Abstract
Background
C-reactive protein (CRP) is a biomarker of inflammation. Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) associated with CRP concentrations and inflammation-related traits such as cardiovascular disease, type 2 diabetes, and obesity. We aimed to replicate previous CRP-SNP associations, assess whether these associations generalize to additional race/ethnicity groups, and evaluate inflammation-related SNPs for a potentially pleiotropic association with CRP.
Methods and Results
We selected and analyzed 16 CRP-associated and 250 inflammation-related GWAS SNPs among 40,473 African American, American Indian, Asian/Pacific Islander, European American, and Hispanic participants from 7 studies collaborating in the Population Architecture using Genomics and Epidemiology (PAGE) study. Fixed-effect meta-analyses combined study-specific race/ethnicity-stratified linear regression estimates to evaluate the association between each SNP and high-sensitivity CRP. Overall, 18 SNPs in 8 loci were significantly associated with CRP (Bonferroni-corrected p<3.1×10−3 for replication, p<2.0×10−4 for pleiotropy): Seven of these were specific to European Americans, while 9 additionally generalized to African Americans (1), Hispanics (5), or both (3); 1 SNP was seen only in African Americans and Hispanics. Two SNPs in the CELSR2/PSRC1/SORT1 locus showed a potentially novel association with CRP: rs599839 (p=2.0×10−6) and rs646776 (p=3.1×10−5).
Conclusions
We replicated 16 SNP-CRP associations, 10 of which generalized to African Americans and/or Hispanics. We also identified potentially novel pleiotropic associations with CRP for two SNPs previously associated with coronary artery disease and LDL cholesterol. These findings demonstrate the benefit of evaluating genotype-phenotype associations in multiple race/ethnicity groups, and of looking for pleiotropic relationships among SNPs previously associated with related phenotypes.
Keywords: genetic epidemiology, inflammation, C-reactive protein, race and ethnicity, single nucleotide polymorphism, pleiotropy
Introduction
C-reactive protein (CRP) is an acute-phase reactant protein produced by the liver. Circulating levels rise sharply following inflammatory stimulation from infection or injury, then fall rapidly following stimulus resolution. In situations of chronic underlying disease, however, CRP remains slightly raised over time, serving as a biomarker to characterize systemic inflammation1, 2. Elevated CRP levels have been associated with a large number of outcomes and traits, such as cardiovascular events1, 2, atherosclerosis3, stroke4, type 2 diabetes (T2D)5, fitness level and body composition6, and cancer7, 8. As such, serum CRP is an important biomarker for the presence or development of disease among apparently healthy individuals1, 2.
Heritability estimates for CRP are between 25 and 56%9–11. Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in 21 loci associated with CRP concentrations12–14. Environmental factors associated with increased CRP levels include female sex9, 15, hormone replacement therapy (HRT) use16, smoking2, and obesity1, 2, while factors associated with decreased CRP levels include exercise17 and the use of aspirin, NSAIDs, and statins1.
CRP levels have previously been shown to vary according to race/ethnicity, and tend to be higher in populations with high rates of diabetes and obesity. Compared to those of European ancestry, those of African15, 18 and American Indian19 ancestry tend to have elevated CRP levels and those of Asian ancestry tend to have lower CRP levels20, 21. Many initial genetic studies of CRP were performed primarily on populations of European descent, and studies examining CRP-associated variants in other race/ethnicity populations have had moderate success in generalizing some, but not all, of these loci18, 22–24. Generalization and fine-mapping of loci in non-European ancestry populations is thus an important step in further elucidating genetic contributions to CRP concentrations in multiple race/ethnicity groups.
Many of the genetic variants associated with serum CRP levels demonstrate pleiotropic effects, and have also been associated with various other outcomes related to inflammation. SNP rs1205 in the gene encoding CRP (CRP), for example, has been associated not only with CRP12, 22, but also with heart rate variability25 and colon cancer7. Given the multiple phenotypes associated with CRP, we hypothesize that genetic variants associated with inflammation-related phenotypes, such as cardiovascular disease, T2D, or obesity, might also be associated with CRP. Identifying additional pleiotropic associations could aid in elucidating shared biological pathways and relationships.
We seek to replicate previously reported associations between SNPs and serum CRP, as well as generalize these associations to four additional race/ethnicity groups: African Americans, Hispanics, Asian/Pacific Islanders, and American Indians. We also evaluate several SNPs previously associated with inflammation-related phenotypes for an association with CRP, both overall and stratified by race/ethnicity groups. The size and diversity of our study population provides an opportunity to replicate previous findings, generalize existing associations to additional race/ethnicity groups, and discover novel pleiotropic associations with this inflammatory biomarker.
Methods
Study description
Data for this analysis were generated as part of the Population Architecture using Genomics and Epidemiology study (PAGE), previously described in detail26. Briefly, PAGE is a consortium of large, well-characterized population-based studies, investigating the epidemiologic architecture of genetic variants associated with complex diseases across various race/ethnicity groups.
Study populations
We included 40,473 participants: 7,228 from Epidemiologic Architecture for Genes Linked to Environment (EAGLE), based on data from three National Health and Nutrition Examination Surveys (NHANES); 1,593 from the Multiethnic Cohort Study (MEC); 9,901 from the Women’s Health Initiative (WHI); and 21,751 from Causal Variants Across the Life Course (CALiCo), itself a consortium of five cohort studies. Participants from CALiCo included: 11,303 from Atherosclerosis Risk in Communities (ARIC); 3,249 from Coronary Artery Risk Development in Young Adults (CARDIA); 4,070 from the Cardiovascular Health Study (CHS); and 3,129 from the Strong Heart Study (SHS).
Studies included one or more of five race/ethnicity groups, totaling 24,958 European American, 8,471 African American, 2,935 Hispanic, 980 Asian/Pacific Islander, and 3,129 American Indian participants. Several of these studies have previously been utilized to evaluate genetic associations with CRP, including participants in ARIC13, CHS13, 27, 28, and CARDIA29. Within EAGLE, we included NHANES 1999–2002 data and excluded NHANES III data (due to non-high-sensitivity CRP measurements). Each study was approved by a local institutional review board, and all participants gave informed consent.
Measurements
Baseline high-sensitivity C-reactive protein (hsCRP) measurements were available in each study as part of previous investigations, with assay method and instrument used to measure CRP varying by study (Supplemental Table 1). Because the distribution of CRP is skewed, the values were natural-log transformed for analyses. Demographic and other epidemiologic information was obtained according to the enrollment protocols of each study (Table 1, Supplemental Table 2).
Table 1.
Demographic and epidemiologic characteristics of study populations used in this analysis, by race/ethnicity group and overall (and by study in Supplemental Table 2).
| Measure | Unit | European American | African American | Hispanic | Asian/Pacific Islander | American Indian | Overall |
|---|---|---|---|---|---|---|---|
| Sample size | n | 24,958 | 8,471 | 2,935 | 980 | 3,129 | 40,473 |
| CRP(mg/dL) | Mean of study-specific medians(range) | 1.9(1.0–2.9) | 2.5(1.3–3.6) | 2.3(1.2–3.1) | 0.8(0.5–1.0) | 3.9(3.9–3.9) | 2.3(0.8–3.9) |
| Age(years) | Mean(SD) | 62.1(12.6) | 56.8(13.5) | 51.3(16.5) | 62.3(8.5) | 39.3(16.4) | 58.5(14.8) |
| Sex | %Female | 66.6% | 66.2% | 62.5% | 67.4% | 61.5% | 65.8% |
| BMI(kg/m^2) | Mean(SD) | 27.9(5.7) | 30.4(6.9) | 28.7(5.7) | 25.1(4.2) | 32.4(7.9) | 28.8(6.3) |
| Smoking | %Current | 13.9% | 20.8% | 16.0% | 7.4% | 34.0% | 16.9% |
| %Former | 40.0% | 29.3% | 26.6% | 35.5% | 23.3% | 35.4% | |
| Hormone Replacement Therapy | %Current | 31.3% | 18.7% | 23.4% | 39.0% | 9.8% | 26.6% |
| %Former | 16.7% | 15.8% | 15.7% | 23.7% | 11.4% | 16.2% |
SNP Selection and Genotyping
In 2008, PAGE investigators identified 266 SNPs previously associated with various phenotypes of interest, focusing on variants related to cardiovascular disease traits, lipids, body mass index (BMI), and T2D. These included: 16 SNPs associated with CRP/inflammation, 21 with cardiovascular disease or myocardial infarction, 26 with BMI/obesity, 51 with T2D or glucose levels, 82 with HDL/LDL/total cholesterol, 29 with triglycerides, 9 with stroke, and 33 with other related phenotypes (Supplemental Table 3; previous trait association refers to the first reported association at the time of SNP selection). Genotyping methods for PAGE have been previously described26. For each SNP, quality control thresholds included SNP and sample call rates >90%, concordance of blinded replicates >98%, and no clear evidence of Hardy-Weinberg disequilibrium (p>0.001).
Due to funding constraints, each PAGE study chose a custom subset of SNPs to genotype depending on the phenotypes they had available. As such, the number of SNPs available varied in each study, with 30 SNPs genotyped in ARIC, 7 in CARDIA, 113 in CHS, 196 in EAGLE, 103 in MEC, 19 in SHS, and 94 in WHI (Table 2, Supplemental Table 3). While this meant that not all SNPs were genotyped in every study, an effort was made to ensure overlap (56% of SNPs were genotyped in more than one study). Since race/ethnicity group availability differed by study, this also meant that not all SNPs were available for analyses in each race/ethnicity group. Nearly all SNPs were available for the European American (n=265), African American (n=266), and Hispanic groups (n=261), while fewer SNPs were available for the Asian/Pacific Islander (n=150) and American Indian groups (n=19). These overlap issues reduced the overall sample size available for any given SNP- and race/ethnicity-specific association, though numbers were still large for most analyses (Table 2, Supplemental Table 4). To avoid small sample size issues in the analysis, for each SNP only study-specific race/ethnicity-specific results with at least 100 genotyped subjects were included in meta-analyses. This restriction meant the exclusion of the WHI American Indian results (≤86 participants for each SNP), which reduced the overall number of SNPs available for analysis in the American Indian group from 96 to 19.
Table 2.
Number of SNPs and participants available for meta-analysis across studies, overall and by race/ethnicity.
| Race/ethnicity group | European American | African American | Hispanic | Asian/ Pacific Islander | American Indian | Overall |
|---|---|---|---|---|---|---|
| SNPs available in each study | ||||||
| WHI | 94 | 92 | 94 | 91 | 94 | |
| EAGLE | 196 | 196 | 196 | 196 | ||
| MEC | 103 | 103 | 103 | 103 | 103 | |
| ARIC | 31 | 31 | 31 | |||
| CARDIA | 7 | 7 | 7 | |||
| CHS | 111 | 118 | 118 | |||
| SHS | 19 | 19 | ||||
| Overall | 265 | 266 | 261 | 150 | 19 | 266 |
| Observations available per SNP | ||||||
| Mean | 7722 | 2367 | 1766 | 617 | 3129 | 12361 |
| Minimum | 233 | 433 | 243 | 388 | 3129 | 802 |
| Maximum | 24319 | 8158 | 2908 | 951 | 3129 | 36299 |
The number of SNPs and observations available are calculated from the number of SNPs genotyped (which varied by study) and the number of participants genotyped in each race/ethnicity group (which varied by study). Each study chose a subset of 266 SNPs to genotype. While these subsets overlapped, not all SNPs were available in all studies. In CHS, seven SNPs were available in African Americans but not in European Americans (rs10010131, rs10938397, rs1501980, rs16890979, rs174547, rs6544713, and rs7679). In WHI, SNPs were excluded from analyses in some race/ethnicity groups with fewer than 100 participants available for that SNP (rs13266634 and rs2383207 for African Americans; rs13266634, rs174547, and rs2383207 for Asian/Pacific Islanders; and all 94 SNPs for American Indians).
Statistical analysis
Each study fit linear regression models to test the association between natural log-transformed CRP and each SNP, coded additively (0/1/2 copies of the coded allele). Separate models were fit for five race/ethnicity groups: European American, African American, Hispanic, Asian/Pacific Islander, and American Indian. MEC performed analyses separately for Hawaiian and Japanese ancestry participants. Race/ethnicity-specific models were adjusted for age, sex, center, and/or principal components, as appropriate for each study. Sampling weights were used in the WHI regression analyses to account for sample selection criteria. Additional models were stratified by sex.
Inverse-variance-weighted fixed effect meta-analyses were used to estimate both overall and race/ethnicity stratified associations for each SNP. Because studies differed in SNPs genotyped and participant race/ethnicity, the overall number of observations available for each SNP-specific meta-analysis varied. Overall combined-race/ethnicity meta-analysis numbers were large for most SNPs (mean number of observations 12,361, range 802–36,299). In the race/ethnicity-stratified meta-analyses, the average number of observations across available SNPs was 7,722 for European Americans, 2,367 for African Americans, 1,766 for Hispanics, 617 for Asian/Pacific Islanders, and 3,129 for American Indians (Table 2, Supplemental Table 4). Additional meta-analyses were stratified by sex. For each SNP, heterogeneity between studies and within race/ethnicity groups was tested using Q and I2 statistics. Meta-regression was used to evaluate the heterogeneity between race/ethnicity groups and sexes. Meta-analyses were performed using Stata version 1230. We used a conservative Bonferroni-corrected p-value to adjust for multiple comparisons, accounting for the maximum number of tested SNPs to determine the statistical significance threshold (0.05/16=3.1×10−3 for replication and 0.05/250=2.0×10−4 for pleiotropy discovery). Since not all SNPs were available in each race/ethnicity group, this correction may have been overly stringent for some groups.
Results
European American and African American were the largest race/ethnicity groups in our analysis (Table 1, Supplemental Table 2). Median CRP concentration varied by study and race/ethnicity: the lowest levels were observed in MEC and Asian/Pacific Islanders, and the highest levels were observed in WHI and American Indians. American Indian participants were on average younger than participants in other groups. Overall there were more women than men (65.8%) due to the focus on women in WHI. Mean BMI values varied by race/ethnicity, with the highest values in the African American and American Indian groups and the lowest values in the Asian/Pacific Islander group. Extensive efforts were made to harmonize variables across PAGE studies, though some study heterogeneity may remain.
In the race/ethnicity-combined analyses, we identified 18 SNPs in 8 loci associated with CRP: 10 of 16 CRP-associated SNPs replicated and 8 of 250 inflammation-related SNPs demonstrated a pleiotropic association with CRP (Table 3). These included: 5 SNPs in the CRP locus; 4 in APOE/APOC1/TOMM40; 2 each in GCKR, HNF4A, and CELSR2/PSRC1/SORT1; and 1 each in HNF1A, IL6R, and LEPR. Several of these SNPs were correlated with each other, with the amount of correlation varying by race/ethnicity (Supplemental Table 5). Of the 18 SNPs (in 8 loci) reaching statistical significance, 15 SNPs (in 7 loci) had previously been associated with CRP, while rs599839 (near PSRC1) and rs646776 (near CELSR2) have not. The SNP rs6857 (in PVRL2) has not itself been previously associated with CRP, though it is in close proximity to the known APOE/APOC1/TOMM40 locus. Since many of these SNPs have previously been associated with other phenotypic traits, the observed associations with CRP suggest that many of these SNPs may have potentially pleiotropic effects.
Table 3.
Association between serum C-reactive protein and SNPs previously associated with inflammation-related phenotypes
| SNP | Allele, position, gene | Within-group | Between- group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Locus | Previously associated trait | Race/ethnicity group | n | # Studies | Coded Allele Frequency | Beta | SE | P-value | p-hetero- geneity | p-hetero- geneity |
| rs1205 | T | European American | 12400 | 2 | 0.33 | −0.17 | 0.01 | 1.03E-31 | 0.02 | |
| CRP | 3′ UTR, CRP | African American | 3669 | 2 | 0.20 | −0.27 | 0.04 | 8.09E-15 | 0.61 | |
| C-reactive Protein | Hispanic | 1842 | 1 | 0.35 | −0.22 | 0.04 | 5.37E-09 | |||
| OVERALL | 17911 | 2 | −0.19 | 0.01 | 1.24E-50 | 0.02 | ||||
| rs1800947 | G | European American | 12404 | 2 | 0.06 | −0.30 | 0.03 | 3.10E-25 | 0.77 | |
| CRP | Exonic, CRP | African American | 3676 | 2 | 0.01 | −0.61 | 0.13 | 1.28E-06 | 0.77 | |
| C-reactive Protein | Hispanic | 1842 | 1 | 0.02 | −0.36 | 0.13 | 6.72E-03 | |||
| OVERALL | 17922 | 2 | −0.32 | 0.03 | 1.13E-30 | 0.05 | ||||
| rs1417938 | A | European American | 3943 | 1 | 0.30 | 0.14 | 0.03 | 5.57E-07 | ||
| CRP | Intronic, CRP | African American | 1328 | 1 | 0.11 | 0.20 | 0.08 | 1.17E-02 | ||
| C-reactive Protein | Hispanic | 1845 | 1 | 0.36 | 0.14 | 0.04 | 2.71E-04* | |||
| OVERALL | 7116 | 1 | 0.15 | 0.02 | 2.97E-11 | 0.79 | ||||
| rs4131568 | T | European American | 11314 | 2 | 0.35 | 0.08 | 0.02 | 1.24E-07 | 0.31 | |
| CRP | Upstream, CRP | African American | 3461 | 2 | 0.07 | −0.08 | 0.06 | 1.54E-01 | 0.96 | |
| C-reactive protein | Hispanic | 1857 | 1 | 0.39 | 0.08 | 0.04 | 3.77E-02 | |||
| OVERALL | 16632 | 2 | 0.07 | 0.01 | 2.45E-07 | 0.02 | ||||
| rs3093058 | T | European American | 12420 | 2 | 0.001 | 0.32 | 0.22 | 1.43E-01 | 0.53 | |
| CRP | Upstream, CRP | African American | 3680 | 2 | 0.17 | 0.48 | 0.04 | 1.40E-40 | 0.87 | |
| C-reactive Protein | Hispanic | 1842 | 1 | 0.01 | 0.67 | 0.20 | 1.02E-03* | |||
| OVERALL | 17942 | 2 | −0.18 | 0.03 | 1.44E-43 | 0.50 | ||||
| rs4420638 | G | European American | 21378 | 4 | 0.18 | −0.24 | 0.02 | 1.01E-56 | 0.12 | |
| APOE/APOC/TOMM40 | Downstream, APOC1 | African American | 6076 | 4 | 0.20 | −0.03 | 0.03 | 2.66E-01 | 0.35 | |
| LDL Cholesterol | Hispanic | 2637 | 2 | 0.10 | −0.18 | 0.05 | 5.58E-04 | 0.04 | ||
| Asian/Pacific Islander | 422 | 1 | 0.12 | −0.06 | 0.14 | 6.76E-01 | ||||
| OVERALL | 30513 | 5 | −0.19 | 0.01 | 4.50E-50 | 1.64E-09 | ||||
| rs2075650 | G | European American | 15926 | 4 | 0.14 | −0.22 | 0.02 | 1.83E-38 | 0.54 | |
| APOE/APOC/TOMM40 | Intronic, TOMM40 | African American | 4729 | 4 | 0.14 | −0.02 | 0.03 | 6.47E-01 | 0.29 | |
| LDL Cholesterol | Hispanic | 2119 | 2 | 0.10 | −0.21 | 0.05 | 1.00E-04 | 0.12 | ||
| Asian/Pacific Islander | 528 | 2 | 0.18 | −0.03 | 0.04 | 5.28E-01 | 0.85 | |||
| OVERALL | 23302 | 5 | −0.16 | 0.01 | 2.59E-32 | 1.60E-09 | ||||
| rs429358 | C | European American | 6993 | 2 | 0.13 | −0.24 | 0.04 | 2.41E-10 | 0.61 | |
| APOE/APOC/TOMM40 | Exonic, APOE | African American | 2220 | 2 | 0.20 | −0.23 | 0.04 | 5.07E-08 | 0.05 | |
| Cardiovascular Disease | Hispanic | 1031 | 2 | 0.11 | −0.36 | 0.07 | 5.08E-08 | 0.02 | ||
| Asian/Pacific Islander | 943 | 2 | 0.11 | −0.19 | 0.05 | 2.25E-04 | 0.84 | |||
| OVERALL | 11187 | 2 | −0.24 | 0.02 | 1.84E-25 | 0.23 | ||||
| rs6857 | T | European American | 3955 | 1 | 0.16 | −0.23 | 0.04 | 2.07E-10 | ||
| APOE/APOC/TOMM40 | 3′ UTR, PVRL2 | African American | 1331 | 1 | 0.06 | −0.23 | 0.11 | 3.04E-02 | ||
| Cardiovascular Disease | Hispanic | 1852 | 1 | 0.10 | −0.28 | 0.06 | 6.12E-06 | |||
| OVERALL | 7138 | 1 | −0.24 | 0.03 | 7.30E-16 | 0.79 | ||||
| rs1260326 | T | European American | 22661 | 5 | 0.42 | 0.10 | 0.01 | 2.38E-17 | 0.16 | |
| GCKR | Exonic, GCKR | African American | 6506 | 5 | 0.15 | 0.06 | 0.03 | 2.06E-02 | 0.42 | |
| Triglycerides | Hispanic | 2873 | 3 | 0.33 | 0.08 | 0.03 | 5.78E-03 | 0.45 | ||
| Asian/Pacific Islander | 951 | 2 | 0.54 | 0.09 | 0.03 | 2.04E-03 | 0.11 | |||
| OVERALL | 32991 | 6 | 0.09 | 0.01 | 5.34E-22 | 0.72 | ||||
| rs780094 | T | European American | 17567 | 5 | 0.40 | 0.10 | 0.01 | 1.53E-16 | 0.32 | |
| GCKR | Intronic, GCKR | African American | 6175 | 5 | 0.19 | 0.03 | 0.03 | 2.26E-01 | 0.23 | |
| C-reactive Protein | Hispanic | 2130 | 2 | 0.34 | 0.07 | 0.03 | 2.62E-02 | 0.26 | ||
| Asian/Pacific Islander | 528 | 1 | 0.54 | 0.05 | 0.03 | 1.51E-01 | 0.63 | |||
| OVERALL | 26400 | 6 | 0.08 | 0.01 | 5.65E-17 | 0.09 | ||||
| rs2650000 | A | European American | 16506 | 5 | 0.35 | −0.12 | 0.01 | 2.62E-23 | 0.18 | |
| HNF1A | Upstream, HNF1A | African American | 5983 | 5 | 0.12 | −0.09 | 0.03 | 5.24E-03 | 0.49 | |
| LDL Cholesterol | Hispanic | 2125 | 2 | 0.36 | −0.11 | 0.03 | 9.45E-04 | 0.08 | ||
| Asian/Pacific Islander | 500 | 1 | 0.45 | −0.03 | 0.03 | 3.61E-01 | 0.25 | |||
| OVERALL | 25114 | 6 | −0.11 | 0.01 | 4.34E-26 | 0.08 | ||||
| rs7310409 | A | European American | 3956 | 1 | 0.40 | −0.18 | 0.03 | 1.57E-10 | ||
| HNF1A | Intronic, HNF1A | African American | 1327 | 1 | 0.32 | −0.15 | 0.06 | 7.94E-03 | ||
| C-reactive protein | Hispanic | 1855 | 1 | 0.41 | −0.13 | 0.04 | 1.13E-03* | |||
| OVERALL | 7138 | 1 | −0.16 | 0.02 | 3.33E-14 | 0.59 | ||||
| rs2228145 | C | European American | 17184 | 4 | 0.40 | −0.10 | 0.01 | 1.47E-18 | 0.09 | |
| IL6R | Exonic, IL6R | African American | 5942 | 4 | 0.14 | −0.13 | 0.03 | 6.72E-05 | 0.27 | |
| C-reactive Protein | Hispanic | 1841 | 1 | 0.52 | −0.18 | 0.04 | 2.18E-06 | |||
| OVERALL | 24967 | 5 | −0.11 | 0.01 | 3.66E-26 | 0.16 | ||||
| rs1892534 | T | European American | 11344 | 2 | 0.39 | −0.08 | 0.02 | 5.80E-08 | 0.13 | |
| LEPR | Downstream, LEPR | African American | 3471 | 2 | 0.46 | −0.08 | 0.03 | 4.45E-03 | 0.49 | |
| C-reactive protein | Hispanic | 1851 | 1 | 0.50 | −0.12 | 0.04 | 1.99E-03* | |||
| OVERALL | 16666 | 2 | −0.09 | 0.01 | 1.02E-11 | 0.67 | ||||
| rs1800961 | T | European American | 17660 | 5 | 0.031 | −0.15 | 0.03 | 4.75E-06 | 0.39 | |
| HNF4A | Exonic, HNF4A | African American | 6222 | 5 | 0.007 | 0.00 | 0.10 | 9.86E-01 | 0.27 | |
| HDL Cholesterol | Hispanic | 2117 | 2 | 0.039 | −0.06 | 0.08 | 4.30E-01 | 0.99 | ||
| Asian/Pacific Islander | 523 | 1 | 0.001 | −0.18 | 0.22 | 4.05E-01 | 0.89 | |||
| OVERALL | 26522 | 6 | −0.13 | 0.03 | 9.39E-06 | 0.45 | ||||
| rs599839 | G | European American | 22427 | 5 | 0.23 | 0.05 | 0.01 | 1.01E-04 | 0.68 | |
| CELSR2/PSRC1/SORT1 | Downstream, PSRC1 | African American | 6596 | 5 | 0.71 | 0.04 | 0.02 | 5.95E-02 | 0.52 | |
| Coronary Artery | Hispanic | 2811 | 3 | 0.22 | 0.06 | 0.03 | 6.96E-02 | 0.25 | ||
| Disease | Asian/Pacific Islander | 946 | 2 | 0.08 | 0.05 | 0.05 | 3.24E-01 | 0.34 | ||
| OVERALL | 32780 | 6 | 0.05 | 0.01 | 1.96E-06 | 0.96 | ||||
| rs646776 | C | European American | 22652 | 5 | 0.22 | 0.05 | 0.01 | 2.02E-04 | 0.66 | |
| CELSR2/PSRC1/SORT1 | Downstream, CELSR2 | African American | 6479 | 5 | 0.35 | 0.03 | 0.02 | 1.97E-01 | 0.65 | |
| LDL Cholesterol | Hispanic | 2889 | 3 | 0.19 | 0.04 | 0.03 | 2.72E-01 | 0.18 | ||
| Asian/Pacific Islander | 951 | 2 | 0.07 | 0.08 | 0.06 | 1.82E-01 | 0.05 | |||
| OVERALL | 32971 | 6 | 0.04 | 0.01 | 3.06E-05 | 0.78 | ||||
CAF=Coded Allele Frequency; SE=Standard Error;
P-values below threshold for replication (p<3.1×10−3) but not pleiotropy discovery (p<2.0×10−4).
In race/ethnicity-stratified analyses, 16 SNPs reached statistical significance in European Americans, along with 5 SNPs in African Americans and 9 SNPs in Hispanics. No SNPs reached statistical significance among the Asian/Pacific Islander or American Indian groups, though not all SNPs were evaluated and power was reduced in these smaller groups. Notably, for the 18 SNPs that were significant in the race/ethnicity-combined analysis, only 9 were available in the Asian/Pacific Islander group and none were available in the American Indian group. Most SNPs showed no evidence of heterogeneity, either within or between race/ethnicity groups. For two SNPs showing some evidence of heterogeneity between race/ethnicity groups (rs2075650 and rs4420638, Figure 1), sensitivity analyses using random-effects meta-analyses provided similar results (Supplemental Figure 1).
Figure 1.
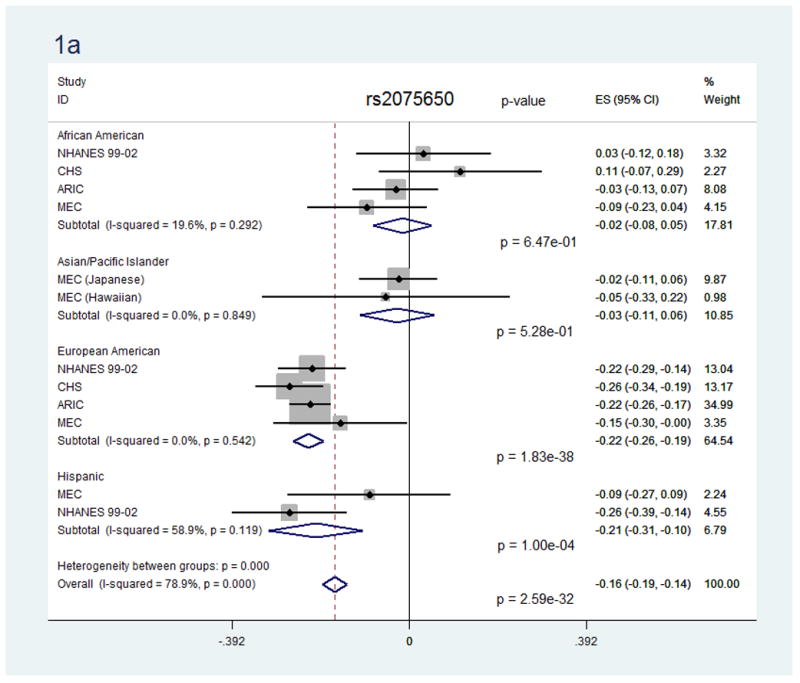
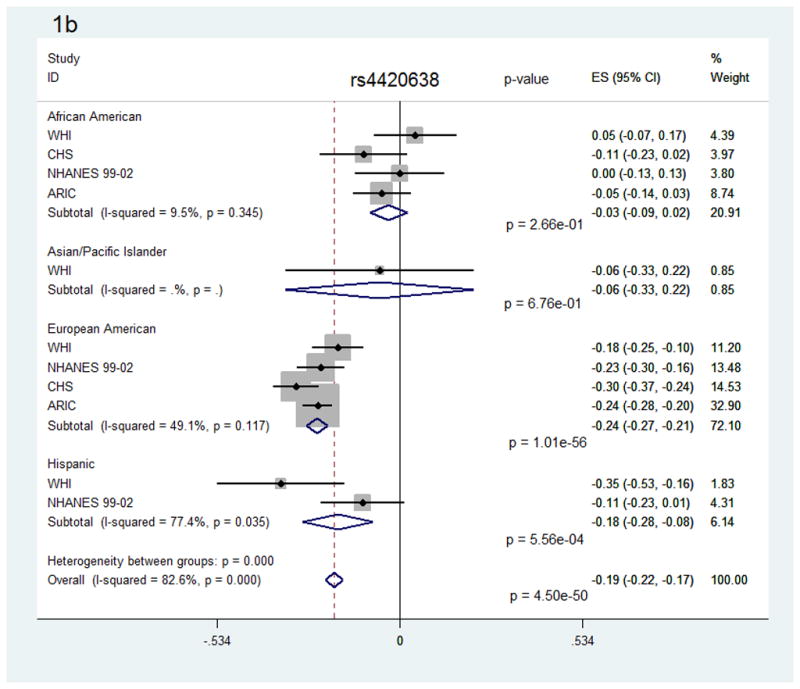
Forest plots for the association of SNPs rs2075650 (a) and rs4420638 (b) with C-reactive protein, overall and by race/ethnicity group.
For generalizability, 7 SNPs were associated only in European Americans, while 10 SNPs in 5 loci demonstrated statistically significant associations with CRP in more than one race/ethnicity group: 3 SNPs in the APOE/APOC1/TOMM40 locus, 4 SNPs in CRP, and 1 each in HNF1A, IL6R, and LEPR (Table 3). For the three SNPs where a statistically significant effect was seen in European Americans, African Americans, and Hispanics, two showed similar effect size estimates across race/ethnicity groups: SNP rs2228145 in IL6R (β=−0.10, −0.13, −0.18, respectively; p-heterogeneity=0.16), and SNP rs429358 in APOE (β=−0.24, −0.23, −0.36, respectively; p-heterogeneity=0.23). Two SNPs also followed this trend in European Americans and Hispanics: SNP rs2075650 in TOMM40 (β=−0.22 and −0.21, respectively; p-heterogeneity=0.76), and SNP rs6857 in PVRL2 (β=−0.23 and −0.28, respectively; p-heterogeneity=0.50). Two SNPs in CRP showed a potential exception to this trend, where a larger effect was seen for African Americans than for European Americans (rs1800947, β=−0.61 vs. −0.30; p-heterogeneity=0.02) or than for European Americans and Hispanics (rs1205, β=−0.27 vs. −0.17 and −0.22, respectively; p-heterogeneity=0.02). In general, however, race/ethnicity-specific effect estimates for a given SNP were in the same direction and of similar magnitude across race/ethnicity groups regardless of the statistical significance of the association. We did observe several exceptions to this trend, where the effect estimate for one group was null or in the opposite direction compared to all other groups, though these exceptions could be due to chance.
We also evaluated whether any of these associations differed by sex (Table 4, Supplemental Table 6). Most of the SNPs which were statistically significant overall remained so in sex-stratified analyses. Two SNPs in GCKR, rs1260326 and rs780094, demonstrated a statistically significant difference in effect between males and females (p-heterogeneity=1.5×10−8), with a stronger effect seen in females than males. Several other SNPs were also suggestive for a potential difference in effect by sex: rs1417938 in CRP (p-heterogeneity=0.03), rs4420638 and rs429358 in APOE/APOC1/TOMM40 (p-heterogeneity=0.03), and rs2650000 in HNF1A (p-heterogeneity=0.01). For these SNPs, the direction of the effect was generally similar in both sexes, while effect estimates were slightly larger and p-values were smaller among the females than the males, as expected given the larger sample size. For the two SNPs in GCKR, the p- values were actually slightly smaller in the female-stratified results than in the overall combined results, despite almost half the sample size.
Table 4.
SNPs showing a potential sex difference in association with C-reactive protein (p-heterogeneity<0.05).
| Male | Female | Between sex | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP(coded allele) | N | Beta | SE | P-value | N | Beta | SE | P-value | P-heterogeneity |
| CRP | rs1417938(A) | 3449 | −0.1 | 0.03 | 1.24E-03* | 3667 | −0.19 | 0.03 | 2.01E-09 | 0.03 |
| APOE/APOC1/TOMM40 | rs4420638(G) | 9234 | −0.23 | 0.02 | 6.99E-27 | 21365 | −0.17 | 0.02 | 2.01E-26 | 0.03 |
| APOE/APOC1/TOMM40 | rs429358(C) | 1012 | −0.16 | 0.04 | 1.18E-04 | 10260 | −0.27 | 0.03 | 2.96E-23 | 0.03 |
| GCKR | rs1260326(T) | 10664 | 0.04 | 0.01 | 2.10E-03 | 22327 | 0.12 | 0.01 | 8.96E-23 | 1.54E-08 |
| GCKR | rs780094(T) | 12095 | 0.04 | 0.01 | 2.85E-03* | 14305 | 0.12 | 0.01 | 4.74E-18 | 1.54E-08 |
| HNF1A | rs2650000(A) | 11545 | −0.13 | 0.01 | 1.48E-18 | 13569 | −0.09 | 0.01 | 1.16E-10 | 0.01 |
P-values below threshold for replication (p<3.1×10−3) but not pleiotropy discovery (p<2.0×10−4).
Discussion
Our multi-ethnic meta-analysis observed 18 SNPs in 8 loci statistically significantly associated with serum CRP concentrations. Ten of these SNPs in 5 loci demonstrated statistically significant associations with CRP in multiple ancestral groups, most of which have not been previously reported in non-European American populations. Among the 18 significant SNPs, 2 SNPs (rs599839 and rs646776, CEU r2=0.9) in 1 locus (CELSR2/PSRC1/SORT1) had not previously been associated with CRP. Three themes emerged from our results: 1) the general consistency of effect at a particular SNP across race/ethnicity groups; 2) variation in association for different SNPs in the same locus can be useful for fine-mapping regions of interest; 3) and the demonstration of pleiotropic effects for specific SNPs.
Generalization
First, for the 10 SNPs which demonstrated a statistically significant association in multiple race/ethnicity groups, the direction and magnitude of the effect was fairly consistent. Several SNPs showed a similar statistically significant effect in European Americans, African Americans, and Hispanics (rs2228145 in IL6R and rs429358 in APOE) or European Americans and Hispanics (rs2075650 in TOMM40 and rs6857 in PVRL2). This trend of similar direction and magnitude across race/ethnicity groups generally held for most SNPs, even where the race/ethnicity-specific associations did not reach statistical significance across all groups (such as rs7310409 in HNF1A and rs599839 in PSRC1). When combined with previous findings, these results suggest a shared genetic influence between race/ethnicity groups at these SNPs.
However, there were several notable exceptions where large differences were seen between race/ethnicity groups. Two SNPs in CRP, for example, showed larger effect sizes in African Americans than in European Americans (rs1800947, rs1205) or Hispanics (rs1205). In other SNPs, an effect was observed in one or two groups but not the others (rs2075650 and rs4420638 in TOMM40, p-heterogeneity<1.6×10−9). Such differences could be indicative of population differences in linkage disequilibrium (LD), since tagging variants in European-ancestry populations may not be representative in other groups18, 31. Taken together, this may suggest that the functional SNP is likely to be within the LD block shared by those race/ethnicity groups with similar effect estimates for that tagSNP, while for the “outlying” race/ethnicity group the tagSNP may not be well correlated with the functional SNP. Alternatively, differences in generalizability could also be due to a reduced ability to detect an association in some race/ethnicity groups in our study due to smaller sample sizes, differences in allele frequency, smaller effect sizes, or lower correlation with functional variants.
Fine-mapping
A related second theme is that differences in SNP generalizability may be informative of the genetic architecture of loci with multiple SNPs associated with CRP. Previous studies have shown that both average CRP levels and SNP-CRP associations can vary by race/ethnicity group18, 23. Additionally, while some polymorphisms have demonstrated an association across race/ethnicity groups (rs120522), others appear specific to particular groups (rs309305818, 22). In our study, some SNPs within the CRP and APOE/APOC1/TOMM40 loci generalized while others did not, providing evidence of potential differences in genetic effect among race/ethnicity groups. Since LD structure also varies by ancestry, we can utilize this information to identify smaller regions that are more likely to be functionally relevant.
We identified three SNPs in the APOE/APOC1/TOMM40 locus associated with CRP that generalized to other race/ethnicity groups. SNPs rs2075650 and rs6857 generalized only to Hispanics, while rs429358 demonstrated an association in European Americans, African Americans and Hispanics. Located in the third exon of the APOE gene, rs429358 is one of two non-synonymous SNPs that define the major ε2, ε3, and ε4 haplotypes of this region32. Our results align with previous studies finding this haplotype associated with serum CRP in European Americans33, African Americans34, Hispanics35, 36, and Asian/Pacific Islanders24. The magnitude and direction of the effect were fairly consistent across race/ethnicity groups, supporting a jointly consistent association across multiple race/ethnicity groups. Previous studies suggest that SNPs at the APOE/APOC1/TOMM40 locus appear to represent a single signal best represented by rs429358, with other SNPs appearing associated due to correlation32, 37. Given the consistency of effect seen in rs429358 across race/ethnicity groups compared to the other SNPs we evaluated in this region, our results appear to support this view, and extend this SNP finding to African Americans and Hispanics.
We also observed race/ethnicity-specific differences in association with CRP among SNPs in the CRP locus. Of the five SNPs demonstrated an overall association, four showed differences in race/ethnicity-specific results. While some of these SNPs may not have generalized to other groups due to smaller sample size, ancestry-related differences in genetic contributions to CRP concentrations at this locus are also possible. For example, for many SNPs in this region the allele frequencies in African Americans were different than other race/ethnicity groups, as previous studies have also highlighted29. In such a situation, investigating SNPs in multiple populations can be useful for evaluating which SNPs might be associated with CRP in multiple populations (rs1205), and which may be race/ethnicity-specific (rs4131568). Other SNPs may have a larger effect in one race/ethnicity group than another (rs1800947). If confirmed, this type of information could potentially be important for ensuring that genetic information is appropriately interpreted and targeted where there are differences in effect by race/ethnicity.
Pleiotropy
The third theme is that our results show evidence for potentially pleiotropic effects. As previously stated, most of the SNPs evaluated in this study have previously been associated with other inflammation-related phenotypes, such as HDL and LDL cholesterol, triglycerides, and coronary artery disease. Studies have also demonstrated associations between some of these SNPs and phenotypes as diverse as colon and rectal cancer (rs12057), cervical cancer survival (rs141793838), and fasting glucose concentration (rs78009439), though only this last trait reached genome-wide significance. Our results support several of these SNPs also being associated with serum CRP concentrations, suggesting that several of these SNPs and loci appear to have pleiotropic effects. This knowledge could be useful for exploring potentially shared disease etiology or mechanisms between phenotypes associated with a given pleiotropic SNP or locus.
We identified a potentially novel pleiotropic association between two correlated SNPs (CEU r2=0.9) in the CELSR2/PSRC1/SORT1 locus and serum CRP levels, as these SNPs have previously been associated with several other inflammation-related phenotypes but not CRP. SNP rs599839 has been associated with LDL cholesterol40–45, coronary artery disease41–43, 46, myocardial infarction47, triglyceride metabolism41, and coronary heart disease45, while SNP rs646776 has been associated with LDL cholesterol40 and progranulin levels48. Variations at rs646776 have also been strongly associated with transcript concentrations of the CELSR2, PSRC1, and SORT1 genes40. Several studies have also found a significant association between variation in rs599839 and sortilin mRNA expression in the liver40, 43, 49. Sortilin, the gene product of SORT1, acts as a multiligand receptor and influences the uptake of LDL particles into cells, with studies suggesting that the G allele of rs599839 offers a protective effect against coronary artery disease mediated through LDL cholesterol lowering43. Our finding that SNPs in this region are also associated with CRP may provide additional insights on the inflammation pathway, and how these SNPs might impact multiple outcomes. Our potentially novel finding in this region also demonstrates the value of evaluating SNPs previously associated with related phenotypes for additional pleiotropic relationships.
Strengths of this study stem from the inclusion of well-characterized study populations with diverse race/ethnicity groups. We had large sample sizes overall, with large numbers for European Americans, African American, and Hispanics. While we had fewer Asian/Pacific Islander and American Indian participants, we were still able to evaluate several SNPs in these groups. While several SNPs in our study demonstrate potential pleiotropic associations with both CRP and inflammation-related traits, we are unable to evaluate whether these effects are truly independent, or if the association with one trait explains the other. Further work is needed to explore these complex relationships.
One limiting factor was that not all participants were genotyped for all SNPs, since SNP panels and race/ethnicity representation varied between participating studies. This reduced the overall sample size available for any given SNP- and race/ethnicity-specific association, though numbers were still large for most analyses, particularly in the European American, African American, and Hispanic groups. Unfortunately, for many SNPs the combination of smaller numbers or unavailable genotype information reduced or eliminated our ability to determine whether CRP associations generalized to the Asian/Pacific Islander or American Indian groups. Race/ethnicity differences in coded allele frequency may have also reduced our ability to detect an association in various groups. For some SNPs, these differences in allele frequency may at least partly explain why an association with CRP in one group did not generalize to another. For these reasons, a lack of generalization in our study should not be interpreted as proof that these SNPs are not associated with CRP in other groups, particularly for the Asian/Pacific Islander or American Indian groups where our ability to detect an association was reduced.
Another potential limiting factor is that we are unable to assess fully whether these potentially pleiotropic associations with CRP are due to correlation with the inflammation-related traits they have previously been associated with. However, previous studies of SNPs in the CRP gene have suggested that their associations with cardiovascular events are only slightly attenuated after adjustment for plasma CRP concentration28. Additionally, a sensitivity analysis in WHI for five of the SNPs in this study showed little effect when adjusting for previously associated traits (coronary heart disease, LDL cholesterol, and triglyceride levels). Together, this suggests that these pleiotropic associations may not be due to statistical correlation between CRP and the other inflammation-related traits, although this cannot be ruled out for all SNPs. Further exploration of the relationships between these traits is warranted.
While our analysis benefited from an a priori selection of interesting SNPs from previous GWAS, it is not a comprehensive analysis across all known loci given the rapid progress made since SNP selection. However, this limitation is lessened by our inclusion of the initial GWAS findings, which tend to have strong effect sizes, and hence tend to explain a larger fraction of the genetic variation than more recent GWAS findings. We applied a Bonferroni-corrected p-value to determine the statistical significance of a given association. While we adjusted for either 16 or 250 independent tests, a potential limitation is that we did not adjust for testing in multiple race/ethnicity groups. However, the Bonferroni adjustment is conservative in nature, and we tested GWAS findings for CRP or related traits. Many, though not all, of our findings greatly surpassed our corrected p-value and reached more stringent genome-wide significance levels (p<5×10−8).
In conclusion, our results support and extend previous observations of associations between CRP and genetic variation in several loci. We generalized several associations between SNPs and serum CRP levels previously identified in European Americans to African Americans or Hispanics, and we also identified a potentially novel CRP locus, previously associated with coronary artery disease and LDL cholesterol. Our findings demonstrate the benefit of evaluating genotype-phenotype associations in multiple race/ethnicity groups, and of looking for pleiotropic relationships among SNPs previously associated with related phenotypes. Additional follow-up and fine-mapping of these loci may lead to better characterization of the functional variants in these regions.
Supplementary Material
Acknowledgments
The PAGE consortium thanks the staff and participants of all PAGE studies for their important contributions.
Funding Sources: The Population Architecture Using Genomics and Epidemiology (PAGE) program is funded by the National Human Genome Research Institute (NHGRI), supported by U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (MEC), U01HG004790 (WHI), and U01HG004801 (Coordinating Center), and their respective NHGRI ARRA supplements. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The complete list of PAGE members can be found at http://www.pagestudy.org.
The data and materials included in this report result from collaboration between the following studies:
“Epidemiologic Architecture for Genes Linked to Environment (EAGLE)” is funded through the NHGRI PAGE program (U01HG004798-01 and its NHGRI ARRA supplement). The study participants derive from the National Health and Nutrition Examination Surveys (NHANES), and these studies are supported by the Centers for Disease Control and Prevention (CDC). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
The Multiethnic Cohort study (MEC) characterization of epidemiological architecture is funded through the NHGRI PAGE program (U01HG004802 and its NHGRI ARRA supplement). The MEC study is funded through the National Cancer Institute (R37CA54281, R01 CA63, P01CA33619, U01CA136792, and U01CA98758).
Funding support for the “Epidemiology of putative genetic variants: The Women’s Health Initiative” study is provided through the NHGRI PAGE program (U01HG004790 and its NHGRI ARRA supplement). The WHI program is funded by the National Heart, Lung, and Blood Institute; NIH; and U.S. Department of Health and Human Services through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf
Funding support for the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) program was provided through the NHGRI PAGE program (U01HG004803 and its NHGRI ARRA supplement). The following studies contributed to this manuscript and are funded by the following agencies: The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute (NHLBI) contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022. The Coronary Artery Risk Development in Young Adults (CARDIA) study is supported by the following National Institutes of Health, NHLBI contracts: N01-HC-95095; N01-HC-48047; N01-HC-48048; N01-HC-48049; N01-HC-48050; N01-HC-45134; N01-HC-05187; and N01-HC-45205. The Cardiovascular Health Study (CHS) is supported by contracts HHSN268201200036C, N01-HC-85239, N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, and grant HL080295 from NHLBI, with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the National Institute on Aging (NIA). The Strong Heart Study (SHS) is supported by NHLBI grants U01 HL65520, U01 HL41642, U01 HL41652, U01 HL41654, and U01 HL65521. The opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Indian Health Service. Assistance with phenotype harmonization, SNP selection and annotation, data cleaning, data management, integration and dissemination, and general study coordination was provided by the PAGE Coordinating Center (U01HG004801-01 and its NHGRI ARRA supplement). The National Institutes of Mental Health also contributes to the support for the Coordinating Center. Funding for work by Jonathan Kocarnik was supported by grant R25CA94880 from NCI. Funding for performing CRP assays in a portion of WHI subjects was provided by NHLBI BAA award HHSN268200960011C.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Ridker PM. C-reactive protein: Eighty years from discovery to emergence as a major risk marker for cardiovascular disease. Clin Chem. 2009;55:209–215. doi: 10.1373/clinchem.2008.119214. [DOI] [PubMed] [Google Scholar]
- 2.Pepys MB, Hirschfield GM. C-reactive protein: A critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ridker PM, Stampfer MJ, Rifai N. Novel risk factors for systemic atherosclerosis: A comparison of C-reactive protein, fibrinogen, homocysteine, lipoprotein(a), and standard cholesterol screening as predictors of peripheral arterial disease. JAMA. 2001;285:2481–2485. doi: 10.1001/jama.285.19.2481. [DOI] [PubMed] [Google Scholar]
- 4.Everett BM, Kurth T, Buring JE, Ridker PM. The relative strength of C-reactive protein and lipid levels as determinants of ischemic stroke compared with coronary heart disease in women. J Am Coll Cardiol. 2006;48:2235–2242. doi: 10.1016/j.jacc.2006.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pradhan AD, Manson JE, Rifai N, Buring JE, Ridker PM. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA. 2001;286:327–334. doi: 10.1001/jama.286.3.327. [DOI] [PubMed] [Google Scholar]
- 6.Mora S, Lee IM, Buring JE, Ridker PM. Association of physical activity and body mass index with novel and traditional cardiovascular biomarkers in women. JAMA. 2006;295:1412–1419. doi: 10.1001/jama.295.12.1412. [DOI] [PubMed] [Google Scholar]
- 7.Slattery ML, Curtin K, Poole EM, Duggan DJ, Samowitz WS, Peters U, et al. Genetic variation in C-reactive protein in relation to colon and rectal cancer risk and survival. Int J Cancer. 2011;128:2726–2734. doi: 10.1002/ijc.25721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groblewska M, Mroczko B, Sosnowska D, Szmitkowski M. Interleukin 6 and C-reactive protein in esophageal cancer. Clin Chim Acta. 2012;413:1583–1590. doi: 10.1016/j.cca.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Wessel J, Moratorio G, Rao F, Mahata M, Zhang L, Greene W, et al. C-reactive protein, an ‘intermediate phenotype’ for inflammation: Human twin studies reveal heritability, association with blood pressure and the metabolic syndrome, and the influence of common polymorphism at catecholaminergic/beta-adrenergic pathway loci. J Hypertens. 2007;25:329–343. doi: 10.1097/HJH.0b013e328011753e. [DOI] [PubMed] [Google Scholar]
- 10.Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, et al. Familial and genetic determinants of systemic markers of inflammation: The NHLBI family heart study. Atherosclerosis. 2001;154:681–689. doi: 10.1016/s0021-9150(00)00586-4. [DOI] [PubMed] [Google Scholar]
- 11.Dupuis J, Larson MG, Vasan RS, Massaro JM, Wilson PW, Lipinska I, et al. Genome scan of systemic biomarkers of vascular inflammation in the framingham heart study: Evidence for susceptibility loci on 1q. Atherosclerosis. 2005;182:307–314. doi: 10.1016/j.atherosclerosis.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Ridker PM, Pare G, Parker A, Zee RY, Danik JS, Buring JE, et al. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: The women’s genome health study. Am J Hum Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiner AP, Beleza S, Franceschini N, Auer PL, Robinson JG, Kooperberg C, et al. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am J Hum Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khera A, McGuire DK, Murphy SA, Stanek HG, Das SR, Vongpatanasin W, et al. Race and gender differences in C-reactive protein levels. J Am Coll Cardiol. 2005;46:464–469. doi: 10.1016/j.jacc.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 16.Dreon DM, Slavin JL, Phinney SD. Oral contraceptive use and increased plasma concentration of C-reactive protein. Life Sci. 2003;73:1245–1252. doi: 10.1016/s0024-3205(03)00425-9. [DOI] [PubMed] [Google Scholar]
- 17.Ford ES. Does exercise reduce inflammation? Physical activity and C-reactive protein among U.S. Adults. Epidemiology. 2002;13:561–568. doi: 10.1097/00001648-200209000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Doumatey AP, Chen G, Tekola Ayele F, Zhou J, Erdos M, Shriner D, et al. C-reactive protein (CRP) promoter polymorphisms influence circulating CRP levels in a genome-wide association study of African Americans. Hum Mol Genet. 2012;21:3063–3072. doi: 10.1093/hmg/dds133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best LG, Zhang Y, Lee ET, Yeh JL, Cowan L, Palmieri V, et al. C-reactive protein as a predictor of cardiovascular risk in a population with a high prevalence of diabetes: The Strong Heart Study. Circulation. 2005;112:1289–1295. doi: 10.1161/CIRCULATIONAHA.104.489260. [DOI] [PubMed] [Google Scholar]
- 20.Saito I, Sato S, Nakamura M, Kokubo Y, Mannami T, Adachi H, et al. A low level of C-reactive protein in Japanese adults and its association with cardiovascular risk factors: The Japan NCVC-Collaborative Inflammation Cohort (JNIC) study. Atherosclerosis. 2007;194:238–244. doi: 10.1016/j.atherosclerosis.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 21.Lakoski SG, Cushman M, Palmas W, Blumenthal R, D’Agostino RB, Jr, Herrington DM. The relationship between blood pressure and C-reactive protein in the Multi-Ethnic Study of Atherosclerosis (MESA) J Am Coll Cardiol. 2005;46:1869–1874. doi: 10.1016/j.jacc.2005.07.050. [DOI] [PubMed] [Google Scholar]
- 22.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, et al. Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation. 2006;114:2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- 23.Okada Y, Takahashi A, Ohmiya H, Kumasaka N, Kamatani Y, Hosono N, et al. Genome-wide association study for C-reactive protein levels identified pleiotropic associations in the IL6 locus. Hum Mol Genet. 2011;20:1224–1231. doi: 10.1093/hmg/ddq551. [DOI] [PubMed] [Google Scholar]
- 24.Wu Y, McDade TW, Kuzawa CW, Borja J, Li Y, Adair LS, et al. Genome-wide association with C-reactive protein levels in CLHNS: Evidence for the CRP and HNF1A loci and their interaction with exposure to a pathogenic environment. Inflammation. 2012;35:574–583. doi: 10.1007/s10753-011-9348-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su S, Lampert R, Zhao J, Bremner JD, Miller A, Snieder H, et al. Pleiotropy of C-reactive protein gene polymorphisms with C-reactive protein levels and heart rate variability in healthy male twins. Am J Cardiol. 2009;104:1748–1754. doi: 10.1016/j.amjcard.2009.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, et al. The next PAGE in understanding complex traits: Design for the analysis of Population Architecture using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol. 2011;174:849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiner AP, Barber MJ, Guan Y, Ridker PM, Lange LA, Chasman DI, et al. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am J Hum Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, et al. Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA. 2006;296:2703–2711. doi: 10.1001/jama.296.22.2703. [DOI] [PubMed] [Google Scholar]
- 29.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, et al. Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet. 2005;77:64–77. doi: 10.1086/431366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.StataCorp. Stata statistical software: Release 12. College Station, TX: StataCorp LP; 2011. [Google Scholar]
- 31.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, et al. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer’s disease: Patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Judson R, Brain C, Dain B, Windemuth A, Ruano G, Reed C. New and confirmatory evidence of an association between APOE genotype and baseline C-reactive protein in dyslipidemic individuals. Atherosclerosis. 2004;177:345–351. doi: 10.1016/j.atherosclerosis.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 34.Chasman DI, Kozlowski P, Zee RY, Kwiatkowski DJ, Ridker PM. Qualitative and quantitative effects of APOE genetic variation on plasma C-reactive protein, LDL-cholesterol, and APOE protein. Genes Immun. 2006;7:211–219. doi: 10.1038/sj.gene.6364289. [DOI] [PubMed] [Google Scholar]
- 35.Aiello AE, Nguyen HO, Haan MN. C-reactive protein mediates the effect of apolipoprotein E on cytomegalovirus infection. J Infect Dis. 2008;197:34–41. doi: 10.1086/524144. [DOI] [PubMed] [Google Scholar]
- 36.Haan MN, Aiello AE, West NA, Jagust WJ. C-reactive protein and rate of dementia in carriers and non carriers of Apolipoprotein APOE4 genotype. Neurobiol Aging. 2008;29:1774–1782. doi: 10.1016/j.neurobiolaging.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curocichin G, Wu Y, McDade TW, Kuzawa CW, Borja JB, Qin L, et al. Single-nucleotide polymorphisms at five loci are associated with C-reactive protein levels in a cohort of Filipino young adults. J Hum Genet. 2011;56:823–827. doi: 10.1038/jhg.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polterauer S, Grimm C, Zeillinger R, Heinze G, Tempfer C, Reinthaller A, et al. Association of C-reactive protein (CRP) gene polymorphisms, serum CRP levels and cervical cancer prognosis. Anticancer Res. 2011;31:2259–2264. [PubMed] [Google Scholar]
- 39.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleber ME, Renner W, Grammer TB, Linsel-Nitschke P, Boehm BO, Winkelmann BR, et al. Association of the single nucleotide polymorphism rs599839 in the vicinity of the sortilin 1 gene with LDL and triglyceride metabolism, coronary heart disease and myocardial infarction. The Ludwigshafen Risk and Cardiovascular Health Study. Atherosclerosis. 2010;209:492–497. doi: 10.1016/j.atherosclerosis.2009.09.068. [DOI] [PubMed] [Google Scholar]
- 42.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linsel-Nitschke P, Heeren J, Aherrahrou Z, Bruse P, Gieger C, Illig T, et al. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 44.Samani NJ, Braund PS, Erdmann J, Gotz A, Tomaszewski M, Linsel-Nitschke P, et al. The novel genetic variant predisposing to coronary artery disease in the region of the PSRC1 and CELSR2 genes on chromosome 1 associates with serum cholesterol. J Mol Med (Berl) 2008;86:1233–1241. doi: 10.1007/s00109-008-0387-2. [DOI] [PubMed] [Google Scholar]
- 45.Angelakopoulou A, Shah T, Sofat R, Shah S, Berry DJ, Cooper J, et al. Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: Cardiovascular Biomarker Genetics Collaboration. Eur Heart J. 2012;33:393–407. doi: 10.1093/eurheartj/ehr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang AZ, Li L, Zhang B, Shen GQ, Wang QK. Association of snp rs17465637 on chromosome 1q41 and rs599839 on 1p13.3 with myocardial infarction in an American caucasian population. Ann Hum Genet. 2011;75:475–482. doi: 10.1111/j.1469-1809.2011.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carrasquillo MM, Nicholson AM, Finch N, Gibbs JR, Baker M, Rutherford NJ, et al. Genome-wide screen identifies rs646776 near sortilin as a regulator of progranulin levels in human plasma. Am J Hum Genet. 2010;87:890–897. doi: 10.1016/j.ajhg.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


