Abstract
Plant auto-inhibited Ca2+-ATPases (ACA) are crucial in defining the shape of calcium transients and therefore in eliciting plant responses to various stimuli. Arabidopsis thaliana genome encodes ten ACA isoforms that can be divided into four clusters based on gene structure and sequence homology. While isoforms from clusters 1, 2 and 4 have been characterized, virtually nothing is known about members of cluster 3 (ACA12 and ACA13). Here we show that a GFP-tagged ACA12 localizes at the plasma membrane and that expression of ACA12 rescues the phenotype of partial male sterility of a null mutant of the plasma membrane isoform ACA9, thus providing genetic evidence that ACA12 is a functional plasma membrane-resident Ca2+-ATPase. By ACA12 expression in yeast and purification by CaM-affinity chromatography, we show that, unlike other ACAs, the activity of ACA12 is not stimulated by CaM. Moreover, full length ACA12 is able to rescue a yeast mutant deficient in calcium pumps. Analysis of single point ACA12 mutants suggests that ACA12 loss of auto-inhibition can be ascribed to the lack of two acidic residues - highly conserved in other ACA isoforms - localized at the cytoplasmic edge of the second and third transmembrane segments. Together, these results support a model in which the calcium pump activity of ACA12 is primarily regulated by increasing or decreasing mRNA expression and/or protein translation and degradation.
Keywords: Ca2+- ATPase, Plasma membrane, Calmodulin, Arabidopsis thaliana
Introduction
Plant sensing of different environmental stimuli such as cold, light, osmotic, salt and drought signals, oxidative stress, plant hormones and pathogens, activate signal transduction systems involving transient increases in cytosolic free Ca2+ concentration. The specificity of the response to the various environmental changes depends on the amplitude, frequency and localization of the cytosolic calcium transient. Transient increases in cytosolic Ca2+ are triggered by the opening of Ca2+ permeable channels that mediate passive ion influx from the apoplast and/or intracellular compartments, while Ca2+ active transporters (Ca2+-H+ antiporters and Ca2+-ATPases) determine the recovery of the resting cytosolic free Ca2+ concentration after the stimulus. Ca2+-ATPases are thought to be crucial in defining the shape of the calcium transient and therefore in eliciting plant specific response to various stimuli (Sanders et al. 2002; McAinsh and Pittman 2009; Dodd et al. 2010; Kudla et al. 2010; Pittman et al. 2011; Spalding and Harper 2011; Bose et al. 2011).
Plant Ca2+-ATPases belong - as those of animals - to subgroup 2 of P-type ATPases super-family, which are characterized by forming a phosphorylated intermediate which changes conformation during the catalytic cycle, and by a number of highly conserved sequence motifs (Møller et al. 1996; Axelsen and Palmgren 1998; Palmgren and Nissen 2011). The genome of higher plants encodes several Ca2+-ATPases which group either with animal sarco-endoplasmic reticulum Ca2+-ATPase in the 2A subgroup of P-type ATPases (ECA, ER-type Ca2+-ATPase) or with animal plasma membrane (PM) Ca2+-ATPase in the 2B subgroup (ACA, auto-inhibited Ca2+-ATPase). Arabidopsis (Arabidopsis thaliana) has four ECA and ten ACA genes, with most cells expressing multiple isoforms (Geisler et al. 2000a; Sze et al. 2000; Bonza and De Michelis 2011; Pedersen et al. 2012).
A distinctive feature of ACAs in comparison to ECAs is an extended cytosolic N-terminal domain. In previously characterized members of the ACA subgroup, the N-terminus has been found to have an auto-inhibitory domain and a partially overlapping high affinity calmodulin (CaM) binding site (Geisler et al. 2000a; Baxter et al. 2003; Kabala and Klobus 2005; Boursiac and Harper 2007; Bonza and De Michelis 2011; Pedersen et al. 2012); a second low affinity CaM-binding site has been recently identified just 8 aa downstream the first one in ACA8, one of the best characterized ACA isoform (Tidow et al. 2012). In all ACA isoforms characterized so far, CaM-binding suppresses the auto-inhibitory action of the N-terminus, increasing Vmax and decreasing K0.5 for free Ca2+.
ACA isoforms can be divided into 4 clusters, based on sequence alignments and intron number and/or position (Baxter et al. 2003): the current evidence suggests that each cluster harbors isoforms with different features, such as sub-cellular localization or regulatory mechanisms (Baxter et al. 2003; Bonza and De Michelis 2011). The abundance of ACA isoforms, together with their co-expression in the same cell types, makes it difficult to define the role of each isoform in the physiology of the plant: however the phenotype of knock out mutants provides evidence for their role in development and/or response to biotic and abiotic stresses (Schiøtt et al. 2004; George et al. 2008; Boursiac et al. 2010; Frei dit Frey et al. 2012). Gaining a deeper knowledge on the biochemical properties and regulation of different ACA isoforms is important to assign them specific functions in vivo and therefore in understanding the biochemical pathways associated to relevant plant responses.
While at least one ACA has been previously characterized from clusters 1, 2 and 4, virtually nothing is known about members of cluster 3, which in Arabidopsis are isoforms ACA12 and ACA13. These isoforms, which are unique in being encoded by intron-less genes, have very low expression level in most cell types under basal conditions, but are dramatically induced upon exposure to a specific stress, such as in response to pathogens or UVB stress [(Boursiac and Harper 2007); http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi (Winter et al. 2007)]. Their N-terminal regions are highly divergent compared to those of other ACAs; moreover they have an Asn (N211 in ACA12) and an Arg (R334 in ACA12) at positions close to transmembrane domain (TM) 2 and 3 respectively, where all other ACAs - as well as animal PM Ca2+-ATPases - have an acidic residue. In different ACA isoforms, as well as in an animal pump isoform, mutation of these acidic residues generates deregulated pumps that show near full activity without further activation by CaM (Curran et al. 2000; Bredeston and Adamo 2004; Fusca et al. 2009).
Here we provide genetic evidence that ACA12 is a functional PM-resident Ca2+-ATPase, and biochemical evidence that ACA12 binds CaM but, unlike other ACAs, is not stimulated by CaM. In addition, a full length ACA12 is able to rescue a yeast mutant deficient in calcium pumps, unlike other well studied ACAs such as ACA8, which only provides a rescue when its auto-inhibitory N-terminus is deleted (Bonza et al. 2004; Baekgaard et al. 2006). Together, this supports a model in which the calcium pump activity provided by ACA12 is not dependent on Ca2+-CaM stimulation, would be therefore primarily regulated by increasing or decreasing mRNA expression and/or protein translation and degradation.
Materials and Methods
Plant lines and growth conditions
Arabidopsis thaliana ecotypes WS or Columbia were used for all plant experiments. For testing the ability of an ACA12 (At3g63380) gene to rescue a loss of function of ACA9 (At3g21180), two WS ecotype-based T-DNA insertion alleles were used, aca9-1 and aca9-2 (Schiøtt et al. 2004). For subcellular localization experiments, a transgene encoding an ACA12-GFP was transformed into A. thaliana ecotype Columbia.
For growing plants, seeds were sown on 0.5× Murashige and Skoog (MS) medium, pH 5.7 and stratified at 4°C for 48 h. Seedlings were grown at room temperature (22°C) under 24 h light for 7–10 days before being transplanted to soil. The soil used was Sunshine SMB-238 supplemented with 10-10-10 fertilizer (Hummert) and Marathon pesticide (Hummert) following the manufacturer's instructions. Plants were grown until maturity in a green house (with light and temperature conditions varying by seasons), or in growth chambers with a photoperiod of 16 h of light at 20°C and 8 h of dark at 18°C.
Plasmid construction
Plasmid construct 35S::ACA12-GFP (plasmid stock ps 391, see Online Resource 1), encodes an ACA12 with a C-terminal GFP followed by a 6His tag, downstream of a 35S promoter in a pBIN101.2 plant expression vector (Bevan 1984), harboring a kanamycin (kanr) resistance marker for bacterial and plant selections. This ACA12 coding sequence was generated by PCR amplification of genomic DNA from A. thaliana (Columbia), and sub-cloning into a pBIN101.2 plant expression vector (Bevan 1984). The ACA12 coding sequence begins with ATGAGGGACCTC and ends with CTCAAGAAACCT. The stop codon was removed to allow an in frame fusion with a GFP. The genomic sequence for ACA12 does not contain any intron. The absence of PCR mistakes was confirmed by DNA sequencing.
Plasmid construct ACA9promoter::ACA12-TAP2(YFP) (ps 688, see Online Resource 1), encodes an ACA12 with a C-terminal YFP downstream of a promoter from a preferentially pollen expressed gene, ACA9 (Schiøtt et al. 2004). This construct was based on a derivative of the pGreenII plant expression vector (Hellens et al. 2000) [kanr in bacteria, hygromycin-resistance (hygr) in plants]. Plasmid construct ACA9promoter::ACA2-TAP2(GFP) (ps 585, see Online Resource 1), is similar to ps 688 except that it encodes an ER-localized ACA2 (Hong et al. 1999) - fused to GFP.
For heterologous expression in yeast, ACA12 and ACA12-GFP coding sequences were amplified by PCR (Robocycler gradient 40, Stratagene) using ps 391 as template and cloned into the pYES2 vector (Online Resource 1). Generation of N211D and R334E mutants of ACA12-GFP was performed by site-directed mutagenesis using the Quickchange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol, using WT ACA12-GFP full length cDNA inserted in the pYES2 as a template (Online Resource 1).
Plant transformation
Plants were transformed with Agrobacterium tumefaciens strain GV3101 using the floral-dip method (Clough and Bent 1998). For the pGreenII vector, the Agrobacterium also included a pSOUP helper plasmid with a tetracycline resistance gene (Hellens et al. 2000). T1 seedlings were grown on 0.5× MS medium (pH 5.7) containing 1% agar, 0.05% MES, and 25 µg/ml hygromycin (pGreenII based constructs) or 50 µg/ml kanamycin (pBIN based vector) to identify stable transgenic lines.
Confocal microscopy
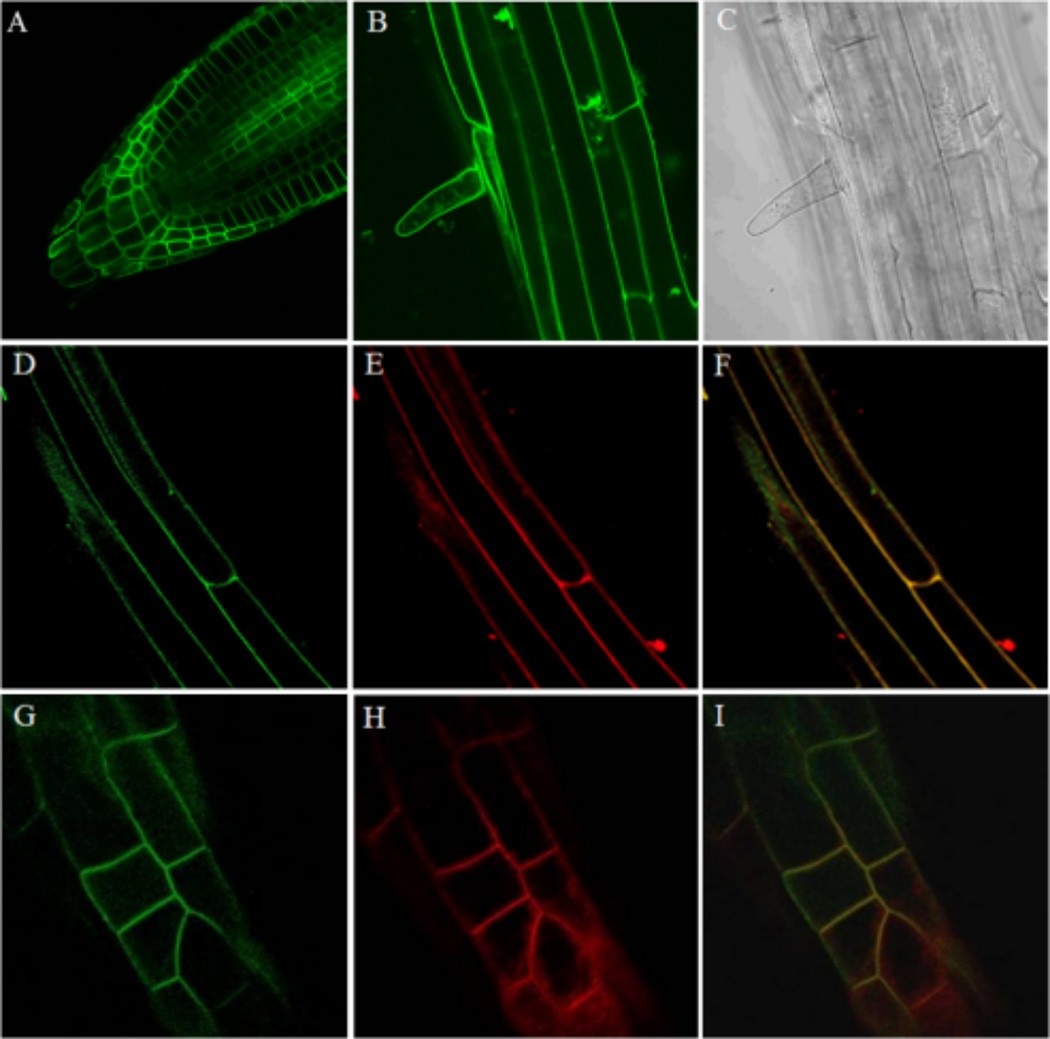
Root images of 7–10 day old dark-grown seedlings expressing ACA12-GFP under the control of the 35S promoter were collected using an Olympus IX81 FV1000 confocal microscope run by the Olympus FluoView 1.07.03.00 software package equipped with a 60× objective (numerical aperture 1.42) (Fig. 1 A–C) or a confocal microscope Leica TCS SP2 AOBS with a 40× oil-immersion objective (Fig.1 D–I). For co-localization experiments seedlings were exposed for 5–10 minutes to 4 µM FM4-64 dye freshly prepared solution (Bolte et al. 2004). Excitation at wavelengths of 488 nm was provided with an Argon-Ion laser. A spectral emission range of 500–600 nm was used for GFP imaging, while a 500–530 nm (GFP) and a 625–665 nm (FM4-64) range were used in co-localization experiments.
Fig. 1.
ACA12-GFP is PM localized. A, B, D and G: GFP fluorescence; C: phase contrast image; E and H: fluorescence of FM4-64 dye; F and I merging. A: root tip; B, C, D, E and F: maturation root zone; G, H and I: elongation root zone.
Yeast Strains, Transformation, and Growth Media
cDNAs coding for ACA12, ACA12-GFP, and ACA12-GFP mutants were in pYES2 vector (Invitrogen) under the control of the GAL1 promoter. Plasmids were used to transform Saccharomyces cerevisiae strain K616 [MATa pmr1::HIS3 pmc1::TRP1 cnb1::LEU2, ade2, ura3 (Cunningham and Fink 1994)] using a lithium acetate/single-stranded DNA/polyethylene glycol protocol and transformants were selected for uracil prototrophy by plating on complete synthetic medium lacking uracil (SC-URA) as described (Bonza et al. 2004). To induce protein expression, the different yeast strains were grown in SC-URA medium containing 2% (w/v) galactose, 1% (w/v) raffinose, 50 mM succinic acid/Tris (pH 5.5), 0.7% (w/v) yeast nitrogen base and 10 mM CaCl2, for 24 h at 20 °C.
Complementation of the Yeast Mutant K616
For complementation experiments, single colonies of the different strains were grown in a SC-URA medium containing 2% (w/v) glucose, 50 mM succinic acid/Tris (pH 5.5), 0.7% (w/v) yeast nitrogen base and 10 mM CaCl2; cultured cells were pelletted, washed and diluted with water to A600 = 1 - 0.3 - 0.1. Five µL of each dilution were spotted on solid SC- URA medium containing 2% (w/v) galactose, 1% (w/v) raffinose, 50 mM succinic acid/Tris (pH 5.5), 0.7% (w/v) yeast nitrogen base supplemented with 10 mM CaCl2 or 5 mM EGTA. As controls, yeast strain K616 transformed with the pYES2 empty vector or expressing full length ACA8 or the Δ74-ACA8 truncated mutant were used (Bonza and Luoni 2010). Growth was recorded after 3 to 8 days of incubation at 20°C.
Isolation of yeast microsomes
Microsomal fraction was isolated as previously reported (Fusca et al. 2009) except that yeast cells were lysed using a Cell Disruptor (Constant Systems Ltd) operating at 27 psi. Protein concentration was determined using the Bio-Rad assay (Bio-Rad) with γ-globulin as standard.
ACA12 and ACA12-GFP Purification by CaM-affinity Chromatography
CaM-affinity purification was performed as described (Fusca et al. 2009) with slight modifications. Proteins (100 mg) from yeast microsomes expressing ACA12 or ACA12-GFP were incubated with n-dodecyl β-D-maltoside (4 mg detergent ml−1: 4 mg protein ml−1) at 25 °C for 30 min in a solubilization medium containing 5% (v/v) glycerol, 50 mM Tris-HCl, pH 7.5, 1 mM p-aminobenzamidine, 2 mM DTT, 1.5 mM ATP, 2 mM CaCl2, 1mM MgSO4, 125 mM NaCl, supplemented with 5 µg ml−1 leupeptin, 5 µg ml−1 pepstatin, 5 µg ml−1 chymostatin immediately before use. The sample was then centrifuged 1 h at 110,000 g at 4°C; the supernatant recovered and incubated overnight under gentle rotation at 4 °C with 0.8 ml of CaM-Sepharose (GeHealthcare) previously equilibrated with solubilization medium supplemented with 37.5 µg ml−1 Brij 58. After removal of the unbound fraction, the resin was washed with 10 ml of washing medium containing 10% (v/v) glycerol, 20 mM Mops-KOH, pH 7.5, 1 mM p-aminobenzamidine, 2 mM DTT, 0.25 mM NaBr, 37.5 µg ml−1 Brij 58, 5 µg ml−1 leupeptin, 5 µg ml−1 pepstatin, 5 µg ml−1 chymostatin, 100 µM CaCl2, 100 µM MgSO4; second and third washes were performed in the same medium but in the absence of CaCl2 and MgSO4 (Bonza et al. 1998; Fusca et al. 2009). CaM-bound proteins were eluted in 2.5 ml of 5 mM Mops-KOH, pH 7.5, 37.5 µg ml−1 Brij 58, 10% (v/v) glycerol and 2 mM EGTA, followed by 1.5 ml of the same solution. The EGTA-eluted fractions were supplemented with 2 mM CaCl2, pooled and concentrated ten to twenty fold on Vivaspin ultrafiltration spin columns, 30 kDa cut-off (Sartorius), to a final concentration of 250–600 µg ml−1. The EGTA-eluted fraction was immediately used for assay of Ca2+-ATPase activity or re-lipidated by overnight incubation at 4°C under gentle rotation with soybean phosphatidylcoline (PC). PC, dissolved in 20 mg ml−1 Brij 58 and then diluted ten times in water to a final concentration of 8.5 mM, was supplied at 500 µM (Gourdon et al. 2011). Re-lipidated ACA12 maintains the same Ca2+-ATPase activity of the freshly purified protein (data not shown).
Electrophoresis and Western Blot analysis
SDS–PAGE and western blotting were performed as previously described (Bonza et al. 2000). ACA12-GFP decoration was performed incubating the nitrocellulose membrane for 1 h at 25 °C with the HisProbe-HRP, a nickel activated derivative of horseradish peroxidase (Thermo Scientific) which can bind to the His tag fused to the C-terminus of the protein. The probe was diluted 1:5000 in 0.05% (w/v) polyoxy-ethylene(20)sorbitan monolaurate, 0.15 M NaCl, 20 mM Tris–HCl, pH 7.4. ECL signal was acquired and quantified by the Fluor-Chem™SP Imaging System and AlphaEaseFC software (Alpha Innotech). Alternatively, gel was stained with silver impregnation (ProteoSilver™ Silver Stain Kit, Sigma Aldrich) or Coomassie blue staining method.
Assays of ACA12 activity
Activity of purified ACA12 (400–600 ng for each sample) was measured as Ca2+-dependent Mg-ATP hydrolysis as previously described (Bonza and Luoni 2010). Ca2+-dependent ATPase activity was evaluated as the difference between activity measured in the presence of 40 µM free Ca2+ and that measured in the absence of Ca2+ in the assay medium. Samples were incubated at 25 °C for 60 min, during which the reaction proceeds linearly. All the assays were performed at least three times, with three replicates.
Results
ACA12 is a functional PM Ca2+-ATPase
To define the intracellular localization of ACA12, transgenic plants expressing GFP-fused ACA12 (ACA12-GFP) under the control of the 35S promoter were generated and analyzed by confocal microscopy. Figure 1 shows that GFP fluorescence localizes at the periphery of the cells, consistent with a PM localization of ACA12 (panels A, B, D, G). To confirm this indication, roots were briefly (5–10 min) exposed to the FM4-64 dye, which under these conditions selectively labels the PM (Bolte et al. 2004): GFP and FM4-64 signals clearly co-localize at cell periphery (panels D–I).
Based on these results we tested whether ACA12 was able to complement the pollen fertility phenotype of an aca9 null mutant. ACA9 is a PM localized isoform primarily expressed in pollen and its deletion has been shown to reduce growth of pollen tubes with high frequency of aborted fertilization leading to a threefold reduction in seed set (Schiøtt et al. 2004). Results in Figure 2 show that the expression of ACA12 rescues the aca9 mutant, while that of ACA2, which encodes for an ACA isoform localized at the endoplasmic reticulum (Hong et al. 1999), is unable to complement the defect. Consistent with this difference in rescue potential, ACA12-YFP was observed to localize at the PM in pollen, whereas ACA2 was limited to the endomembranes (Online Resource 2). Together, these results indicate that ACA12 is a functional PM Ca2+ pump.
Fig. 2.
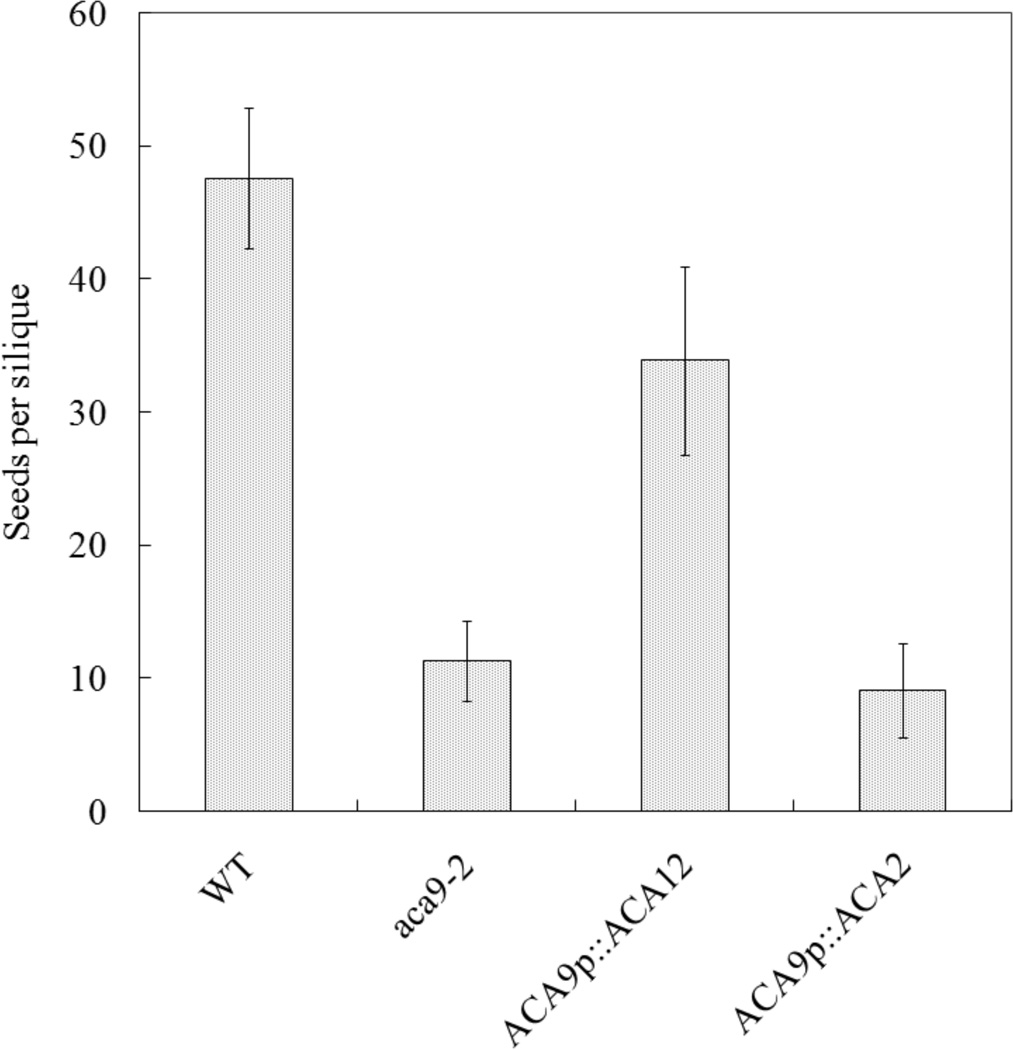
ACA12 rescues the reduced fertility phenotype of aca9 null mutants. Average seed set per silique was determined for plants grown under green house conditions. All wild type (WT), aca9-2, and transgenic lines were created using an A. thaliana ecotype WS background. Seeds were counted from 10 consecutive siliques on the primary bolt. Multiple plants were used for WT (N = 3), and aca9-2 (N = 4), and one each for 20 independent transgenic plant lines harboring an ER control ACA2 construct (ps 585), and 26 independent transgenic plants lines harboring an ACA12 construct (ps 688). Using another knockout line (aca9-1) transformed with the same ACA12 construct, an equivalent level of restored seed set was visually confirmed in a separate experiment. Four representative transgenic lines for ACA12-TAP2(YFP) in aca9-2 are ss 2099–2102, and ss 2103 to 2106 for the control ACA2-GFP in aca9-2.
Expression of ACA12 in yeast and protein purification
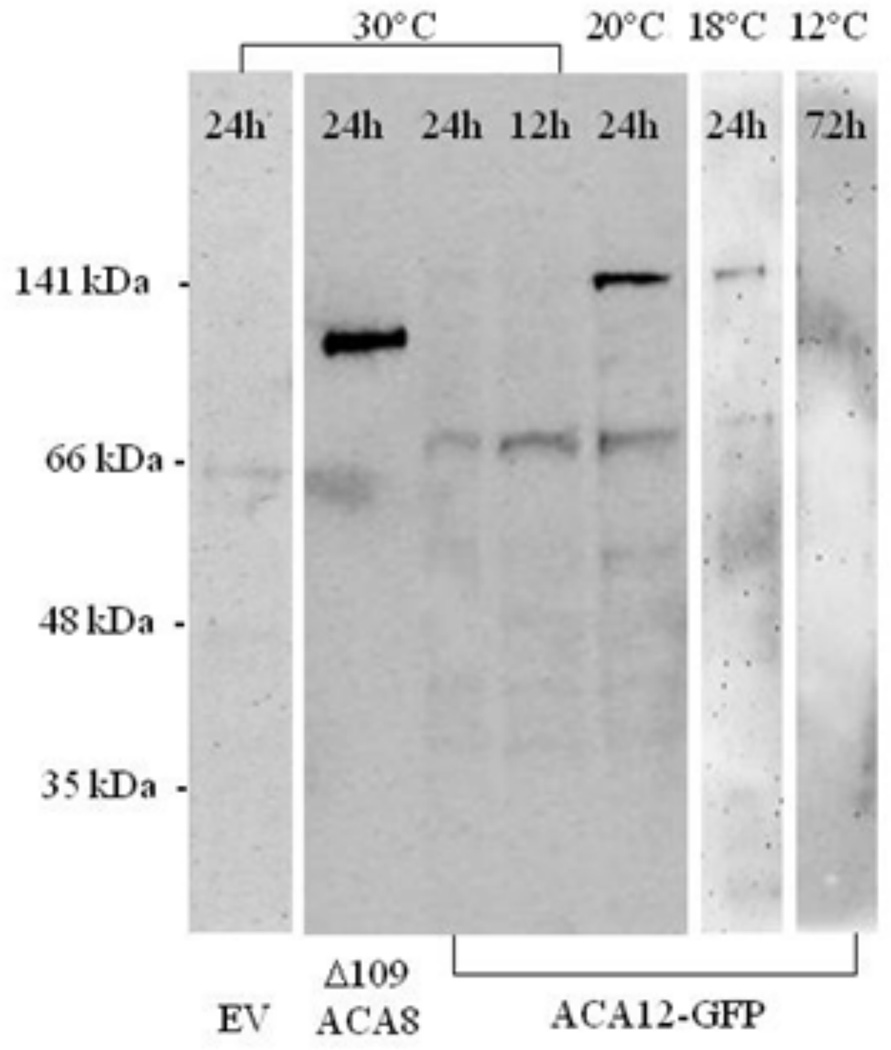
To biochemically characterize ACA12, plasmids encoding ACA12 and ACA12-GFP (the latter contains also a 6His tag) under the control of the GAL1 promoter were used to transform Saccharomyces cerevisiae strain K616, which lacks endogenous Ca2+-ATPases (Cunningham and Fink 1994). After induction of the yeast strain carrying ACA12-GFP, microsomes were isolated and solubilized microsomal proteins were subjected to SDS-PAGE, blotted and decorated with the HisProbe-HPR. After standard induction conditions (24 h at 30 °C) the probe recognizes only a very faint band of the expected molecular mass (141 kDa). However, an increase in accumulation was observed if the temperature during induction was lowered to 20 °C. Nevertheless, even in this condition, ACA12-GFP expression levels were very low compared to that of another ACA isoform: the signal in the lane containing a His-tagged Δ109-ACA8 is much stronger than that of ACA12-GFP, although hundred fold more microsomal proteins were loaded in the latter case; moreover also under this conditions the HisProbe-HPR labels several smaller bands (the most prominent of about 70 kDa) indicative of ACA12-GFP degradation. Attempts to improve ACA12-GFP expression by further lowering the temperature of induction till 12 °C (Fig. 3), by lowering Ca2+ concentration in the medium during induction to possibly select cells efficiently expressing the heterologous Ca2+-ATPase (Cunningham and Fink 1994), or by changing promoter were unsuccessful (data not shown).
Fig. 3.
Expression of ACA12-GFP in yeast strain K616. K616 yeast transformed with an empty pYES2 vector (EV), with Δ109-ACA8-6His or with ACA12-GFP was galactose-induced at the specified temperatures for the indicated times; solubilized microsomal proteins (30 µg for EV and ACA12-GFP and 0.3 µg for Δ109-ACA8) were subjected to SDS-PAGE, blotted and decorated with the HisProbe-HPR.
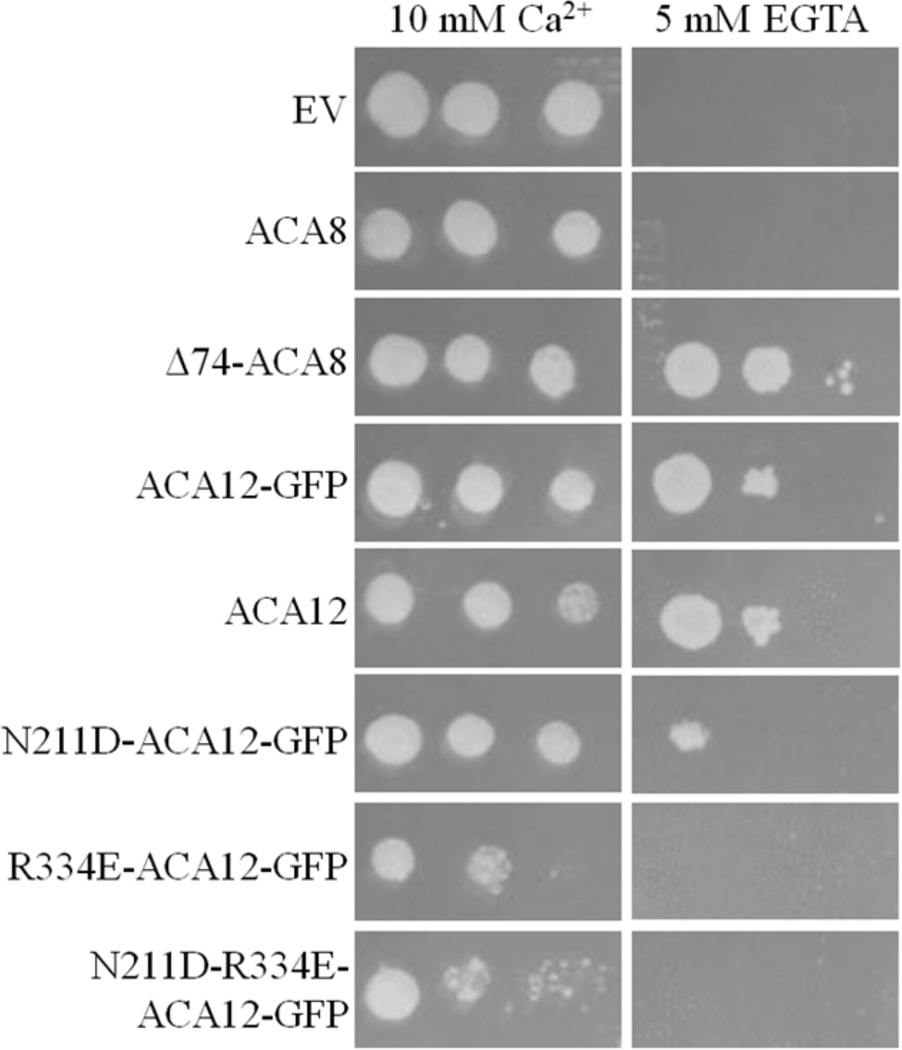
K616 yeast strain, being devoid of endogenous Ca2+-ATPases, is unable to grow in Ca2+-depleted media unless it expresses a deregulated form of ACA (Curran et al. 2000; Bonza et al. 2004; Baekgaard et al. 2006). We checked whether at 20 °C ACA12-GFP allows K616 growth in a Ca2+-depleted medium: Figure 4 shows that K616 yeast expressing ACA12-GFP grows in the presence of 5 mM EGTA nearly as well as that expressing the deregulated Δ74-ACA8 mutant, while that expressing WT ACA8 is unable to grow under these conditions (Bonza et al. 2004). This result indicates that the yeast expressed ACA12-GFP protein is functional and not auto-inhibited. A rescue was also obtained by K616 transformation with an untagged ACA12, indicating that the WT protein is also expressed in a functional form and that the addition of the GFP did not somehow cause the tagged pump to become de-regulated.
Fig. 4.
ACA12 complements the phenotype of Ca2+-ATPase deficient yeast K616 strain. Precultures of K616 transformed with the specified constructs grown in synthetic complete medium lacking uracil (SC-URA), 2% (w/v) glucose, 10 mM CaCl2 were pelletted, washed and diluted with water. Five µl of cells at A600 = 1.0 - or serial 1:3 dilutions - were spotted on SC-URA, 2% (w/v) galactose, 1% (w/v) raffinose, supplemented with 10 mM CaCl2 or 5 mM EGTA. All media were supplemented with 50 mM succinic acid/Tris (pH 5.5), 0.7% (w/v) yeast nitrogen base. Plates were incubated for 8 days at 20 °C. Results are from one experiment representative of three.
Given the low expression level of ACA12-GFP (and probably also of ACA12, see below) it is extremely unlikely that yeast growth under selective conditions is enabled by some ACA12 fragment which has lost the auto-inhibitory domain. However, to evaluate this possibility, we purified ACA12 from yeast using CaM-affinity chromatography (Bonza et al. 1998; Bonza et al. 2000; Fusca et al. 2009; Bonza and Luoni 2010), and tested its biochemical properties.
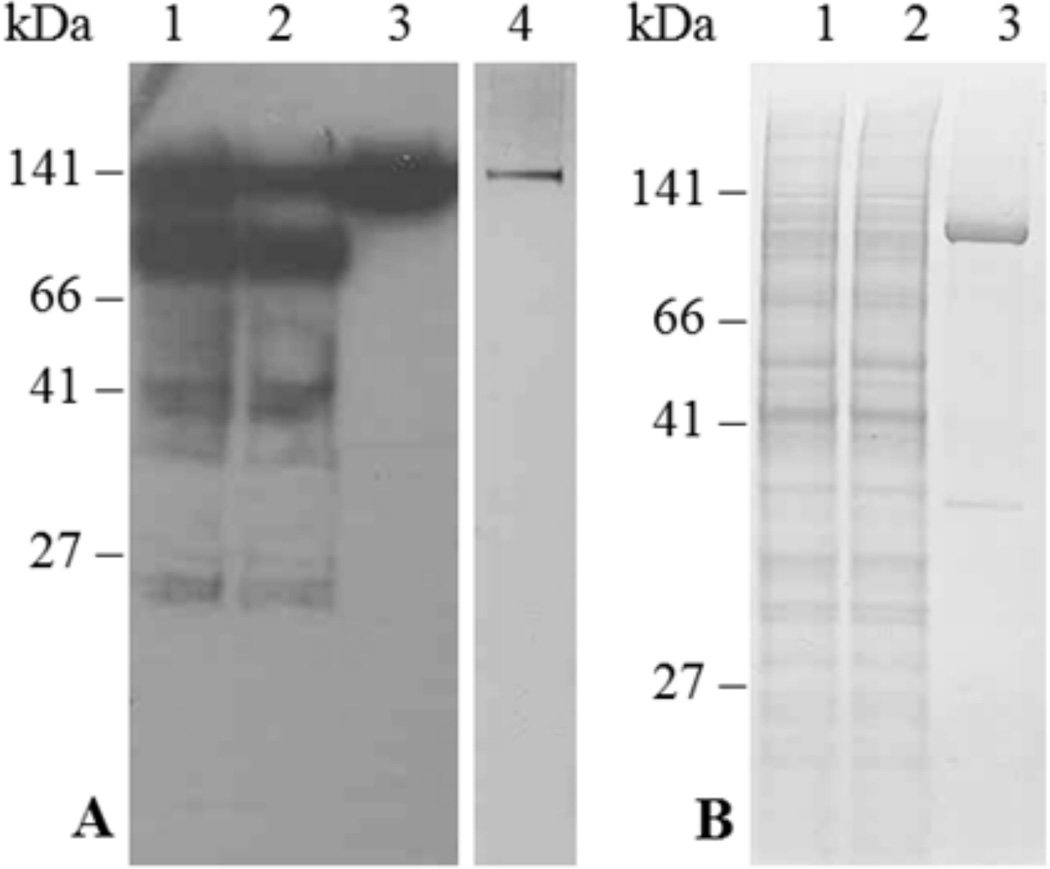
Panel A in Fig. 5 shows the main fractions obtained from purification of ACA12-GFP, which can be easily monitored thanks to the His tag. Upon overnight incubation with CaM-Sepharose of the microsomal proteins solubilized from K616 cells expressing ACA12-GFP, a relevant portion of the protein remains associated to the resin (compare lane 1 with lane 2, on which comparable amounts of the solubilized microsomal fraction and of the unbound fraction were loaded). The intact ACA12-GFP protein (141 kDa) is eluted upon Ca2+-chelation with 2 mM EGTA (lane 3), while the His-tagged lower molecular weight bands are undetectable in the EGTA-eluted fraction. Moreover, the 141 kDa band is by far the most prominent after SDS-PAGE and silver staining of the EGTA-eluted fraction (lane 5), indicating that this procedure allows the recovery of quite pure ACA12-GFP protein, albeit in tiny amounts (ca. 1 µg protein per mg of microsomal protein). Upon application of the same purification procedure to microsomes from K616 cells expressing WT ACA12 (panel B in Fig. 5), a band of 113 kDa is prominent in the EGTA-eluted fraction (lane 3), indicating that CaM-affinity also allows purification of the untagged ACA12 protein, albeit still with low yield (ca. 1 µg ACA12 per mg of microsomal protein).
Fig. 5.
Purification of ACA12-GFP (panel A) and ACA12 (panel B) by CaM-Sepharose affinity. Yeast microsomal proteins (100 mg), solubilized with n-dodecyl β-d-maltoside, were incubated overnight with CaM-Sepharose; after collection of the unbound protein fraction, the resin was washed as described in the Materials and Methods and CaM-bound proteins were eluted with 4 ml of elution buffer containing 2 mM EGTA. Panel A: lane 1, 10 µl of solubilized microsomal proteins (40 µg); lane 2, 10 µl of the unbound protein fraction; lane 3, 9 µl of the EGTA-eluted fraction; lane 4, 1 µl of the EGTA-eluted fraction concentrated ten-fold. Fractions were subjected to SDS-PAGE, blotted and decorated with the HisProbe-HPR (lanes 1–3) or subjected to SDS-PAGE and silver-stained (lane 5). Panel B: lane 1, 10 µl of solubilized microsomal proteins (40 µg); lane 2, 10 µl of the unbound protein fraction; lane 3, 3.5 µl of the EGTA-eluted fraction concentrated twenty-fold. Fractions were subjected to SDS-PAGE and stained with Comassie blue.
The purified ACA12 protein has Ca2+-ATPase activity insensitive to CaM even when supplied at 10 µM concentration (Table 1). ACA12 activity is inhibited by vanadate, a well known inhibitor of P-type ATPases, but in contrast with other ACAs [(Bonza and De Michelis 2011) and references therein] is virtually unable to use ITP as an alternative substrate (Table 1). As with other ACAs, ACA12 is inhibited by micromolar concentrations of eosin Y (Fig. 6), a fluorescein derivative which acts as a competitive inhibitor with respect to the nucleoside triphosphate substrate (De Michelis et al. 1993); however, the effective concentrations are about 2 order of magnitude higher that those required to inhibit ACA8 (Bonza et al. 2004).
Table 1. Activity of CaM-affinity purified ACA12.
Ca2+-ATPase activity of ACA12 was measured as described in the Materials and Methods. Results (plus or minus standard error of the mean) are from three experiments, each with three replicates, performed on different purified fractions.
| Assay conditions | Ca2+-ATPase activity (nmol Pi min−1 mg−1 protein) |
|---|---|
| ATP | 289 ± 27 |
| ATP + 1 µM CaM | 303 ± 31 |
| ATP + 10 µM CaM | 309 ± 29 |
| ATP + 200 µM vanadate | 3 ± 1 |
| ITP | 25 ± 3 |
Fig. 6.
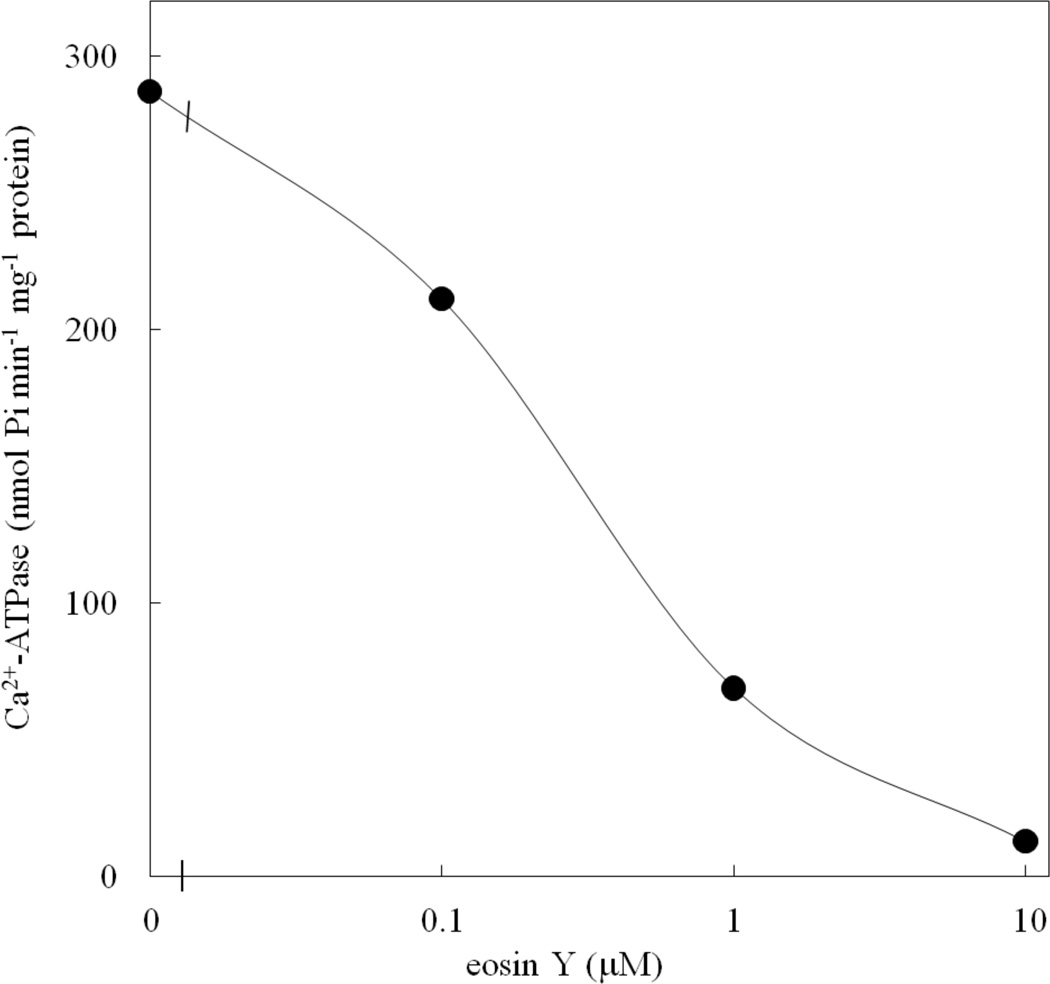
Effect of eosin Y on ACA12 activity. Ca2+-dependent ATPase activity of ACA12 was measured as described in the Materials and Methods, in the presence of the specified eosin Y concentrations. Results are from one experiment, representative of three.
The specific activity of the purified ACA12 is low as compared to the CaM-stimulated activity of ACA8 purified under the same conditions which ranges between 2 and 3 µmol min−1mg−1 protein [(Bonza and Luoni 2010); unpublished data from the authors laboratory]. This low activity is very unlikely to be an intrinsic characteristic of ACA12, since it is able to complement the K616 phenotype despite its very low expression level. Rather, it is probably due to protein damage during purification: indeed, the activity of purified ACA12 decreases by 50 to 80 % upon freezing and storage at −80 °C (data not shown) and can be preserved overnight only by maintaining the protein at 4 °C in the presence of phosphatidylcholine (100 fold molar excess). We tried to improve ACA12 activity by adding phospholipids during the solubilization and/or purification procedure, without success. The finding that the expression level of ACA12-GFP in mature leaves of transgenic plants expressing ACA12-GFP under the control of the 35S promoter is fairly low (data not shown) suggests that protein instability might be an intrinsic feature of ACA12.
Single point mutations of ACA12 suppress its ability to complement the phenotype of the K616 yeast strain
Alignment of ACA12 (Fig. 7) with the other ACAs of Arabidopsis shows that two residues crucial for auto-inhibition of type 2B Ca2+-ATPases and conserved in all members of the other three clusters, are variant in ACA12 and ACA13: ACA12 N211 aligns with an Asp, and R334 with a Glu in ACAs belonging to the other three clusters. In studies on other ACA isoforms as well as on a PMCA, single point mutations of each of these two acidic residues, respectively localized at the edge of TM2 and of TM3, generates constitutively active proteins, insensitive to CaM (Curran et al. 2000; Bredeston and Adamo 2004; Fusca et al. 2009). We generated ACA12 mutants in which N211 was mutated to Asp and/or R334 to Glu: all mutants are expressed in K616 yeast at levels similar to that of the WT protein (Online Resource 3). While the N211D-ACA12 only partially impairs the ability of the construct to rescue the growth phenotype (Fig. 4), there was no rescue with a single substitution of R334E-ACA12 or a double N211D-R334E substitution.
Fig. 7.

Alignment of Arabidopsis ACA isoforms. Top panel: TM2 (underlined in ACA12) and beginning of the small cytoplasmic loop, with the variant N211 of ACA12 highlighted (white on black background); bottom panel: end of the small cytoplasmic loop and TM3 (underlined in ACA12) with the variant R344 of ACA12 highlighted (white on black background). Sequence alignment was performed using ClustalW.
Discussion
In this paper we perform a characterization of ACA12 and we show that it has biochemical and regulatory features distinct from other characterized ACAs, potentially providing a unique biological function for plant cells. First, we demonstrate that it is localized at the PM: this is shown by confocal microscopy of plants expressing the ACA12-GFP or –YFP fusion proteins (Figure 1 and Online Resource 2), which co-localizes with the PM dye FM4-64 (Bolte et al. 2004), in agreement with the observations of Frei dit Frey et al (2012), and by the ability of ACA12 – expressed under the ACA9 promoter – to rescue the phenotype of a null mutant of ACA9 (Figure 2), a PM isoform primarily expressed in pollen (Schiøtt et al. 2004). The latter result also provides the first demonstration that ACA12 is a functional Ca2+-ATPase.
ACA12 and 13 are 40 to 50% similar to the other eight ACA isoforms in Arabidopsis, which together can be divided into 4 different clusters. However, their sequence is highly divergent in the regulatory N-terminus which contains the auto-inhibitory domain and the CaM-binding sites: in this region similarity between ACA12 and ACA13 is only 18% and their similarity to other ACAs range between 9 and 16%. In particular only one of the two anchor residues in the high affinity CaM-binding site (W28 of ACA12) is conserved. Nevertheless, ACA12 binds CaM and this has allowed its purification by CaM-affinity chromatography (Fig. 5).
ACA12 differs from the other ACAs characterized so far for being virtually unable to use ITP as a substrate alternative to ATP, and for being less sensitive to inhibition by the fluorescein derivative eosin Y (Table 1, Fig. 6). Since fluorescein and its derivatives are competitive inhibitors of P-type ATPases, which act by binding to a conserved Lys residue in the nucleotide-binding domain, these results suggest that the conformation of this domain on ACA12 is more similar to that of other P-type ATPase – which are ATP-specific and less sensitive to inhibition by fluorescein derivatives – than to that of ACA isoforms pertaining to the other three clusters (De Michelis et al. 1993; Bonza et al. 2004; Bonza and De Michelis 2011).
The activity of the purified ACA12 protein could not be activated by CaM (Table 1). Moreover, despite its low expression level, ACA12 was able to complement the K616 phenotype, making it able to grow in the presence of 5 mM EGTA (Fig. 4), as only deregulated mutants of other ACA isoforms are able to do (Harper et al. 1998; Chung et al. 2000; Geisler et al. 2000b; Hwang et al. 2000; Sze et al. 2000; Bonza et al. 2004; Kabala and Klobus 2005; Schiøtt and Palmgren 2005; Baekgaard et al. 2006; Lee et al. 2007). Taken together, these results clearly indicate that ACA12 is a deregulated isoform of Arabidopsis type 2B Ca2+-ATPase. This property may be connected to the lack of two acidic residues, conserved in other subgroups of ACAs as well as in animal type 2B Ca2+-ATPases, at the edge of TM2 and of TM3 respectively. Residue N211 of ACA12 aligns with a conserved Asp and residue R334 with an equally conserved Glu (Fig. 7): in different ACA isoforms, as well as in an animal pump isoform, mutation of these acidic residues generates a deregulated pump almost insensitive to further activation by CaM (Curran et al. 2000; Bredeston and Adamo 2004; Fusca et al. 2009). The N211D-ACA12 mutant allows some growth of the K616 yeast strain in low Ca2+ medium, while R334E-ACA12 as well as the double mutant N211D-R334E-ACA12 are unable to complement the K616 phenotype (Fig. 4); since the mutant proteins are expressed to levels similar to that of WT ACA12, the simplest interpretation of these results is that introduction of a negatively charged residue at one, or better both, of these positions restore in ACA12 the auto-inhibited conformation typical of other type 2B Ca2+-ATPases in the absence of CaM. However, since the activity of the R334E mutant purified with the same protocol used for the WT protein was barely detectable also in the presence of CaM (data not shown), the possibility exists - albeit unlikely since most ACA isoforms have a Glu at this position - that the mutation generates a non functional protein.
The question of the role that a deregulated ACA isoform as ACA12 can play in the physiology of the plant remains open. A careful analysis of gene knockouts for ACA12 has not yet been reported. While our study did not detect any obvious phenotype resulting from the expression of ACA12 under a 35S or ACA9 promoter, we did not conduct a systematic analysis of different growth conditions. However, expression profiling through microarrays shows that, in contrast to the relatively constant and redundant expression of most ACA isoforms, under normal growth conditions ACA12 transcripts are barely detectable throughout plant development. Nevertheless, ACA12 might be expressed under normal conditions in some specific cell type, such as guard cells [http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi (Winter et al. 2007)].
One potential hint on a possible role of ACA12 in plant development comes from the observation that, when transiently co-expressed in N. benthamiana leaves, ACA12 interacts at the PM with the brassinosteroid receptor BRI1 and with CLV1, which plays a central role in the control of the size of the central stem cell pool in the shoot apical meristem (Gish and Clark 2011; Frei dit Frey et al. 2012). However the same ability is shared by ACA8 (Frei dit Frey et al. 2012), a widely expressed PM isoform (Cerana et al. 2006). Since both BRI1 and CLV1 are members of the large family of receptor-like kinases (RLK) (Gish and Clark 2011), the two pumps might be substrate of their kinase activity. In the case of ACA8 it is known that this pump is phosphorylated in vivo at several Ser residues in the regulatory N-terminus and that phosphorylation can affect both auto-inhibition and CaM-binding (Giacometti et al. 2012 and references therein). Although most of these Ser residues are not conserved in ACA12, we cannot exclude that phospho- dephosphorylation might also impact ACA12 activity, and that the yeast expressed enzyme might have properties different from that expressed in planta.
RLKs constitute a large gene family in plants, with more than 600 members in Arabidopsis, and are involved in diverse ligand-mediated signaling pathways (Shiu and Bleecker 2001). In particular, several RLKs have been shown to be receptor proteins involved in perception of pathogen-associated molecular patterns (Boller and Felix 2009; Ranf et al. 2011); one of the first events in defense signaling is a transient increase of cytosolic free Ca2+ concentration (Ranf et al. 2011). Interestingly, interactomics screening by the split ubiquitin test has identified four orphan RLKs as ACA12 interactors (http://cas-biodb.cas.unt.edu/project/mind/search.php). Moreover, ACA12 expression is dramatically induced upon exposure to pathogens [(Boursiac and Harper 2007; http://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi; Winter et al. 2007)], suggesting that it may be involved in the adaptive response to these stress signals: expression of a deregulated PM Ca2+-ATPase may impact on the shape of pathogen-induced Ca2+ transient (Boller and Felix 2009; Ranf et al. 2011) and consequently on the response of the plant to a new pathogen attack.
Supplementary Material
Acknowledgments
This work was supported by grants to JFH from NSF DBI-0420033 for stress-dependent phenotype screens, NIH 1RO1 GM070813-01 for studies on pollen tube tip growth, and DOE DE-FG03-94ER20152 for studies on membrane biogenesis and genetic analyses of ACAs. Confocal microscopy was made possible by support at UNR from NIH COBRE grant RR024210. We thank Prof. Anna Moroni, Dipartimento di Bioscienze, Università degli Studi di Milano (Italy), for her generous support during completion of this work, Woo Sik Chung for assistance in cloning ACA12 from plant genomic DNA and Kathy Troung for assistance in plant rescue experiments.
LITERATURE CITED
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46(1):84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Baekgaard L, Luoni L, De Michelis MI, Palmgren MG. The plant plasma membrane Ca2+ pump ACA8 contains overlapping as well as physically separated auto-inhibitory and calmodulin-binding domains. J Biol Chem. 2006;281(2):1058–1065. doi: 10.1074/jbc.M508299200. [DOI] [PubMed] [Google Scholar]
- Baxter I, Tchieu J, Sussman MR, Boutry M, Palmgren MG, Gribskov M, Harper JF, Axelsen KB. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003;132(2):618–628. doi: 10.1104/pp.103.021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic acids research. 1984;12(22):8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A Renaissance of Elicitors: Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B. FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. Journal of microscopy. 2004;214(Pt 2):159–173. doi: 10.1111/j.0022-2720.2004.01348.x. [DOI] [PubMed] [Google Scholar]
- Bonza C, Carnelli A, Ida De Michelis M, Rasi-Caldogno F. Purification of the Plasma Membrane Ca2+-ATPase from Radish Seedlings by Calmodulin-Agarose Affinity Chromatography. Plant Physiol. 1998;116(2):845–851. doi: 10.1104/pp.116.2.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza MC, De Michelis MI. The plant Ca2+-ATPases repertoire: biochemical features and physiological functions. Plant Biol. 2011;13:421–430. doi: 10.1111/j.1438-8677.2010.00405.x. [DOI] [PubMed] [Google Scholar]
- Bonza MC, Luoni L. Plant and animal type 2B Ca2+-ATPases: evidence for a common auto-inhibitory mechanism. FEBS Lett. 2010;584(23):4783–4788. doi: 10.1016/j.febslet.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Bonza MC, Luoni L, De Michelis MI. Functional expression in yeast of an N-deleted form of At-ACA8, a plasma membrane Ca2+-ATPase of Arabidopsis thaliana, and characterization of a hyperactive mutant. Planta. 2004;218(5):814–823. doi: 10.1007/s00425-003-1160-y. [DOI] [PubMed] [Google Scholar]
- Bonza MC, Morandini P, Luoni L, Geisler M, Palmgren MG, De Michelis MI. At-ACA8. encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol. 2000;123(4):1495–1506. doi: 10.1104/pp.123.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose J, Pottosin II, Shabala SS, Palmgren MG, Shabala S. Calcium efflux systems in stress signaling and adaptation in plants. Front Plant Sci. 2011;2:85. doi: 10.3389/fpls.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursiac Y, Harper JF. The origin and function of calmodulin regulated Ca2+ pumps in plants. J Bioenerg Biomembr. 2007;39(5–6):409–414. doi: 10.1007/s10863-007-9104-z. [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Lee SM, Romanowsky S, Blank R, Sladek C, Chung WS, Harper JF. Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 2010;154(3):1158–1171. doi: 10.1104/pp.110.159038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredeston LM, Adamo HP. Loss of autoinhibition of the plasma membrane Ca2+ pump by substitution of aspartic 170 by asparagin. Activation of plasma membrane calcium ATPase 4 without disruption of the interaction between the catalytic core and the C-terminal regulatory domain. J Biol Chem. 2004;279(40):41619–41625. doi: 10.1074/jbc.M403116200. [DOI] [PubMed] [Google Scholar]
- Cerana M, Bonza MC, Harris R, Sanders D, De Michelis MI. Abscisic acid stimulates the expression of two isoforms of plasma membrane Ca2+-ATPase in Arabidopsis thaliana seedlings. Plant Biol (Stuttg) 2006;8(5):572–578. doi: 10.1055/s-2006-924111. [DOI] [PubMed] [Google Scholar]
- Chung WS, Lee SH, Kim JC, Heo WD, Kim MC, Park CY, Park HC, Lim CO, Kim WB, Harper JF, Cho MJ. Identification of a calmodulin-regulated soybean Ca2+-ATPase (SCA1) that is located in the plasma membrane. Plant Cell. 2000;12(8):1393–1407. doi: 10.1105/tpc.12.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124(3):351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran AC, Hwang I, Corbin J, Martinez S, Rayle D, Sze H, Harper JF. Autoinhibition of a calmodulin-dependent calcium pump involves a structure in the stalk that connects the transmembrane domain to the ATPase catalytic domain. J Biol Chem. 2000;275(39):30301–30308. doi: 10.1074/jbc.M002047200. [DOI] [PubMed] [Google Scholar]
- De Michelis MI, Carnelli A, Rasi-Caldogno F. The Ca2+ pump of the plasma membrane of Arabidopsis thaliana- characteristics and sensitivity to fluorescein derivatives. Bot Acta. 1993;106(1):20–25. [Google Scholar]
- Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620. doi: 10.1146/annurev-arplant-070109-104628. [DOI] [PubMed] [Google Scholar]
- Frei dit Frey N, Mbengue M, Kwaaitaal M, Nitsch L, Altenbach D, Häweker H, Lozano-Duran R, Njo MF, Beeckman T, Huettel B, Borst JW, Panstruga R, Robatzek S. Plasma membrane calcium ATPases are important components of receptor-mediated signaling in plant immune responses and development. Plant Physiol. 2012;159(2):798–809. doi: 10.1104/pp.111.192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusca T, Bonza MC, Luoni L, Meneghelli S, Marrano CA, De Michelis MI. Single point mutations in the small cytoplasmic loop of ACA8, a plasma membrane Ca2+-ATPase of Arabidopsis thaliana, generate partially deregulated pumps. J Biol Chem. 2009;284(45):30881–30888. doi: 10.1074/jbc.M109.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Axelsen KB, Harper JF, Palmgren MG. Molecular aspects of higher plant P-type Ca2+-ATPases. Biochim Biophys Acta. 2000a;1465(1–2):52–78. doi: 10.1016/s0005-2736(00)00131-0. [DOI] [PubMed] [Google Scholar]
- Geisler M, Frangne N, Gomes E, Martinoia E, Palmgren MG. The ACA4 gene of Arabidopsis encodes a vacuolar membrane calcium pump that improves salt tolerance in yeast. Plant Physiol. 2000b;124(4):1814–1827. doi: 10.1104/pp.124.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L, Romanowsky SM, Harper JF, Sharrock RA. The ACA10 Ca2+-ATPase regulates adult vegetative development and inflorescence architecture in Arabidopsis. Plant Physiol. 2008;146(2):716–728. doi: 10.1104/pp.107.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti S, Marrano CA, Bonza MC, Luoni L, Limonta M, De Michelis MI. Phosphorylation of serine residues in the N-terminus modulates the activity of ACA8, a plasma membrane Ca2+-ATPase of Arabidopsis thaliana. J Exp Bot. 2012;63(3):1215–1224. doi: 10.1093/jxb/err346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish LA, Clark SE. The RLK/Pelle family of kinases. Plant Journal. 2011;66(1):117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourdon P, Andersen JL, Hein KL, Bublitz M, Pedersen BP, Liu XY, Yatime L, Nyblom M, Nielsen TT, Olesen C, Møller JV, Nissen P, Morth JP. HiLiDe-Systematic Approach to Membrane Protein Crystallization in Lipid and Detergent. Cryst Growth Des. 2011;11(6):2098–2106. [Google Scholar]
- Harper JF, Hong B, Hwang I, Guo HQ, Stoddard R, Huang JF, Palmgren MG, Sze H. A novel calmodulin-regulated Ca2+-ATPase (ACA2) from Arabidopsis with an N-terminal autoinhibitory domain. J Biol Chem. 1998;273(2):1099–1106. doi: 10.1074/jbc.273.2.1099. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant molecular biology. 2000;42(6):819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- Hong B, Ichida A, Wang Y, Gens JS, Pickard BG, Harper JF. Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum. Plant Physiol. 1999;119(4):1165–1176. doi: 10.1104/pp.119.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Harper JF, Liang F, Sze H. Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal autoinhibitory domain. Plant Physiol. 2000;122(1):157–168. doi: 10.1104/pp.122.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabala K, Klobus G. Plant Ca2+ -ATPases Acta Physiol Plant. 2005;27:559–574. [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22(3):541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Kim HS, Han HJ, Moon BC, Kim CY, Harper JF, Chung WS. Identification of a calmodulin-regulated autoinhibited Ca2+-ATPase (ACA11) that is localized to vacuole membranes in Arabidopsis. FEBS Lett. 2007;581(21):3943–3949. doi: 10.1016/j.febslet.2007.07.023. [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Pittman JK. Shaping the calcium signature. New Phytol. 2009;181(2):275–294. doi: 10.1111/j.1469-8137.2008.02682.x. [DOI] [PubMed] [Google Scholar]
- Møller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286(1):1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P. P-type ATPases. Annu Rev Biophys. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- Pedersen CN, Axelsen KB, Harper JF, Palmgren MG. Evolution of plant p-type ATPases. Front Plant Sci. 2012;3:31. doi: 10.3389/fpls.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK, Bonza MC, De Michelis MI. Ca2+ pumps and Ca2+ antiporters in plant development. In: Geisler M, Venema K, editors. Transporters and Pumps in Plant Signalling. Berlin, Heidelberg, D: Springer-Verlag; 2011. pp. 133–161. [Google Scholar]
- Ranf S, Eschen-Lippold L, Pecher P, Lee J, Scheel D. Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant Journal. 2011;68(1):100–113. doi: 10.1111/j.1365-313X.2011.04671.x. [DOI] [PubMed] [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. Calcium at the crossroads of signaling. Plant Cell. 2002;14(Suppl):S401–S417. doi: 10.1105/tpc.002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiøtt M, Palmgren MG. Two plant Ca2+ pumps expressed in stomatal guard cells show opposite expression patterns during cold stress. Physiol Plant. 2005;124(2):278–283. [Google Scholar]
- Schiøtt M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc Natl Acad Sci USA. 2004;101(25):9502–9507. doi: 10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding EP, Harper JF. The ins and outs of cellular Ca2+ transport. Curr Opin Plant Biol. 2011;14(6):715–720. doi: 10.1016/j.pbi.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze H, Liang F, Hwang I, Curran AC, Harper JF. Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:433–462. doi: 10.1146/annurev.arplant.51.1.433. [DOI] [PubMed] [Google Scholar]
- Tidow H, Poulsen LR, Andreeva A, Knudsen M, Hein KL, Wiuf C, Palmgren MG, Nissen P. A bimodular mechanism of calcium control in eukaryotes. Nature. 2012;491(7424):468–472. doi: 10.1038/nature11539. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An "Electronic Fluorescent Pictograph" browser for exploring and analyzing large-scale biological data sets. PloS One. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.