Abstract
The ability to quantify the local electrostatic environment of proteins and protein/peptide assemblies is key to yielding a microscopic understanding of many biological interactions and processes. Herein, we show that the ester carbonyl stretching vibration of two non-natural amino acids, L-aspartic acid 4-methyl ester and L-glutamic acid 5-methyl ester, is a convenient and sensitive probe in this regard since its frequency correlates linearly with the local electrostatic field for both hydrogen-bonding and non-hydrogen-bonding environments. We expect that the resultant frequency-electric field map will find use in various applications. In addition, we show that, when situated in a non-hydrogen bonding environment, this probe can also be used to measure the local dielectric constant (ε). For example, applying it to amyloid fibrils formed by Aβ16-22 reveals that the interior of such β-sheet assemblies has a ε of ~5.6.
Keywords: IR Probe, Protein Electrostatics, Hydrogen Bonding
Electrostatic interactions are ubiquitous in biological molecules and, in many cases, play a key role in molecular association and enzymatic reactions.[1] However, quantifying the local electric field or how it changes inside a protein, especially in a site-specific manner and/or as a function of time, still remains a challenging task. One promising method in this regard is vibrational Stark spectroscopy,[2] which capitalizes on the fact that vibrational transitions have an intrinsic dependence on local electrostatic environment and uses an infrared (IR) probe that has a well-defined, localized vibrational mode to sense local electric field amplitude through the response of the frequency.[3] For example, the vibrational Stark effect has been used to determine the local electric field at protein interfaces and to monitor protein conformational transitions and dynamics.[4] While the theoretical underpinning of this methodology is straightforward, in practice the application of vibrational Stark spectroscopy to biological systems is currently limited by the availability of suitable vibrational probes. Herein, we show, using linear and nonlinear IR measurements and molecular dynamics (MD) simulations, that the ester carbonyl vibration in two non-natural amino acids can be used to quantitatively and site-specifically probe the electric fields of proteins, including those arising from hydrogen-bond (H-bond) interactions.
The utility of a vibrational probe to reliably and conveniently measure local electric fields in proteins is evaluated by how well it meets several criteria. First and foremost, its frequency must show a sensitive and quantifiable dependence on the local electric field. Also, a chemical or biological method must exist to incorporate the probe into a protein. Furthermore, it must minimally perturb the native chemical and structural environment of interest. Finally, its vibration must be a localized mode having a large cross-section and, ideally, be located in an uncongested region of the IR spectrum of proteins (e.g., 1700–2400 cm-1).
For naturally occurring proteins, the amide I vibration arising from backbone amide units offers the largest IR intensity (molar extinction coefficient ~800 M-1cm-1) and shows a strong dependence on the local electrostatic environment, such as hydration, and, as a result, has been widely used to investigate protein conformational transitions.[5] However, the amide I transition is generally delocalized and also contains contributions from other vibrational modes, thus making it rather challenging to serve as a standalone probe of local electric field. One viable strategy to overcome this limitation is to incorporate a single carbonyl group (C=O) into an amino acid sidechain. The computational study of Cho and coworkers has predicted that the stretching mode of such a carbonyl is not only localized, but its frequency also varies linearly with the electrostatic field for both H-bonding and non-H-bonding environments,[6] thus making it an ideal candidate for the aforementioned applications. Indeed, Boxer and coworkers[7] have recently shown that the C=O stretching frequency of p-acetyl-L-phenylalanine (p-Ac-Phe) can serve as a reporter of the local electrostatic field of proteins. However, this vibrational transition (at ~1673 cm–1) overlaps with the protein amide I band and, thus its application requires careful background subtraction using the wild-type protein. To circumvent this inconvenience, we propose to use the C=O stretching vibration of an ester moiety as an alternative. Previous studies[8] have shown that the ester carbonyl absorbs in a spectral region (1700-1800 cm–1) where no other protein IR bands are present at neutral pH,[9] except those arising from protonated carboxylic groups.[10] Specifically, we test the utility of two ester containing non-natural amino acids, L-aspartic acid 4-methyl ester (hereafter referred to as DM) and L-glutamic acid 5-methyl ester (hereafter referred to as EM). While, to the best of our knowledge, DM and EM have not been previously introduced in proteins, we expect that this would not be a challenging task, as other ester containing sidechains have been successfully incorporated into proteins via genetic methods.[11]
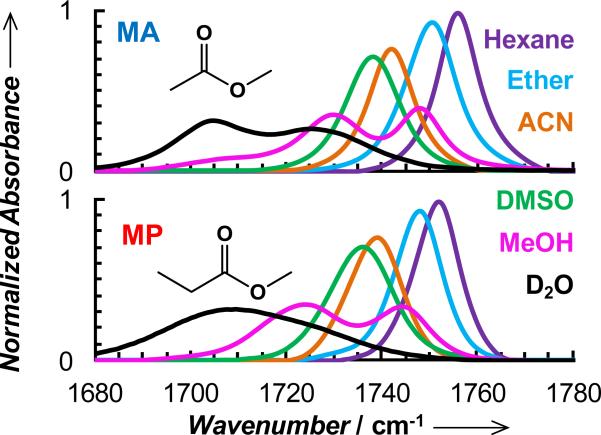
To provide a quantitative assessment of the electric field dependence of the ester carbonyl stretching vibrations of DM and EM, we first performed detailed vibrational solvatochromism studies on their respective sidechain mimics, methyl acetate (MA) and methyl propionate (MP). We chose MA and MP in these experiments, instead of the non-natural amino acids, because the model compounds are soluble in a wide range of polar and nonpolar solvents. As shown (Figure 1 and Table S1 in Supporting Information), the ester carbonyl stretching frequencies of MA and MP exhibit a strong dependence on the chosen solvents, ranging from hexane, an aprotic solvent with a very low dielectric constant (1.89 at 20°C), to water. For example, upon changing the solvent from hexane to DMSO, the center frequency of MA is red-shifted by 18.1 cm-1, compared to the 14.4 cm-1 shift observed for p-Ac-Phe.[7] Thus, these results substantiate the utility of these ester carbonyl stretching vibrations as sensitive probes of the local electric field, provided that a quantitative relationship between the electric field and frequency can be determined.
Figure 1.
Normalized FTIR spectra of MA and MP obtained in different solvents, as indicated. The concentration of the solute in each case was 20 mM and normalization is based on the integrated area of the band obtained in hexane (i.e., the spectra collected in other solvents were scaled so that their integrated areas are equal to that obtained in hexane). For MA in hexane, the peak absorbance was measured to be 0.0715, which gives rise to a molar extinction coefficient of 650 M-1 cm-1.
Interestingly, while in aprotic solvents the ester carbonyl stretching vibration results in a single absorption band, in protic solvents, where H-bonding between the vibrator and solvent is possible, the linear IR spectra contain more than one resolvable feature, suggesting that differently solvated or H-bonded species are present. Such spectral features have also been observed for nitrile and amide modes in protic solvents such as methanol.[12] For example, in D2O the IR spectrum of MA gives rise to two distinct peaks, centered at 1703.6 and 1727.0 cm-1, which is consistent with the two dimensional IR (2D IR) study of Righini and coworkers.[13] In agreement with the study of Tominaga and coworkers,[14] the ester carbonyl stretching band of MA in methanol consists of three resolvable spectral features, centered at 1748.1, 1729.6 and 1708.1 cm-1. For MP, however, the spectrum obtained in D2O is broad and almost featureless. To help better discern the underlying spectral contributions, we further carried out 2D IR measurements on MP in D2O and methanol. As shown (Figure S1 in Supporting Information), the 2D IR spectrum indicates that under the linear IR profile of MP in D2O, two peaks, at 1703.1 and 1721.1 cm-1, are resolvable, representing two distinct species.
With this information at hand, we then tried to determine how the ester C=O stretching frequencies of MA and MP vary with local electric field. As shown (Figure S2 in Supporting Information), the commonly used Onsager reaction field model[15] works fairly well for describing the trend obtained with aprotic solvents, consistent with the study of Asbury and coworkers,[16] but fails to predict the frequency shifts induced by protic solvents (Figure S3 in Supporting Information).
Therefore, following the work of Boxer and coworkers,[17] we used MD simulations to directly quantify the electric field experienced by the ester carbonyl vibration and, additionally, to help assign the two C=O stretching bands observed in protic solvents (see details and Figures S4 and S5 in the Supporting Information). Briefly, for aprotic solvents the electric field was directly calculated by averaging the values obtained from ~20,000 frames from each MD simulation, whereas for protic solvents, because of the possibility of different H-bonding patterns, we first divided the frames from each MD simulation into different clusters, according to a set of geometric criteria for H-bond formation (see details in the Supporting Information), and then the average electric field for each cluster was calculated. As shown (Figure S5, Supporting Information), for MA in water, the majority of the carbonyls are H-bonded to water, forming either 1 (51%) or 2 H-bonds (44%). Thus, we propose that the two peaks observed in the linear IR spectrum arise predominantly from these two species. Because H-bonding of water to a carbonyl induces a red-shift in its stretching vibration, we attribute the lower-frequency component to the doubly H-bonded species and the higher-frequency peak to the singly H-bonded species. In addition, the percentages of the lower- and higher-frequency components of MA in D2O (Figure 1), calculated based on their integrated areas, are 55% and 45%, which provides further evidence supporting the above assignment. Furthermore, MD simulations revealed the existence of less populated but differently H-bonded species, which were assumed to contribute to the broad width of the spectrum. A similar observation and assignment was made for MP in water. For both compounds in methanol, the majority of the carbonyls were found to be either non-H-bonded or singly H-bonded, with relative percentages in agreement with the ratio of the integrated areas of the two IR peaks. Thus, we assigned the two major IR bands to these species, with the H-bonded carbonyls vibrating at a lower frequency. For MA, a minor band at ~1710 cm-1 is clearly observable, which, based on MD simulations, was attributed to doubly H-bonded carbonyls. Finally, the width of the calculated electric field distribution for each differently solvated species shows a correlation with the corresponding spectral width of the C=O stretching vibration (Figure S6, Supporting Information), suggesting that the MD simulations are able to exhaustively sample the heterogeneous electrostatic environments of the probe.
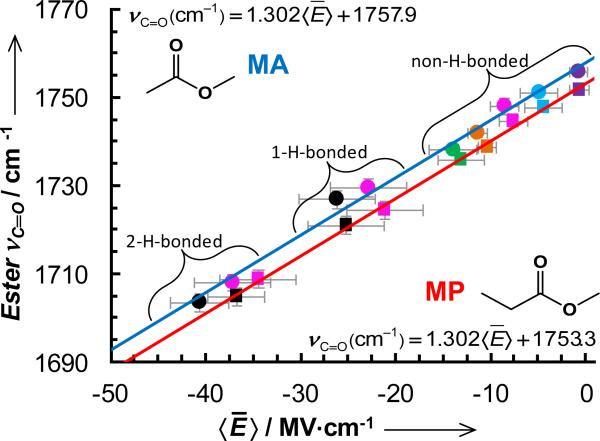
As shown (Figure 2), the center frequencies of the ester carbonyl stretching vibrations of MA and MP show a linear dependence on the calculated electric field for both protic and aprotic solvents, indicating that an ester moiety, such as that in DM and EM, could be used to quantitatively determine the local electrostatic field of proteins, using the frequency-field map shown in Figure 2.
Figure 2.
Center frequencies of the carbonyl stretching vibrations of MA (circles) and MP (squares) versus calculated local electric field for different solvents (represented by the same colors as those used in Figure 1). The solid lines are the best fits of these data to the linear equations indicated in the figure.
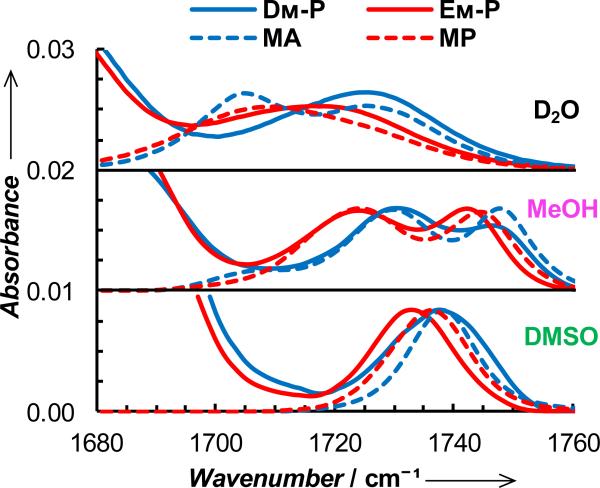
To demonstrate the utility of this ester vibrational mode in biological applications, we first used DM and EM to probe the local electrostatic and/or hydration environment of two short peptides, Ac-YDMK-NH2 (hereafter referred to as DM-P) and Ac-YEMK-NH2 (hereafter referred to as EM-P). As shown (Figure 3), the ester carbonyl stretching bands of these peptides in D2O indicate, when compared to those of MA and MP, that the population of the 2 H-bonded species (i.e., the spectral intensity at ~1705 cm-1) is significantly decreased. This result is not surprising as, in comparison to their respective model compounds, the sidechains of DM and EM are expected to be situated in a more crowded environment, thus limiting the accessibility of water molecules to the ester carbonyl and hence decreasing the probability of forming two H-bonds. In addition, and perhaps more convincing, EM-P, the ester carbonyl of which is expected to be further extended into the solvent than that of DM-P, shows a smaller decrease in this regard. Thus, these results provide further validation of the sensitivity of the C=O stretching vibration of the ester moiety to its local electrostatic environment. In support of this notion, Xie and coworkers have shown that the native structural analogs of DM and EM, i.e., the protonated carboxylic acid sidechains of Asp and Glu, can be used to sense H-bond formation in proteins.[10] The spectra of these two peptides in DMSO and methanol also support this notion. For example, in DMSO the center frequency of the ester C=O stretching band of EM-P is red-shifted by 3.4 cm-1 from that of MP, whereas that of DM-P is similar to that of MA. This red-shift results from the addition of the peptide environment around the vibrational probe. Besides the solvent-induced electric field, the ester carbonyl group will also experience electrostatic forces arising from the peptide backbone and other amino acid sidechains, thus making the vibrational frequency position dependent. In other words, the red-shift observed for EM-P is most likely due to the closer (compared to DM-P) proximity between the ester carbonyl and the polar amine group of the lysine sidechain. As indicated (Figure 3), the non-hydrogen-bonded peaks of these peptides obtained in methanol also corroborate this picture. Thus, these results demonstrate the ability of the ester carbonyl stretching vibration to sense minimal changes in its local electrostatic environment.
Figure 3.
Offset FTIR spectra in the ester C=O stretching region of DM-P, MA, EM-P, and MP in different solvents, as indicated. The peptide concentrations were 2 mM and, in each case, the spectrum of the model compound has been normalized with respect to that of the corresponding peptide.
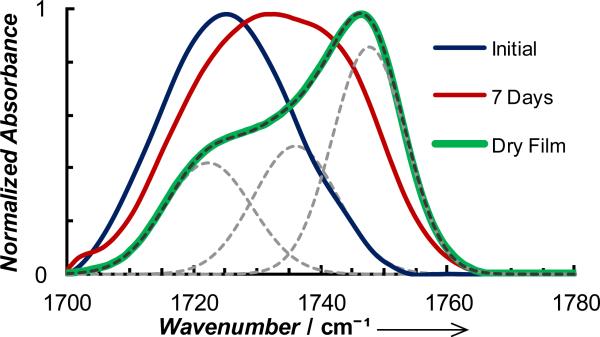
In the second study, we used DM to quantify the electrostatic environment in amyloid fibrils formed by a short segment of the Alzheimer's β-amyloid peptide, KLVFFAE (i.e., Aβ16-22). While it is generally assumed that the interior of amyloid has a low dielectric constant, to the best of our knowledge, no experiments have been attempted to directly measure the electrostatic properties of such β-sheet assemblies. Here, we mutated the leucine residue of Aβ16-22 to DM (the resultant peptide is referred to as Aβ-DM) since the sidechains are similar in size. As shown (Figure 4), prior to full onset of peptide aggregation (as judged by the amide I′ band at 1625 cm-1), the ester band has a peak at ~1725 cm-1, indicating that the DM sidechain is mostly hydrated and forms 1 H-bond with water, as expected. Upon further aggregation, the ester band becomes broader and blue-shifted, indicating that the population of the H-bonded ester carbonyls decreases. A further analysis indicates that this band can be decomposed into two Gaussians with center frequencies at 1727.2 and 1743.7 cm-1, respectively (Figure S8, Supporting Information). This is consistent with the formation of β-sheet fibrils, which, according to the Aβ16-22 fibrillar structures determined by Eisenberg and coworkers,[18] would lead to the creation of two distinct environments for the DM sidechain: one where the sidechain is sequestered in a dehydrated interface and the other where it is exposed to solvent (Figure S7, Supporting Information). However, as revealed by AFM measurements (Figure S9, Supporting Information), the fibrils/aggregates thus formed are rather heterogeneous, which not only leads to a broad ester C=O stretching band, but also prevents a more quantitative assessment of the structural features of the fibrils based on their IR signals.
Figure 4.
Normalized stretching vibrational bands of the ester carbonyl of 15 mM Aβ-DM obtained immediately after the sample was prepared, after 7 days of incubation in D2O, and also in the form of a dried film (dried under a flow of nitrogen for 7 days), as indicated. The band obtained with the dry film can be decomposed into 3 Gaussians (grey dashed lines) with the center frequencies given in the text.
The structural model of Eisenberg and coworkers[18] indicates that the β-strands stack in an anti-parallel fashion, with waters confined in the core of the fibrils. To probe these water molecules and also to form more homogeneous fibrils, we placed an aliquot of the abovementioned aggregated Aβ-DM sample on the surface of a germanium crystal of an attenuated total reflectance (ATR) unit and allowed it to dry under a gentle flow of N2 for 7 days. This drying procedure should remove most, if not all, of the bulk water. The resulting ester C=O stretching spectrum (Figure 4), shows three well-resolved peaks at 1722.3, 1736.0 and 1747.6 cm-1. The lowest frequency peak (i.e., 1722.3 cm-1) coincides with that arising from singly H-bonded ester carbonyls (Figure 1), indicating that water is indeed present in the amyloid fibrils. On the other hand, the 1747.6 cm-1 peak must arise from non-H-bonded ester carbonyls, or those situated at the aforementioned dry interfaces (Figures S7 and S8, Supporting Information), whereas the 1736.0 cm-1 peak most likely corresponds to outward facing DM sidechains that are H-bonded with water prior to sample drying, and its frequency reflects the local electrostatic environment of the dry air/fibril interfaces. Supporting these assignments is the fact that the relative percentages of these peaks, determined from their integrated areas, are 28% (1722.3 cm-1), 31% (1736.0 cm-1) and 41% (1747.6 cm-1), which are similar to those (i.e., 25%, 25% and 50%) calculated based on the structural model of Eisenberg and coworkers (Figure S7, Supporting Information).[18]
Using the frequency-field relationship obtained for MA (Figure 2) the three peaks (in order of increasing frequency) give rise to the following local electric fields: -26.3, -16.2, and -8.0 MV·cm-1. What is more important is that we can use the experimentally determined frequency-Onsager field relationship for this probe (Supporting Information) to estimate the dielectric constant to be 5.6 for the dry interior of the well-packed fibrils. While it is well known that a low dielectric constant environment would increase the strength of H-bonding and other types of electrostatic interactions, it is challenging to quantitatively assess the dielectric constants of proteins and peptides, especially in a site-specific manner. Thus, it is our belief that the methodology demonstrated here holds utility in relevant biological studies.
In conclusion, we have established that the ester carbonyl stretching frequencies of two non-natural amino acids, L-aspartic acid 4-methyl ester (DM) and L-glutamic acid 5-methyl ester (EM), show a linear dependence on the local electric field and, thus, can quantify, in a site-specific manner, the local electrostatic environment of proteins. In comparison to commonly used nitrile-, azide-, or CD-based IR probes,[19] the ester carbonyl stretching vibration offers one distinct advantage: its large dynamic range makes it more useful to probe small changes in the local electric field. For example, it is sensitive enough to probe the difference in the electric fields between two points in a peptide environment that are separated by a single methylene unit. In addition, the size of DM is similar to that of asparagine, aspartic acid, and leucine, while EM is similar to glutamine and glutamic acid. Taken together, these attributes of DM and EM suggest that they are two of the most promising local electrostatic IR probes of proteins. In addition, we have devised a new method that allows one to determine the local dielectric constant of proteins. Applying this method to amyloid fibrils formed by an Aβ-peptide fragment indicates that their interiors have a ε of 5.6 ± 1.5. Because the C=O stretching vibration of esters is also Raman active,[20] we expect that the frequency-field relationships devised here can also be used to study relevant biochemical and biophysical problems in conjunction with Raman based techniques.[21]
Experimental Section
Methyl acetate (acetic acid methyl ester) (MA) and methyl propionate (propanoic acid methyl ester) (MP) were purchased from Acros Organics (Fair Lawn, NJ) and used as received. Fmoc-L-Asp(Me)-OH was purchased from Chem-Impex International Inc. (Wood Dale, IL) and F-moc-L-Glu(Me)-OH was purchased from Santa Cruz Biotechnology Inc. Peptides were synthesized using standard Fmoc solid-phase methods on an PS3 peptide synthesizer from Protein Technologies (Tucson, AZ). All linear IR spectra, except the one for the Aβ-DM dry film, which was obtained using a Horizon ATR unit from Harrick (Pleasantville, NY), were collected on a Nicolet Magna-IR 860 FTIR spectrometer at 1 cm-1 resolution using a home-made CaF2 sample holder. More details about the linear and 2D IR instrumentations can be found elsewhere[22] or in the Supporting Information. AFM images were acquired on a Bruker Dimension Icon AFM (Santa Barbara, CA). The details of MD simulations and electric field calculations are given in the Supporting Information.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (GM-065978) for funding.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Ileana M. Pazos, Department of Chemistry, University of Pennsylvania 231 S. 34th Street, Philadelphia, PA 19104, United States
Ayanjeet Ghosh, Department of Chemistry, University of Pennsylvania 231 S. 34th Street, Philadelphia, PA 19104, United States.
Matthew J. Tucker, Department of Chemistry, University of Nevada 1664 N. Virginia Street, Reno, Nevada 89557, United States.
Feng Gai, Department of Chemistry, University of Pennsylvania 231 S. 34th Street, Philadelphia, PA 19104, United States.
References
- 1.a Suydam IT, Snow CD, Pande VS, Boxer SG. Science. 2006;313:200–204. doi: 10.1126/science.1127159. [DOI] [PubMed] [Google Scholar]; b Ragain CM, Newberry RW, Ritchie AW, Webb LJ. J. Phys. Chem. B. 2012;116:9326–9336. doi: 10.1021/jp303272y. [DOI] [PubMed] [Google Scholar]
- 2.Boxer SG. J. Phys. Chem. B. 2009;113:2972–2983. doi: 10.1021/jp8067393. [DOI] [PubMed] [Google Scholar]
- 3.Kim H, Cho M. Chem. Rev. 2013:5817–5847. doi: 10.1021/cr3005185. [DOI] [PubMed] [Google Scholar]
- 4.a Volk M, Kholodenko Y, Lu HSM, Gooding EA, DeGrado WF, Hochstrasser RM. J. Phys. Chem. B. 1997;101:8607–8616. [Google Scholar]; b Schkolnik G, Utesch T, Salewski J, Tenger K, Millo D, Kranich A, Zebger I, Schulz C, Zimanyi L, Rakhely G, Mroginski MA, Hildebrandt P. Chem. Commun. 2012;48:70–72. doi: 10.1039/c1cc13186a. [DOI] [PubMed] [Google Scholar]; c Chung JK, Thielges MC, Fayer MD. J. Am. Chem. Soc. 2012;134:12118–12124. doi: 10.1021/ja303017d. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Walker DM, Hayes EC, Webb LJ. Phys. Chem. Chem. Phys. 2013;15:12241–12252. doi: 10.1039/c3cp51284c. [DOI] [PubMed] [Google Scholar]
- 5.a Dyer RB, Gai F, Woodruff WH. Acc. Chem. Res. 1998;31:709–716. [Google Scholar]; b Woutersen S, Pfister R, Hamm P, Mu YG, Kosov DS, Stock G. J. Chem. Phys. 2002;117:6833–6840. [Google Scholar]; c Demirdoven N, Cheatum CM, Chung HS, Khalil M, Knoester J, Tokmakoff A. J. Am. Chem. Soc. 2004;126:7981–7990. doi: 10.1021/ja049811j. [DOI] [PubMed] [Google Scholar]; d Waegele MM, Culik RM, Gai F. J. Phys. Chem. Lett. 2011;2:2598–2609. doi: 10.1021/jz201161b. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Serrano AL, Waegele MM, Gai F. Protein Sci. 2012;21:157–170. doi: 10.1002/pro.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J-H, Cho M. J. Chem. Phys. 2011;134:54513. doi: 10.1063/1.3580776. [DOI] [PubMed] [Google Scholar]
- 7.Fried SD, Bagchi S, Boxer SG. J. Am. Chem. Soc. 2013;135:11181–11192. doi: 10.1021/ja403917z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a George WO, Houston TE, Harris WC. Spectrochim. Acta, Part A. A. 1974;30:1035–1057. [Google Scholar]; b Patel KB, Eaton G, Symons MCR. J. Chem. Soc., Faraday Trans. 1985;81:2775–2786. [Google Scholar]
- 9.Barth A, Zscherp C. Q. Rev. Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 10.Nie B, Stutzman J, Xie A. Biophys. J. 2005;88:2833–2847. doi: 10.1529/biophysj.104.047639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a Wang J, Xie J, Schultz PG. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]; b Takimoto JK, Xiang Z, Kang J-Y, Wang L. Chembiochem. 2010;11:2268–2272. doi: 10.1002/cbic.201000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Kim YS, Hochstrasser RM. J. Phys. Chem. B. 2007;111:9697–9701. doi: 10.1021/jp074267x. [DOI] [PubMed] [Google Scholar]; b Ghosh A, Hochstrasser RM. Chem. Phys. 2011;390:1–13. doi: 10.1016/j.chemphys.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Candelaresi M, Pagliai M, Lima M, Righini R. J. Phys. Chem. A. 2009;113:12783–12790. doi: 10.1021/jp906072w. [DOI] [PubMed] [Google Scholar]
- 14.Banno M, Ohta K, Yamaguchi S, Hirai S, Tominaga K. Acc. Chem. Res. 2009;42:1259–1269. doi: 10.1021/ar9000229. [DOI] [PubMed] [Google Scholar]
- 15.Onsager L. J. Am. Chem. Soc. 1936;58:1486–1493. [Google Scholar]
- 16.Pensack RD, Banyas KM, Asbury JB. Phys. Chem. Chem. Phys. 2010;12:14144–14152. doi: 10.1039/c0cp00971g. [DOI] [PubMed] [Google Scholar]
- 17.Fried SD, Wang L-P, Boxer SG, Ren P, Pande VS. J. Phys. Chem. B. 2013;117:16236–16248. doi: 10.1021/jp410720y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colletier J-P, Laganowsky A, Landau M, Zhao M, Soriaga AB, Goldschmidt L, Flot D, Cascio D, Sawaya MR, Eisenberg D. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16938–16943. doi: 10.1073/pnas.1112600108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. J. Am. Chem. Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]; b Maienschein-Cline MG, Londergan CH. J. Phys. Chem. A. 2007;111:10020–10025. doi: 10.1021/jp0761158. [DOI] [PubMed] [Google Scholar]; c Oh K-I, Choi J-H, Lee J-H, Han J-B, Lee H, Cho M. J. Chem. Phys. 2008:128. [Google Scholar]; d Oh K-I, Lee J-H, Joo C, Han H, Cho M. J. Phys. Chem. B. 2008;112:10352–10357. doi: 10.1021/jp801558k. [DOI] [PubMed] [Google Scholar]; e Lindquist BA, Furse KE, Corcelli SA. Phys. Chem. Chem. Phys. 2009;11:8119–8132. doi: 10.1039/b908588b. [DOI] [PubMed] [Google Scholar]; f Bunagan MR, Gao J, Kelly JW, Gai F. J. Am. Chem. Soc. 2009;131:7470–7476. doi: 10.1021/ja901860f. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Waegele MM, Tucker MJ, Gai F. Chem. Phys. Lett. 2009;478:249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Marek P, Mukherjee S, Zanni MT, Raleigh DP. J. Mol. Biol. 2010;400:878–888. doi: 10.1016/j.jmb.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Ye S, Zaitseva E, Caltabiano G, Schertler GFX, Sakmar TP, Deupi X, Vogel R. Nature. 2010;464:1386–1389. doi: 10.1038/nature08948. [DOI] [PubMed] [Google Scholar]; j Zimmermann J. r., Thielges MC, Yu W, Dawson PE, Romesberg FE. J. Phys. Chem. Lett. 2011;2:412–416. [Google Scholar]; k Zimmermann J, Thielges MC, Seo YJ, Dawson PE, Romesberg FE. Angew. Chem. Int. Ed. 2011;50:8333–8337. doi: 10.1002/anie.201101016. [DOI] [PubMed] [Google Scholar]; l Tucker MJ, Gai XS, Fenlon EE, Brewer SH, Hochstrasser RM. Phys. Chem. Chem. Phys. 2011;13:2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]; m Dutta S, Rock W, Cook RJ, Kohen A, Cheatum CM. J. Chem. Phys. 2011:135. doi: 10.1063/1.3623418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dybal J, Stokr J, Schneider B. Collect. Czech. Chem. Commun. 1982;47:2027–2036. [Google Scholar]
- 21.Nafie LA, Raman Spectrosc J. 2013;44:1629–1648. [Google Scholar]
- 22.a Huang CY, Getahun Z, Zhu YJ, Klemke JW, DeGrado WF, Gai F. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2788–2793. doi: 10.1073/pnas.052700099. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Kim YS, Hochstrasser RM. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11185–11190. doi: 10.1073/pnas.0504865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.