Abstract
Glioblastoma multiforme (GBM) is a class of primary brain tumours characterized by their ability to rapidly proliferate and diffusely infiltrate surrounding brain tissue. The aggressive growth of GBM leads to the development of regions of low oxygenation (hypoxia), which can be clinically assessed through [18F]-fluoromisonidazole (FMISO) positron emission tomography (PET) imaging. Building upon the success of our previous mathematical modelling efforts, we have expanded our model to include the tumour microenvironment, specifically incorporating hypoxia, necrosis and angiogenesis. A pharmacokinetic model for the FMISO-PET tracer is applied at each spatial location throughout the brain and an analytical simulator for the image acquisition and reconstruction methods is applied to the resultant tracer activity map. The combination of our anatomical model with one for FMISO tracer dynamics and PET image reconstruction is able to produce a patient-specific virtual PET image that reproduces the image characteristics of the clinical PET scan as well as shows no statistical difference in the distribution of hypoxia within the tumour. This work establishes proof of principle for a link between anatomical (magnetic resonance image [MRI]) and molecular (PET) imaging on a patient-specific basis as well as address otherwise untenable questions in molecular imaging, such as determining the effect on tracer activity from cellular density. Although further investigation is necessary to establish the predictive value of this technique, this unique tool provides a better dynamic understanding of the biological connection between anatomical changes seen on MRI and biochemical activity seen on PET of GBM in vivo.
Keywords: Glioma, GBM, mathematical modelling, MRI, FMISO-PET, hypoxia
1. Introduction
Glioblastoma multiforme (GBM) is a primary brain tumour characterized by excessive proliferation and diffuse invasion of the brain parenchyma while remaining localized in the brain (Kleihues et al., 2002; Louis et al., 2007). Although GBM patients receive extensive treatment including surgical resection, radiotherapy and chemotherapy, these tumours prove to be uniformly fatal with a median survival of 11 months (Burnet et al., 2007). GBMs inevitably recur despite aggressive treatment due to the extensive invasion of tumour cells invisible to medical imaging, which can vary from patient to patient. In addition to a poor prognosis, anatomical and metabolic traits of GBM manifest differently across patients, suggesting that treatment could benefit from information on tumour growth dynamics on an individual patient level. The process of interpreting clinical images for treatment planning is a relatively qualitative process because of the lack of quantitative tools, making GBM an ideal medical condition for patient-specific mathematical modelling. In this report, we demonstrate a proof-of-principle system that builds upon previously established methods of GBM modelling, positron emission tomography (PET) radio-tracer kinetics and PET imaging simulation to form an integrated method of generating a patient-specific FMISO-PET simulation. This approach is both novel and clinically relevant because it provides a link between anatomical (magnetic resonance image [MRI]) and molecular (PET) image modalities.
1.1. Patient-specific mathematical modelling of GBM: proliferation–invasion and proliferation-invasion-hypoxia–necrosis–angiogenesis models
Patient-specific mathematical modelling of glioma growth and invasion began in the 1990s using crude information regarding tumour volume provided by the patients’ computerized tomography (CT) images (Tracqui et al., 1995). Since then, glioma modelling has developed to incorporate realistic heterogeneity and anisotropy of the brain anatomy informed by routine MRIs (Harpold et al., 2007; Jbabdi et al., 2005) in the form of the proliferation–invasion (PI) model. This model quantifies glioma growth in terms of net rates of proliferation (ρ) and invasion (D) and is represented as a reaction–diffusion equation as follows:
| (1) |
In words, the rate of change of glioma cell density equals the sum of the net dispersal of glioma cells and the net proliferation of glioma cells. This model, studied by Swanson et al. (2008) (Harpold et al., 2007), has proven to be an accurate predictor of anatomical changes that can be imaged by MRI in individual patients (Wang et al., 2009; Szeto et al., 2009; Swanson et al., 2003, 2008; Rockne et al., 2009). Further, patient-specific PI model parameters for biological aggressiveness (D and ρ) can be estimated from routinely available pre-treatment MRIs (Harpold et al., 2007). These patient-specific proliferation and invasion kinetic rates are prognostic of survival (Wang et al., 2009) and response to therapy (Swanson et al., 2003, 2008; Rockne et al., 2009) in individual patients.
Although the PI model has been validated on both the population and patient-specific levels, it provides limited insight with regard to tumour activity at the molecular level. Despite the PI model’s focus on macroscopic down-stream net behaviour of GBM, we have recently reported that the PI model-defined metrics of biological aggressiveness (ρ/D) correlated closely with the tumour’s hypoxic burden imageable on 18F-fluoromisonidazole (FMISO) PET in GBM Szeto et al. (2009). This connection suggested a quantitative link between the dynamics of anatomical growth (seen on MRI) and the molecular characteristics of the tumour (hypoxia imaged on FMISO-PET) thus providing a grounding upon which to build a more sophisticated model that included molecular information regarding the tumour.
Our previous successes with the PI model combined with the need for more information on the molecular level dynamics of tumour growth has led us to explore an expanded model, which we will refer to as the proliferation-invasion-hypoxia–necrosis–angiogenesis (PIHNA) model (Swanson et al., 2011). The PIHNA model characterizes GBM evolution by partitioning the malignant tumour cells into subpopulations based on fundamental histological characteristics of GBM including normoxic GBM cells, hypoxic GBM cells, necrotic tissue, neo-angiogenic vasculature and angiogenic factors ((5)–(10)). The PIHNA model provides insight into the biological interplay of metabolic changes that take place within and between cells, which can be validated through functional imaging, such as PET on a patient-specific level. Here, we explore our ability to simulate maps of hypoxia using the PIHNA model to compare with patient FMISO-PET images. However, to realistically compare model predictions with clinical imaging data, we must incorporate the dynamic nature and implicit noise characteristics of PET imaging in our virtual FMISO-PET images.
1.2. Simulating patient-specific FMISO-PET images
From a histological perspective, GBMs are characterized by central or focal necrosis and hypoxia, which results from the rapid depletion of nutrients that corresponds with aberrant growth (Rajendran et al., 2004; Fischer et al., 2005; Louis et al., 2007). In a clinical setting, hypoxia is measured in GBM patients in vivo using FMISO-PET. FMISO covalently binds to macromolecules within hypoxic cells and remains sequestered within them (Vallabhajosula, 2007). Our aim is to transform PIHNA model predictions of tumour-induced hypoxia so that it can be directly compared with clinical FMISO-PET data. To this end, we aim to construct a virtual FMISO-PET by simulating FMISO tracer dynamics combining PIHNA-predicted hypoxia with a pharmacokinetic (PK) model together with clinical-scale PET image acquisition and reconstruction algorithms.
In this investigation, we consider a patient with histologically diagnosed GBM for a comprehensive assessment of our ability to predict hypoxic burden using anatomic information derived solely from pre-treatment MRI characteristics using our PIHNA model. Figure 1 provides an overview of this process. Using the BrainWeb atlas (Cocosco et al., 1997), a virtual tumour is simulated using the PIHNA model in two spatial dimensions using a point-source initial concentration of tumour cells until the simulated tumour matched the size by area of the actual tumour observed on the patient’s T2-weighted MRI (see Section 2.4). To simulate an FMISO-PET scan, a PK model (Thorwarth et al., 2005; Wang et al., 2009) was used to determine the activity of the FMISO tracer within the brain as it acts between the vascular tissue and various tissue subpopulations of the brain over the time interval of virtual PET imaging. Finally, a summed tracer activity map underwent image reconstruction using an analytic simulator, (ASIM; Alessio et al., 2005), of the image acquisition and reconstruction procedures associated with the tomograph when obtaining the actual patient FMISO-PET. The resultant simulated FMISO-PET image is constructed to mimic the noise characteristics and acquisition artefacts representative of the actual clinical FMISO-PET image for the patient.
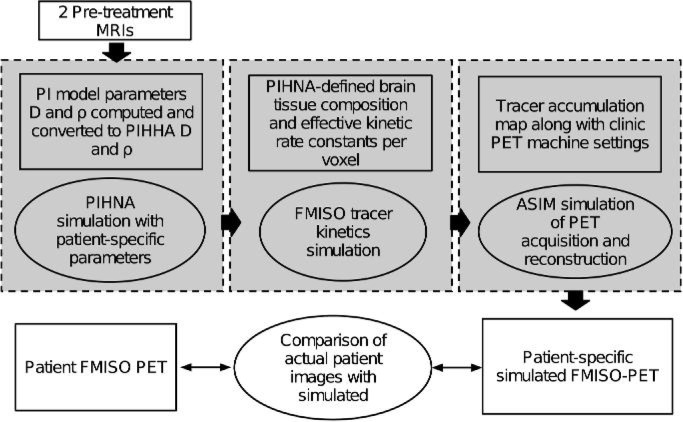
Fig. 1.
Flow diagram outlining overall methodology for developing simulated FMISO-PETs. Boxes represent tangible inputs, outputs and parameters. Ellipsoids correspond to different simulation and in silico processing steps. Grey boxes denote the three major components derived from previous works in generating simulated FMISO-PET.
2. Materials and methods
2.1. Numerical simulation of PIHNA model on 2D anatomically accurate brain domain
A fractional step method, also known as operator splitting, was utilized to implement the PIHNA model on a 2D grid with cerebral spinal fluid (CSF), grey and white matter concentrations defined at each grid point by the virtual brain atlas BrainWeb (Cocosco et al., 1997). For a reaction–diffusion equation of the form: ut (t) = D(u, x, t) + R(u, x, t), where D represents the diffusion operator and R represents the reaction operator, the fractional step method allows the two operators to be treated separately. Specifically, the conjugate gradient method was used for the diffusion operator and a two stage TRBDF2 method was used for the reaction terms along with Newton’s method for solving the non-linear equations. The numerical time step (k) was chosen to meet numerical stability requirements, with the spatial grid determined by the BrainWeb atlas resolution (dx = dy = 1mm). All methods were implemented in MATLAB. PIHNA model simulations rely essentially upon two patient-specific parameters, diffusion (Dc) and proliferation (ρ) since all other rates of diffusion, proliferation, conversion are either taken from the literature or defined relative to the normoxic cell population. These characteristic parameters are computed from their PI model equivalents shown in Table 2 and Fig. 8, which can be calculated using only two preoperative MRI observations (Harpold et al., 2007; Szeto et al., 2009; Wang et al., 2009) and then scaled to the D and ρ domain of the PIHNA model.
Table 2.
PIHNA model parameters with references. Patient-specific parameters Dc and ρ were calculated from tumour volumes of two preoperative MRI, with velocity of radial expansion of 6.7 mm/year measured on T1Gd. To account for the differing rates of diffusion between grey and white matter, the following relationship was applied to all diffusion coefficients in the PIHNA model simulation: Dw = 10Dg
| Parameter | Definition | Value | Reference |
|---|---|---|---|
| Dc,Dh | Max net rate of dispersal of glioma cells | 1.87 mm2/year | Patient specific |
| ρ | Max net rate of proliferation of glioma cells | 4.61/year | Patient specific |
| K | Carrying capacity of the tissue | 2.39 × 108 cells/cm3 | Assuming average cell radius of 10 μm |
| γ | Maximum rate of conversion of hypoxic to normoxic (when vascular density is high) | 0.5 (1/day) | A fast rate of hypoxic to normoxic conversion in the presence of sufficient vasculature |
| β | Maximum rate of conversion of normoxic to hypoxic (when no vasculature available) | ρ/10 (1/day) | Assumed to be proportional to the metabolic rate of the glioma cells (e.g. ρ) |
| αh | Rate of hypoxic cell necrosis | ρ/20 (1/day) | Assumed to be proportional to the metabolic rate of the glioma cells (e.g. ρ) |
| Dν | Random motility of endothelial cells | 5 × 106 (cm2/day) | Sherratt & Murray (1990); Levine et al. (2001) |
| μ | Maximal net proliferation rate of endothelial cells | log(2)/15 (1/day) | Xiu et al. (2006) |
| Da | Diffusion of TAF (e.g., VEGF) | 8.64 × 10−5 (cm2/day) | Levine et al. (2001) |
| δc | Rate of production of TAF by normoxic cells | 7.59 × 10−16 (μmol/cell/day) | Calibrated from Michaelis-Menten term |
| δh | Rate of production of TAF by hypoxic cells | 1.43 × 10−12 (μmol/cell/day) | Calibrated from Michaelis–Menten term |
| KM | Michaelis–Menten half-max of response of ECs to angiogenic factors | 5.75 × 10−7 (μmol/cell/day) | Mac Gabhann & Popel (2004) |
| Q | TAF consumption per EC proliferation | 1 × 105 (molecules/cell) | Estimated from number of VEGF receptors per cell, given in Mac Gabhann & Popel (2004) |
| w | TAF washout by vessels | 2.17 × 10−6 (1/cell/day) | Set this so that the background 3% vessels washout TAF at the same speed as the decay rate |
| αn | Necrosis contact death | log(2)/50/(0.1*K) (1/day) | Chose so that 10% necrosis gives tissue a 50-day half-life. |
| λa | TAF half-life | log(2)/64 (1/min) | Serini et al. (2003) |
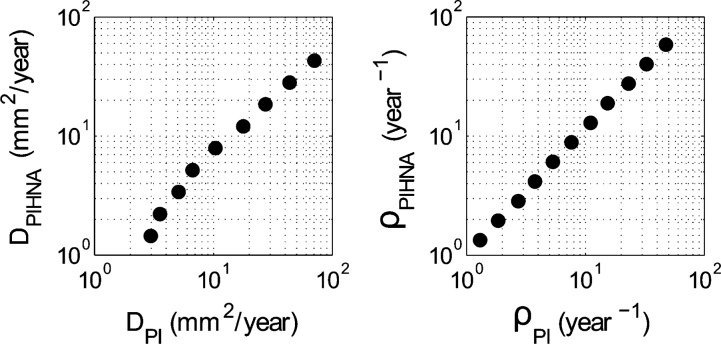
Fig. 8.
A one-to-one mapping exists for the patient-specific estimates of DPI and ρPI associated with the PI model derived from serial pre-treatment MRIs and the patient-specific estimates of DPIHNA and ρPIHNA to be used in the PIHNA model simulations to generate the dynamics seen on the patients MRIs.
2.2. PK model for FMISO tracer activity
In generating a simulated FMISO-PET, we considered a three-compartment PK model (Wang et al., 2009) to define the FMISO tracer activity within each voxel of the virtual brain. Voxel resolution was 1 mm by 1 mm. Outputs from the PIHNA model describe the evolution of the different types of tumour cell populations and brain tissue (c, h, ν, n) within each voxel within a 2D plane of the virtual brain over time. The PK model was used to simulate the injection, uptake, activity and washout of the FMISO tracer. The three-compartment PK model was taken from literature (Wang et al., 2009) and is schematically illustrated in Fig. 2. The first compartment (Vb) in the PK model represents the concentration of FMISO delivered to a voxel of brain through the blood in the vascular compartment. The second compartment (A) represents the diffusing component of FMISO existing in the interstitial space between the blood and brain tissue. The third compartment (B) represents the concentration of sequestered FMISO within the brain tissue of the voxel, which is predominately determined by its concentration of hypoxic tissue (Wang et al., 2009). Because the PK model for the tracer activity is not stiff, it was solved using the ode23 routine in MATLAB, with a relative error tolerance of 10−3 and an absolute error tolerance of 10−6. The final radiolabel kinetics solution described tracer concentration within each voxel for the different tissue compartments over the span of 180 min after injection. Kinetic rates, derived from literature (Wang et al., 2009) and listed in Table 1, determined how FMISO diffuses from vasculature (compartment Vb) into tissue (compartment A), and the rate at which FMISO tracer from compartment A is metabolized and sequestered by hypoxic cell populations (compartment B). A virtual FMISO injection, determined directly by the actual dosage given to the patient was introduced into the vascular tissue of the virtual patient brain. Directional blood flow through the brain and tumour was not modelled. The PK model was solved at each voxel, with the rate of tracer diffusion and uptake determined by the proportions of hypoxic tumour, normoxic tumour, necrotic tumour and normal brain cells at that location (Table 1). The result is a spatialtemporal map of tracer activity during the FMISO-PET imaging time-frame, which was approximately 3 h. To generate a standardized uptake value (SUV) image to compare with the clinical scan, the tracer activity map was summed from 120 to 140 min after injection, as done in the clinic, and reconstructed using a filtered backprojection algorithm (see Section 2.3).
Fig. 2.
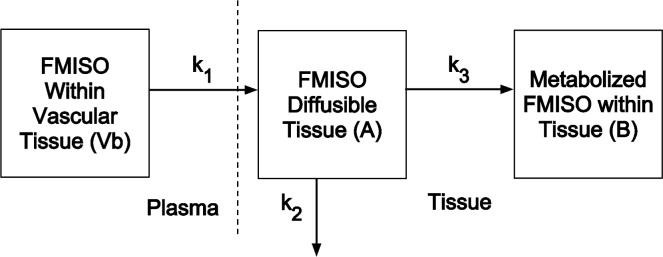
PK model rate constants (k1, k2 and k3) characterize the FMISO kinetics between compartments: k1 specifies the rate of tracer diffusion into tissue and interstitial space from the vasculature, k2 specifies the rate of tracer loss back to the vasculature and k3 specifies the rate of FMISO sequestering by the voxel of brain tissue. Table 1 outlines the various kinetic rates for the different brain tissue types.
Table 1.
Kinetic rates in PK model (Wang et al., 2009) for different tissue types defined by the PIHNA model and BrainWeb atlas
| Constant | Hypoxic tumour | Necrotic tumour | Normoxic tumour | Normal tissue |
|---|---|---|---|---|
| k1 (min−1) | 0.30 | 0.09 | 0.30 | 0.15 |
| k2 (min−1) | 0.45 | 0.15 | 0.45 | 0.20 |
| k3 (min−1) | 8 × 10−3 | 3 × 10−3 | 4 × 10−3 | 4 × 10−3 |
Applying the PK model to each voxel of brain tissue resulted in a unique PK solution per voxel determined by the varying concentrations of tissue types predicted by the PIHNA model and the BrainWeb atlas. The composition of tissue types adjusts the kinetic rate constants that characterize the FMISO action within that particular voxel. For instance, a tissue voxel comprising of half normoxic and half hypoxic cancer cells is assumed to have an intermediate PK behaviour with FMISO. The model is solved for each voxel with a system of ordinary differential equations:
| (2) |
| (3) |
The blood activity of FMISO (Vb), was the forcing function of the compartmental model. Vb was fitted to a typical patient FMISO blood activity curve seen from other studies and matched with the case study patient’s total dosage during actual PET imaging (Bruehlmeier et al., 2004). To generate a virtual tracer map that describes the total FMISO activity over time for each voxel, each compartment in the solution was summed in the following manner:
| (4) |
Thus, in generating the tracer map, Vb is scaled by the proportion of vasculature (ν from the PIHNA simulation) in the voxel (where K is the tumour cell carrying capacity of the voxel) and compartments A and B are scaled by the proportion of non-vascular tissue within the voxel
2.3. Simulation of the PET imaging process
The PET imaging of the tracer distribution within the brain determined by the three-compartment PK model was simulated with an ASIM (Comtat et al., 1999) that models the effects of attenuation, scattered and random coincidences, detector blurring and photon counting noise. Other effects can be modelled (e.g. detector efficiency variations) but were not in this case. Since ASIM is not a photon tracking simulator (e.g. SimSET; Lewellen et al., 1998), it cannot exactly reproduce the biases found in PET raw data after the corrections for photon scatter are applied, as these are scanner and object dependent. Thus, data corrections that are exact in the mean were applied, which produced accurate statistical and resolution properties in the simulated raw PET data. To model the patient raw data used for the case study, the estimated tracer intensity was summed between 120 and 140 min post injection as is done clinically at the University of Washington. This summed image was forward projected into essentially noiseless PET raw data with a sampling corresponding to that of the GE Advance scanner used for the patient study (Degrado et al., 1994). Detector blurring due to inter-crystal penetration and scattering was applied based on measured point spread function data (Alessio et al., 2005). The effects of photon attenuation were modelled and mean value of 30% scattered and random coincidences then added. Poisson noise was then added based on a total of 2.5×106 coincident photon events per image slice. Next, the noisy raw data were corrected accurately in the mean for attenuation and scattered and random coincidences to remove the bias of these effects but leave characteristic noise in the data (Comtat et al., 1999). Finally, the data were reconstructed into images using the filtered backprojection algorithm (Kak & Slaney, 2001) with a Hanning window matched to that used for the patient images.
2.4. Image analysis
In order to comparatively study functional and anatomical imaging, spatial registration was performed using PMOD to establish a common spatial coordinate system for each patient image set (FMISOPET, T1Gd and T2 MRI) relative to the BrainWeb atlas (Cocosco et al., 1997; Kwan et al., 1996, 1999; Collins et al., 1998). After image registration, 2D tumour areas were computed for the patient for both T1Gd and T2 MRI using a semi-automatic program developed in-house using tools available within MATLAB (Swanson et al., 2009). Following our previous work (Swanson et al., 2009), several regions of interest were compared between clinical and virtual MRI and FMISO-PET images. Regions of necrosis, denoted T0 were defined as the non-contrasting tumour core in the clinical T1Gd image or a cell density of >99% the cell carrying capacity (K) of the tissue in the PIHNA model output. The region of T1Gd enhancement was defined as regions in the simulated tumour with cell density of ⩾80% of the tissue cell carrying capacity in the PIHNA model output. Similarly, the T2 region was defined as the region of cell density ⩾16% of the tissue cell carrying capacity. These cut-off values were determined experimentally through previous work (Harpold et al., 2007). An additional region, denoted T2 Buff., was defined as the T2 abnormality with the addition of a uniform 2 cm margin or ‘buffer’ around its outermost edge for both clinical and simulated images. The T2 Buff. region was included because of its use as a target region for radiation therapy (Rockne et al., 2009). To account for the differences in voxel intensity between the clinical and the virtual FMISO, the simulated PET image was normalized to the background voxel activity of the clinical PET by matching the voxel intensities of a region of interest away from the tumour abnormality.
2.5. Case study
We consider a 43-year-old male patient diagnosed with a right fronto-temporal GBM. This patient received two pre-operative imaging observations consisting of both T1-weighted, gadolinium-enhanced and T2-weighted MRI in addition to a single FMISO-PET just prior to a subtotal resection of the bulk tumour mass. The patient was subsequently treated with radiation and chemo therapies. The FMISOPET scan was performed on an GE Medical Systems Advance Tomograph (Waukesha, WI) operating in 2D high sensitivity mode with 35 imaging planes covering a 15-cm axial field of view (Degrado et al., 1994) prior to treatment. All patient FMISO-PET images were reconstructed with a Hanning filter after scatter correction, resulting in a reconstructed spatial resolution of ∼12 mm (Degrado et al., 1994). Further details of the MRI and PET imaging protocols are given by Rajendran et al. (2003), Swanson et al. (2009) and in Section 2.4.
3. Results and discussion
The ability of our extended PIHNA model to produce patient-specific FMISO-PET images using only serial MRIs as inputs supports our previous results and successes with our PI model of GBM growth in modelling patient-specific tumour dynamics (Harpold et al., 2007). As an indicator of tumour resistance to many forms of treatment (Rockwell et al., 2009), hypoxia is an important and defining characteristic of GBM, and the impact a patient-specific predictive model could have on treatment planning is immense. We have made every effort to apply our model to clinical situations and maximize their utility in such circumstances.
3.1. PIHNA model output
Simulations of the PIHNA model result in continuous mappings of tumour cell concentration for normoxic (c), hypoxic (h) and necrotic (n) cell populations as well as vasculature (ν) and tumour angiogenic factors (a). MRI regions of interest (T0, T1Gd and T2) were defined using tumour cell density thresholds (Harpold et al., 2007), where the total tumour cell population (T) includes normoxia, hypoxia, vasularity and necrosis (T = n + h + c + ν; Swanson et al., 2003; Harpold et al., 2007). Plotting these regions on the corresponding T1Gd- or T2-simulated BrainWeb MRI allows for direct comparisons of PIHNA model predictions with clinical images. Furthermore, the hypoxic cell density from the PIHNA output is compared with the clinical FMISO-PET. Figure 3 shows a summary comparison of the PIHNA-derived tumour boundaries with the corresponding actual patient images. This demonstrates capabilities of the PIHNA model to produce predictive patient-specific realizations of tumour growth as visualized on clinical MRI. The hypoxia map shows the distribution of hypoxic tumour cells that may give clinicians greater insight in deciding on the course of therapy. However, it is important to note the PIHNA model predictions are still limited predominantly by the anatomical mismatch between the BrainWeb human brain atlas used for simulation purposes and the patient’s actual brain geometry.
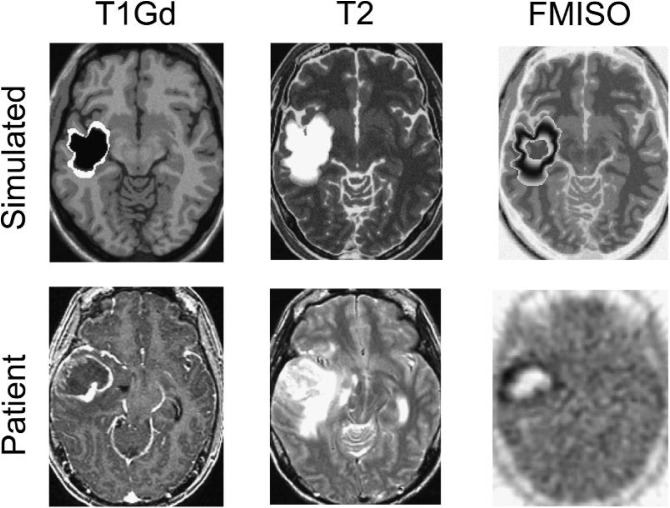
Fig. 3.
Comparison of clinical MRI and FMISO-PET images with PIHNA simulated MRI regions and mapping of hypoxic tumour cells. The virtual tumour was grown until the T2-weighted MRI area matched that of the actual patient’s. Simulated T1Gd abnormality was defined by 80% of the total carrying capacity (Harpold et al., 2007), shown in white with necrosis defined by the necrotic cell population (n) shown in black. Simulated T2 MRI was defined as 16% of the carrying capacity and is shown in white. A contour plot of hypoxic cell density (h) is shown, which informs the tracer activity model (PK) and simulated PET scan (ASIM). BrainWeb atlas MRIs were used as a background for the simulated tumour images.
While the MRI simulations can and have been validated by comparison with clinical MRI scans (Szeto et al., 2009), the hypoxia mapping cannot be directly compared to clinical FMISO-PET due to the lack of FMISO tracer kinetics and PET acquisition scatter in the PIHNA model output. In order to generate a more robust virtual PET, we integrated hypoxia mapping from the PIHNA output with a PK model and ASIM.
3.2. Simulated FMISO tracer activity
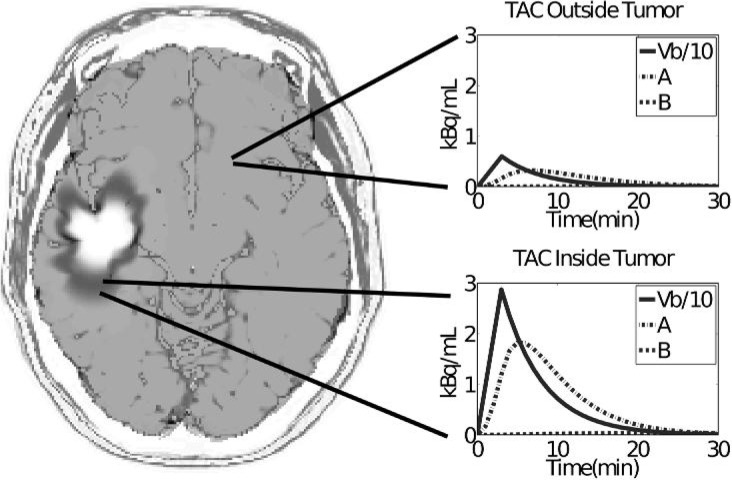
Using the PIHNA output of the different cell subpopulations, the PK model determined voxel-by-voxel solutions for the kinetic action of FMISO. Each voxel is characterized by a different cell subpopulation profile, depending on the outputs of the BrainWeb and PIHNA simulations. In the 2D image, modelling PKs of the FMISO tracer allowed us to better map the regions that would sequester the tracer. For example, if a voxel was 30% hypoxic and 70% normoxic, it would uptake less tracer than a region that was 100% hypoxic and 0% normoxic. Further, areas of the brain not containing tumour would have the FMISO tracer diffuse in and diffuse out without localization. Clinically, a static summed imaged of the FMISO-PET is utilized for detection of hypoxia, where the summed image is taken by integrating the amount of tracer in voxels from 120 to 140 min post injection of the tracer. We followed a similar protocol to generate the virtual FMISO by integrating the FMISO tracer activity from 120 to 140 min post virtual injection (Fig. 4). Representative voxels were chosen in normal (Fig. 4, top right) and hypoxic (Fig. 4, bottom right) brain regions to display the differences in tracer kinetics. Kinetics within the hypoxic tissue show faster uptake of tracer due to increased vascularity, as expected from the angiogenic cascade. This leads to a more pronounced effect of Vb in hypoxic tissue with greater diffusivity of the tracer and sequestering of FMISO as compared to the kinetics of a voxel within normal brain region. Sequestering of FMISO is expected in the hypoxic regions due to the hypoxic cells’ higher affinity for the tracer as modelled by equations governing compartment B ((2) and (3)). Tracer activity in the skin and CSF was added after the voxel kinetic modelling at relative intensity levels that approximated clinical scans. Modelling the tracer kinetics within the CSF and skin was beyond the scope of this work.
Fig. 4.
Summed FMISO tracer activity curves with representative voxels from normal brain tissue and hypoxic tumour regions outside and within the tumour region, respectively, shown from 0 to 30 min post injection. Although the summed image is generated from tracer activity in the voxel 120–140 min. post injection, the tracer dynamics are visually more evident at earlier time points.
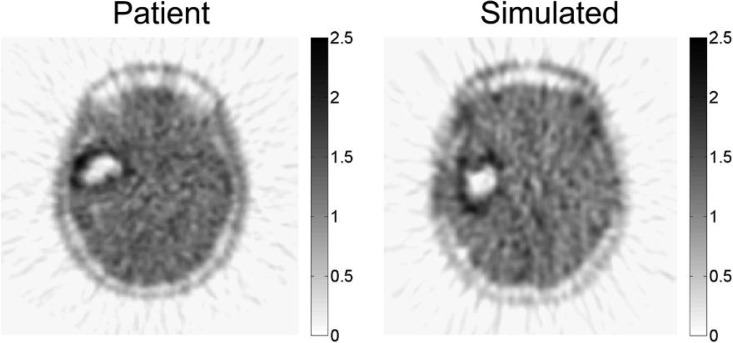
After applying the PK model to each voxel of the PIHNA output, we applied ASIM to the summed tracer map in order to mirror PET acquisition, producing an image that can be directly compared with the corresponding clinical FMISO-PET (Fig. 5). Radial streaking artefacts characteristic of filtered back-projection image reconstruction are visible in the simulated acquisitions (Fig. 5), comparable with those found in the clinical scans (Figs. 3 and 5).
Fig. 5.
Simulated FMISO-PET image produced using ASIM from PHINA model-predicted distribution of hypoxia (Fig. 3 top right) compared to the patient’s actual clinical FMISO-PET. Intensity was measured in SUV units.
In clinical FMISO-PETs, the intensity of the FMISO tracer indicated hypoxic activity. However, a higher intensity can indicate either more cells that can be denoted hypoxic, a group of cells that has a higher relative hypoxia than neighbouring cells or a combination of both. There is no established mapping of hypoxic cell density to FMISO tracer intensity. In order to compare the original PIHNA model output of hypoxia in cells/cc of tissue to FMISO tracer intensity, the voxel intensity was scaled after applying the PK model and ASIM to the average background brain signal in the clinical FMISOPET. This allows for the comparison between hypoxic cell density and virtual FMISO tracer activity. Finally, the virtual FMISO-PET can be qualitatively and quantitatively compared with the real FMISOPET (Fig. 5).
To a casual observer, while the morphological differences between the clinical and simulated images may seem significant, the limitation of the model to represent the actual tumour shape stems from the inherent differences between neuroanatomy of the patient and that of the BrainWeb idealized brain phantom. As the tumour grows, various parts of the brain anatomy put pressure on the tumour and deforms it, which has yet to be accounted for in the PIHNA model. However, the novel concept presented in this work is the methodology to predict patient-specific FMISO distribution relative to MRI-defined regions of interest. By first building the connection between gross tumour shape viewable under MRI and metabolic information offered through FMISO-PET, potential new avenues for interpreting MRI/PET images and simulation of treatment plan outcomes are established. Overall, this is an integrated system to produce a virtual FMISO-PET image with image acquisition and reconstruction techniques that are taken directly from clinical methods. This method then allows for validating mathematical models of biochemical activity in human tumours by directly translating model results to clinically relevant formats.
3.3. Comparing regions of actual and simulated hypoxia
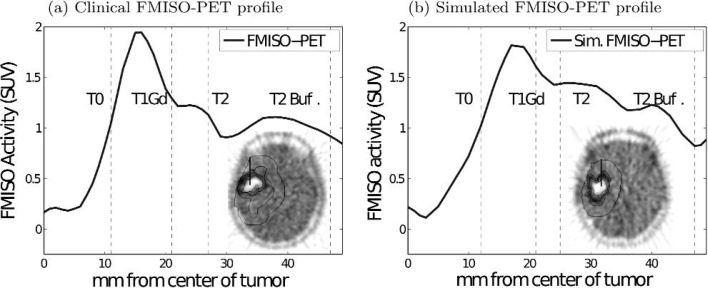
In order to compare the tumour tracer activity profile of clinical and simulated FMISO-PET, 1D radial plots were generated by choosing a representative line segment originating at the centre of the tumour and ending at the edge of the T2-Buff region of interest (Fig. 6). The insets in Fig. 6 show the line segment chosen in both the real (Fig. 6a) and simulated (Fig. 6b) case. These line segments spanned MRI-defined regions, including T0, T1Gd, T2 and T2-Buff, to obtain plots of FMISO intensity and distinguish regions of peak tracer activity relative to the MRI-defined regions of interest. The maximum FMISO activity lies within the T1Gd region for both the clinical patient PET and the PIHNA/ASIM-generated virtual PET, which is consistent with results from a previous study (Swanson et al., 2009).
Fig. 6.
Comparison of 1D profiles of FMISO activity of both simulated and clinical FMISO-PET images. Insets show the orientation of the 1D profile within the 2D field of view as well as the boundaries of MRI-defined regions of interest.
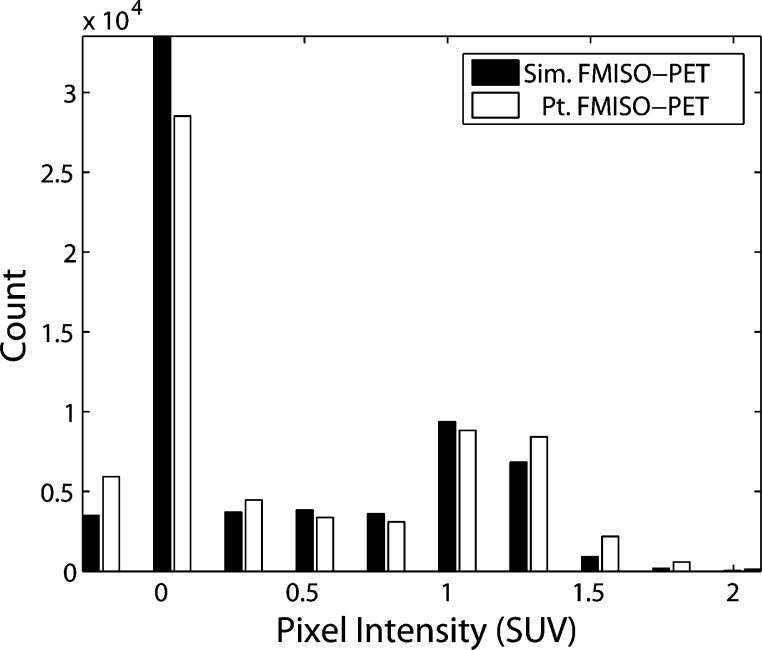
To assess the overall distribution of hypoxia and tracer accumulation in the actual FMISO-PET and simulated FMISO-PET relative to the MRI-defined regions of interest, we compared histograms of voxel intensity. The distributions were compared for regions of interest that included: the entire PET field of view, head (Fig. 7), T2-Buff., T2, T1Gd and T0 regions. Voxel intensity histograms showed statistically significant agreement between simulated and actual images for all regions of interest. A two-sample Kolomogorov–Smirnov (K–S) test was used to quantitatively assess the differences in distributions of the clinical and simulated FMISO-PET images within the different regions of interest. The null hypothesis for the K–S test was that the two distributions were sampled from the same population and the alternative hypothesis was that the two distributions were from two statistically different populations. The null hypotheses could not be rejected in any of the comparisons within the regions of interest using a significance level of 0.05. Figure 7 shows the distribution matching of real and simulated voxel values for the entire head space which passed the K–S test with a p value of 0.975.
Fig. 7.
Histograms of real compared to simulated FMISO-PET voxel intensities within the simulation domain (head space). Kolmogorov-Smirnov (K-S) test determined that the null hypothesis that the two distributions are not the same could not be rejected with 97.5% certainty. Negative voxel intensity indicates noise related to the imaging process.
Further, comparisons of the FMISO intensity regions in all the MRI-defined regions (T0, T1Gd, T2 and T2-Buff) showed similar results. This result suggests that the relationship between FMISO-tracer distribution relative to MRI-defined regions of interest can be described by the sim-FMISO-PET potentially providing a tool for further understanding and interpreting hypoxia and other FMISO-based information through sequential MRI scans. This method is a first step towards establishing and quantifying the relationship between anatomical (MRI) and functional (FMISO-PET) imaging techniques in human gliomas in vivo, which may provide greater insight to the patient disease state than either imaging modality can provide alone.
3.4. Clinical implications for virtual FMISO-PET images
The ability to produce patient-specific predictive FMISO-PET images based on measured MR images would provide quantitative anatomic, molecular and cellular information that would aid in our understanding of the spatiotemporal dynamics of tumour growth. Specifically, hypoxia is an indicator of tumour resistance to many forms of treatment (Rockwell et al., 2009). An accurate and predictive map of tumour hypoxia may allow clinicians to use more quantitative and systematic methods, compared to the relatively qualitative techniques currently used in treating patients, to improve patient survival and quality of life. Further, such a predictive tool provides novel dynamic insight into the connection between tumour cell phenotype, measured in terms of net rates of proliferation and invasion, and the spatiotemporal pattern of molecular changes seen in these tumour giving insight into the dynamics of tumours otherwise unavailable using standard experimental and clinical investigative means.
Although we have already shown that there is a strong connection between biological aggressiveness (D and ρ) assessed from serial MRIs and hypoxic burden assessed on FMISO-PET (Szeto et al., 2009), this work serves as a further validation of the connection between our anatomic PI model based on gross tumour dynamics and net downstream behaviours and molecular activity in tumours. In other words, this set of patient-specific models (PI and PIHNA) can be used to provide unique insight into the connection between changes in the molecular activity of tumours and the biological aggressiveness of that tumour imaged as changes on serial MRI. Current analysis of human FMISO-PET images relies on the relatively arbitrary choice of a single intensity threshold to differentiate regions of hypoxia from the rest of the brain. However, it is likely that this fixed cutoff neglects significant information in the FMISO-PET image.
This work provides a proof of principle for a tool for quantifying the information that is lost as a function of the imaging process, thus allowing for the design of more sophisticated image acquisition and reconstruction techniques. Further evaluations are needed to determine how accurately simulation results quantitatively predict clinical PET imaging data. Furthermore, this work allows for the improved interpretation of patient images and provides a way of exploring unanswered biological questions in silico, such as differentiating intensity of FMISO activity versus the density of hypoxic cells. Ultimately, this approach creates a path for better understanding of not only the biology of brain tumours by elucidating the connection between MRI-based abnormalities and molecular activity presented on PET imaging but also reveal their limitations.
4. Conclusions
The results of this study indicate that a spatial model for angiogeneiss in gliomas (PIHNA) combined with a PK model for FMISO activity and PET image reconstruction techniques (ASIM) were able to operate together to produce realistic MRI and FMISO-PET simulations. A two-sample Kolmogorov-Smirnov (K-S) statistical test verified that there was no statistically significant difference in the distributions of FMISO-defined hypoxia or pixel intensities relative to the MRI abnormality between the virtual and the clinical FMISO-PET image.
Current analysis of human FMISO-PET images relies on the relatively arbitrary choice of a single intensity threshold to differentiate regions of hypoxia from the rest of the brain (Swanson et al., 2009). However, it is likely that this fixed cutoff neglects significant information in the FMISO-PET image. This work provides a tool for quantifying the information that is lost as a function of the imaging process, which allows for the design of better image acquisition and reconstruction techniques.
A limitation of this modelling approach includes fundamental differences in the morphology of the MRI-defined tumour abnormality between simulated and actual due to mass effect, which is not included in the PIHNA model presented here. For instance, the MRI images of the patient show distinct deviation of the brain midline and insula region due to herniation. Future work implementing the force component of tumour growth may provide more accurate simulations of the tumour shape (Chen et al., 2001; Gevertz et al., 2008). Further work in accounting for biomechanical forces of tumour growth within the brain may improve the ability to predict tumour shape and anatomical evolution. In addition, using a patient-specific tissue classification map instead of a generic atlas may yield more accurate results.
Further evaluations of this modelling tool are needed to determine how accurately simulation results quantitatively predict clinical PET imaging data in glioma patients. However, the ability to produce simulated PET images from MRI data using the PIHNA model suggests a way to link functional metabolic information provided by PET with anatomical information provided by MRI on a patient-specific basis. This result can impact current clinical GBM treatment by providing greater quantitative insight on the regions of treatment resistance and the in vivo biochemical dynamics of the disease within individual patients. Furthermore, this modelling system may improve GBM treatment by providing a novel means for distinguishing hyper activity from hyper density of hypoxic cells on a FMISO-PET image.
Appendix. PIHNA model
The PIHNA model is a continuous reaction–diffusion model and is a logical extension of our previous PI model. It was designed in order to consider the complex interplay of essential histological characteristics of GBM and has already provided insights into the grading of GBM (Swanson et al., 2011). The PIHNA model partitions the tumour cell populations into three classes: well-oxygenated ‘normoxic’ tumour cells (c), hypoxic tumour cells (h) and dead or necrotic tumour cells (n) along with angiogenic factors (a), density of vasculature (ν) and total tumour cell density (T). The interplay between these variables represent the ‘angiogenic cascade,’ which we define using five coupled partial differential equations:
Normoxia
| (A.1) |
Hypoxia
| (A.2) |
Tumour angiogenic factors (TAFs)
| (A.3) |
Vasculature
| (A.4) |
Necrosis
| (A.5) |
Diffusion coefficients
| (A.6) |
Briefly, the rate of change of normoxic cell density over time depends on the cell’s net diffusion and proliferation rates and is coupled to the hypoxic model compartment such that normoxic cells turn hypoxic when vasculature is limited (A.1). The rate of change of hypoxic cell density is based upon the diffusion and vasculature dependence of these cells to remain hypoxic or turn normoxic or necrotic (A.2). TAFs released by both normoxic and hypoxic cells is regulated by a third equation involving modest secretion of these factors by normoxic cells and significant production by hypoxic cells (A.3). Vasculature is defined by a fourth equation that incorporates diffusion (A.4). Finally, the last equation governs growth of the necrotic core as a result of insufficient vasculature and death of hypoxic cells (A.5). Besides controlling for growth and death of the different cell populations and metabolic factors, the model follows our previous work and considers differing rates of cell migration within the brain by distinguishing between grey and white matter such that all cell populations diffuse faster in white matter than in grey, Dw = 10Dg (Swanson et al., 2003; Jbabdi et al., 2005). Interactions between cell populations and TAFs are governed by Michaelis–Menten kinetics. Parameter values and references are listed in Table 2. Further discussion regarding the form and justification of the equations can be found elsewhere (Swanson et al., 2011).
Patient-specific PI model parameters (DPI, ρPI) were determined from pre-treatment MRI tumour volumes (Harpold et al., 2007; Szeto et al., 2009; Wang et al., 2009) and were converted using a one-toone mapping of PIHNA model parameters Dc and ρ (Fig. 8). While the travelling wave dynamics of the PI model are well understood (Fisher, 1937) and the same qualitative phenomenon are observed in the PIHNA model, a precise analytical relationship between PIHNA model parameters and shape and speed of the travelling wave solution has not been articulated, necessitating the one-to-one mapping between the PI model parameter space and PIHNA.
References
- Alessio A, Kinahan P, Harrison R, Lewellen T. Measured spatially variant system response for pet image reconstruction. IEEE Nucl Sci Symp Med Imaging Conf. 2005;4:1986–1990. [Google Scholar]
- Bruehlmeier M, Roelcke U, Schubiger PA, Ametamey SM. Assessment of hypoxia and perfusion in human brain tumors using pet with 18F-Fluoromisonidazole and 15O-H2O. J. Nucl. Med. 2004;45:1851–1859. [PubMed] [Google Scholar]
- Burnet NG, Lynch AG, Jefferies SJ, Price SJ, Jones PH, Antoun NM, Xuereb JH, Pohl U. High grade glioma: imaging combined with pathological grade defines management and predicts prognosis. Radiother: Oncol. 2007;85:371–378. doi: 10.1016/j.radonc.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Chen CY, Byrne HM, King JR. The influence of growth-induced stress from the surrounding medium on the development of multicell spheroids. J Math Biol. 2001;43:191–220. doi: 10.1007/s002850100091. [DOI] [PubMed] [Google Scholar]
- Cocosco CA, Kollokian V, Kwan RKS, Evans AC. Neuroimage, Proceedings of the Third International Conference on the Functional Mapping of the Human Brain. part 2/4. Vol. 5. Copenhagen: 1997. Brainweb: online interface to a 3D MRI simulated brain database; p. S425. [Google Scholar]
- Collins DL, Zijdenbos AP, Kollokian V, Sled JG, Kabani NJ, Holmes CJ, Evans AC. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17:463–468. doi: 10.1109/42.712135. [DOI] [PubMed] [Google Scholar]
- Comtat C, Kinahan PE, Defrise M, Michel C, Lartizien C, Townsend DW. M7-83 simulating whole-body pet scanning with rapid analytical methods. In: Britton C, editor. IEEE Nuclear Science and Medical Imaging Symposium; 26–30 October; Seattle, WA: IEEE; 1999. pp. 1260–1264. Series: IEEE Transactions on Nuclear Science, vol. 47. [Google Scholar]
- Degrado TR, Turkington TG, Williams JJ, Stearns CW. Performance characteristics of a whole-body pet scanner. J Nucl Med. 1994;35:1398–1406. [PubMed] [Google Scholar]
- Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The wave of advance of advantageous genes. Ann Eugenics. 1937;7:353–369. [Google Scholar]
- Gevertz JL, Gillies GT, Torquato S. Simulating tumor growth in confined heterogeneous environments. Phys Biol. 2008;5:036010. doi: 10.1088/1478-3975/5/3/036010. [DOI] [PubMed] [Google Scholar]
- Harpold HL, Alvord EC, Jr, Swanson KR. The evolution of mathematical modeling of glioma proliferation and invasion. J Neuropathol Exp Neurol. 2007;66:1–9. doi: 10.1097/nen.0b013e31802d9000. [DOI] [PubMed] [Google Scholar]
- Jbabdi S, Mandonnet E, Duffau H, Capelle L, Swanson KR, Pelegrini-Issac M, Guillevin R, Benali H. Simulation of anisotropic growth of low-grade gliomas using diffusion tensor imaging. Mag Res Med. 2005;54:616–624. doi: 10.1002/mrm.20625. [DOI] [PubMed] [Google Scholar]
- Kak AC, Slaney M. Principles of Computerized Tomographic Imaging. Philadelphia, PA: Society of Industrial and Applied Mathematics; 2001. [Google Scholar]
- Kleihues P, Louis DN, Scheithauer BW, Rorke LB, Reifenberger G, Burger PC, Cavenee WK. The who classification of tumors of the nervous system. J Neuropathol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- Kwan RKS, Evans AC, Pike GB. An extensible MRI simulator for post-processing evaluation. Vis. Biomed. Comput. (VBC’96) 1996:135–140. Lecture Notes in Computer Science, vol 1131. [Google Scholar]
- Kwan RKS, Evans AC, Pike GB. MRI simulation-based evaluation of image-processing and classification methods. IEEE Trans Med Imag. 1999;18:1085–1097. doi: 10.1109/42.816072. [DOI] [PubMed] [Google Scholar]
- Levine HA, Pamuk S, Sleeman BD, Nilsen-Hamilton M. Mathematical modeling of capillary formation and development in tumor angiogenesis: penetration into the stroma. Bull Math Biol. 2001;63:801–863. doi: 10.1006/bulm.2001.0240. [DOI] [PubMed] [Google Scholar]
- Lewellen TK, Harrison RL, Vannoy S. The SimSET Program in Monte Carlo Calculations in Nuclear Medicine. Vol. 3. Philadelphia, PA: Institute of Physics Publishing; 1998. pp. 77–92. [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 who classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mac Gabhann F, Popel AS. Model of competitive binding of vascular endothelial growth factor and placental growth factor to VEGF receptors on endothelial cells. Am J Physiol Heart Circ Physiol. 2004;286:H153–H164. doi: 10.1152/ajpheart.00254.2003. [DOI] [PubMed] [Google Scholar]
- Rajendran JG, Mankoff DA, O’Sullivan F, Peterson LM, Schwartz DL, Conrad EU, Spence AM, Muzi M, Farwell DJ, Krohn KA. Hypoxia and glucose metabolism in malignant tumors: evaluation by [18F]fluoromisonidazole and [18F]fluorodeoxyglucose positron emission tomography imaging. Clin Cancer Res. 2004;10:2245–2252. doi: 10.1158/1078-0432.ccr-0688-3. [DOI] [PubMed] [Google Scholar]
- Rajendran JG, Wilson DC, Conrad EU, Peterson LM, Bruckner JD, Rasey JS, Chin LK, Hofstrand PD, Grierson JR, Eary JF, Krohn KA. [(18)F]FMISO and [(18)F]FDG PET imaging in soft tissue sarcomas: correlation of hypoxia, metabolism and VEGF expression. Eur J Nucl Med Mol Imaging. 2003;30:695–704. doi: 10.1007/s00259-002-1096-7. [DOI] [PubMed] [Google Scholar]
- Rockne R, Alvord EC, Jr, Rockhill JK, Swanson KR. A mathematical model for brain tumor response to radiation therapy. J Math Biol. 2009;58:561–578. doi: 10.1007/s00285-008-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell S, Dobrucki IT, Kim EY, Marrison ST, Vu VT. Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr Mol Med. 2009;9:442–458. doi: 10.2174/156652409788167087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini G, Ambrosi D, Giraudo E, Gamba A, Preziosi L, Bussolino F. Modeling the early stages of vascular network assembly. EMBO J. 2003;22:1771–1779. doi: 10.1093/emboj/cdg176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt JA, Murray JD. Models of epidermal wound healing. Proc R Soc Lond B. 1990;241:29–36. doi: 10.1098/rspb.1990.0061. [DOI] [PubMed] [Google Scholar]
- Swanson KR, Bridge C, Murray JD, Alvord EC., Jr Virtual and real brain tumors: using mathematical modeling to quantify glioma growth and invasion. J Neurol Sci. 2003;216:1–10. doi: 10.1016/j.jns.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Swanson KR, Chakraborty G, Wang CH, Rockne R, Harpold HL, Muzi M, Adamsen TC, Krohn KA, Spence AM. Complementary but distinct roles for MRI and 18f-fluoromisonidazole PET in the assessment of human glioblastomas. J Nucleic Med. 2009;50:36–44. doi: 10.2967/jnumed.108.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KR, Rockne RC, Claridge J, Chaplain MA, Alvord EC, Jr, Anderson ARA. Quantifying the role of angiogenesis in malignant progression of gliomas: in silico modeling integrates imaging and histology. Cancer Res. 2011;71:7366–7375. doi: 10.1158/0008-5472.CAN-11-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KR, Rostomily RC, Alvord EC., Jr A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: a proof of principle. Br J Cancer. 2008;98:113–119. doi: 10.1038/sj.bjc.6604125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto MD, Chakraborty G, Hadley J, Rockne R, Muzi M, Alvord EC, Krohn KA, Spence AM, Swanson KR. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 2009;69:4502–4509. doi: 10.1158/0008-5472.CAN-08-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorwarth D, Eschmann SM, Paulsen F, Alber M. A kinetic model for dynamic [18F]-FMISO PET data to analyse tumour hypoxia. Phys Med Biol. 2005;50:2209–2224. doi: 10.1088/0031-9155/50/10/002. [DOI] [PubMed] [Google Scholar]
- Tracqui P, Cruywagen GC, Woodward DE, Bartoo GT, Murray JD, Alvord EC., Jr A mathematical model of glioma growth: the effect of chemotherapy on spatio-temporal growth. Cell Prolif. 1995;28:17–31. doi: 10.1111/j.1365-2184.1995.tb00036.x. [DOI] [PubMed] [Google Scholar]
- Vallabhajosula S. (18)F-labeled positron emission tomographic radiopharmaceuticals in oncology: an overview of radiochemistry and mechanisms of tumor localization. Sem Nucleic Med. 2007;37:400–419. doi: 10.1053/j.semnuclmed.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Wang W, Georgi JC, Nehmeh S, Narayanan M, Paulus T, Bal M, O Donoghue J, Zanzonico P, Schmidtlein C, Lee N. Evaluation of a compartmental model for estimating tumor hypoxia via FMISO dynamic pet imaging. Phys Med Biol. 2009;54:3083–3100. doi: 10.1088/0031-9155/54/10/008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Rockhill JK, Mrugala M, Peacock DL, Lai A, Jusenius K, Wardlaw JM, Cloughesy T, Spence AM, Rockne R, Alvord EC, Jr, Swanson KR. Prognostic significance of growth kinetics in newly diagnosed glioblastomas revealed by combining serial imaging with a novel bio-mathematical model. Cancer Res. 2009;69:9133–9140. doi: 10.1158/0008-5472.CAN-08-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu M, Turner SM, Busch R, Gee TA, Hellerstein MK. Measurement of endothelial cell proliferation rate in vivo using 2H2O labeling: a kinetics biomarker of angiogenesis. FASEB J. 2006;20:A718. [Google Scholar]