 10.1021/bi201676y. [DOI] [PubMed] [Google Scholar]
- 18.Palczewski K, Kumasaka T, Hori T, Behnke CA, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 19.Audet M, Bouvier M. Restructuring G-protein-coupled receptor activation. Cell. 2012;151:14–23. doi: 10.1016/j.cell.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Wald G. The receptors of human color vision. Science. 1964;145:1007–16. doi: 10.1126/science.145.3636.1007. [DOI] [PubMed] [Google Scholar]
- 21.Kim JE, Tauber MJ, Mathies RA. Wavelength dependent cis–trans isomerization in vision. Biochemistry. 2001;40:13774–8. doi: 10.1021/bi0116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palczewski K. G protein-coupled receptor rhodopsin. Annu Rev Biochem. 2006;75:743–67. doi: 10.1146/annurev.biochem.75.103004.142743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koenig D, Hofer H. The absolute threshold of cone vision. J Vision. 2011;11 doi: 10.1167/11.1.21. pii: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanishi K. Recent bioorganic studies on rhodopsin and visual transduction. Chem Pharm Bull. 2000;48:1399–409. doi: 10.1248/cpb.48.1399. [DOI] [PubMed] [Google Scholar]
- 25.Saari JC. Vitamin A metabolism in rod and cone visual cycles. In: Cousins RJ, editor. Annual Review of Nutrition. Vol. 32. Palo Alto: Annual Reviews; 2012. pp. 125–46. [DOI] [PubMed] [Google Scholar]
- 26.Luecke H, Schobert B, Richter HT, Cartailler JP, et al. Structure of bacteriorhodopsin at 1.55 angstrom resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 27.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 angstrom resolution. Science. 2000;288:1390–6. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki D, Irieda H, Homma M, Kawagishi I, et al. Phototactic and chemotactic signal transduction by transmembrane receptors and transducers in microorganisms. Sensors. 2010;10:4010–39. doi: 10.3390/s100404010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu H-Y, Lin Y-C, Chang Y-N, Tseng H, et al. A novel six-rhodopsin system in a single archaeon. J Bacteriol. 2010;192:5866–73. doi: 10.1128/JB.00642-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neutze R, Pebay-Peyroula E, Edman K, Royant A, et al. Bacteriorhodopsin: a high-resolution structural view of vectorial proton transport. Biochim Biophys Acta. 2002;1565:144–67. doi: 10.1016/s0005-2736(02)00566-7. [DOI] [PubMed] [Google Scholar]
- 31.Gai F, Hasson KC, McDonald JC, Anfinrud PA. Chemical dynamics in proteins: the photoisomerization of retinal in bacteriorhodopsin. Science. 1998;279:1886–91. doi: 10.1126/science.279.5358.1886. [DOI] [PubMed] [Google Scholar]
- 32.Pfisterer C, Gruia A, Fischer S. The mechanism of photo-energy storage in the halorhodopsin chloride pump. J Biol Chem. 2009;284:13562–9. doi: 10.1074/jbc.M808787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.del Rosario RCH, Oppawsky C, Tittor J, Oesterhelt D. Modeling the membrane potential generation of bacteriorhodopsin. Math Biosci. 2010;225:68–80. doi: 10.1016/j.mbs.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Klare JP, Bordignon E, Engelhard M, Steinhoff H-J. Transmembrane signal transduction in archaeal phototaxis: the sensory rhodopsin II-transducer complex studied by electron paramagnetic resonance spectroscopy. Eur J Cell Biol. 2011;90:731–9. doi: 10.1016/j.ejcb.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Choe H-W, Kim YJ, Park JH, Morizumi T, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–5. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 36.Zhu SS, Brown MF, Feller SE. Retinal conformation governs pK(a) of protonated Schiff base in rhodopsin activation. J Am Chem Soc. 2013;135:9391–8. doi: 10.1021/ja4002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakanishi K, Baloghnair V, Arnaboldi M, Tsujimoto K, et al. An external point-charge model for bacteriorhodopsin to account for its purple color. J Am Chem Soc. 1980;102:7945–7. [Google Scholar]
- 38.Rajput J, Rahbek DB, Andersen LH, Hirshfeld A, et al. Probing and modeling the absorption of retinal protein chromophores in vacuo. Angew Chem Int Ed Engl. 2010;49:1790–3. doi: 10.1002/anie.200905061. [DOI] [PubMed] [Google Scholar]
- 39.Sekharan S, Sugihara M, Buss V. Origin of spectral tuning in rhodopsin – it is not the binding pocket. Angew Chem Int Ed Engl. 2007;46:269–71. doi: 10.1002/anie.200603306. [DOI] [PubMed] [Google Scholar]
- 40.Kloppmann E, Becker T, Ullmann GM. Electrostatic potential at the retinal of three archaeal rhodopsins: implications for their different absorption spectra. Proteins. 2005;61:953–65. doi: 10.1002/prot.20744. [DOI] [PubMed] [Google Scholar]
- 41.Motto MG, Sheves M, Tsujimoto K, Baloghnair V, et al. Opsin shifts in bovine rhodopsin and bacteriorhodopsin – comparison of 2 external point-charge models. J Am Chem Soc. 1980;102:7947–9. [Google Scholar]
- 42.Rajamani R, Lin YL, Gao JL. The opsin shift and mechanism of spectral tuning in rhodopsin. J Comput Chem. 2011;32:854–65. doi: 10.1002/jcc.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathies R, Stryer L. Retinal has a highly dipolar vertically excited single-state – implications for vision. Proc Natl Acad Sci USA. 1976;73:2169–73. doi: 10.1073/pnas.73.7.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ujwal R, Bowie JU. Crystallizing membrane proteins using lipidic bicelles. Methods. 2011;55:337–41. doi: 10.1016/j.ymeth.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–7. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- 46.Nathans J, Thomas D, Hogness DS. Molecular-genetics of human color vision – the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 47.Stenkamp RE, Filipek S, Driessen CA, Teller DC, et al. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim Biophys Acta. 2002;1565:168–82. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 48.Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–4. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 49.Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–8. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 50.Chan T, Lee M, Sakmar TP. Introduction of hydroxyl-bearing amino-acids causes bathochromic spectral shifts in rhodopsin – aminoacid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 1992;267:9478–80. [PubMed] [Google Scholar]
- 51.Luecke H, Schobert B, Lanyi JK, Spudich EN, et al. Crystal structure of sensory rhodopsin II at 2.4 angstroms: insights into color tuning and transducer interaction. Science. 2001;293:1499–503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimono K, Ikeura Y, Sudo Y, Iwamoto M, et al. Environment around the chromophore in pharaonis phoborhodopsin: mutation analysis of the retinal binding site. Biochim Biophys Acta. 2001;1515:92–100. doi: 10.1016/s0005-2736(01)00394-7. [DOI] [PubMed] [Google Scholar]
- 53.Shimono K, Iwamoto M, Sumi M, Kamo N. Effects of three characteristic amino acid residues of Pharaonis phoborhodopsin on the absorption maximum. Photochem Photobiol. 2000;72:141–5. doi: 10.1562/0031-8655(2000)072<0141:eotcaa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 54.Ren L, Martin CH, Wise KJ, Gillespie NB, et al. Molecular mechanism of spectral tuning in sensory rhodopsin II. Biochemistry. 2001;40:13906–14. doi: 10.1021/bi0116487. [DOI] [PubMed] [Google Scholar]
- 55.Wada A, Tsutsumi M, Inatomi Y, Imai H, et al. Retinoids and related compounds. Part 26. Synthesis of (11 Z)-8,18-propano- and methano-retinals and conformational study of the rhodopsin chromophore. J Chem Soc. 2001;1:2430–9. [Google Scholar]
- 56.Baasov T, Sheves M. Alteration of PKA of the bacteriorhodopsin protonated Schiff-base – a study with model compounds. Biochemistry. 1986;25:5249–58. [Google Scholar]
- 57.Livnah N, Sheves M. Model compounds can mimic spectroscopic properties of bovine rhodopsin. J Am Chem Soc. 1993;115:351–3. [Google Scholar]
- 58.Sakmar TP, Franke RR, Khorana HG. The role of the retinylidene Schiff-base counterion in rhodopsin in determining wavelength absorbency and Schiff-base PKA. Proc Natl Acad Sci USA. 1991;88:3079–83. doi: 10.1073/pnas.88.8.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honig B, Greenberg AD, Dinur U, Ebrey TG. Visual-pigment spectra – implications of protonation of retinal Schiff-base. Biochemistry. 1976;15:4593–9. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- 60.Andersen LH, Nielsen IB, Kristensen MB, El Ghazaly MOA, et al. Absorption of Schiff-base retinal chromophores in vacuo. J Am Chem Soc. 2005;127:12347–50. doi: 10.1021/ja051638j. [DOI] [PubMed] [Google Scholar]
- 61.Sinicropi A, Basosi R, Olivucci M. Recent applications of a QM/MM scheme at the CASPT2//CASSCF/AMBER (or CHARMM) level of theory in photochemistry and photobiology. J Phys Conf Ser. 2008;101:012001. [Google Scholar]
- 62.Wanko M, Hoffmann M, Strodel P, Koslowski A, et al. Calculating absorption shifts for retinal proteins: Computational challenges. J Phys Chem B. 2005;109:3606–15. doi: 10.1021/jp0463060. [DOI] [PubMed] [Google Scholar]
- 63.Valsson O, Angeli C, Filippi C. Excitation energies of retinal chromophores: critical role of the structural model. Phys Chem Chem Phys. 2012;14:11015–20. doi: 10.1039/c2cp41387f. [DOI] [PubMed] [Google Scholar]
- 64.Soederhjelm P, Husberg C, Strambi A, Olivucci M, et al. Protein influence on electronic spectra modeled by multipoles and polarizabilities. J Chem Theory Comput. 2009;5:649–58. doi: 10.1021/ct800459t. [DOI] [PubMed] [Google Scholar]
- 65.Yan B, Spudich JL, Mazur P, Vunnam S, et al. Spectral tuning in bacteriorhodopsin in the absence of counterion and coplanarization effects. J Biol Chem. 1995;270:29668–70. doi: 10.1074/jbc.270.50.29668. [DOI] [PubMed] [Google Scholar]
- 66.Wang WJ, Nossoni Z, Berbasova T, Watson CT, et al. Tuning the electronic absorption of protein-embedded all-trans-retinal. Science. 2012;338:1340–3. doi: 10.1126/science.1226135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleywegt GJ, Bergfors T, Senn H, Lemotte P, et al. Crystal-structures of cellular retinoic acid-binding protein-I and protein-Ii in complex with all-trans-retinoic acid and a synthetic retinoid. Structure. 1994;2:1241–58. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 68.Winter NS, Bratt JM, Banaszak LJ. Crystal-structures of holo and apo-cellular retinol-binding protein-Ii. J Mol Biol. 1993;230:1247–59. doi: 10.1006/jmbi.1993.1239. [DOI] [PubMed] [Google Scholar]
- 69.Crist RM, Vasileiou C, Rabago-Smith M, Geiger JH, et al. Engineering a rhodopsin protein mimic. J Am Chem Soc. 2006;128:4522–3. doi: 10.1021/ja058591m. [DOI] [PubMed] [Google Scholar]
- 70.Vasileiou C, Vaezeslami S, Crist RM, Rabago-Smith M, et al. Protein design: reengineering cellular retinoic acid binding protein II into a rhodopsin protein mimic. J Am Chem Soc. 2007;129:6140–8. doi: 10.1021/ja067546r. [DOI] [PubMed] [Google Scholar]
- 71.Vasileiou C, Wang WJ, Jia XF, Lee KSS, et al. Elucidating the exact role of engineered CRABPII residues for the formation of a retinal protonated Schiff base. Proteins. 2009;77:812–22. doi: 10.1002/prot.22495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dwyer JJ, Gittis AG, Karp DA, Lattman EE, et al. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys J. 2000;79:1610–20. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee KSS, Berbasova T, Vasileiou C, Jia XF, et al. Probing wavelength regulation with an engineered rhodopsin mimic and a C15-retinal analogue. ChemPlusChem. 2012;77:273–6. [Google Scholar]
- 74.Honig B, Dinur U, Nakanishi K, Baloghnair V, et al. External point-charge model for wavelength regulation in visual pigments. J Am Chem Soc. 1979;101:7084–6. [Google Scholar]
- 75.Sakmar TP. Redder than red. Science. 2012;338:1299–300. doi: 10.1126/science.1231589. [DOI] [PubMed] [Google Scholar]
- 76.Mei Y, Zhang F. Molecular tools and approaches for optogenetics. Biol Psychiat. 2012;71:1033–8. doi: 10.1016/j.biopsych.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–92. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 78.Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PLoS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–74. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]