Abstract
The Vpu protein of human immunodeficiency virus type 1 (HIV-1) is known to enhance virion release from certain cell types. To accomplish this function, Vpu interacts with the restriction factor known as bone marrow stromal cell antigen 2 (BST-2)/tetherin. In this study, we analyzed whether the Vpu protein is associated with microdomains known as lipid or membrane rafts. Our results indicate that Vpu partially partitions into detergent resistant membrane (DRM) fractions when expressed alone or in the context of simian-human immunodeficiency virus (SHIV) infection. The ability to be partitioned into rafts was observed with both subtype B and C Vpu proteins. The use of cholesterol lowering lovastatin/M-β-cyclodextrin and co-patching experiments confirmed that Vpu can be detected in cholesterol rich regions of membranes. Finally, we present data showing that raft association-defective transmembrane mutants of Vpu have impaired enhanced virus release function, but still maintain the ability to down-regulate CD4.
INTRODUCTION
Viral protein U (Vpu) encoded by human immunodeficiency virus type I (HIV-1) augments viral pathogenesis by down-modulating CD4 molecules from the surface of infected cells and enhancing virion release (Fujita et al., 1997; Klimkait et al., 1990; reviewed in Ruiz et al., 2010; Schubert et al., 1998). Membrane association is critical for both activities although an earlier study indicated that the primary structure of the transmembrane domain was irrelevant for CD4 down-modulation (Schubert et al., 1996). The ability of Vpu to enhance virion release from some cell types has been attributed to antagonism of the cellular protein bone marrow stromal antigen 2 (BST-2; also known as CD317, HM1.24 and tetherin) (Neil et al., 2008; Van Damme et al., 2008). BST-2 localizes to the sites of HIV-1 budding and provides a physical, protease-sensitive link between the cellular and viral membranes (Perez-Caballero et al., 2009). BST-2 is a type II integral membrane protein that is anchored into the cell membrane by an amino terminal transmembrane domain and by a glycophosphatidylinositol (GPI) anchor at the carboxyl terminal region. GPI anchored proteins are often found in membrane microdomains known as membrane rafts and fully processed BST-2 has been shown to partition to these domains (Kupzig et al., 2003). A more recent study documented a punctate distribution of BST-2 on the surface of HIV-1 infected cells and that removal of the GPI anchor resulted in BST-2 being exclusively associated with sites of assembly suggesting that BST-2 may partition to multiple types of microdomains with distinct membrane compositions (Perez-Caballero et al., 2009). As it is documented that the subtype B Vpu is neither incorporated into virions nor is it present at the sites of HIV-1 assembly and maturation, the location of Vpu mediated antagonism of BST-2 has remained in question. However, since BST-2 may partition to multiple, compositionally distinct rafts during intracellular processing, it may interact with Vpu in rafts distinct from those essential to HIV-1 maturation and egress. The mechanism(s) by which Vpu counteracts BST-2 are under investigation but the down-regulation of BST-2 from the cellular surface, increased degradation of BST-2 and sequestration at alternate intracellular sites are all potentially dependent co-partitioning to similar membrane rafts.
Membrane rafts are essential to HIV-1 replication as the assembly and budding of virions is a dynamic process that is dependent upon the association of viral proteins with these microdomains (Ono and Freed, 2001). HIV-1 proteins Gag, Env and Nef have been identified as membrane raft associated proteins and the partitioning to these microdomains is essential for assembly, budding and enhancement of viral infectivity (Nguyen and Hildreth, 2000; Fournier et al., 2002). Membrane rafts are enriched in cholesterol and sphingolipids forming tightly packed, highly ordered regions within the membrane. The presence of cholesterol and sphingolipids in rafts confers resistance to solubilization by some non-ionic detergents such as Triton X-100 at low temperatures. Due to their insolubility, membrane rafts are often referred to as detergent resistant membranes (DRMs). Because the isolation of DRMs is accomplished at 4C and isolated DRMs are 0.1 to 1μm vesicles, some investigators have questioned their existence in live cells. However, more recent studies have shown that DRMs can also be isolated by extraction at physiological temperature (Chen et al., 2009). Additionally, morphological approaches (confocal microscopy; atomic force microscopy, and fluorescence resonance energy transfer) have been used to study in situ localization of membrane rafts (Pralle et al., 2000; Prior et al., 2003; Sharma et al., 2004; Kusumi et al., 2005).
In this study, we employed both biochemical and morphological approaches to determine whether Vpu associates with membrane rafts and further characterized the potential for this association to affect both functions of Vpu. We report here that Vpu partially partitions to detergent resistant membrane microdomains in a cholesterol dependent manner. We also demonstrate the involvement of the transmembrane domain in this partitioning and identify targeted mutations within this domain that abolish membrane raft association. Furthermore, membrane raft association of the Vpu protein correlates with the enhancement of virion release function, but not CD4 surface down-regulation. Taken together, these results establish that separate Vpu functions require distinct membrane localization patterns, implicate specific regions of the transmembrane domain as targets for disrupting Vpu function and provide further evidence for the importance of membrane rafts in HIV-1 pathogenesis.
RESULTS
The subtype B Vpu protein partially partitions to the detergent resistant membrane fractions
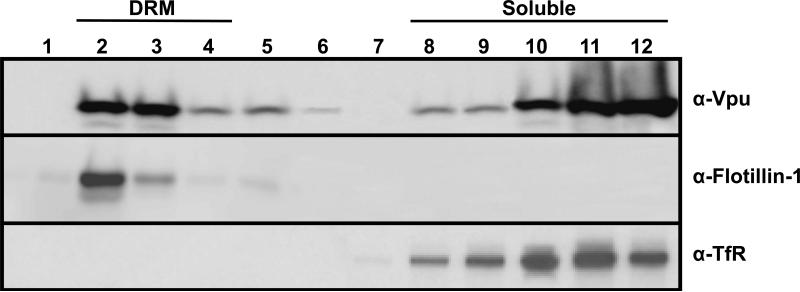
We transfected 293 cells with a vector expressing a codon-optimized version of the subtype B Vpu (Vphu) and at 48 hours the DRMs were isolated on sucrose gradients using ultracentrifugation. Fractions were collected from the top of the gradients, the proteins concentrated and analyzed by Western blot analysis. Vphu was detected predominantly in the soluble fractions at the bottom of the gradient (Fig. 1; upper panel) where a non-raft protein (transferrin receptor) was located (Fig. 1; lower panel). However, the Vphu protein was also detected at fractions at the top of the gradient that corresponded to the DRMs. The DRM fractions were identified by the presence of flotillin 1 (a raft protein) (Fig. 1; middle panel). All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments. The results provide evidence that Vpu is partially partitioned into membrane raft proteins.
Figure 1.
Vpu partitions into detergent resistant membrane (DRM) fractions. 293 cells were transfected with a plasmid expressing Vphu protein. At 48 hours, cells were lysed in ice cold DRM buffer containing 1% Triton X-100, and raft proteins separated from non-raft proteins on discontinuous sucrose gradients by ultracentrifugation as described in the Materials and Methods section. Fractions were collected from the top of the gradient, and Vpu proteins detected by Western blot analysis using a rabbit anti-Vpu serum (Upper panel) or stripped and reprobed with either an antibody directed against raft protein flotillin-1 (Middle panel) or the non-raft protein transferrin receptor (Lower panel). Fraction 1 is the top of the gradient and fraction 12 the bottom of the gradient.
Vpu is detected in the DRM fractions isolated from virus infected cells
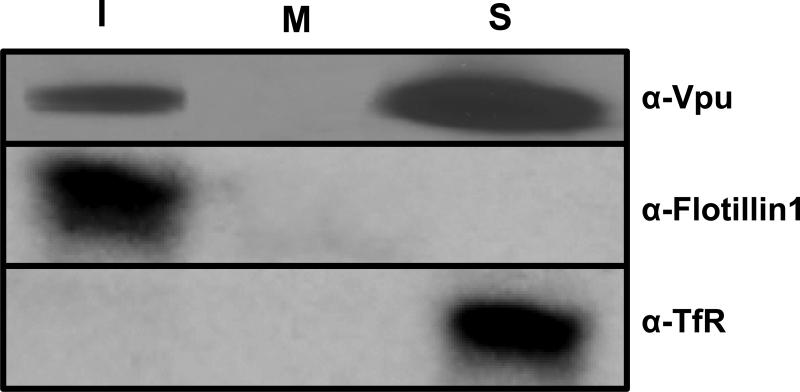
We next determined if the Vpu could be detected in DRM fractions from virus-infected cells. C8166 cells were inoculated with 104 TCID50 of simian-human immunodeficiency virus expressing an intact Vpu protein (SHIVKU-1bMC33) or no Vpu protein (novpuSHIVKU-1bMC33) and incubated for 5 days. At 5 days, cells were starved and radiolabeled with 35S-methionine/cysteine for 2 hours followed by DRM extraction and isolation by ultracentrifugation. Fractions containing the DRMs (fractions 1-3; I), the middle of the gradient (fraction 6-7; M), and the soluble non-raft proteins (Fractions 10-12;S) were each pooled, the Vpu proteins were immunoprecipitated using a rabbit anti-Vpu antibody, and the proteins separated by SDS-PAGE. Aliquots were analyzed by Western blot for flotillin-1 or transferrin receptor. Vpu could be detected in both the DRM (top of gradient) and soluble fractions, but very little was detected in fractions collected from the middle of the gradient (Fig. 2). All experiments were performed at least twice, and most three times. These results indicate that Vpu could also be detected in DRMs isolated from virus-infected T cells.
Figure 2.
Vpu expressed in virus-infected cells also partitions into DRM fractions. C8166 cells were inoculated with 100 ng of SHIVKU-1bMC33 or novpuSHIVKU-1bMC33 and incubated for 5 days. At this time cells were starved for methionine/cysteine for 2 hours and radiolabeled for 2 hours with 1 mCi of 35S-methionine/cysteine. The cells were harvested by low speed centrifugation, washed, and lysed in ice cold DRM buffer containing 1% Triton X-100. Raft proteins were separated from non-raft proteins using discontinuous sucrose gradients by ultracentrifugation as described in the Materials and Methods section. Fractions 1-3, 6-7 and 10-12 were pooled and the Vpu proteins immunoprecipitated using a rabbit anti-Vpu serum and immunoprecipitates collected on Protein A Sepharose beads. The remaining supernatants were analyzed by Western blot for flotillin-1 and transferrin receptor. The proteins were separated using SDS-PAGE and proteins visualized using standard autoradiographic techniques.
Vpu fusion proteins also partition to DRMs
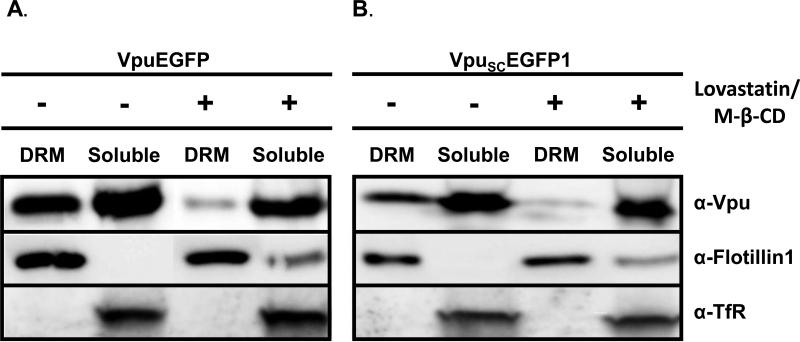
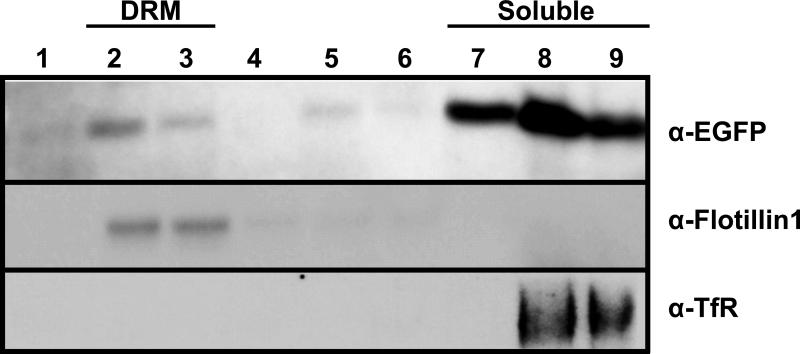
To facilitate the detection of Vpu in different compartments of the cell, we previously developed a vector that expressed Vpu fused to the protein enhanced green fluorescent protein (VpuEGFP) (Singh et al., 2003; Pacyniak et al., 2005). We determined if the VpuEGFP protein would also partition to the DRM fractions. Cells were transfected with vectors expressing either the subtype B fusion protein (VpuEGFP) or the subtype C Vpu protein (VpuSCEGFP1) and fractionated as described above. Both the VpuEGFP and VpuSCEGFP1 were detected in the DRM fractions, indicating that these tagged proteins could be used to study raft association (Fig. 3A-B). All experiments were performed at least twice, and most three times.
Figure 3.
Vpu fusion proteins also partition into detergent resistant membranes and cholesterol depletion reduces the amount of VpuEGFP and VpuSCEGFP1 in DRM fractions. Left side of panels A and B. 293 cells were transfected with vectors expressing either VpuEGFP (panel A) or VpuSCEGFP1 (panel B) proteins. At 48 hours, cells were lysed in ice cold DRM buffer containing 1% Triton X-100, and raft proteins separated from non-raft proteins on discontinuous sucrose gradients by ultracentrifugation as described in the Materials and Methods section. Fractions 1-3 (I), fractions 6-7 (M) and 10-12 (S) were pooled, concentrated and the Vpu proteins detected by Western blot analysis using a rabbit anti-EGFP serum. Right side of panels A and B. 293 cells were transfected with vectors expressing either VpuEGFP (Panel A) or VpuSCEGFP1 (Panel B) proteins. Following transfection, cells were incubated in the presence of 4 μM lovastatin for 48 hours. Thirty minutes prior to lysis, M-β-CD was added to a final concentration of 10mg/ml. Cells were processed as described for the untreated samples. Panels C-D. Micrographs of untreated cultures (Panel C) or those treated with M-β-CD (Panel D).
Cholesterol depletion abolishes the ability of VpuEGFP and VpuSCEGFP1 to be partitioned to the DRM fractions
As membrane rafts are rich in cholesterol, compounds that reduce cholesterol levels in cells such as M-β-cyclodextrin (M-β-CD) or statins disrupt membrane raft formation (Keller and Simons, 1998; Scheiffele et al., 1999). We examined if lovastatin/M-β-CD treatment would reduce the level of Vpu in DRM fractions. 293 cells were transfected with vectors expressing VpuEGFP or VpuSCEGFP1, treated with 4 μM lovastatin for 48 hours and with M-β-CD for the last 30 minutes prior to cell lysis. The cells were lysed as described above, DRM and soluble fractions separated by ultracentrifugation and fractions collected as described above. All experiments were performed at least twice, and most three times. The results clearly indicate that there were reduced levels of Vpu B and C fusion proteins in the DRM fractions following treatment with lovastatin and M-β-CD, indicating that Vpu resistance to Triton-X-100 is cholesterol dependent and likely correlates with raft association (Fig. 3A-B). The observation of flotillin-1 in rafts of lovastatin/ M-β-CD treated cells and may relate to the ability of flotillin-1 to occupy detergent-resistant, buoyant complexes that do not rely on cholesterol for their integrity and has been previously reported (Browman et al., 2006; Gkantiragas et al., 2001). Companion monolayers of 293 cells treated with lovastatin and M-β-CD showed no toxicity/cell death compared to untreated cells using microscopy (Fig. 3C-D) and trypan blue staining (data not shown).
Vpu partially partitions into membrane rafts isolated at physiological temperature
Classical methods of the isolation of DRMs involve solubilization at 4C, which has been challenged by some investigators. Therefore, we also examined Vpu partitioning into membrane raft fractions when the extraction was performed at 37C. 293 cells were transfected and at 48 hours post-transfection, were lysed in DRM37 buffer plus 1% Triton-X-100 and rafts isolated as described in the Materials and Methods section (Chen et al., 2009). Rafts were separated from non-rafts by ultracentrifugation and fractions collected. The fractions were analyzed using Western blots for the presence of Vpu, flotillin, and transferrin receptor. The raft (flotillin 1) and non-raft (transferrin receptor) markers were predominantly localized at the top and bottom of the gradients, respectively (Fig. 4). Vpu was detected in both raft and non-raft fractions, similar to what we observed with solubilization of cells in the presence of 1% Triton-X-100 at 4C. All experiments were performed at least twice, and most three times. These results show the Vpu could be detected in DRMs at physiological temperature.
Figure 4.
Vpu partitions into membrane rafts isolated at physiological temperature. 293 cells were transfected with a vector expressing VpuEGFP and incubated at 37C. At 48 hours post-transfection, cells solubilized in DRM37 buffer with 1 % Triton-X-100 and raft proteins separated from non-raft proteins using discontinuous flotation sucrose gradients and ultracentrifugation as described in the Materials and Methods section. Fractions were collected and VpuEGFP, flotillin or transferrin receptor detected by Western blot analysis using antibodies described in the Materials and Methods. Panel A. Detection of VpuEGFP. Panel B. Detection of flotillin-1. Panel C. Detection of transferrin receptor. Fraction 1 is the top of the gradient and fraction 9 the bottom of the gradient.
The subtype C Vpu protein co-localizes with a patched membrane raft protein
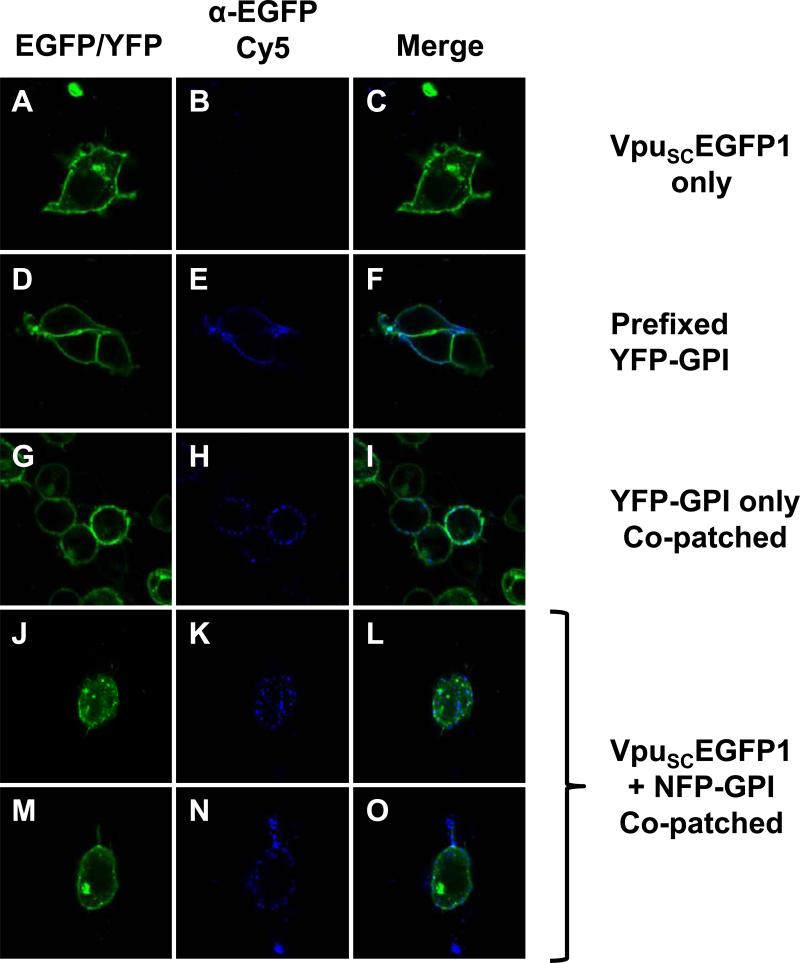
To further verify that Vpu is found in membrane rafts, we employed co-patching techniques for aggregating membrane rafts to a size visible by confocal microscopy and then examined if VpuSCEGFP1 co-localized with a known membrane raft protein. We used plasmids expressing either a fluorescent or non-fluorescent form of YFP with an ER translocation signal and a GPI anchor sequence. This protein is recognized by a mouse anti-GFP antibody (Clontech) in live cells, as the YFP portion of the protein is found extracellularly. While all known Vpu proteins are found in small amounts at the cell surface, most are prominently retained in the Golgi apparatus, making visualization of raft association difficult in live cells. Most live cell methods of raft detection involve surface staining or aggregation of rafts by cholera toxin or antibody co-patching. To increase the likelihood of finding Vpu in a raft by these methods, we utilized a Vpu protein which is predominantly found at the cell surface, VpuSC(Pacyniak et al., 2005). We used several controls to validate this approach. First, cells were transfected with the plasmid expressing VpuSCEGFP1 and live stained with anti-GFP as described in the Materials and Methods (Fig. 5A-C). As expected, these cells show no staining, since the EGFP tag of VpuSCEGFP1 is cytoplasmic. We next transfected cells with the plasmid expressing YFP-GPI, fixed and then stained with mouse anti-GFP (Fig. 5D-F). The antibody staining shows nearly complete co-localization at the cell surface, indicative of antibody specificity to the extracellular YFP. Finally, we transfected cells with the plasmid expressing YFP-GPI, then stained the live cells at 37C as described in Materials and Methods (Fig. 5G-I). This induces aggregation of the membrane rafts, making them visible by standard microscopy. Note the punctate staining of the co-patched YFP-GPI compared to the pre-fixed sample. To determine if VpuSCEGFP1 would co-localize with this membrane raft protein, we used a plasmid expressing a non-fluorescent version of the same YFP-GPI construct (NFP-GPI), which produces no background (data not shown). This allowed us to use the VpuSCEGFP1 protein without having overlapping fluorescent signals. Cells were co-transfected with plasmids expressing NFP-GPI and VpuSCEGFP1, then stained at 37C (Fig. 5 J-O). VpuSCEGFP1 partially co-localized with the patches of NFP-GPI, indicating that Vpu is found in at least some GPI-anchored protein containing membrane rafts. This provides additional evidence that Vpu is a membrane raft protein.
Figure 5.
Co-patching experiments reveal that VpuSCEGFP1 partially co-localizes with NFP-GPI. Panels A, D, G, J, and M are fluorescent micrographs using a filter for YFP/EGFP . Panels B, E, H, K, aand N are fluorescent micrographs for Cy5. Panels C, F, I, L, and O are a merge of the two fluorescent micrographs to the left. Panels A-C. 293 cells were transfected with vector expressing VpuSCEGFP1. Note the punctuate fluorescence in Panel C. Panels D-F. 293 cells were transfected with vector expressing YFP-GPI and pre-fixed. Panels G-I. 293 cells were transfected with a vector expressing YFP-GPI and co-patched. Panels J-O. 293 cells were transfected with both VpuSCEGFP1 and NFP-GPI.
The transmembrane domain of Vpu is involved in raft association
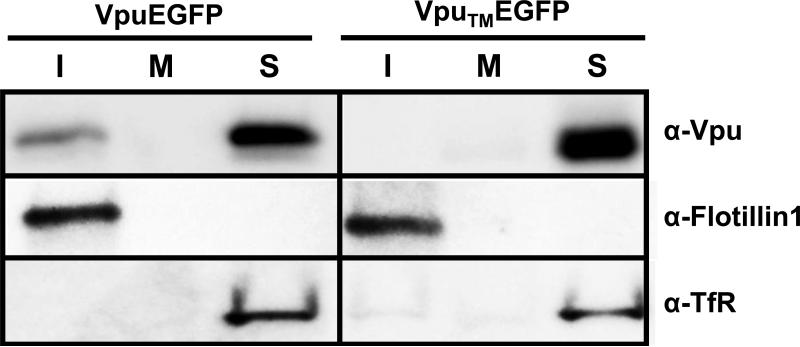
We next determined if amino acids within the transmembrane domain were involved in the transport of VpuEGFP to membrane rafts. Previous studies have shown that scrambling the hydrophobic amino acids of the transmembrane domain (VpuTMEGFP) results in a protein that is transported to similar compartments within the cell but unable to enhance virus release or down-regulate CD4 expression from the cell surface (Schubert et al., 1996; Hout et al., 2005). We determined if this Vpu fused to EGFP (VpuTMEGFP) would partition to raft fractions. The results shown in Fig. 6 indicate that VpuTMEGFP partitioned exclusively to the detergent soluble fractions of the gradient. All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments. Appropriate controls for raft isolation and purity were performed and are shown. These results provide genetic evidence for the specificity of Vpu partitioning to the DRM fractions.
Figure 6.
Scrambling the transmembrane domain prevents VpuEGFP raft association. 293 cells were transfected with vectors expressing either VpuEGFP or VpuTMEGFP proteins. At 48 hours, cells were lysed in ice cold DRM buffer containing 1% Triton X-100, and raft proteins separated from non-raft proteins on discontinuous sucrose gradients by ultracentrifugation as described in the Materials and Methods section. Fractions were collected from the top of the gradient, and fractions from the top (1-3; I) middle (6-7; M) and bottom (10-12;S) were analyzed for the presence of Vpu, Flotillin-1 and transferrin receptor proteins by Western blot analysis using a antibodies described in the Materials and Methods.
Characterization of Vpu transmembrane mutants
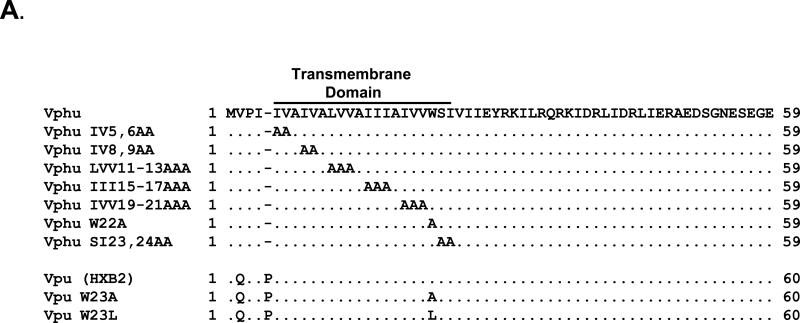
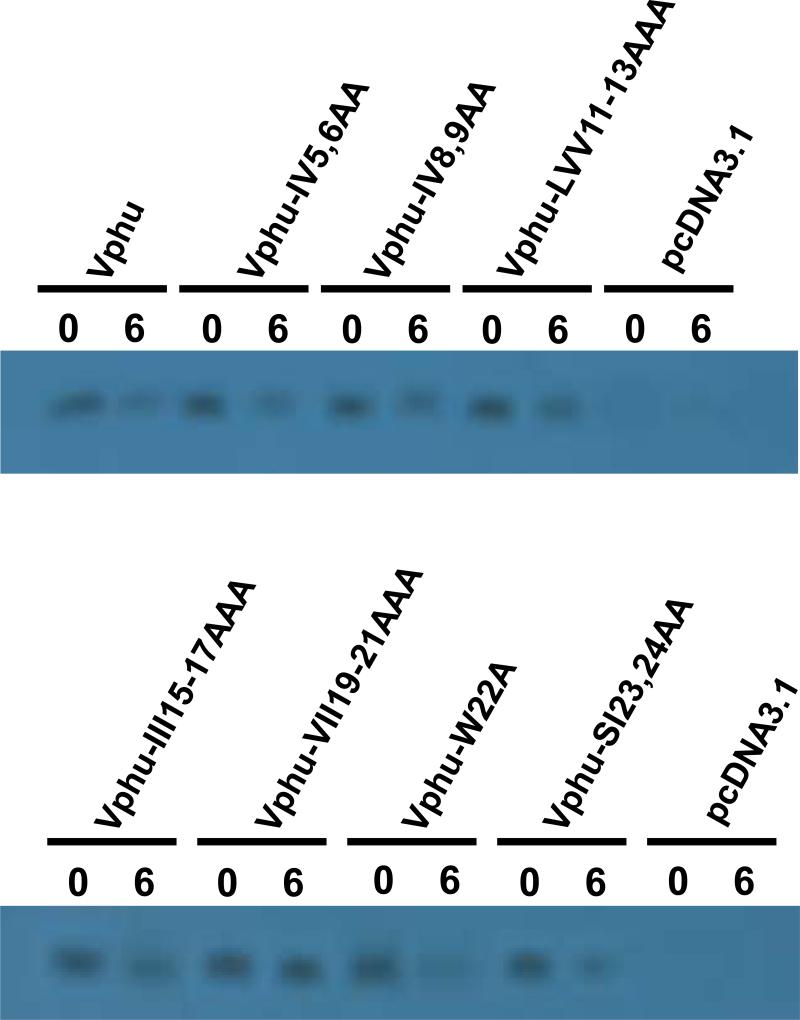
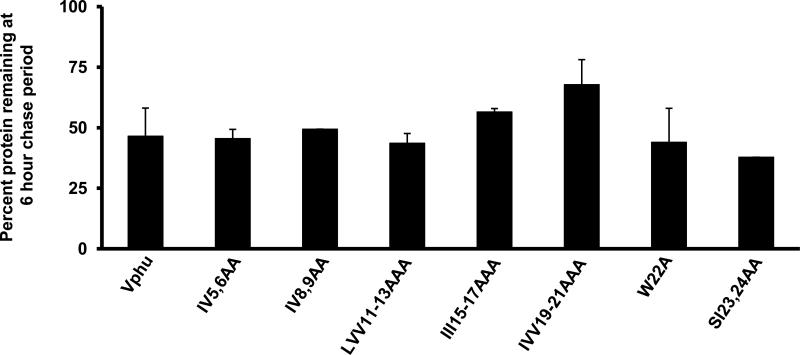
Based on the above results suggesting the involvement of the transmembrane domain in raft association, we constructed a series of vectors expressing Vphu proteins with 1-3 amino acid changes in the transmembrane domain (Fig. 7A). We first analyzed the stability of these mutants using pulse-chase analysis. 293 cells were transfected with each mutant or the unmodified Vphu. At 48 hours, the cells were starved and radiolabeled with 35S-methionine/cysteine and then the radiolabel chased in cold excess methionine/cysteine for 0 and 6 hours. All conditions were run in duplicate and the average percent protein remaining and standard deviation calculated. The results indicate that the majority of the Vphu mutants had a similar stability or were more stable than the unmodified Vphu, which had 47% remaining at the 6 hour chase period (Fig. 7B-C). Vphu mutants W22A and SI23,24AA had less protein remaining at the 6-hour chase period (28% and 38%, respectively).
Figure 7.
Characterization of Vphu transmembrane mutants. Panel A. A series of transmembrane mutants were constructed in which 1-3 non-alanine residues were changed to alanines. Panel B. Pulse chase analysis of the Vphu transmembrane mutants. 293 cells were transfected with either the unmodified Vphu or the Vphu TM mutants. At 48 hours, cells were starved for methionine/cysteine and radiolabled for 1 hour as described in the Materials and Methods section. The radiolabel was removed, cells washed and incubated in excess cold methionine/cysteine for 0 and 6 hours. The cells were lysed and processed for immunoprecipitation assays using a rabbit anti-Vpu serum. The immunoprecipitates were collected on Protein A Sepharose, boiled and visualized by SDS PAGE (12% gel) and standard autoradiographic techniques. The numbers above each lane represent the length of time chased in cold medium.
Identification of critical amino acids in the transmembrane domain of Vpu that are required for membrane raft association
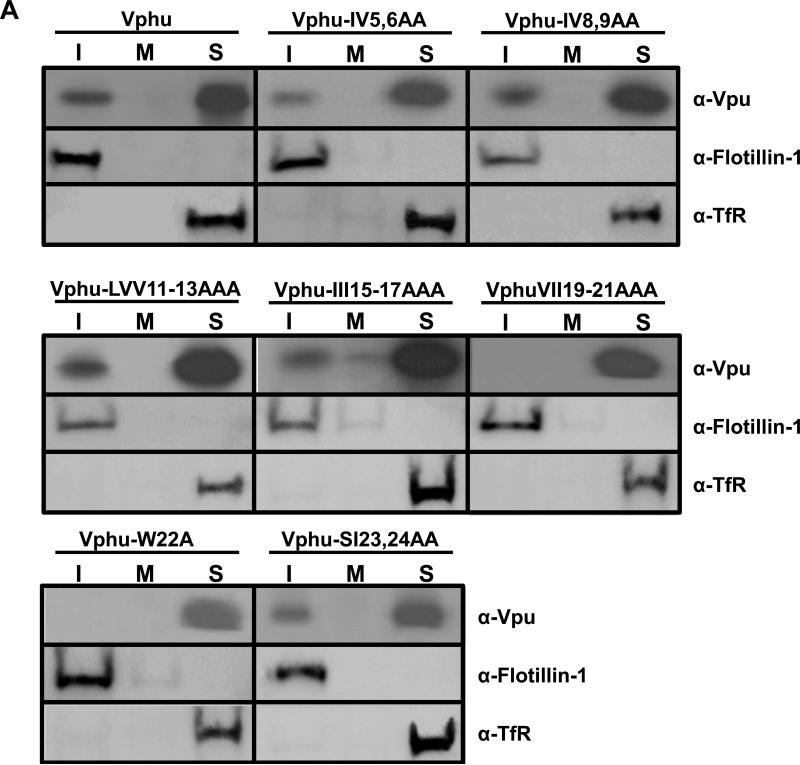
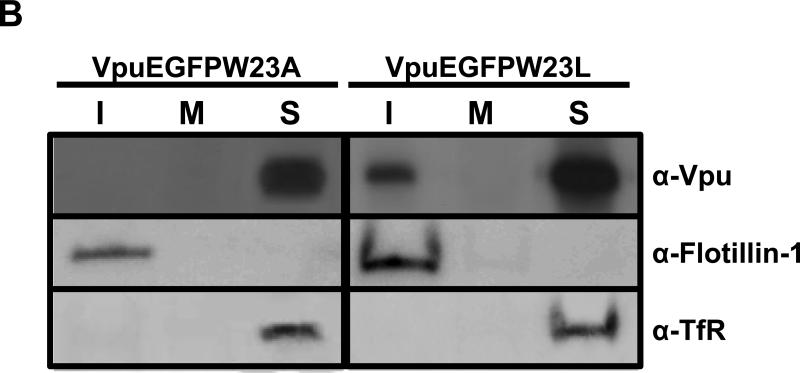
We determined if one or more amino acid residues within the transmembrane domain were critical for membrane raft association. Vectors expressing these mutant Vphu proteins were transfected into 293 cells and at 36 hours the DRMs were isolated as described above. All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments. Appropriate controls for raft isolation and purity were performed and are shown. We found that two mutants, W22A and IVV19-21AAA had no detectable Vpu in the DRM fractions (Fig. 8A). Additionally, substitution of the W22 with the more hydrophobic leucine did not affect raft association (Fig. 8B). Taken together, these results suggest that membrane raft association can be manipulated by substituting specific amino acids in the TM domain.
Figure 8.
Identification of Vphu transmembrane mutants that are not associated with membrane rafts. Raft association of individual transmembrane mutants. 293 cells were transfected with vectors expressing each of the raft mutants described in Figure 7A. At 48hours, DRMs were extracted and isolated as described in the Materials and Methods section. Fractions 1-3 (rafts; I), 5-6 (middle of the gradient; M) and 10-12 (non-raft; S) were pooled and analyzed for the presence of Vpu, flotillin-1, and transferrin receptor. Panel A. 293 cells transfected with vectors expressing each of the Vphu mutants described in Figure 7A. Panel B. 293 cells transfected with vectors expressing VpuEGFPW23A and VpuEGFPW23L mutants described in Fig. 7A.
The W22A mutant is localized to the same compartments as the unmodified VpuEGFP
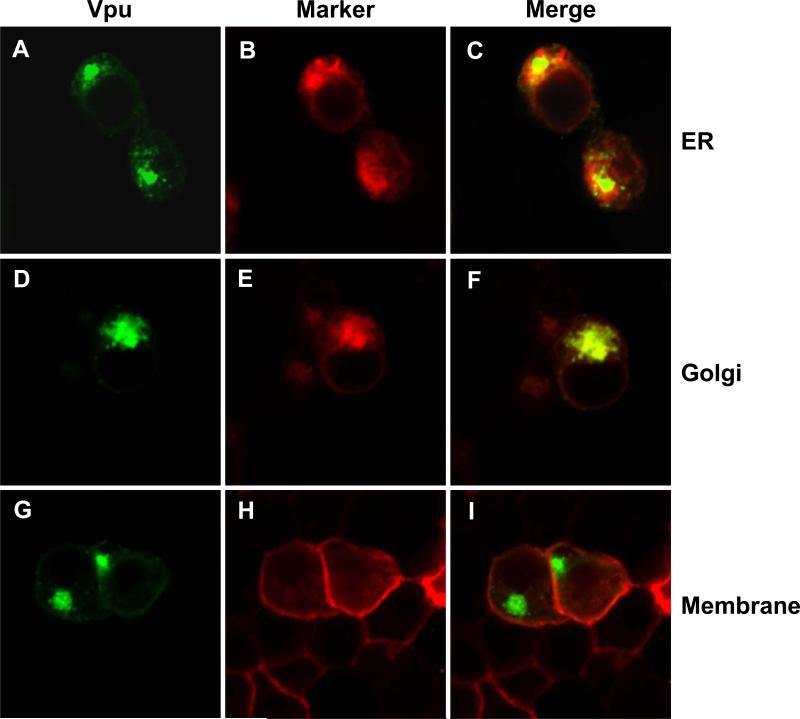
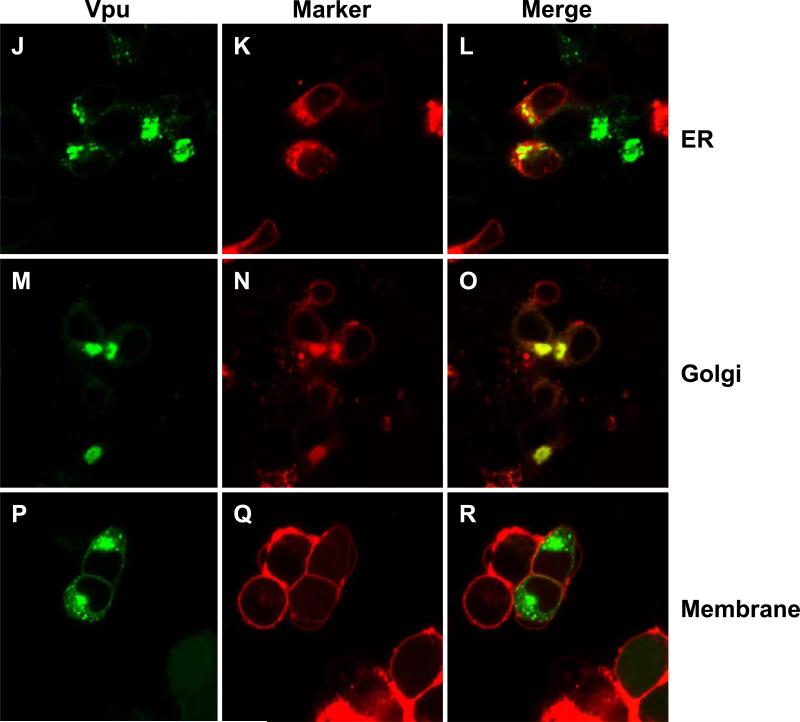
As the W22A mutant appeared to have the greatest effect on raft association, we determined if this mutant was mislocalized in the cell. 293 cells were transfected with vectors expressing either VpuEGFP or VpuEGFPW23A and ER, Golgi or membrane markers. The results indicate that the VpuEGFP (Fig. 9A-I) and the VpuEGFPW23A (Fig. 9J-R) were localized to similar compartments, suggesting that transport to the intracellular compartments was not the reason for the lack of raft association by the W22A/W23A mutants.
Figure 9.
VpuEGFPW23A and VpuEGFP are localized to similar compartments. 293 cells were transfected with a vectors expressing either VpuEGFP or VpuEGFPW23A and either ER-DsRed2, Golgi-DsRed2 or Membrane-DsRed2 as described in the Materials and Methods section. At 48 hours, cells were processed for confocal microscopy. Figure 9A-I. 293 cells transfected with VpuEGFP. Panels A, D, and G are fluorescent micrographs showing expression of VpuEGFP. Panels B, E, H are fluorescent micrographs showing expression of DsRed2 proteins. Panels C, F, and I are a merge of the two panels to the left. Panels A-C. 293 cells transfected with VpuEGFP and ER-DsRed2. Panels D-F. 293 cells transfected with VpuEGFP and Golgi-DsRed2. Panels G-I. 293 cells transfected with VpuEGFP and Mem-DsRed2. Figures 9J-R. 293 cells transfected with VpuEGFPW23A. Panels J, M, and P are fluorescent micrographs showing expression of VpuEGFPW23A. Panels K, N, and Q are fluorescent micrographs showing expression of DsRed2 proteins. Panels L, O, and R are a merge of the two panels to the left. Panels J-L. 293 cells transfected with VpuEGFPW23A and ER-DsRed2. Panels N-O. 293 cells transfected with VpuEGFPW23A and Golgi-DsRed2. Panels P-R. 293 cells transfected with VpuEGFPW23A and Mem-DsRed2.
Membrane raft association correlates with enhanced virus release
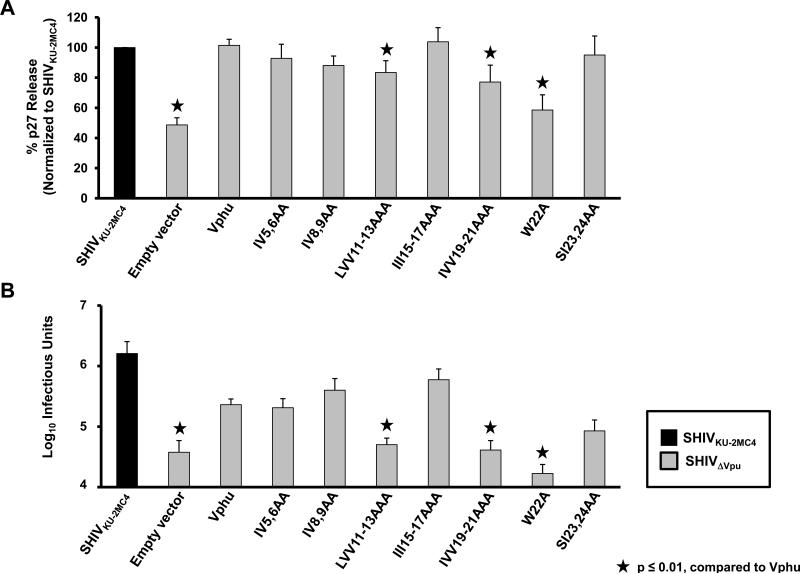
We examined these same mutants for the ability to enhance virus release from infected cells. For these experiments, we used HeLa cells, which express bone marrow stromal antigen 2 (BST-2). Vpu has been shown to interact with BST-2 to permit enhanced virus release from infected cells (Douglas et al., 2009). HeLa cells were co-transfected with plasmids expressing the Vphu mutants described above and SHIVΔVpu and assessed for p27 release. All conditions were run at least four separate times and the average percent p27 release and standard error calculated. The results indicate that Vpu mutants W22A and IVV19-21AAA and to a lesser extent LVV11-13AAA showed a significant decrease in p27 release, which correlated well with the lack of association with membrane rafts (Fig. 10A). Analysis of infectious virus released also showed the same general pattern when compared to the p27 assays (Fig. 10B). Significance of the p27 and infectious virus release for each virus mutant was assessed by comparison to the Vphu control with a p<0.01 considered significant.
Figure 10.
Raft association correlates with enhanced virus release. The same mutants described in Figure 7A were used to determine the level of virus release in the presence of human BST-2. HeLa cells were co-transfected with SHIVΔVpu and vectors expressing Vphu or the different transmembrane mutants. At 48 hours, the culture medium was collected and assayed for p27 and infectious virus released from cells. Panel A. The level of p27 released from transfected cells. Significance in the restriction of p27 release was calculated with respect to the SHIVKU-2MC4 in the presence of each respective BST-2 using a Student's t-test with p< 0.01 considered significant (★) .Panel B. The level of infectious virus released as determined by infection of TZM-bl cells and staining for the presence of β-galactosidase activity at 48 hours post-inoculation as described in the Materials and Methods section. Significance in the restriction of infectivity was determined with respect to the unmodified Vphu using a Student's t-test with p<0.01 considered significant (★).
Membrane raft association is not required for CD4 down-regulation
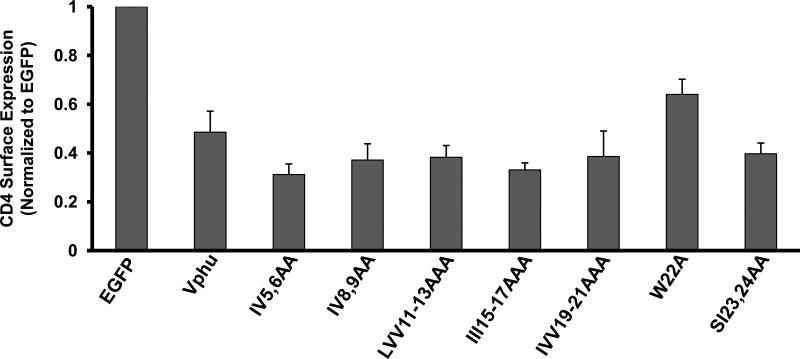
As the other major function of Vpu is shunting of CD4 to the proteasome for degradation, we also examined surface expression of CD4 in the presence of these Vpu mutants. HeLaCD4+ cells were transfected with vectors expressing Vphu or the mutants described above and EGFP. Cells were analyzed for CD4 surface expression by flow cytometry as previously described (Hill et al., 2008, 2009). All conditions were run three separate times and the average CD4 surface expression and standard error calculated. This Vpu function was not impaired with the majority of these mutants (Fig. 11). The two Vphu mutants that were not detected in rafts, Vphu-IVV19-21AAA and W22A, had normal (VphuIVV19-21AAA) or slightly impaired (W22A) surface CD4 down-modulation compared to the unmodified Vphu protein. Taken together, these results indicate that raft association was not a requirement for Vpu-mediated down-modulation of CD4.
Figure 11.
CD4 down-regulation by Vphu transmembrane mutants. HeLa CD4+ cells were transfected with plasmids expressing each of the unmodified Vphu or mutant Vphu proteins and one expressing EGFP. At 48 hours, live cells were immunostained for CD4. Cells were assessed for CD4 surface expression using flow cytometry. A mean fluorescent intensity (MFI) ratio of transfected (EGFP positive) to untransfected cells (EGFP negative)was calculated for each sample with the EGFP transfection control normalized to 1.0. Normalized ratios from three separate experiments were averaged and the standard error calculated.
DISCUSSION
Many enveloped viruses such as vaccinia virus, the orthomyxoviruses (influenza), paramyxoviruses (measles, RSV), filoviruses (Ebola, Marburg) herpesviruses (EBV, HSV-1, pseudorabies), flaviviruses (West Nile virus), rhabdoviruses (VSV), and retroviruses (MLV) use membrane rafts as portals for entry and/or egress from cells (Barman et al., 2000; Bavari et al., 2002; Bender et al., 2003; Brown and Lyles, 2003; Chung et al., 2005; Keller and Simons, 1998; Li et al., 2002; Manie et al., 2000; Marty et al., 2004; Medigeshi et al., 2008; Scheiffele et al., 1999). In addition to the viruses listed above, HIV-1 also exploits membrane rafts at several steps in its replication cycle. The HIV-1 envelope is known to be enriched in cholesterol (Aloia et al., 1993; Brugger et al., 2005). Both Gag and Env were found in detergent resistant membranes (Nyugen and Hildreth, 2000). These investigators found that GPI-anchored proteins such as Thy1 and CD59 as well as GM-1 ganglioside were present but that a non-raft protein, CD45, was largely absent from viral particles. It was subsequently shown that membrane rafts were critical for virus assembly and release (Ono and Freed, 2001). These investigators found that the myristylated N-terminal domain of Gag and the I domain of the nucleocapsid protein were critical for raft association and that the cholesterol depleting compound, M-β-CD, reduced the efficiency of virus release. The palmitoylation of Env gp41 was initially shown to be important for raft targeting and infectivity but later studies showed that substitution of the cysteines in the cytoplasmic domain of gp41 (thus resulting in no palmitoylation) only partially affected infectivity or had no effect (Ruoso et al., 2000; Bhattacharya et al., 2004; Chan et al., 2005). It was later shown that mutations in the Gag p17 protein would prevent Env incorporation into rafts and virions, suggesting that Gag regulates Env incorporation into rafts (Bhattacharya et al., 2006; Patil et al., 2010). Other investigators have shown that the HIV-1 Env has a cholesterol recognition/interaction amino acid consensus (CRAC) domain that is responsible for raft association (Epand et. al. 2006; Vishwanathan et al., 2008).
In this study, we have determined that the Vpu protein partially associates with membrane rafts. We have shown this using biochemical fractionation, through the use of cholesterol reducing drugs and co-patching experiments. In the results presented here, Vpu expressed either exogenously in the absence of other viral proteins or in virus-infected T cells (C8166 cell line), partitions into DRMs. Our studies used Triton-X-100 as it is the most stringent with respect to extraction of DRMs (Schuck et al., 2003; Shogomori and Brown, 2003). We also showed that Vpu partially partitions into DRMs when extracted at physiological temperature as rafts isolated at 37C are thought to be more similar to rafts in live cells. In addition to showing that Vpu partitions into DRMs, it was necessary to show that disruption of membrane rafts by cholesterol depletion would prevent Vpu from partitioning into DRMs. Drugs that reduce cholesterol levels in cells, either by inhibiting an HMGCoA reductase in the cholesterol synthesis pathway (lovastatin) or by extracting cholesterol from cell membranes (cyclodextrin; M-β-CD), are known to disrupt membrane rafts. We determined if a combination of lovastatin and M-β-CD would eliminate Vpu from DRMs, which would indicate that Vpu resistance to Triton-X-100 is a specific property of raft inclusion. This treatment significantly reduced the level of Vpu in membrane rafts. Finally, Vpu association with lipid rafts was confirmed by co-localization of VpuSCEGFP with a raft marker in live cells. As the subtype B Vpu used in most of the experiments in this study is found predominantly in the Golgi, ER, and endosomes, and only small amounts are found at the surface, we used VpuSC, which is efficiently transported to the cell surface for this assay. While it seems likely that Vpu is associated with rafts internally, these rafts are, as yet, difficult to visualize. Most studies of rafts in Golgi bodies or endosomes have been done using biochemical lipidomic techniques rather than live cell imaging. Taken together, these experiments provide strong evidence that Vpu is partially localized to membrane rafts and at least some rafts containing GPI-anchored proteins.
As the transmembrane or membrane proximal domains are most likely to be involved in membrane raft targeting, we examined the detergent resistance of a Vpu fusion protein with a scrambled TM domain (VpuTMEGFP). This protein was not associated with DRM fractions, indicating that the TM domain may be involved in membrane raft association. However, since this scrambled TM domain has a total of 15 amino acid changes, it is possible that these substitutions may have caused an alteration of the spatial orientation of the protein in the membrane, the flexible linker region following the TM domain or the first α-helical region proximal to the membrane. Previous modeling studies have indicated that changes in the Vpu TM domain have the ability to cause secondary structure changes downstream (Candler et al., 2005; Sramala et al., 2003). In order to address this concern, we used Vpu mutants with more targeted mutations. Our results indicate that Vpu mutants Vphu-IVV19-21AAA and Vphu-W22A were no longer associated with membrane raft fractions. While these results show that amino acid substitutions in the TM domain can affect incorporation into rafts, they do not necessarily rule out that other amino acids in the TM domain (the alanine residues) or residues in the cytoplasmic domain may also be involved. Recent computer modeling studies have suggested that the transmembrane domain of Vpu is flexible in adapting to different lipid environments (Kruger and Fischer, 2008). When Vpu was simulated moving through various lipid environments representative of the Golgi apparatus, Vpu exhibited no particular preference for lipid thickness or composition. Rather, the tilt angle and kink around the region I17 to S23 adjust to the membrane. This modeling data correlates well with the loss of DRM association of IVV19-21AAA and W22A. If these residues are important to the structural flexibility of the Vpu transmembrane domain, substitution with alanines could reduce the ability of the protein to adapt to a changing lipid environment, thus excluding it from membrane rafts. Substitution of the W23 with a longer, more hydrophobic leucine resulted in a protein which was still found in the DRMs. As this protein was more similar to wild-type Vpu in efficiency of CD4 down-modulation than W23A (data not shown), this further suggests that structural changes are involved in raft inclusion and exclusion of Vpu.
To demonstrate that membrane raft association was relevant to Vpu functions and to virus replication, we assayed the various Vpu TM mutants for the ability to down-modulate CD4 surface expression and enhance virus release. Our results showed that all of the Vpu TM mutants down-modulated CD4 surface expression although Vphu-W22A was slightly impaired compared to the other mutants. We measured enhanced virus release by two methods, release of p27 antigen from cells and by quantifying levels of infectious virus released from cells. The two assays were in agreement and indicate that virus release was significantly reduced by the LVV11-13AAA, IVV19-21AAA and W22A mutants with the W22A consistently having the most reduction in particle release. Of these three mutants, both LVV19-21AA and W22A were not incorporated into rafts, implying a correlation between membrane raft association and Vpu-mediated virus release. One question that arises is, “Why did LVV11-13AAA have impaired release since it was clearly observed in rafts?” While the answer is presently unknown, it is possible that substitution of three hydrophobic residues with less hydrophobic alanines may have altered the structure/ flexibility of the protein or protein-protein interactions (such as with BST-2) of the domain although it had no effect on CD4 down-modulation. As is a caveat of all mutagenesis based studies, it is unknown whether the results observed in this study were based on structural alterations of the protein or roles for specific amino acids. Further studies will be needed to determine the affects the mutations introduced in this study have on the overall protein structure as well as any protein-protein interactions. However, regardless of the mechanisms by which the membrane raft association and enhanced virion release function of the Vpu proteins were altered these results demonstrate the ability of exogenous forces to disrupt function. This introduces the potential for therapeutic molecules designed to alter the spatial orientation of the TMD such that membrane raft association and/or protein-protein interactions (such as BST-2) would be disrupted.
Recently, Vpu was shown to antagonize the activity of a molecule known as bone marrow stromal cell antigen 2 (BST-2) or tetherin (Neil et al., 2008; Van Damme et al., 2008). BST-2 is thought to work by “tethering” particles at the cell surface, which may account for the observed maturation of HIV-1Δvpu (adherence of particles to the cell plasma membrane with a common observation of several virus particles in the “string of pearls” arrangement and the observation of virus particles being taken or maturing into vesicles). BST-2 has been shown to associate with membrane rafts, including the sites of HIV-1 maturation and release (Kupzig et al., 2003; Perez-Caballero et al., 2009; Rollason et al., 2007). In the absence of Vpu, BST-2 may become associated with rafts that are ultimately involved in virus assembly and release from cells. This is supported by findings that HIV-1 Gag and BST-2 co-localize in intracellular compartments of the cell (Neil et al., 2008; Van Damme et al., 2008). Vpu is not incorporated into virions suggesting that Vpu may associate with membrane rafts that are not involved in virus assembly and release. It is known that membrane rafts are diverse in both lipid and protein composition and it has been shown that distinct rafts can be isolated using immune selection procedures (Drevot et al., 2002; Knorr et al., 2009). One group of investigators showed that wild type BST-2 was found in several different clusters on the plasma membrane and that removal of the GPI anchor resulted in a BST-2 exclusively associated with sites of budding (Perez-Caballero et al., 2009). This implies that wild type BST-2 is found in more than one type of membrane raft. Thus, it is conceivable that Vpu could possibly interact with BST-2 before or after transport to the Golgi complex and become associated with a subpopulation of membrane rafts not associated with virus assembly and release at the cell surface. Membrane rafts were originally shown to initially form in the Golgi complex (Simons and van Meer, 1988) and to be enriched in the trans Golgi network. However, more recent studies have shown that the ER and cis Golgi are also sites for sorting of proteins and lipids (Alfalah et al., 2005; Browman et al., 2006). BST-2 was found in membrane rafts only after its N-linked carbohydrate chains were fully processed, suggesting that raft association of this protein occurs in the Golgi complex (Kupzig et al., 2003). Recent data has been presented that suggests the presence of Vpu in the trans Golgi is important for Vpu antagonism of BST-2 (Dube et al., 2009). It will be of interest to determine if Vpu and BST-2 can be co-immunoprecipitated from similar rafts and if so, where does this association occur? Finally, identification of Vpu mutants that are functional for either CD4 down-modulation or enhanced virus release will be useful in determining if one function is more important for pathogenesis using the SHIV/macaque model.
MATERIALS AND METHODS
Cells, viruses, and plasmids
The 293 and HeLa cell lines were maintained in Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum, gentamicin (5 μg per ml) and penicillin/streptomycin (100U per ml and 100 μg per ml). TZM-bl cells expressing luciferase and β-galactosidase genes under the control of an HIV-1 promoter were maintained in DMEM containing 10% fetal bovine serum with antibiotics. HeLa CD4+ cells were maintained in DMEM containing 10% fetal bovine serum antibioics and 1 mg/ml of G-418. The derivation and pathogenicity of SHIVKU-1bMC33 and novpuSHIVKU-1bMC33 have been described (McCormick-Davis et al., 2000; Singh et al., 2003; Stephens et al., 2002). The derivation of the SHIVKU-2MC4Δvpu plasmid has been described (Ruiz et al., 2010). Vectors expressing the subtype B (pcvpuegfp) and C Vpu (pcvpuscegfp1) proteins fused to enhanced green fluorescent protein (eGFP) and the VpuTMEGFP (pcvpuTMegfp) mutant have been previously described (Singh et al., 2003; Gomez et al., 2005; Pacyniak et al., 2005; Ruiz et al., 2008). The vector expressing the codon optimized (human) Vpu protein (Vphu) was kindly provided by the NIH AIDS Reference and Reagents Program (Nguyen et al., 2004). The YFP-GPI and NFP-GPI constructs were kindly provided by Dr. Akira Ono. The YFP-GPI construct has the ER translocation signal from rabbit phlorizin hydrolase fused to the N-terminus of Venus YFP and a GPI anchor sequence from CD59 fused to the C-terminus. The non-fluorescent version (NFP-YFP) contains the mutation Y67C that abolishes fluorophore formation. The vectors expressing ER-DsRed2, Golgi-Red2, and Mem-DsRed2 were obtained from Clontech.
Isolation of detergent resistant membranes (DRMs)
293 cells were cultured in 60 mm well dishes for 24 hours, then transfected using branched polyethylenimine (PEI; Sigma). At 48 hours post-transfection, cells were lysed in ice cold DRM buffer (25 mM Tris pH 7.5, 150mM NaCl, 10mM EDTA) containing 1% Triton-X-100. Cells remained on ice in lysis buffer for 20 minutes before being pushed through a 22 gauge needle at least 7 times. The lysate was then mixed with an equal volume of 80% sucrose (w/v) in DRM buffer to a final concentration of 40% sucrose. The lysate was placed in the bottom of a SW41 ultracentrifuge tube and overlaid with 8 ml 30% sucrose and 2 ml 5% sucrose. Gradients were spun to equilibrium at 38,000 × rpm (247,000 × g) for 18 hours (SW41 rotor, Beckman Coulter). One ml fractions were taken from the top, concentrated, and analyzed by Western blot using a rabbit anti-EGFP antibody or rabbit anti-Vpu antibody (NIH AIDS Research and Reference Reagents Program). Controls include the raft protein flotillin-1 (mouse anti-flotillin-1, BD Biosciences) and the non-raft protein, transferrin receptor (mouse anti-transferrin receptor, BD Biosciences). All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments.
For isolation of DRMs from virus infected cells, C8166 cells were inoculated with 104 TCID50 of either SHIVKU-1bMC33 or novpuSHIVKU-1bMC33 for 4 hours. At this time, cells were washed twice and incubated in fresh medium for 5 days at 37C. Cultures were starved and radiolabled with 1 mCi of 35S-methionine/cysteine for 2 hours. The cells were centrifuged at low speed (800 × g) to pellet the cells, washed three times in medium without serum and processed as above. One ml fractions were taken from the top and raft (fractions 1-3), middle (fractions 6-7), and non-raft (fractions 10-12) fractions were analyzed by immunoprecipitation using a rabbit anti-Vpu antibody and by Western blot for raft and non-raft control proteins.
Isolation of detergent resistant membranes (DRMs) at physiological temperature
For these experiments, we used the procedure of Chen et al., (2009). 293 cells were cultured in 12 well dishes for 24 hours and then transfected using PEI. At 36-48 hours post-transfection, cells were lysed in DRM37 (10mM HEPES pH 7.0, 50mM KOAc, 1mM Mg(OAc)2, 1mM EDTA, 200mM sucrose). The lysates were pushed through a 22 gauge needle at least 7 times, then incubated at 37C for 5 minutes. Lysates were then mixed with an equal volume of 2% Triton X-100 in DRM37 and incubated at 37C for 5 minutes. The lysates were mixed with an equal volume of 80% sucrose in DRM37, layered at the bottom of an ultracentrifuge tube and overlaid with 7 ml 30% sucrose/DRM37 and 2 ml DRM37 (6.5% sucrose). Gradients were spun to equilibrium at 38,000 × rpm overnight (SW41 rotor, Beckman Coulter) at 4C. One ml fractions were taken from the top and analyzed for various proteins by Western blot. All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments. Appropriate controls for raft isolation and purity were performed and are shown.
Cholesterol depletion experiments
293 cells were transfected with a plasmid expressing VpuEGFP or VpuSCEGFP1 and treated with 4 μM lovastatin for 48 hours. Thirty minutes prior to lysis of cells, the cultures were incubated with M-β-cyclodextrin (M-β-CD; 10 mg/ml). Lysates were prepared, and subjected to ultracentrifugation as described above. One ml fractions were collected from the top of the gradient. Fractions 1-3, 6-7, and 10-12 were each pooled, methanol precipitated and resuspended in 1X sample reducing buffer. The samples were then analyzed for the presence of VpuEGFP or VpuSCEGFP1 by Western blot using an antibody directed against EGFP. All experiments were performed at least twice, and most three times. Blots shown are representative of all experiments. Appropriate controls for raft isolation and purity were performed and are shown.
Co-patching experiments
293 cells cultured on cover slips were transfected with YFP-GPI or NFP-GPI and VpuSCEGFP1 using PEI. At 48 hours post-transfection, cover slips were washed three times in 1X PBS, then incubated in primary antibody (mouse anti-GFP, Clontech) for 30 minutes at 37C. Unbound antibody was removed by washing three times in 1X PBS followed by reaction with a secondary antibody (goat anti-mouse-Cy5, Molecular Probes) at 37C for 30 minutes. Unbound antibody was removed by washing three times in 1X PBS and cells fixed in 2% paraformaldehyde/PBS for 15 minutes. Cover slips were mounted in SlowFade Antifade solution A (Molecular Probes). A Nikon A1 confocal microscope was used to collect 100X images with a 2X digital zoom, using EZ-C1 software. The pinhole was set to large (100nm) for all wavelengths. EGFP was excited using an argon 488 nm laser and viewed through the FITC filter (525/25nm) and Cy5 was excited at 638 nm and viewed through a Cy5 filter (700/38 nm).
Site-directed mutagenesis
Mutations introduced into all plasmids were accomplished using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. All plasmid inserts were sequenced to ensure the validity of the mutations and that no other mutations were introduced during the cloning process.
Pulse-chase analysis of Vphu TMD mutant proteins
293 cells were transfected with vectors expressing each mutant protein. At 48 hours post-transfection, the medium was removed and cells were incubated in methionine/cysteine-free medium for 2 hours. The cells were then radiolabeled with 200 μCi of 35S-Translabel (methionine and cysteine, MP Biomedical) for 1 hour. The radiolabel was chased in DMEM containing 100X unlabeled methionine/cysteine medium for 0 and 6 hours. Vphu proteins were immunoprecipitated using a rabbit anti-Vpu serum and collected on Protein A-Sepharose beads on a rotator for 18 hours. Non-transfected 293 cells, starved, radiolabeled and chased for 0 hours served as a negative control. Beads were washed three times with 1X radioimmunoprecipitation buffer (RIPA: (50 mM TrisBHCl, pH 7.5; 50 mM NaCl; 0.5% deoxycholate; 0.2% SDS; 10 mM EDTA), and the samples resuspended in sample reducing buffer. Samples were boiled and the Vphu proteins separated by SDS-PAGE (12% gel). Proteins were then visualized using standard autoradiographic techniques. All conditions were run in duplicate, the pixel densities of each band determined using ImageJ software, normalized to the hour 0 sample, and the average percent protein remaining and standard deviation calculated.
Confocal microscopy studies
293 cells were cultured on coverslips one day prior to being transiently transfected with plasmids expressing either VpuEGFP or VpuEGFPW23A and one of the intracellular marker proteins (ER-DsRed2, Golgi-DsRed2 or Membrane-DsRed2) using branched polyethylenimine (Sigma). Cultures were maintained for 36-48 hours before being washed three times in PBS, and fixed in 2% paraformaldehyde/PBS. Cover slips were mounted in glycerol containing mounting media (Slowfade Antifade solution A). A Nikon A1 confocal microscope was used to collect 100X images with a 2X digital zoom, using EZ-C1 software. The pinhole was set to large (100nm) for all wavelengths. EGFP was excited using an argon 488nm laser and viewed through the FITC filter (525/25nm). DsRed2 were excited using a 561 DPSS laser and viewed through the Texas Red filter (595/50nm).
Virion release assays
Hela cells (105) were seeded into each well of a 24-well tissue culture plate 24 hours prior to transfection. Cells were transfected as described above with 1 μg of plasmid expressing full length SHIV proviral DNA (either SHIVKU-2MC4 or SHIVΔVpu) and 200 ng of plasmid expressing various mutant vphu genes. Cells were incubated at 37C in 5% CO2 atmosphere for 48 hours. Supernatants were collected and cellular debris removed through low speed centrifugation. Cells were lysed in 250 μl of 1X RIPA buffer and the nuclei removed through high speed centrifugation. The amount of p27 present within the supernatant and the cell lysates was determined using a commercially available p27 ELISA kit (Zeptometrix Incorporated) and the percent of p27 release calculated. All conditions were run at least four separate times and the average percent p27 release and standard error calculated. Significance with respect to the unmodified Vphu was calculated using a Student's t-test with a p<0.01 considered significant.
Infectious virus release assays
TZM-bl cells (104) expressing luciferase and β-galactosidase genes under the control of an HIV-1 promoter were seeded into each well of a 96-well tissue culture plate 24 hours prior to infection. Supernatants collected from HeLa cells co-transfected with SHIV and Vphu expressing plasmids as described above were added to the TZM-bl cells and serially diluted. At 48 hours post-infection, cells were washed twice in 1X PBS and incubated in a fixative solution (0.25% glutaraldehyde, 0.8% formaldehyde in phosphate buffered saline) for 5 minutes at room temperature. The cells were washed three times in 1X PBS and covered in staining solution (400 μg/ml X-gal, 4 mM MgCl2, 4 mM K3Fe(CN)6, 4 mM K4Fe(CN)6-3H2O in phosphate buffered saline) and incubated for 2 hours at 37C. Cells were washed once in 1X PBS and then covered in 1X PBS during counting. The TCID50 for each supernatant was calculated based on wells containing cells expressing β-galactosidase. All conditions were run at least four times and the TCID50 calculated. The average TCID50 and standard error were calculated. Significance in the restriction of infectivity was determined with respect to the unmodified Vphu using a Student's t-test with p<0.01 considered significant.
Assessment of CD4 cell surface expression
For analysis of cell surface CD4 expression in the presence of the various Vphu mutant proteins, HeLa CD4+ cells were seeded into 6-well tissue culture plates 1 day prior to transfection such that 24 hours later they would be 60-70% confluent. Cells were co-transfected with plasmids expressing one of the various Vphu mutant proteins and EGFP. Cells transfected with EGFP only were used as the control. At 48 hours post-transfection, cells were removed from the plate using Ca2+/Mg2+-free PBS containing 10 mM EDTA and stained with PE-Cy5 conjugated anti-CD4 (BD Bioscience). Cells were analyzed using an LSRII flow cytometer and the mean fluorescence intensity (MFI) of PE-Cy5 for transfected cells (EGFP positive) and non-transfected (EGFP negative) was calculated. An MFI ratio was calculated for each sample with the EGFP control (no Vphu) normalized to 1.0. Normalized ratios from three separate experiments were averaged and the standard error calculated. All groups were compared to the EGFP control as well as the Vphu only control using a Student's t-test (unpaired, p<0.01 significant).
ACKNOWLEDGMENTS
The work reported here is supported by grant NIH AI51981 to E.B.S. We thank members of the KUMC Biotechnology Support Facility and Northwestern University for their assistance with the sequence analysis and oligonucleotide synthesis. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Anti-Bst-2 (cat# 11722) from Drs. Klaus Strebel and Amy Andrew; and TZM-bl from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc.; the pcDNAVphu (catalog #10076) from Drs. Stephen Bour and Klaus Strebel.; the HelaCD4+ cells (catalog # 154) from Dr. Richard Axel. We thank the laboratory Dr. Akira Ono at the University of Michigan for the YFP-GPI (originally by Kai Simons, Max Planck Institute of Molecular Cell Biology and Genetics, Germany) and NFP-GPI constructed by Ian Hogue in Dr. Ono's laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alfalah M, Wetzel G, Fischer I, Busche R, Sterchi EE, Zimmer KP, Sallmann HP, Naim HY. A novel type of detergent-resistant membranes may contribute to an early protein sorting event in epithelial cells. J. Biol. Chem. 2005;280:42636–42643. doi: 10.1074/jbc.M505924200. [DOI] [PubMed] [Google Scholar]
- Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl. Acad. Sci. U S A. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S, Nayak DP. Analysis of the transmembrane domain of influenza virus neuraminidase, a type II transmembrane glycoprotein, for apical sorting and raft association. J. Virol. 2000;74:6538–6545. doi: 10.1128/jvi.74.14.6538-6545.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender FC, Whitbeck JC, Ponce de Leon M, Lou H, Eisenberg RJ, Cohen GH. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J. Virol. 2003;77:9542–9552. doi: 10.1128/JVI.77.17.9542-9552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Peters PJ, Clapham PR. Human immunodeficiency virus type 1 envelope glycoproteins that lack cytoplasmic domain cysteines: impact on association with membrane lipid rafts and incorporation onto budding virus particles. J. Virol. 2004;78:5500–5506. doi: 10.1128/JVI.78.10.5500-5506.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. J. Virol. 2006;80:5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman DT, Resek ME, Zajchowski LD, Robbins SM. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006;119:3149–3160. doi: 10.1242/jcs.03060. [DOI] [PubMed] [Google Scholar]
- Brown EL, Lyles DS. Organization of the vesicular stomatitis virus glycoprotein into membrane microdomains occurs independently of intracellular viral components. J. Virol. 2003;77:3985–3992. doi: 10.1128/JVI.77.7.3985-3992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich H-G. The HIV lipidome: A raft with an unusual composition. Proc Natl. Acad. Sci., USA. 2005;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candler A, Featherstone M, Ali R, Maloney L, Watts A, Fischer WB. Computational analysis of mutations in the transmembrane region of Vpu from HIV-1. Biochim. Biophys. Acta. 2005;1716:1–10. doi: 10.1016/j.bbamem.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Chan WE, Lin HH, Chen SS. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J Virol. 2005;79:8374–8387. doi: 10.1128/JVI.79.13.8374-8387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Jen A, Warley A, Lawrence MJ, Quinn PJ, Morris RJ. Isolation at physiological temperature of detergent-resistant membranes with properties expected of lipid rafts: the influence of buffer composition. Biochem J. 2009;417:525–533. doi: 10.1042/BJ20081385. [DOI] [PubMed] [Google Scholar]
- Chung CS, Huang CY, Chang W. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J Virol. 2005;79:1623–1634. doi: 10.1128/JVI.79.3.1623-1634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Früh K, Moses AV. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a β-TrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, Cohen EA. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J. Virol. 2009;83:4574–4590. doi: 10.1128/JVI.01800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevot P, Langlet C, Guo XJ, Bernard AM, Colard O, Chauvin JP, Lasserre R, He HT. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 2002;21:1899–1908. doi: 10.1093/emboj/21.8.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epand RF, Thomas A, Brasseur R, Vishwanathan SA, Hunter E, Epand RM. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45:6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K, Omura S, Silver J. Rapid degradation of CD4 in cells expressing human immunodeficiency virus type 1 Env and Vpu is blocked by proteasome inhibitors. J.Gen.Virol. 1997;78:619–625. doi: 10.1099/0022-1317-78-3-619. [DOI] [PubMed] [Google Scholar]
- Gkantiragas I, Brügger B, Stüven E, Kaloyanova D, Li XY, Löhr K, Lottspeich F, Wieland FT, Helms JB. Sphingomyelin-enriched microdomains at the Golgi complex. Mol. Biol. Cell. 2001;12:1819–1833. doi: 10.1091/mbc.12.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez LM, Pacyniak E, Mulcahy ER, Flick M, Gomez M, Nerrient E, Ayouda A, Santiago M, Hahn B, Stephens EB. Vpu mediated CD4 down-regulation and degradation is conserved among highly divergent SIVcpz strains. Virology. 2005;335:46–60. doi: 10.1016/j.virol.2005.01.049. [DOI] [PubMed] [Google Scholar]
- Hout DR, Gomez M, Pacyniak E, Gomez L, Mulchay ER, Culley N, Pinson DM, Powers M, Wong SW, Stephens EB. Scrambling of the amino acids within the transmembrane domain of Vpu results in simian-human immunodeficiency virus (SHIVTM) that is less pathogenic for pig-tailed macaques. Virology. 2005;339:56–69. doi: 10.1016/j.virol.2005.04.038. [DOI] [PubMed] [Google Scholar]
- Keller P, Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J. Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM. The human immunodeficiency virus type 1-specific protein Vpu is required for efficient virus maturation and release. J. Virol. 1990;64:621–629. doi: 10.1128/jvi.64.2.621-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr R, Karacsonyi C, Lindner RJ. Endocytosis of MHC molecules by distinct membrane rafts. Cell Sci. 2009;122:1584–94. doi: 10.1242/jcs.039727. [DOI] [PubMed] [Google Scholar]
- Kruger J, Fischer WB. Exploring the Conformational Space of Vpu from HIV-1: A Versatile Adaptable Protein. J. Comput. Chem. 2008;29:2416–2124. doi: 10.1002/jcc.20986. [DOI] [PubMed] [Google Scholar]
- Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Suzuki K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim Biophys Acta. 2005;1746:234–251. doi: 10.1016/j.bbamcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Li M, Yang C, Tong S, Weidmann A, Compans RW. Palmitoylation of the murine leukemia virus envelope protein is critical for lipid raft association and surface expression. J Virol. 2002;76:11845–11852. doi: 10.1128/JVI.76.23.11845-11852.2002. Li M, Yang C, Tong S, Weidmann A, Compans RW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manié SN, de Breyne S, Vincent S, Gerlier D. Measles virus structural components are enriched intolipid raft microdomains: a potential cellular location for virus assembly. J Virol. 2000;74:305–311. doi: 10.1128/jvi.74.1.305-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty A, Meanger J, Mills J, Shields B, Ghildyal R. Association of matrix protein of respiratory syncytial virus with the host cell membrane of infected cells. Arch. Virol. 2004;149:199–210. doi: 10.1007/s00705-003-0183-9. [DOI] [PubMed] [Google Scholar]
- McCormick-Davis C, Mukherjee S, Dalton SB, Singh DK, Pinson DM, Berman NEJ, Foresman L, Stephens EB. A molecular clone of simian-human immunodeficiency virus (SHIVKU-1bMC33) with a truncated, non-membrane bound Vpu results is not required for rapid CD4+ T cell loss and neuroAIDS in pig-tailed macaques. Virology. 2000;272:112–126. doi: 10.1006/viro.2000.0333. [DOI] [PubMed] [Google Scholar]
- Medigeshi GR, Hirsch AJ, Streblow DN, Nikolich-Zugich J, Nelson JA. West Nile virus entry requires cholesterol-rich membrane microdomains and is independent of alphavbeta3 integrin. J Virol. 2008;82:5212–5219. doi: 10.1128/JVI.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Nguyen DH, Hildreth JE. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74:3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K-L, Llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, Bour S. Codon optimization of the HIV-1 vpu and vif genes stabilizes their messenger RNA and allows for highly efficient Rev-independent expression. Virology. 2004;319:163–175. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Ono A, Freed E,O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak E, Gomez M, Gomez L, Mulcahy E, Jackson M, Flick M, Hout DR, Stephens EB. Identification of a region within the cytoplasmic domain of the subtype B Vpu protein of human immunodeficiency virus type 1 (HIV-1) that responsible for retention in the Golgi complex and its absence in subtype C Vpu proteins. AIDS Res. Hum. Retro. 2005;21:379–394. doi: 10.1089/aid.2005.21.379. [DOI] [PubMed] [Google Scholar]
- Patil A, Gautam A, Bhattacharya J. Evidence that Gag facilitates HIV-1 envelope association both in GPI-enriched plasma membrane and detergent resistant membranes and facilitates envelope incorporation onto virions in primary CD4+ T cells. Virol. J. 2010;7:3. doi: 10.1186/1743-422X-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell. 2009;139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G. Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J. Cell Sci. 2007;120:3850–3858. doi: 10.1242/jcs.003343. [DOI] [PubMed] [Google Scholar]
- Ruiz A, Hill MS, Schmitt K, Guatelli J, Stephens EB. Requirements of membrane proximal tyrosine and dileucine sorting signals for efficient transport of the subtype C Vpu proteins to the plasma membrane and virus release. Virology. 2008;378:58–68. doi: 10.1016/j.virol.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Guatelli JC, Stephens EB. The Vpu protein: new concepts in virus release and CD4 down-modulation. Curr. HIV-1 Res. 2010;8:240–252. doi: 10.2174/157016210791111124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouso I, Mixon MB, Chen BK, Kim PS. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. U S A. 2000;97:13523–13525. doi: 10.1073/pnas.240459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Bour S, Ferrer-Montiel AV, Montal M, Maldarell F, Strebel K. The two biological activities of human immunodeficiency virus type 1 Vpu protein involve two separable structural domains. J. Virol. 1996;70:809–819. doi: 10.1128/jvi.70.2.809-819.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Bacik I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J. Virol. 1998;72:2280–2288. doi: 10.1128/jvi.72.3.2280-2288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U S A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Shogomori H, Brown DA. Use of detergents to study membrane rafts: the good, the bad, and the ugly. J. Biol Chem. 2003;384:1259–1263. doi: 10.1515/BC.2003.139. [DOI] [PubMed] [Google Scholar]
- Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- Singh DK, Griffin DM, Pacyniak E, Jackson M, Werle M, Wisdom B, Sun F, Hout DR, Pinson DM, Gunderson RS, Powers MF, Wong SW, Stephens EB. The presence of the casein kinase II phosphorylation sites of Vpu enhances the CD4+ T cell loss caused by a simian human immunodeficiency virus (SHIVKU-1bMC33) in pig-tailed macaques. Virology. 2003;313:435–451. doi: 10.1016/s0042-6822(03)00339-8. [DOI] [PubMed] [Google Scholar]
- Sramala I, Lemaitre V, Faraldo-Gomez JD, Vincent S, Watts A, Fischer WB. Molecular dynamics simulations on the first two helices of Vpu from HIV-1. Biophys J. 2003;84:3276–3284. doi: 10.1016/S0006-3495(03)70052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens EB, McCormick C, Pacyniak E, Griffin D, Sun F, Pinson DM, Gunderson RS, Wong SW, Berman NEJ, Singh DK. A chimeric simian-human immunodeficiency virus with the vpu sequences removed prior to envelope glycoprotein gene (novpuSHIVKU1bMC33) results in enhanced Env biosynthesis in lymphocytes but is less pathogenic in pig-tailed macaques. Virology. 2002;293:252–261. doi: 10.1006/viro.2001.1244. [DOI] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Martin MA. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science. 1988;241:1221–1223. doi: 10.1126/science.3261888. [DOI] [PubMed] [Google Scholar]
- Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli JC. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan SA, Thomas A, Brasseur R, Epand RF, Hunter E, Epand RM. Hydrophobic substitutions in the first residue of the CRAC segment of the gp41 protein of HIV. Biochemistry. 2008;47:124–30. doi: 10.1021/bi7018892. [DOI] [PubMed] [Google Scholar]