Abstract
Pathogenic simian-human immunodeficiency viruses (SHIV)contain HIV-1 Vpu and SIV Nef, both shown to counteract BST-2 (HM1.24; CD317; tetherin) inhibition of virus release in a species-specific manner. We show that human and pig-tailed BST-2 (ptBST-2) restrict SHIV. We found that sequential “humanize” of the transmembrane domain (TMD) of the pig-tailed BST-2 (ptBST-2) protein resulted in a fluctuation in sensitivity to HIV-1 Vpu. Our results also show that the length of the TMD in human and ptBST-2 proteins is important for BST-2 restriction and susceptibility to Vpu. Taken together, our results emphasize the importance of tertiary structure in BST-2 antagonism and suggests that the HIV-1 Vpu transmembrane domain may have additional functions in vivo unrelated to BST-2 antagonism.
INTRODUCTION
Human bone marrow stromal cell antigen 2 (BST-2; HM1.24; CD317 or tetherin) was recently identified as a potent inhibitor of the release of multiple enveloped viruses including human immunodeficiency virus type I (HIV-1) and simian immunodeficiency virus (SIV) (Jouvenet et al., 2009; Kaletsky et al., 2009; Neil et al., 2008; Van Damme et al., 2008). While the exact cellular function of human BST-2 (hBST-2) in the human host is unclear, it has been shown to play a role in regulating the growth and development of B cells, to be involved in the organization of the subapical actin cytoskeleton in polarized epithelial cells (Rollason et al., 2009), and most recently to function as a ligand for ILT7, a receptor that inhibits IFN production from plasmacytoid dendritic cells (Cao et al., 2009). BST-2 is an interferon-induced, lipid raft-associated, type II integral membrane protein with an unusual topology that is similar in membrane orientation to a neuro-pathogenic form of the prion protein (PrP) (Kupzig et al., 2003). It contains a short cytoplasmic N-terminus followed by a transmembrane domain (TMD), a central extracellular domain predicted to form a coiled-coil structure, and a C-terminal cleavage site predicted to form a glycosyl-phosphatidylinositol (GPI) anchor (Ishikawa et al., 1995; Kupzig et al., 2003). This topology supports membrane-spanning models of restriction of virion release, in which the protein would form dimers providing a physical, protease-sensitive link between the cellular and viral membranes (Perez-Caballero et al., 2009). This model is supported by evidence that hBST-2 is incorporated into nascent virions and that the formation of cysteine-linked dimers is required for hBST-2 restriction of HIV-1 virion release (Ali et al., 2010; Andrew et al., 2009; Fitzpatrick et al., 2010; Habermann et al., 2010; Perez-Caballero et al., 2009).
Several studies have provided evidence that viruses including HIV-1 and SIV have evolved to acquire activity against the antiviral properties of BST-2. The HIV-1 group M Vpu proteins counteract specifically the restriction of human, chimpanzee and gorilla BST-2 proteins, with the exception of Vpu from strains JR-CSF and YU-2, which also antagonized BST-2 proteins derived from Greater-spot nosed and African green monkeys (Jouvenet et al., 2009; Neil et al., 2008; Sauter et al., 2009; Van Damme et al., 2008). The Nef proteins isolated from SIVmac, SIVcpz, SIVgor, and SIVagm also antagonize the effects of certain non-human primate BST-2 proteins, including those isolated from rhesus (rhBST-2) and pig-tailed (ptBST-2) macaques (Jia et al., 2009; Sauter et al., 2009; Zhang et al., 2009). Similar to other known intrinsic viral restriction factors such as the APOBEC3 and TRIM families of proteins, the ability of specific viral proteins to counteract BST-2 restriction is species-specific (Gupta et al., 2009; Jia et al., 2009; McNatt et al., 2009; Zhang et al., 2009). Specific domains and amino acids within these proteins mediate this specificity, allowing small evolutionary changes to confer susceptibility to cross-species infection (Gupta et al., 2009; Jia et al., 2009; McNatt et al., 2009;Yap et al., 2005; Zhang et al., 2009). The interaction of Vpu with hBST-2 involves the TMDs of both proteins (Douglas et al., 2009; McNatt et al., 2009; Rong et al., 2009). Several studies have identified amino acids within the human, rhesus and tantalus monkey BST-2 proteins required for Vpu sensitivity/resistance. These studies found several combinations of substitutions within the transmembrane domain of hBST-2 that reduced susceptibility to Vpu, including delGI,T45I (where glycine and isoleucine residues at positions 25 and 26 were deleted in combination with the substitution of a threonine at position 45 with an isoleucine) and delGI, I33V, I36L (where the glycine and isoleucine residues at positions 25 and 26 were deleted in combination with the substitution of the isoleucine at position 33 with a valine and mutation of the isoleucine at position 36 to a leucine). These studies also noted the importance of the proline residue at position 40 in sensitivity to Vpu (McNatt et al., 2009). More recent studies focused on the specific role of amino acids within the BST-2 proteins from non-human primates in the susceptibility and resistance to SIVmac Nef proteins. Several groups obtained similar results that identified an amino acid motif that is present within the cytoplasmic domain (D/GIWK14-17) of pig-tailed and rhBST-2 as important for SIV Nef susceptibility. These investigators showed that this motif could be inserted into the hBST-2 protein and impart susceptibility to the SIV Nef protein (Jia et al., 2009; Zhang et al., 2009).
Simian-human immunodeficiency viruses (SHIVs) have been used extensively for in vivo studies to understand the role of HIV-1 Vpu and Env proteins as well as other SIV proteins in viral pathogenesis. These chimeric viruses express both the HIV-1 Vpu and SIVmac Nef proteins shown to counteract BST-2 orthologues from human and non-human primates and possibly represent a useful virus to study these interactions. In this study, we examined the characteristics and anti-viral activities of BST-2 proteins from humans and pig-tailed macaques with specific amino acid exchanges in the context of SHIVs lacking the HIV-1 vpu gene, the SIV nef gene, or both. Our results indicate that the transmembrane domain is a determinant of HIV-1 Vpu susceptibility, however, the length of the domain may play more of a role in this rather than specific amino acids and or combinations of residues.
RESULTS
Comparison of the sequence of pig-tailed and rhesus bst-2 genes
We amplified and sequenced the bst-2 genes from five rhesus macaques (CX54, AH64, AS05, AS34 and AS89) and seven pig-tailed macaques (CC8X, W004, W005, W006, W007, W013, and W018). The genes isolated from the rhesus macaques exhibited considerable variability compared to genes isolated from pig-tailed macaques, most notably in the N-terminal regions (Fig. 1). We observed a 50:50 distribution of arginine and cysteine at position 9, a 60:40 ratio of aspartic acid to glycine at position 14, a 70:30 distribution of valine to isoleucine at position 29 and finally an 80:20 ratio of leucine to proline at position 43. In contrast the pig-tailed sequences did not show significant variability with the exception of one sequence that had a leucine instead of a methionine at position 13. Based on the sequence analysis, we cloned the ptbst-2 gene for expression studies.
Figure 1.
Sequence analysis of BST-2 genes amplified from rhesus (A) and pig-tailed (B) macaques. RNA was isolated from either spleen tissue or PBMC from five rhesus macaques and seven pig-tailed macaques. The bst-2 gene was amplified from each of these samples and bulk sequenced. The sequences obtained from the rhesus macaques are presented along with the sequences of rhBST-2 from the genomic database. The BST-2 proteins used in this study for both rhesus and pig-tailed macaques were used as references and the dashes represent similar identity.
Analysis of human and ptBST-2 mutants
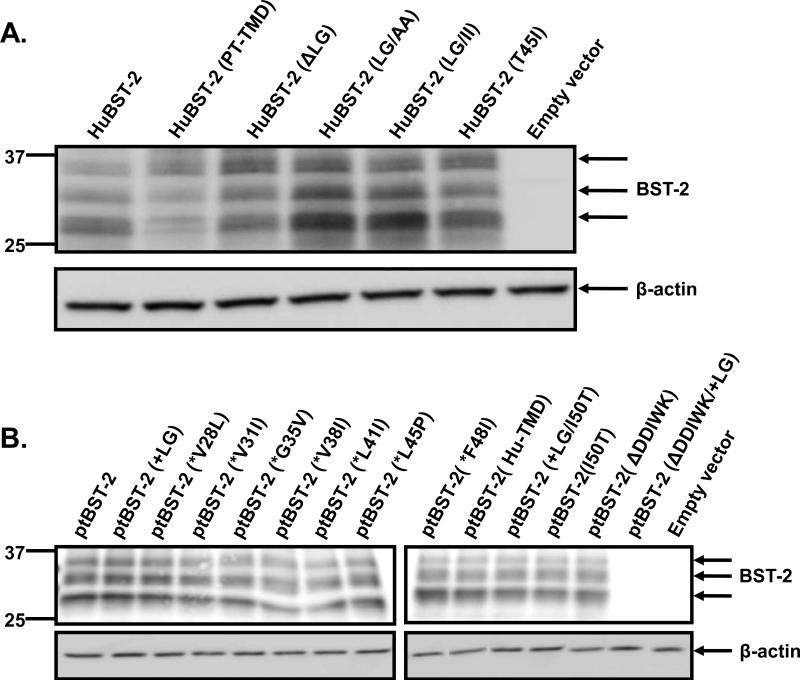
We constructed a series of mutations in the human (hBST-2) and pig-tailed macaque (ptBST-2) bst-2 genes. These are presented in Fig. 2. We first examined the expression of each mutant BST-2 protein by transfection of 293 cells with vectors expressing each mutant protein and subsequent Western blot analysis of the cell lysates (Fig. 3). Substitution of the leucine and glycine residues at positions 24 and 25 in huBST-2 with alanines or isoleucines (hBST-2LG/AA or hBST-2LG/II) consistently resulted in higher levels of expression, suggesting that these mutations may increase the half-life of these proteins. The only other protein with altered expression was the ptBST-2(ΔDDWIK/+LG). This amino acid substitution resulted in decreased protein expression, which could be due to altered stability of the protein and was not further studied.
Figure 2.
The amino acid sequence of the N-terminal region of human and pig-tailed BST-2 proteins, and the BST-2 mutants analyzed in this study.
Figure 3.
Expression of parental and mutant BST-2 proteins analyzed in this study. 293 cells were transfected with vectors expressing each of the BST-2 proteins. At 48 hours, cell lysates were collected and the nuclei removed through centrifugation. The lysates were boiled in sample reducing buffer and protein expression was examined through Western blot analysis using a rabbit polyclonal anti-BST-2 antibody (NIH). Panel A. 293 cells transfected with vectors expressing human parental and mutant BST-2 proteins. Panel B. 293 cells transfected with vectors expressing pig-tailed parental and mutant BST-2 proteins.
SHIV can serve as models for studying BST-2 mediated restriction
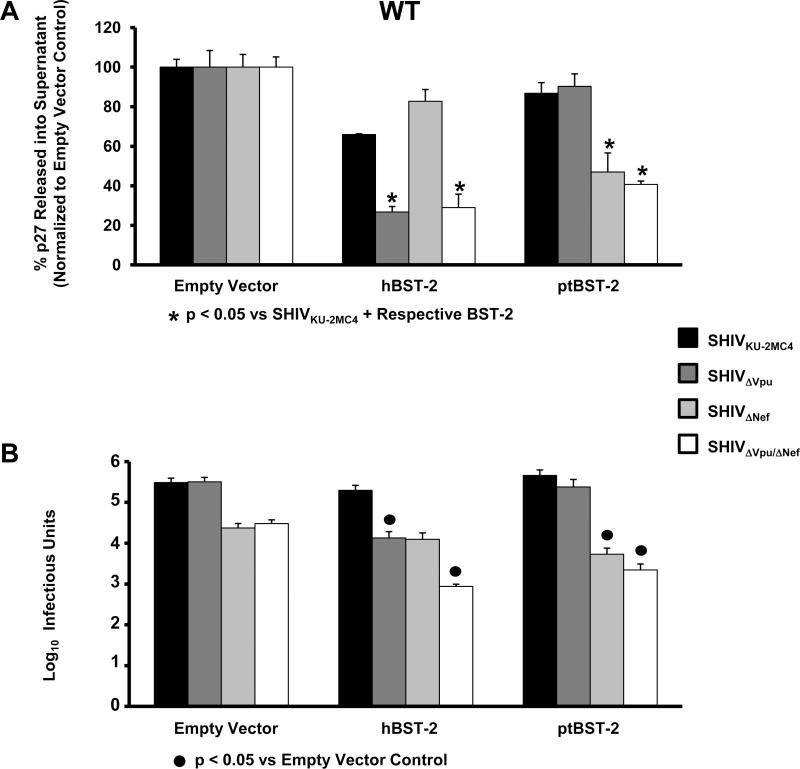
In order to examine the ability of the wild-type hBST-2 and ptBST-2 proteins to restrict virus release, we used parental SHIVKU-2MC4 to construct three SHIVs that did not express either HIV-1 Vpu (SHIVΔVpu), SIV Nef (SHIVΔNef) or both proteins (SHIVΔVpu/ΔNef). HEK 293 cells were co-transfected with plasmids expressing one of the four SHIVs and each of the BST-2 constructs. At 48 hours post-transfection, the supernatants were collected and cleared by centrifugation, the cells were lysed and the nuclei and cellular debris cleared by centrifugation. The supernatants and cell lysates were used to quantify the percent p27 antigen release from cells and the supernatants were also evaluated for the number of infectious doses (TCID50) (Fig. 4). The percent release of p27 in the context of each SHIV in the presence and absence of each BST-2 protein is shown in Fig. 4A. All samples were normalized to their respective SHIV empty vector controls. Significance in the restriction of p27 release for the SHIVKU-2MC4 samples was determined with respect to the parental SHIVKU-2MC4 empty vector control using a Student's t-test (▲). Significance in the restriction of p27 release for the SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef samples was calculated with respect to the SHIVKU-2MC4 in the presence of each respective BST-2 using a Student's t-test (*). The level of infectious virus released was determined using TZM-bl indicator cells (Fig. 4B). The results are represented as the average log10 infectious units. Significance in the restriction of infectious units released was determined with respect to the appropriate empty vector control (#25Cf) since Nef has significant effects on virion infectivity that are not associated with BST-2 incorporation (Aiken et al., 1995; Chowers et al., 1994). All four SHIVs exhibited similar p27 release in the absence of any BST-2 protein. In the presence of hBST-2, SHIVΔVpu and SHIVΔVpu/ΔNef viruses exhibited a significant decrease in p27 release from cells while SHIVΔNef had no decrease in p27 release. Similar results were observed in the infectious units assay. In the presence of ptBST-2 protein, only SHIVΔNef and SHIVΔVpu/ΔNef showed a decrease in p27 release as well as infectious units released. These results are in congruence with those published by other investigators that found that hBST-2 is susceptible to HIV-1 Vpu and ptBST-2 is sensitive to the SIV Nef (Neil et al., 2008; Van Damme et al., 2008; Zhang et al., 2009), indicating that these SHIVs can serve as models for studying the individual and additive effects of different BST-2 proteins.
Figure 4.
BST-2 dependent down-regulation of SHIV virion release from cells transfected with hBST-2 and ptBST-2. Panel A. p27 release assay. 293 cells were co-transfected with vectors expressing proviral DNA from one of four SHIV (SHIVKU-2MC4, SHIVΔVpu, SHIVΔNef or SHIVΔVpu/ΔNef) and a vector expressing either the hBST-2 or ptBST-2 protein. At 48 hours post-transfection, the supernatants and cell lysates were collected and cleared of cellular debris and nuclei by centrifugation. The p27 content of both the supernatant and cell lysate from each sample was quantified using a p27 antigen capture assay (Zeptometrix) and the percent p27 release calculated. All conditions were run at least three separate times, normalized to their respective SHIV empty vector controls, and the average percent p27 release and standard error calculated. Significance in the restriction of p27 release for the SHIVKU-2MC4 samples was determined with respect to the parental SHIVKU-2MC4 empty vector control using a Student's t-test (▲). Significance in the restriction of p27 release for the SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef samples was calculated with respect to the SHIVKU-2MC4 in the presence of each respective BST-2 using a Student's t-test (*). Panel B. Infectious units assay using TZM-bl indicator cells. Supernatants from transfections described above were added to the TZM-bl cells and serially diluted. At 48hrs post-infection, cells were washed, fixed, and stained for 2 hours. The TCID50 for each supernatant was calculated based on wells containing cells expressing β-galactosidase. All conditions were run at least three times and the TCID50 calculated. The average TCID50 and standard error were calculated. Significance in the restriction of infectivity was determined with respect to the parental SHIV empty vector control using a Student's t-test with p<0.05 considered significant (●).
Mutant ptBST-2 antagonism of SHIV release
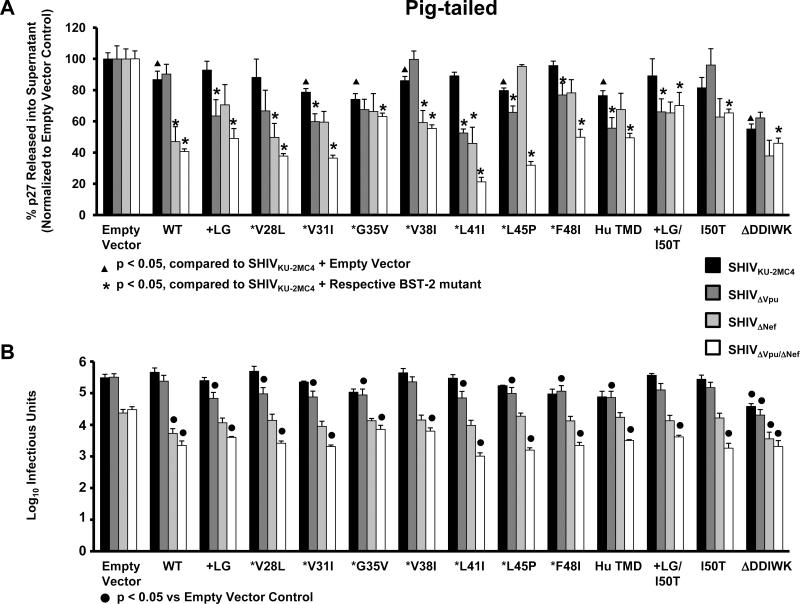
Previous studies showed that the transmembrane domain of the hBST-2 protein dictates its interaction with and susceptibility to HIV-1 Vpu (McNatt et al., 2009; Gupta et al., 2009; Rong et al., 2009). We determined if cumulative consecutive amino acid substitutions in the transmembrane domain of the ptBST-2 would result in a gradual increase in sensitivity to Vpu. We generated a library of ptBST-2 constructs with successive Ahumanizing@amino acid substitutions starting from the amino terminal end of the TMD with an insertion of two amino acids, a leucine and a glycine, at positions 29 and 30. We transfected each construct into 293 cells in conjunction with one of four different simian-human immunodeficiency virus expressing plasmids. The supernatants and cell lysates were used to quantify the percent p27 antigen release from cells and the supernatants were also evaluated for the number of infectious doses (TCID50) (Fig 5). The percent release of p27 in the context of each SHIV in the presence and absence of each BST-2 protein is shown in Fig. 5A. Results and significance are expressed and calculated similar to Fig. 4A. The level of infectious virus released was determined using TZM-bl indicator cells (Fig. 5B). Again, the results and significance are expressed and calculated as described for Fig. 4B. Surprisingly, a gradual increase in sensitivity to Vpu was not observed. We observed that most mutations resulted in BST-2 sensitivity to HIV-1 Vpu but the level was random in distribution. Mutations that significantly increased sensitivity to Vpu, including substitution of the leucine at position 41 with an isoleucine, did not retain this phenotype with additional “humanizing” amino acid substitutions. This indicates that the substitutions were not sufficient for the interaction with and antagonism by HIV-1 Vpu and suggests that their effect on the spatial orientation of the TMD within the membrane is also important.
Figure 5.
BST-2 dependent down-regulation of SHIV virion release from cells transfected with vectors expressing ptBST-2 mutants. Panel A. p27 release assay. 293 cells were co-transfected with vectors expressing proviral DNA from one of four SHIV (SHIVKU-2MC4, SHIVΔVpu, SHIVΔNef or SHIVΔVpu/ΔNef) and a vector expressing either the hBST-2 or ptBST-2 protein. At 48 hours post-transfection, the supernatants and cell lysates were collected and cleared of cellular debris and nuclei by centrifugation. The p27 content of both the supernatant and cell lysate from each sample was quantified using a p27 antigen capture assay (Zeptometrix) and the percent p27 release calculated. All conditions were run at least three separate times, normalized to their respective SHIV empty vector controls, and the average percent p27 release and standard error calculated. Significance in the restriction of p27 release for the SHIVKU-2MC4 samples was determined with respect to the parental SHIVKU-2MC4 empty vector control using a Student's t-test (▲). Significance in the restriction of p27 release for the SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef samples was calculated with respect to the SHIVKU-2MC4 in the presence of each respecitve BST-2 using a Student's t-test (*). Panel B. Infectious units assay using TZM-bl indicator cells. Supernatants from transfections described above were added to the TZM-bl cells and serially diluted. At 48hrs post-infection, cells were washed, fixed, and stained for 2 hours. The TCID50 for each supernatant was calculated based on wells containing cells expressing β-galactosidase. All conditions were run at least three times and the TCID50 calculated. The average TCID50 and standard error were calculated. Significance in the restriction of infectivity was determined with respect to the parental SHIV empty vector control using a Student's t-test with p<0.05 considered significant (●).
The length of the transmembrane domain is important for hBST-2 susceptibility to Vpu
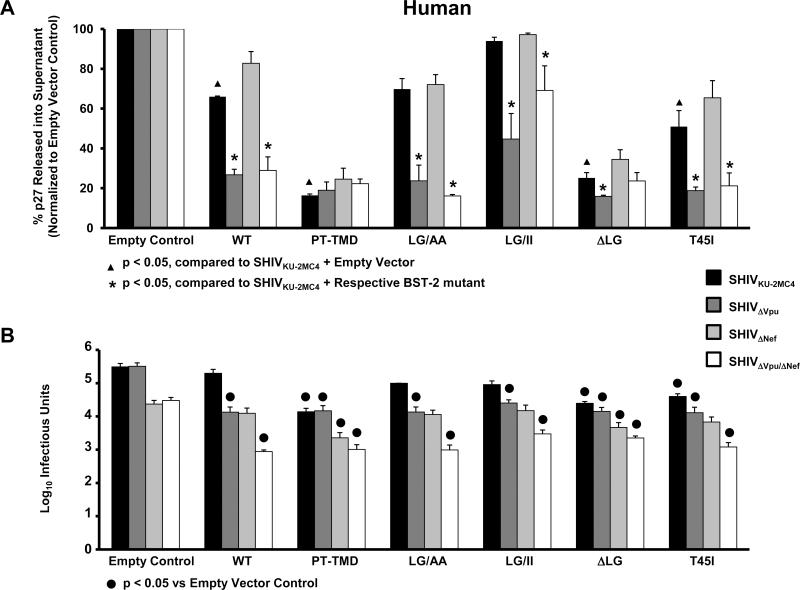
Our results shown in Fig. 5 indicated that insertion of the two residues in the transmembrane domain of ptBST-2 alone significantly increased the susceptibility to HIV-1 Vpu. These results were confirmed by quantifying the level of infectious virus released using TZM-bl indicator cells (Fig. 5B). These results suggested that the length of the transmembrane domain may be important for Vpu antagonism. To test this hypothesis, we generated additional hBST-2 constructs with different amino acid substitutions in the transmembrane domain. These constructs included hBST-2 proteins in which the TMD was replaced with the TMD from ptBST-2 (hBST-2/PT-TMD), the leucine and glycine residues at positions 24 and 25 were substituted with alanines (hBST-2LG/AA), the residues at positions 24 and 25 were substituted with hydrophobic, isoleucine residues (hBST-2LG/II), or the two residues were deleted (hBST-2ΔLG). These plasmids were used in the same virion release assay described above to determine the percent virion release (Fig. 6A). The hBST-2/PT-TMD protein was no longer susceptible to any viral protein, similar to results that have been published previously (McNatt et al., 2009; Douglas et al., 2009). The hBST-2ΔLG protein exhibited similar results to hBST-2/PT-TMD, while the hBST-2LG/AA and hBST-2LG/II proteins produced results similar to the wild-type protein. These data suggest that alteration of the length of the transmembrane domain and not necessarily the hydrophobic nature of the amino acid residues at these positions contributed to the resistance to HIV-1 Vpu. These results were confirmed by quantifying the levels of infectious virus released using TZM-bl indicator cells (Fig. 6B). The results and significance for both assays are expressed and calculated as described for Fig. 4.
Figure 6.
BST-2 dependent down-regulation of SHIV virion release from cells transfected with vectors expressing hBST-2 mutants. Panel A. p27 release assay. 293 cells were co-transfected with vectors expressing proviral DNA from one of four SHIV (SHIVKU-2MC4, SHIVΔVpu, SHIVΔNef or SHIVΔVpu/ΔNef) and a vector expressing either the hBST-2 or ptBST-2 protein. At 48 hours post-transfection, the supernatants and cell lysates were collected and cleared of cellular debris and nuclei by centrifugation. The p27 content of both the supernatant and cell lysate from each sample was quantified using a p27 antigen capture assay (Zeptometrix) and the percent p27 release calculated. All conditions were run at least three separate times, normalized to their respective SHIV empty vector controls, and the average percent p27 release and standard error calculated. Significance in the restriction of p27 release for the SHIVKU-2MC4 samples was determined with respect to the parental SHIVKU-2MC4 empty vector control using a Student's t-test (▲). Significance in the restriction of p27 release for the SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef samples was calculated with respect to the SHIVKU-2MC4 in the presence of each respective BST-2 using a Student's t-test (*). Panel B. Infectious units assay using TZM-bl indicator cells. Supernatants from transfections described above were added to the TZM-bl cells and serially diluted. At 48hrs post-infection, cells were washed, fixed, and stained for 2 hours. The TCID50 for each supernatant was calculated based on wells containing cells expressing β-galactosidase. All conditions were run at least three times and the TCID50 calculated. The average TCID50 and standard error were calculated. Significance in the restriction of infectivity was determined with respect to the parental SHIV empty vector control using a Student's t-test with p<0.05 considered significant (●).
Deletion of residues 13-17 in the ptBST-2 decreases sensitivity to SHIV Nef
In order to study the susceptibility of the ptBST-2 mutants to the SIVmac Nef protein encoded in our SHIVs, we generated a construct that expressed a protein with a deletion in the region previously shown to be sufficient for antagonism by Nef. Deletion of residues 13-17 within this region (ptBST-2ΔDDIWK) decreased susceptibility of the protein to SIVmac Nef and resulted in a decrease in p27 released from cells transfected with all four SHIV constructs (Fig.5A). These results are in accordance with those published previously that residues 14-17 within the rhesus BST-2 protein determine susceptibility to SIVmac Nef. Again, these results were confirmed using TZM-bl cell assays that indicated a similar level of infectious virus (Fig. 5B).
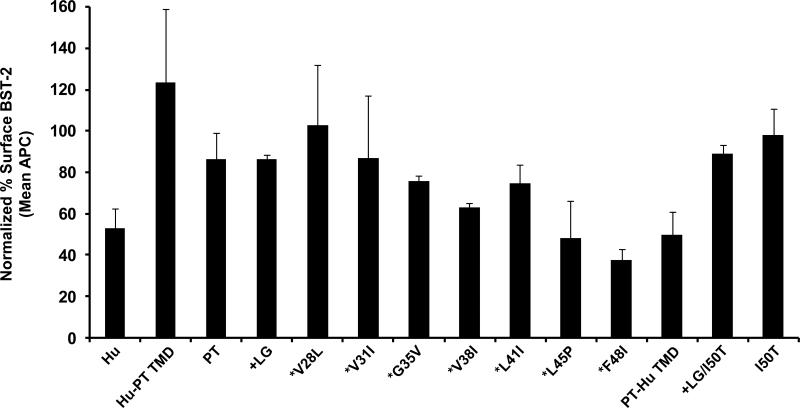
Down-regulation of ptBST-2 mutant cell surface expression by Vpu and Nef
We analyzed the ability of Vpu to down-regulate cell surface expression of hBST-2, ptBST-2 and the ptBST-2 mutants. The results in Fig. 7 show that Vpu caused a down-regulation of hBST-2 and ptBST-2 surface expression of 47% and 14%, respectively. Replacement of the hBST-2 TMD with that of ptBST-2 protein (hBST-2PT-TMD) resulted in an increase in cell surface expression in the presence of Vpu. Our results indicate that the insertion of a leucine and glycine at positions 29 and 30 (ptBST-2+LG) did not have a substantial effect on surface down-regulation by Vpu when compared to ptBST-2. We also did not observe an increase in Vpu-mediated surface down-regulation with *V28L and V31I mutants. However, continued humanization of the ptTMD led to a gradual increase in Vpu responsiveness. Exchange of the ptBST-2 TMD resulted in surface expression levels of 50%, similar to the hBST-2 in the presence of Vpu. We also analyzed the ability of SHIV Nef protein to down-regulate the surface expression of ptBST-2 and hBST-2. In our hands, the SIV Nef did not significantly down-regulate the surface expression of either hBST-2 or ptBST-2 proteins (data not shown).
Figure 7.
The down-regulation of cell surface expression of ptBST-2 mutants by Vpu. To measure surface down-regulation of BST-2 mutants in the presence of HIV-1 Vpu, 293 cells were seeded on a 12-well format and transfected using Lipofectamine 2000. For each transfection, 0.25 μg of pCG-GFP, 0.5 μg of pVphu or mock DNA plasmid, and 0.06 μg of each BST-2 noted above were used. At 24 hours, the cells were reacted with a mouse anti-HM1.24 antibody, then with an APC-conjugated goat anti-mouse antibody, followed by fixation. The cells were analyzed by two-color flow cytometry. For each BST-2 mutant, the APC mean fluorescent intensity (MFI) of high GFP expressing cells in the presence of Vphu was normalized to the MFI of cells without Vphu. The data are represented as percent BST-2 remaining on the surface of transfected cells in the presence of Vphu. All conditions were run at least twice and the standard deviation calculated.
DISCUSSION
Previous studies have shown that BST-2 restricts HIV-1 and SIVmac in a species-specific manner with these effects being counteracted by HIV-1 Vpu and SIV Nef (Jia et al., 2009; McNatt et al., 2009; Neil et al., 2008; Van Damme et al., 2008; Zhang et al., 2009;). SHIVs that express both of these proteins have been used extensively to study the effects of these and other viral proteins in vivo. Thus, one goal of these studies was to determine if SHIVs would counter human and pig-tailed BST-2 proteins and if specific mutations in these primate BST-2 proteins would increase or decrease sensitivity to Vpu and Nef. We approached this by characterizing BST-2-mediated restriction of the unmodified SHIVKU-2MC4 and three additional SHIVs that did not express Vpu, Nef or both proteins. Our results demonstrate that SHIVs display similar properties as HIV-1 and SIV with respect to human and pig-tailed BST-2. That is, SHIVΔVpu was restricted by hBST-2 while SHIVΔNef was inhibited by ptBST-2. This suggests that the amino acid substitutions in the Nef protein during adaptation of SHIVKU-2MC4 to cause disease in rhesus and pig-tailed macaques did not result in a loss of Nef antagonism against ptBST-2.
The resistance of the ptBST-2 to SHIV Vpu and its sensitivity to SHIV Nef allowed us to characterize the role of individual residues within the transmembrane domain of the ptBST-2 associated with these properties. While previous studies presented data on the role of the individual residues of the hBST-2 TMD, we used a series of sequential amino acid substitutions to “humanize” the ptBST-2 TMD (Gupta et al., 2009; McNatt et al., 2009; Rong et al., 2009). Our results are in accordance with these groups in that the two amino acid deletion in the ptBST-2 TMD (L29 and G30) are key residues involved in the susceptibility to HIV-1 Vpu. However, our analysis of a series of proteins with a progressive “humanize” of the TMD revealed a fluctuation in sensitivity to HIV-1 Vpu. All of the ptBST-2 transmembrane domain mutants displayed susceptibility to either HIV-1 Vpu, SIV Nef or both. However, the four mutations that displayed the most interesting effects were ptBST-2(+LG), ptBST-2(*G35V), ptBST-2(*V38I) and ptBST-2(*L41I). Insertion of the leucine and glycine at positions 29 and 30 clearly increased the sensitivity of the protein to HIV-1 Vpu without significantly altering the susceptibility to SIV Nef. Substitution of the glycine at position 35 with a valine eliminated the sensitivity to SHIV Vpu gained with the preceding mutations. This substitution also seemed to eliminate sensitivity to SHIV Nef. Mutation of the valine at position 38 to an isoleucine resulted in a complete loss of the sensitivity to HIV-1 Vpu gained with mutation of the preceding residues, but again without alteration of SHIV Nef susceptibility. Finally, substitution of the leucine at position 41 with an isoleucine resulted in an increased sensitivity to HIV-1 Vpu and an increase in the additive effects of Vpu and Nef. Any alterations in Nef susceptibility observed with the transmembrane domain mutations are most likely due to an alteration in the positioning of the cytoplasmic tail along the membrane. These results suggest that the overall structure, the spatial orientation of amino acids in the membrane, or possibly the tilt angle of the TMD in the membrane may be important factors. Insertion of the leucine and glycine at positions 29 and 30 into the ptBST-2 resulted in the most observable effect on the antagonism by Vpu leading us to hypothesize that the length of the transmembrane domain was important for this interaction. We tested this hypothesis by constructing three mutants of hBST-2 with either deletion of the LG (ΔLG), or substitution with amino acids with either a small hydrophobic side group (LG/AA) or a larger hydrophobic side group (LG/II). The hBST-2ΔLG deletion restricted particle release of all four SHIVs tested, while the LG/AA and LG/II mutants remained highly sensitive to Vpu and resistant to Nef. These results indicate that the length and not the degree of hydrophobicity of these amino acids is critical in defining susceptibility to HIV-1 Vpu. For these “humanize” ptBST-2 mutants, down-regulation of surface expression by HIV-1 Vpu generally correlated with the relief of restriction, with some exceptions. For example, wt ptBST-2 was minimally down-regulated by Vpu, but when ptBST-2 contained the TMD of huBST-2 it was efficiently down-regulated. Conversely, the huBST-2 mutant that contained the TMD of ptBST-2 was not down-regulated by Vpu. These results are consistent with the data on p27 release. A gradual increase in Vpu-mediated surface down-regulation was observed with sequential humanization of the ptTMD, and a reduction of ptBST-2 surface expression to levels similar to those of huBST-2 in response to Vpu was observed when the TMD was exchanged with that of huBST-2. These results generally correlated with p27 release detected with the SHIVcVpu/Nef. However, the surface down-regulation data did not perfectly correlate with the results of p27 release. For example, the effect of Vpu on the surface expression of the ptBST(*L41I) mutant is less than expected based on the p27 release data. These results suggest that the huBST-2 transmembrane domain is involved in Vpu-mediated surface down-regulation, however it may not be sufficient to fully explain the anti-viral properties or gain in susceptibility of the ptBST-2 mutants.
The level of surface expression of hBST-2 compared to ptBST-2 is unknown and it is possible that differences in these levels may help explain the possible differences in viral restriction efficiency as well as mechanisms of antagonism by viral proteins. Differences in surface expression could be due to differences in protein expression, internalization rates and/or turnover rates of each species of protein. Studies examining these properties are necessary in order to further elucidate the mechanisms by which different species of BST-2 proteins restrict virion release and how they are antagonized by different viral proteins. Studies within our laboratory suggest that there are differences in expression levels of the hBST-2 compared to the ptBST-2 and present a possible explanation for the differences in restriction efficiency observed in this study (unpublished data). However, the studies presented herein reflect intraspecies analysis and do not indicate cross-species examination.
This study introduces SHIV as an in vitro model for studying BST-2 mediated restriction laying a basis for future studies that could lead to an in vivo model. These results also emphasize the necessity for studies focused on the structural components of BST-2 and their contributions to the antiviral properties of the protein. These studies support the findings by Perez-Caballero and colleagues (2009) that the amino acid identity of the protein is less significant than the structural properties in virion restriction. Taken together, these results provide a basis for the development of new small molecule inhibitors designed to mimic the effects of BST-2 on multiple enveloped viruses.
The physiological relevance of the anti-HIV activity of BST-2 has not yet been demostrated in vivo. Previously published studies from our laboratory demonstrate that a functional Vpu protein is important for pathogenicity of SHIV in the pig-tailed macaque model, which expresses a Vpu resistant BST-2, suggesting a role for other functions of Vpu in viral pathogenesis (Hout et al., 2005; McCormick-Davis et al., 2000; Singh et al., 2001; Singh et al., 2003; Stephens et al., 2002). Of note are our studies using a SHIV expressing the VpuM2 protein (SHIVM2). This protein has been shown to down-regulate CD4 from the surface of CD4+ cells, however it is unable to rescue virion release from cells expressing high levels of BST-2 (data not shown). SHIVM2 caused a loss of CD4+ T cells within one month, high viral loads and histological lesions in infected macaques (Hout, et al., 2006). These studies provide evidence for additional functions of the Vpu transmembrane domain in vivo unrelated to BST-2 antagonism, which are potentially related to the ion channel properties exhibited by Vpu. Just as the BST-2 TM domain has amino acids/structures that are critical for interactions with Vpu, there are most likely amino acids within the Vpu TM domain that are also critical for these interactions. It will be of interest to identify these residues and determine if they are conserved in different subtypes of HIV-1.
MATERIALS AND METHODS
Plasmid Construction
The human bst-2 (hBST-2) gene was amplified from a plasmid expressing a full length cDNA of hBST-2 (Origene) using oligonucleotides containing 5’ BamHI and 3’ XhoI sites. The fragment was ligated into a pcDNA3.1(+) expression vector (Promega) digested with BamHI and XhoI restriction enzymes. The plasmid was sequenced to ensure no mutations were introduced during the cloning process. The pig-tailed bst-2 (ptBST-2) and rhesus bst-2 (rhBST-2) genes were amplified from cDNA generated from PBMC isolated from uninfected juvenile pig-tailed and rhesus macaques using a reverse-transcriptase polymerase chain reaction (RT-PCR). Oligonucleotides used contained 5’ BamHI and 3’ NotI restriction sites. The resulting fragment was ligated into a pcDNA3.1(+) expression vector (Promega) digested with BamHI and NotI restriction enzymes. Mutations introduced into all plasmids were accomplished using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. All plasmids were sequenced to ensure the validity of the mutations and that no other mutations were introduced during the cloning process. The pcVphu construct was kindly provided by the NIH AIDS Reference and reagent Program.
Sequence analysis of ptBST-2 and rhBST-2 variants
The bst-2 gene was amplified from five and seven different rhesus and pig-tailed macaque spleen tissue or PBMC, respectively, using a one-tube Titan reverse transcriptase kit (Roche). The resulting fragments were purified using a Qiaquick Gel Extraction kit (Qiagen) and subjected to sequence analysis.
Proviral DNA Plasmid Construction
The construction of molecular clone SHIVKU-2MC4 has been described previously (Liu et al., 1999). In order to construct SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef another molecular clone ΔvpuΔnefSHIVPPc was used and the construction of this clone has been described previously (Joag et al., 1998). A 1444 base pair fragment was removed from SHIVKU-2MC4 using restriction enzymes SphI and NheI. This fragment was replaced with the corresponding 1382 basepair fragment of ΔvpuΔnefSHIVPPc. The resulting construct expressed a Vpu protein with a 62 base pair deletion and an inactive start codon (SHIVΔVpu). A 525 base pair fragment was removed from SHIVKU-2MC4 using restriction enzymes BamHI and NcoI. This fragment which encoded for part of the gp41 and nef genes was replaced with the corresponding 321 base pair fragment of ΔvpuΔnefSHIVPPc. The resulting construct expressed a nef gene with a 204 base pair deletion that included the start codon (SHIVΔNef). A 525 base pair fragment was removed from SHIVΔVpu using restriction enzymes BamHI and NcoI. This fragment which encoded for part of the gp41 and nef genes was replaced with the corresponding 321 base pair fragment of ΔvpuΔnef SHIVPPc. The resulting construct expressed a vpu gene with a 62 base pair deletion and an inactive start codon and a nef gene with a 204 base pair deletion that included the start codon (SHIVΔVpu/ΔNef).
Cell lines and transfections
The HEK 293 and TZM-bl cell lines were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum (FBS), gentamicin (5μg per ml) and penicillin/streptomycin (100U/100μg per ml). Both cell lines were transfected using a cationic polymer (polyethylenimine) transfection reagent (ExGen™ 500, MBI Fermentas) using the manufacturer's protocol. 293 cells used for surface expression studies were maintained in Eagle's minimal essential medium (Quality Biological) supplemented with 10% FBS and 1× L-glutamine and were transfected using Lipofetamine 2000 (Invitrogen) according to manufacturer-provided instructions.
Western blot assays
Human 293 cells (105) were seeded into a 24-well tissue culture plate 24 hours prior to transfection. Cells were transfected as described above with 1μg of plasmid expressing full length SHIV proviral DNA (either SHIVKU-2MC4, SHIVΔVpu, SHIVΔNef, and/or SHIVΔVpu/ΔNef) and empty vector or 10ng of plasmid expressing various untagged bst-2 genes. Cells were incubated at 37°C in 5% CO2 atmosphere for 48hrs. Cells were lysed in 200μl of 1X RIPA buffer (50 mM TrisBHCl, pH 7.5; 50 mM NaCl; 0.5% deoxycholate; 0.2% SDS; 10 mM EDTA) and the nuclei removed through high speed centrifugation. Cell lysates were made 1X with sample reducing buffer (100mM Tris-HCl pH 6.8, 30% glycerol, 2% SDS, 10% 2-mercaptoethanol, 0.05% bromophenol blue) and boiled for 5 minutes. Samples were separated on SDS-PAGE gels and transferred to PVDF membrane. BST-2 proteins were detected using a rabbit polyclonal anti-BST-2 primary antibody kindly provided by the NIH AIDS Reagent and Reference Program (1:1000 dilution). The secondary antibody used was an alkaline phosphatase-conjugated goat anti-rabbit IgG (whole molecule) (Sigma). Alkaline phosphatase substrate used was CDP-Star (Sigma) for chemiluminescent detection and an LAS-4000 Imager (Fujifilm) was used for visualization and analysis of proteins.
Virion release assays
293 cells (105) were seeded into a 24-well tissue culture plate 24 hours prior to transfection. Cells were transfected as described above with 1μg of plasmid expressing full length SHIV proviral DNA (either SHIVKU-2MC4, SHIVΔVpu, SHIVΔNef, or SHIVΔVpu/ΔNef) and 10ng of plasmid expressing various untagged bst-2 genes. Cells were incubated at 37°C in 5% CO2 atmosphere for 48 hrs. Supernatants were collected and cellular debris removed through low speed centrifugation. Cells were lysed in 200μl of 1X RIPA buffer (50 mM TrisBHCl, pH 7.5; 50 mM NaCl; 0.5% deoxycholate; 0.2% SDS; 10 mM EDTA) and the nuclei removed through high speed centrifugation. The amount of p27 present within the virion containing supernatant and the cell lysates was determined using a commercially available p27 ELISA kit (Zeptometrix Incorporated) and percent p27 release calculated. All conditions were run at least three separate times, normalized to the empty vector control, and the average percent p27 release and standard error calculated. Significance in the restriction of p27 release for the SHIVKU-2MC4 samples was determined with respect to the parental SHIVKU-2MC4 empty vector control using a Student's t-test with p<0.05 considered significant (▲). Significance in the restriction of p27 release for the SHIVΔVpu, SHIVΔNef, and SHIVΔVpu/ΔNef samples was calculated with respect to the SHIVKU-2MC4 in the presence of each respective BST-2 using a Student's t-test with p<0.05 considered significant (*).
Infectious particle release assays
TZM-bl cells (104) expressing luciferase and β-galactosidase genes under the control of an HIV-1 promoter were seeded into a 96-well tissue culture plate 24 hours prior to infection. Supernatants collected from either the 293 cells co-transfected with SHIV and BST-2 expressing plasmids as described above were added to the TZM-bl cells and serially diluted. At 48hrs post-infection, cells were washed twice in 1X PBS and incubated in a fixative solution (0.25% glutaraldehyde, 0.8% formaldehyde in phosphate buffered saline) for 5 min. at room temperature. The cells were washed three times in 1X PBS and covered in staining solution (400μg/ml X-gal, 4mM MgCl2,4mM K3Fe(CN)6, 4mM K4Fe(CN)6-3H2O in phosphate buffered saline) and incubated for 2 hrs. at 37C. Cells were washed once in 1X PBS and then covered in 1X PBS during counting. The TCID50 for each supernatant was calculated based on wells containing cells expressing β-galactosidase. All conditions were run at least three times and the TCID50 calculated. The average TCID50 and standard error were calculated. Significance in the restriction of infectivity was determined with respect to the parental SHIV empty vector control using a Student's t-test with p<0.05 considered significant (#25Cf).
BST-2 surface expression assays
For surface down-regulation of BST-2 mutants in the presence of HIV-1 Vpu, 106 293 cells were seeded on a 12-well format and transfected using Lipofectamine 2000. For each transfection, 0.25 μg of pCG-GFP, 0.5 μg of pVphu or mock DNA plasmid, and 0.06 μg of each BST-2 noted above were used. After 24 hours, the cells were stained with mouse anti-HM1.24 antibody (Chugai) at a concentration of 10 μg/ml, at 4°C in phosphate-buffered saline (PBS) with 0.1% sodium azide and 2% fetal bovine serum, followed by APC-conjugated goat anti-Mouse antibody (Biolegend) at 10 μg/ml in the same staining buffer, then fixed with 1% paraformaldehyde. The cells were analyzed by two-color flow cytometry. For each BST-2 mutant, the APC mean fluorescent intensity (MFI) of high GFP expressing cells in the presence of Vphu was normalized to the MFI of cells without Vphu. The data are represented as percent BST-2 remaining on the surface of transfected cells in the presence of Vphu. All conditions were run at least twice and the standard deviation calculated.
ACKNOWLEDGMENTS
The work reported here is supported by grant NIH grant AI51981 to E.B.S. and AI081688 to J.C.G. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Anti-Bst-2 (cat# 11722) from Drs. Klaus Strebel and Amy Andrew; and TZM-bl from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc.; and the pcDNAVphu (catalog #10076) from Drs. Stephen Bour and Klaus Strebel. The murine monoclonal antibody to BST-2 used for the flow cytometry was a gift from Chugai Pharmaceutical Co, Kanagawa, Japan. We also would like to thank members of the KUMC Biotechnology Support Facility and the Northwestern University Genomics Core Facility for their assistance with the oligonucleotide synthesis and sequence analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69(8):5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MS, Hammonds J, Ding L, Spearman P. CAML does not modulate tetherin-mediated restriction of HIV-1 particle release. PLos One. 2010;5(2):e9005. doi: 10.1371/journal.pone.0009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AJ, Miyagi E, Kao S, Strebel K. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology. 2009;6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Bover L, Cho M, Wen X, Hanabuchi S, Bao M, Rosen DB, Wang YH, Shaw JL, Du Q, Li C, Arai N, Yao Z, Lanier LL, Liu YJ. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 2009;206(7):1603–1604. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowers MY, Spina CA, Kwoh NJ, Fitch NJ, Richman DD, Guatelli JC. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68(5):2906–2014. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarrol MN, Gustin JK, Fruh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a β-TrCP-dependent mechanism. J. Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLos Pathog. 2010;6(3):e1000701. doi: 10.1371/journal.ppat.1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RK, Hue S, Schaller T, Verschoor E, Pillay D, Towers GJ. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 2009;5:1000443. doi: 10.1371/journal.ppat.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A, Krijnse Locker, J., Oberwinkler H, Eckhardt M, Homann S, Andrew A, Strebel K, Krausslich HG. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J. Virol. 2010;84:4646–4658. doi: 10.1128/JVI.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Fegley B, Mulcahy ER, Hill MS, Culley N, Pinson DM, Nothnick W, Powers MF, Wong SW, Stephens EB. Substitution of the transmembrane domain of Vpu in simian-human immunodeficiency virus (SHIVKU1bMC33) with that of M2 of influenza A results in a virus that is sensitive to inhibitors of the M2 ion channel and is pathogenic for pig-tailed macaques. Virology. 2006;344:541–559. doi: 10.1016/j.virol.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Hout DR, Gomez ML, Pacyniak E, Gomez LM, Inbody SH, Mulcahy ER, Culley N, Pinson DM, Powers MF, Wong SW, Stephens EB. Scrambling of the amino acids within the transmembrane domain of Vpu results in a simian-human immunodeficiency virus (SHIV™) that is less pathogenic for pig-tailed macaques. Virology. 2005;339:56–69. doi: 10.1016/j.virol.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Ishikawa J, Kaisho T, Tomizawa H, Lee BO, Kobune Y, Inazawa J, Oritani K, Itoh M, Ochi T, Ishihara K, Hirano T. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST-2, that may be involved in pre-B-cell growth. Genomics. 1995;26:527–534. doi: 10.1016/0888-7543(95)80171-h. [DOI] [PubMed] [Google Scholar]
- Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST-2. PLoS Pathog. 2009;5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joag SV, Liu ZQ, Stephens EB, Smith MS, Kumar A, Li Z, Wang C, Sheffer D, Jia F, Foresman L, Adany I, Lifson J, McClure HM, Narayan O. Oral immunization of macaques with attenuated vaccine virus induces protection against vaginally transmitted AIDS. J. Virol. 1998;72:9069–9078. doi: 10.1128/jvi.72.11.9069-9078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniesz PD. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 2009;83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc. Natl. Acad. Sci., USA. 2009;106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. BST-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Liu ZQ, Muhkerjee S, Sahni M, McCormick-Davis C, Leung K, Li Z, Gattone VJ, II, Tian C, Doms RW, Hoffman TL, Raghavan R, Narayan O, Stephens EB. Derivation and biological characterization of a molecular clone of SHIVKU-2 that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology. 1999;260:295–307. doi: 10.1006/viro.1999.9812. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Dalton SB, Hout DR, Singh DK, Berman NEJ, Yong C, Pinson DM, Foresman L, Stephens EB. A molecular clone of simian-human immunodeficiency virus (DvpuSHIVKU1bMC33) with a truncated, non-membrane-bound Vpu results in rapid CD4+ T cell loss and neuro-AIDS in pig-tailed macaques. Virology. 2000;272:112–126. doi: 10.1006/viro.2000.0333. [DOI] [PubMed] [Google Scholar]
- McNatt MW, Zang T, Hatziioannou T, Bartlett M, Fofana IB, Johnson WE, Neil SJ, Bieniasz PD. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS Pathog. 2009;5:e1000300. doi: 10.1371/journal.ppat.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollason R, Korolchuk V, Hamilton C, Jepson M, Banting G. A CD317/tetherin-RICH2 complex plays a critical role in the organization of the subapical actin cytoskeleton in polarized epithelial cells. J. Cell Biol. 2009;184:721–736. doi: 10.1083/jcb.200804154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J. Virol. 2009;83:7536–7546. doi: 10.1128/JVI.00620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma T, Noda T, Urata S, Kawaoka Y, Yasuda J. Inhibition of Lassa and Marburg virus production by tetherin. J. Virol. 2009;83:2382–2385. doi: 10.1128/JVI.01607-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D, Schindler M, Specht A, Landford WN, Munch J, Kim KA, Votteler J, Schubert U, Bibollet-Ruche F, Keele BF, Takehisa J, Ogando Y, Ochsenbauer C, Kappes JC, Ayouba A, Peeters M, Learn GH, Shaw G, Sharp PM, Bieniasz PD, Hahn BH, Hatziioannou T, Kirchoff F. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe. 2009;6:409–421. doi: 10.1016/j.chom.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DK, McCormick C, Pacyniak E, C. Lawrence K, Dalton SB, Pinson DM, Sun F, Berman NEJ, Calvert M, Wong SW, Stephens EB. A simian human immunodeficiency virus with a non-functional vpu (ΔvpuSHIV KU-1bMC33) isolated from a macaque with neuroAIDS has selected for mutations in env and nef that contribute to its pathogenic phenotype. Virology. 2001;282:123–140. doi: 10.1006/viro.2000.0821. [DOI] [PubMed] [Google Scholar]
- Singh DK, Griffin DM, Pacyniak E, Jackson M, Werle MJ, Wisdom B, Sun F, Hout DR, Pinson DM, Gunderson RS, Powers MF, Wong SW, Stephens EB. The presence of the casein kinase II phosphorylation sites of Vpu enhances the CD4+ T cell loss caused by the simian-human immunodeficiency virus SHIVKU-lbMC33 in pig-tailed macaques. Virology. 2003;313:435–451. doi: 10.1016/s0042-6822(03)00339-8. [DOI] [PubMed] [Google Scholar]
- Stephens EB, McCormick C, Pacyniak E, Griffin D, Pinson DM, Sun F, Nothnick W, Wong SW, Gunderson R, Berman NE, Singh DK. Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology. 2002;293:252–261. doi: 10.1006/viro.2001.1244. [DOI] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Stoye JP. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr Biol. 2005;15:73–78. doi: 10.1016/j.cub.2004.12.042. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchoff F, Bieniasz PD, Hatziioannou T. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe. 2009;6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]