Abstract
The pentein superfamily is a mechanistically diverse superfamily encompassing both noncatalytic proteins and enzymes that catalyze hydrolase, dihydrolase and amidinotransfer reactions on guanidine substrates. Despite generally low sequence identity, they possess a conserved structural fold and display common mechanistic themes in catalysis. The structurally characterized catalytic penteins possess a conserved core of residues that include a Cys, His and two polar, guanidine-binding residues. All known catalytic penteins use the core Cys to attack the substrate’s guanidine moiety to form a covalent thiouronium adduct and all cleave one or more of the guanidine C–N bonds. The mechanistic information compiled to date supports the hypothesis that this superfamily may have evolved divergently from a catalytically promiscuous ancestor.
Keywords: Pentein, Guanidine, Arginine deiminase, Dimethylarginine dimethylaminohydrolase, Agmatine deiminase, Peptidylarginine deiminase, Arginine:glycine amidinotransferase, Arginine:inosamine phosphate, amidinotransferase, Nα-succinylarginine dihydrolase, Guanidino-modifying superfamily
1. Introduction
The pentein superfamily consists of functionally diverse proteins grouped together based on their similar β/α propeller structural fold. This superfamily contains numerous enzymes that modify guanidines, including hydrolases, dihydrolases and amidinotransferases. They fulfill diverse biological roles in gene regulation, translation, cell–cell signaling, metabolism, natural product biosynthesis and cell survival, in organisms ranging from bacteria to humans.
Assigning structure and function to penteins has proven difficult because many of them do not share significant sequence similarity to one another. It was believed for a time that this superfamily contained only enzymes that modify guanidines, but X-ray crystallography has unexpectedly revealed that the catalytic penteins contain the same overall fold as the ribosome anti-association factors aIF6 and eIF6 [1], despite very poor sequence identity [2]. The catalytic members of the superfamily have collectively been referred to as the guanidino-modifying superfamily. However, the broader term pentein can be used to refer to all proteins, both noncatalytic and catalytic, that contain the unifying pentein structural fold [3,4].
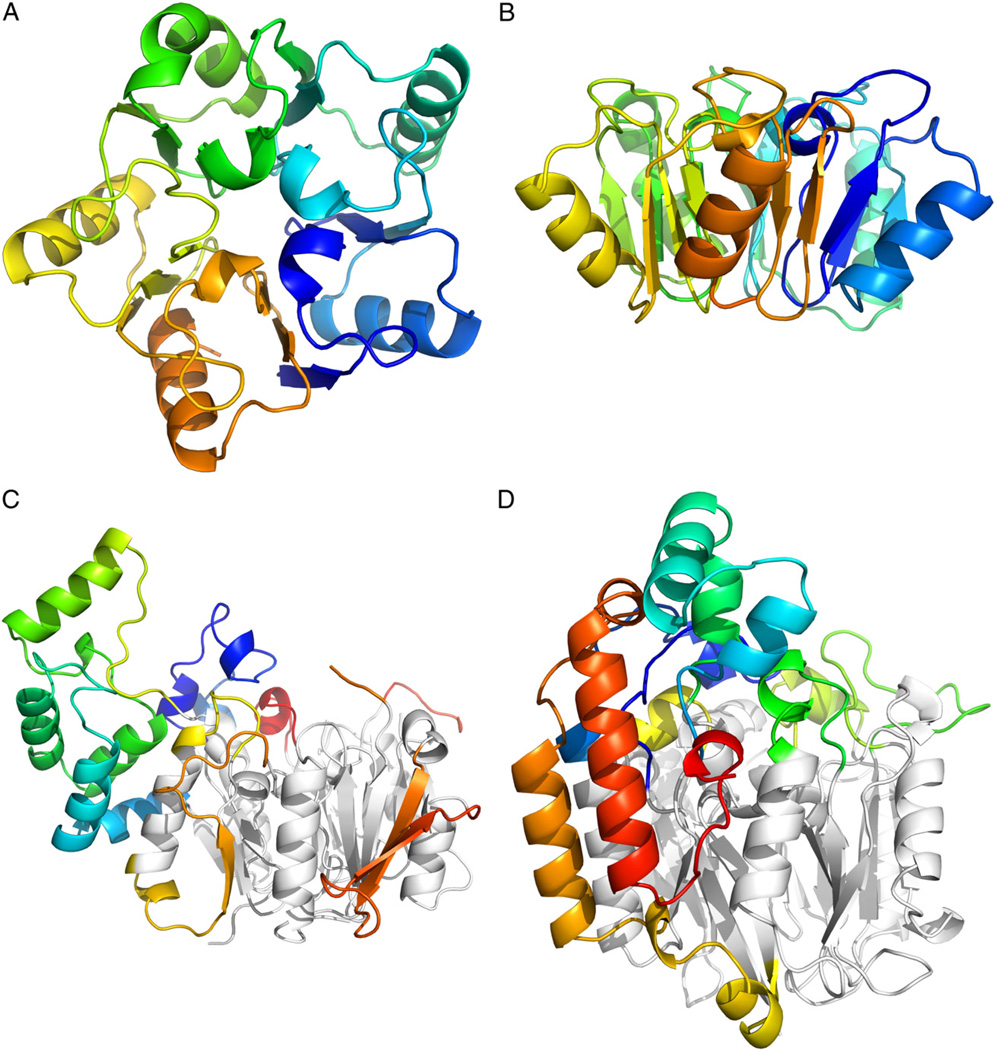
The fold of the non-catalytic proteins aIF6 and eIF6 is the simplest known form of the pentein fold. The structures contain five “quasiidentical” α/β subdomains which are positioned about a five-fold axis of pseudosymmetry forming a β/α propeller [5]. A large hollow is present where the domains intersect in the center of the protein. In the case of eIF6, small loops surrounding this central hollow facilitate binding to the ribosomal interaction partner rpL23 [6]. Catalytic members of the pentein superfamily are typically decorated with more elaborate loop inserts and extensions [7] (Fig. 1).
Fig. 1.
The pentein fold consists of five αββαβ subdomains symmetrically arranged around afivefold axis of pseudosymmetry. A) Top-down viewofribosome anti-association factor eIF6 (PDB ID: 1G61). B) Side view of eIF6. C) Side view of arginine deiminase (PDB ID: 2A9G). D) Side view of succinylarginine dihydrolase (PDB ID: 1YNI). E) Side view of arginine: glycine amidinotransferase (PDB ID: 4JDW). F) Side view of PAD4 (PDB ID: 1WDA). In C–F, the conserved pentein core is colored gray, while additional inserts and loops are colored by amino acid residue number.
Part of the central hollow is used to bind substrates in the catalytic penteins. These enzymes share a similar active site, with changes in the extra loops altering the substrate specificity of these enzymes to recognize a variety of natural guanidines, including l-arginine [8], Nω-methylated arginine [9,10],Nα-succinylarginine [11,12], l-canavanine [13], agmatine [14], N-amidino scyllo-inosamine phosphate [13] and peptidylarginine [15]. Although most catalytic residues are conserved throughout the superfamily, the guanidino-modifying penteins catalyze different reactions. Three functional classes of guanidino-modifying penteins are known: hydrolases, which convert guanidines to urea derivatives; amidinotransferases, which transfer an amidine group from one amine to another; and dihydrolases, which formally hydrolyze guanidines twice to produce carbon dioxide or bicarbonate, ammonia and a primary amine.
This review will explore the common and divergent features of substrate binding and catalysis present in the pentein superfamily. The common activesite scaffold is modified slightly in each enzyme to allow recognition of a wide variety of substrates while maintaining a similar catalytic core. It has also become increasingly clear that the catalytic penteins are marked by a unifying mechanistic thread, a similar first reaction intermediate. These observations suggest that the penteins may have undergone divergent evolution from a common ancestor, and that changes to substrate specificity and intermediate partitioning may have provided these enzymes with a path for the evolution of new functions.
2. Pentein catalytic families
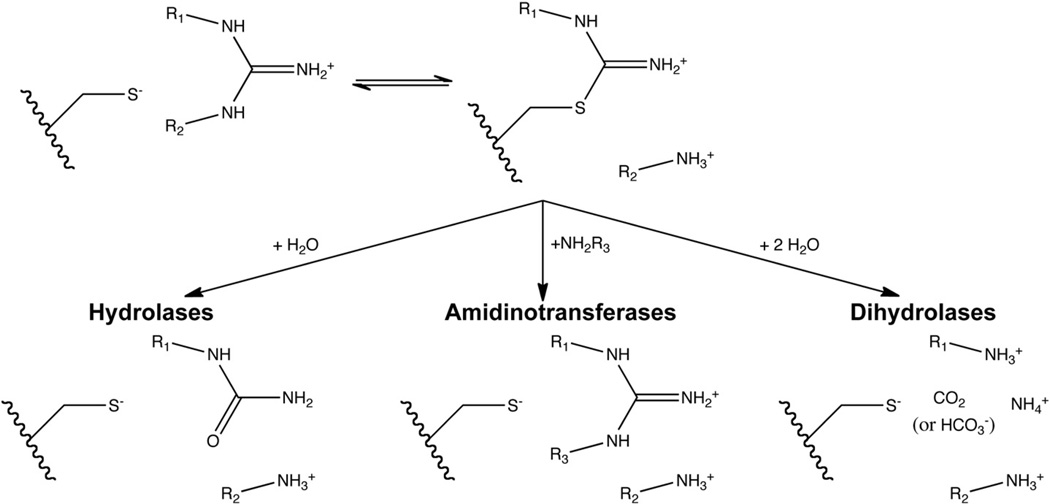
The catalytic penteins have been subdivided into three families according to the reaction type [7] (Fig. 2). Hydrolases catalyze the attack of a guanidine by water to form a urea derivative. The most well-characterized hydrolases include arginine deiminase (ADI), which converts l-arginine to citrulline and ammonia; dimethylarginine dimethylaminohydrolase (DDAH), which converts Nω, Nω-methyl-l-arginine to citrulline and dimethylamine; peptidylarginine deiminase (PAD), which converts arginine residues in peptides and proteins to citrulline residues and ammonia; and agmatine deiminase (AgD), which converts agmatine to N-carbamoyl putrescine.
Fig. 2.
Reactions catalyzed by penteins. The guanidine substrate binds and reacts with the core Cys residue to form a thiouronium intermediate, which is used in the hydrolase, amidinotransferase and dihydrolase reactions. It is assumed in this figure that the active-site Cys is deprotonated prior to formation of the alkylthiouronium adduct. However, the protonation model at work is unclear (see Fig. 6).
The second family, amidinotranferases, catalyzes the transfer of a one carbon amidine (–C(=NH)NH2) from a guanidine compound donor to an amine acceptor. The mechanistically characterized amidinotransferases include arginine:glycine amidinotransferase (AGAT), which transfers the amidino group from l-arginine to glycine forming l-ornithine and guanidinoacetate; arginine:inosamine-phosphate amidinotransferase (AIAT), which uses l-arginine as an amidine donor and various amino compounds as acceptors to form l-ornithine and a corresponding guanidine; and arginine:lysine amidinotransferase, which uses l-arginine to donate an amidine to l-lysine to form l-ornithine and l-homoarginine.
The third family, dihydrolases, formally catalyzes two hydrolysis reactions on a guanidine, breaking all three C–N bonds and freeing the guanidine carbon as CO2 (or HCO3). No dihydrolase has been mechanistically characterized, although the structure of Nα-succinylarginine dihydrolase (SADH, also labeled AstB by genetic studies) has provided some mechanistic insight. This enzyme converts Nα-succinylarginine to Nα-succinylornithine, two ammonia molecules and one carbon dioxide or bicarbonate molecule. No other dihydrolases have been purified or characterized, but several have been functionally annotated based on the substitution of an Asn residue for one of the active-site carboxylate residues conserved in the hydrolases and amidinotransferases (see below).
3. Substrate recognition
The interactions of penteins with their cognate substrates include those that directly bind the guanidino group and those that bind its substituents. A set of core residues are mostly conserved and facilitate binding of the guanidino group, while residues responsible for binding its substituents are more diverse. Several conserved and semiconserved binding motifs in each enzyme subfamily account for the binding and exclusion of ligands.
3.1. Specificity is governed by non-guanidine-binding residues
Most guanidine-containing substrates of the pentein superfamily members are derivatives of arginine, with substitutions possible at either the guanidine or amino acid moieties. The exception is AIAT, which uses arginine as an amidine donor and a variety of substrates that include scyllo-inosamine phosphate and glycine as acceptors. Since arginine is the most common pentein substrate and is recognized across all three catalytic families, we will discuss it as an exemplar substrate.
The aliphatic chain portion of the substrate arginine side chain is accommodated by a hydrophobic region. In arginine deiminase and dimethylarginine dimethylaminohydrolase, the key residue in this region is a Phe [16–20], and in peptidyl-arginine deiminase 4, agmatine deiminase and succinylarginine dihydrolase, this residue is a Trp [12,14,15,21,22].
There are three distinct modes for recognizing substrate carboxyl or carbonyl groups [7]. In the first, used by ADI and DDAH, Arg residues on the second and third subdomain repeats make mono- and bidentate interactions, respectively, with the substrate carboxyl group. In the second, used by PAD, the peptide substrate carbonyl forms a hydrogen bond directly with one Arg residue on the second subdomain repeat and indirectly through a water molecule hydrogen bonded to a second Arg residue on the second subdomain. In both of these cases, the substrate carbonyl or carboxyl is oriented between the second and third subdomains. Arg:gly amidinotransferase, in contrast, binds its arginine substrate with the carboxyl group oriented between the fourth and fifth subdomains, representing a shift of approximately 120° pivoted around the guanidino carbon relative to the hydrolases such as ADI and DDAH. The carboxyl group forms a bidentate interaction with an Arg residue on the fourth subdomain. The catalytic implications of this rotated binding mode will be discussed further in Section 4.3.
AgD is specific to agmatine, the decarboxylated form of arginine. The crystal structure of AgD demonstrates the use of a bulky residue to exclude carboxyl-containing substrates. The active-site cleft where the substrate’s carboxyl or carbonyl binds in other penteins is narrowed in AgD [14], presumably preventing the binding of free and peptidyl arginine substrates. One residue in particular, Trp106, likely excludes l-arginine and peptidylarginine substrates by introducing additive steric bulk at this position. This is in contrast to PAD4, which uses a wider active site pocket to bind its larger peptidyl arginine substrates. AIAT also makes use of a larger active-site cleft to bind a wide variety of guanidine and amino substrates as amidine donors and acceptors, respectively, conferring the enzyme with promiscuous activity. l-arginine, l-canavanine, streptidine 6-phosphate, 2-deoxystreptidine-6-phosphate, and N-amidino-scyllo-inosamine 4-phosphate function can serve as amidino donor substrates for AIAT, while scyllo-inosamine 4-phosphate, N1-amidinosreptamine 6-phosphate, 3-amino-3-deoxy-scyllo-inosamine 4-phosphate, 2-amino-2-deoxy-neo-inositol 5-phosphate, l-ornithine, l-canaline, glycylglycine, 1,4-diaminobutyl-1-phosphonate and hydroxylamine can serve as acceptors [13].
Penteins have two different binding motifs for recognizing the substrate’s α nitrogen, one for substrates containing N-amides and another for those with free amino groups. Free amino substrates, such as l-arginine, l-canavanine, Nω, Nω-dimethyl-l-arginine and agmatine, are recognized by an Asn or Asp residue in addition to two backbone carbonyl groups which provide hydrogen bond connections to the positively charged substrate amine. N-amide-linked pentein substrates, such as peptidyl arginine and Nα-succinylarginine, are recognized by hydrogen bonds with a His residue (SADH) or a backbone carbonyl, accompanied by expansion of the active-site pocket to accommodate the larger substrate molecule.
The hydrolases contain a small channel that overlaps with the binding site of the arginine’s alkyl side chain in the amidinotransferases (see Section 5). The channel’s size and properties may help protect the reactive intermediate from unwanted side reactions. Amine nucleophiles have been shown to be excluded in a size- and sterics-dependent manner from the channel in DDAH [23], demonstrating that the channel may allow small nucleophiles such as water to enter the active site and react, while isolating the reaction from larger more reactive nucleophiles. The water molecule used in the second half reaction of the hydrolases is oriented through a hydrogen bond the conserved active-site His. Another residue, located in the channel near the substrate, is also believed to orient the nucleophilic water through a hydrogen bond. ADI [19,20], DDAH [16,17,24] and AgD from Helicobacter pylori [14] use a Ser or Thr in this position, but other hydrolases differ. For example, the residue is an Glu in PAD4 [15], an Asp in AgD from Enterococcus faecalis [21], and an Asn in AgD from Arabidopsis thaliana [14]. Nα-succinylarginine dihydrolase has a Val residue in this position, which cannot form hydrogen bonds to water, but may still interact with reaction intermediates. However, the function of this residue has not been experimentally established.
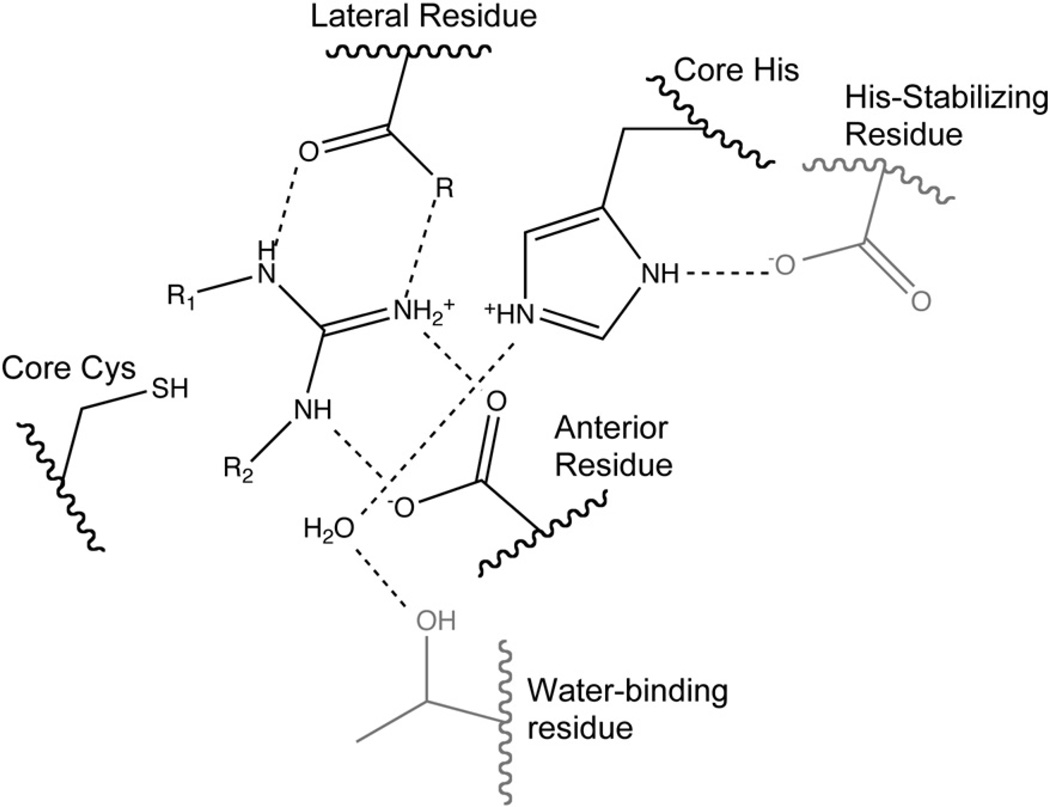
3.2. Core binding residues
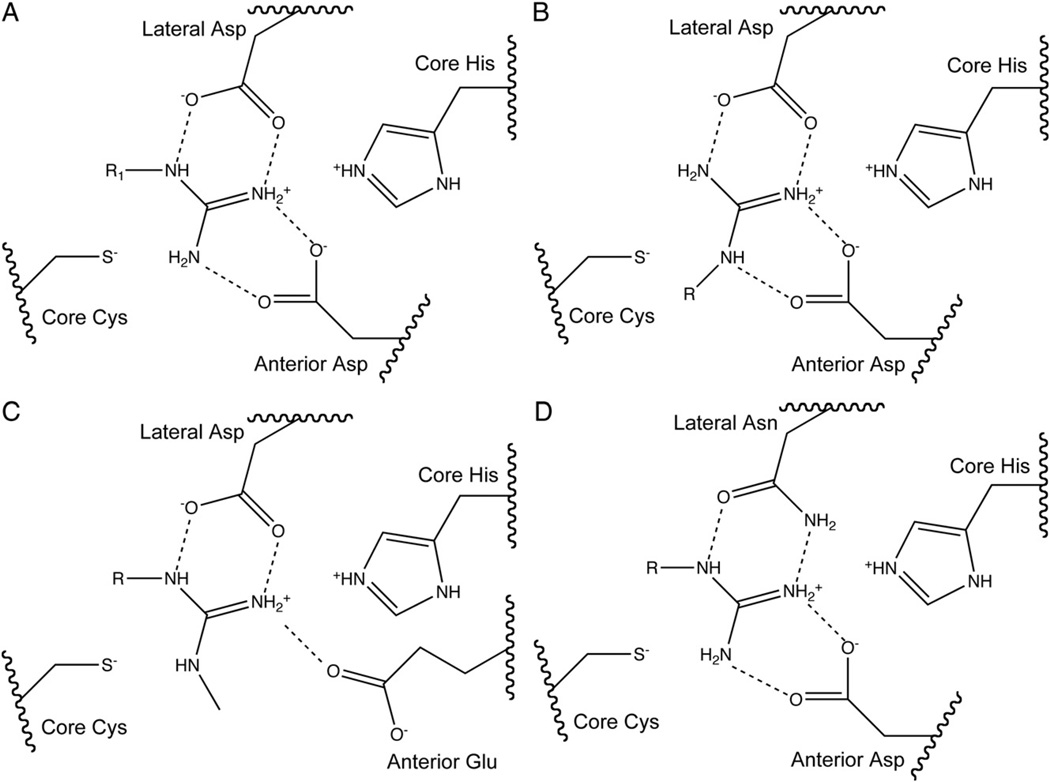
The substrate’s guanidine is held in place in a very polar environment bounded by four residues. Most penteins, including AGAT, AgD, PAD and ADI, bind the guanidine moiety through bidentate interactions with the side chain carboxylates of two conserved acidic residues. Typically an Asp in the hydrolases or Asn residue in the dihydrolases is located lateral to the axis formed by the Nδ–Cζ bond of arginine substrates and binds to Nδ and Nω. The carboxylate side chain of a second residue positioned anterior to the Nδ–Cζ bond binds to Nω and Nω′. Since the amidinotransferases use a rotated binding mode relative to the other penteins, the lateral residue in these enzymes interacts with Nω and Nω′ of arginine amidinotransferase substrates and the anterior residue interacts with Nδ and Nω. These two residues have been proposed to play a large electrostatic role in ADI catalysis [8]. DDAH and SADH, though, bind the guanidine using different means. DDAH uses a Glu residue anterior to the Nδ–Cζ bond to make a monodentate interaction with the unsubstituted Nω of the Nω-methyl-l-arginine or asymmetric Nω,Nω-dimethyl-l-arginine substrates and an Asp residue in the lateral position, similar to other penteins [18]. DDAH also contains a Lys residue which forms an ion pair with one oxygen of the anterior Glu. The aliphatic carbon side chain of this Lys may facilitate binding of the methylated substrates by providing a hydrophobic pocket. In contrast, SADH possesses the anterior Asp residue but has an Asn residue located lateral to the Nδ–Cζ bond. Conserved Cys and His residues sandwich the planar guanidine’s si and re faces, and are essential for catalysis. In most penteins (DDAH, AgD, ADI, AGAT and SADH), an Asp or Glu residue stabilizes the core His, but in PAD4, a Ser is present in this location. To avoid confusion based on differences in amino acid numbering across the superfamily, the two polar active-site guanidine-binding residues will be referred to as the “lateral” and “anterior” residues, as defined using the orientation of hydrolase substrates. The conserved, catalytic active-site Cys and His residues will be referred to as the “core” Cys and His. The locations of major guanidine-binding residues are shown generically in Fig. 3 and specifically in Fig. 4. Important substrate-binding residues are listed in Table 1.
Fig. 3.
Generic active-site diagram. Residues not conserved are shown in gray. R1=Nδ of arginine substrate in hydrolases, R2=Nδ of arginine substrate in amidinotransferases.
Fig. 4.
Active-site diagram showing core catalytic residues for A) ADI, AgD and PAD, B) AGAT and AIAT, C) DDAH and D) SADH. Note that the terms “lateral” and “anterior” refer to the position of the residue relative to the substrate arginine’s Nδ—Cζ bond in the hydrolases and dihydrolases.
Table 1.
Residues at the key active-site positions for known penteins.
| Enzyme | Organism | PDBID | Core Cys | Core His | Lateral | Anterior | His-stabilizing | Water-binding |
|---|---|---|---|---|---|---|---|---|
| DDAH | Pseudomonas aeruginosa | 1H70 | Cys249 | His162 | Asp66 | Glu65 | Glu114 | Thr165 |
| DDAH | Human | 2JAI | Cys273 | His172 | Asp78 | Glu77 | Asp126 | Ser175 |
| DDAH | Bovine | 2C6Z | Cys273 | His172 | Asp78 | Glu77 | Asp126 | Ser175 |
| AgD | Helicobacter pylori | 3HVM | Cys324 | His202 | Asp84 | Asp204 | Glu140 | Thr205 |
| AgD | Arabidopsis thaliana | 3H7K | Cys366 | His224 | Asp94 | Asp226 | Glu162 | Asn227 |
| AgD | Enterococcus faecalis | 2JER | Cys357 | His218 | Asp96 | Asp220 | Glu157 | Asp221 |
| PAD4 | Human | 1WDA | Cys645 | His471 | Asp350 | Asp473 | Ser406 | Glu474 |
| ADI | Pseudomonas aeruginosa | 2A9G | Cys406 | His278 | Asp166 | Asp280 | Glu224 | Thr281 |
| ADI | Mycoplasma arginini | 1LXY | Cys398 | His269 | Asp161 | Asp271 | Glu213 | Thr272 |
| AGAT | Human | 8JDW | Cys407 | His303 | Asp170 | Asp305 | Asp254 | Ala306 |
| AIAT | Streptomyces griseus | 1BWD | Cys332 | His227 | Asp108 | Asp229 | Asp179 | Ala230 |
| SADH | Escherichia coli | 1YNI | Cys365 | His248 | Asn110 | Asp250 | Glu174 | Val251 |
Annotations of important pentein active-site residues: Lateral residue: The residue positioned lateral to the axis formed by the arginine substrate’s Nδ–Cζ bond as oriented in the hydrolases and dihydrolases. Anterior residue: The residue positioned anterior to the axis formed by the arginine substrate’s Nδ–Cζ bond as oriented in the hydrolases and dihydrolases. Core His: The conserved catalytic active-site His residue. Core Cys: the conserved catalytic active-site Cys residue. Water-binding residue: The residue in hydrolases, which, in concert with the core His, forms a hydrogen bond to the nucleophilic water molecule, presumably to orient it for attack. His-stabilizing residue: The residue near the core His which may help orient and stabilize it for catalysis.
4. Pentein catalysis
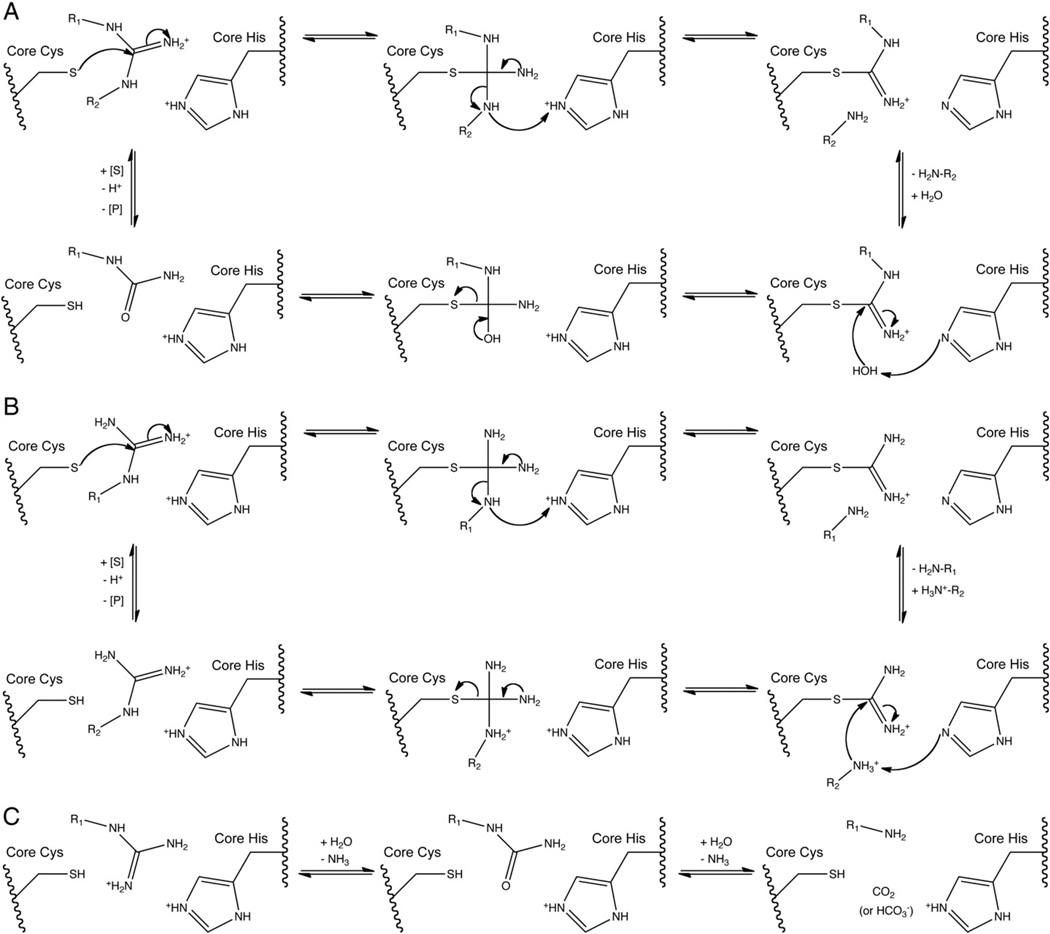
The pentein hydrolases, dihyrolases and amidinotransferases all use similar catalytic machinery and form a thiouronium covalent adducts in their first respective half reactions. Depending on the catalysis, this reactive intermediate then partitions into hydrolysis, dihydrolysis, or amidinotransfer reaction pathways. The proposed mechanisms for the different families of penteins discussed in this section are summarized in Fig. 5.
Fig. 5.
Proposed reaction mechanisms for A) hydrolases, B) amidinotransferases, C) dihydrolases.
4.1. Formation of S-alkyl thiouronium adduct
In all known catalytic penteins, the first step of catalysis is generally proposed to be nucleophilic attack by the active-site Cys to form a tetrahedral adduct, followed by cleavage of a C—N bond of the guanidine to form an S-alkyl thiouronium adduct. When the substrate binds, the active site positions the substrate such that the core Cys and His are on opposite faces of the bound guanidine. Electrostatic interactions with the guanidine-binding pocket of the active site stabilize the guanidine in its cationic form to enhance the electrophilicity of Cζ for attack by the core Cys. Presumably, a tetrahedral adduct results from this attack. Donation of a proton by the core His residue, assisted by electrostatic interactions with the guanidine-binding pocket, is proposed to stabilize the tetrahedral intermediate and subsequent leaving group, promoting cleavage of the scissile C—N bond and loss of an amino compound to form a planar thiouronium adduct. After this point in the catalytic mechanism, the different families of catalytic penteins diverge.
The proposed nucleophilic role of the core Cys in the hydrolases is supported by numerous X-ray crystallography, site-directed mutagenesis and thiol-modification studies in ADI [8,19,20,25,26], DDAH [9,10,23,27], PAD [15,28–30] and AgD [14,31]. One possible exception is A. thaliana AgD, where a core Cys mutation reduces activity by less than 10-fold, which suggests a possible alternative role for the core Cys or a different nucleophile [32]. The role of the core His as an acid-base catalyst is also supported by structural and mutagenic studies in ADI [8,20,26], DDAH [9,23], PAD4 [15] and AGAT [33] Solvent isotope and pH rate profiling studies with PAD4 inactivators have suggested that proton donation is important for stabilization of the tetrahedral intermediate during inactivation, implying that a similar mechanism could occur during normal catalysis [34].
Computational studies with ADI have suggested that the catalytically active state of the enzyme with substrate bound consists of the Cys thiolate anion and His imidazolium cation [35]. However, there is evidence in some penteins, notably DDAH and ADI, that the core Cys is predominantly protonated in the resting state of the unliganded enzyme [27,35,36]. This Cys has an elevated pKa value of 9.6 in ADI [35], and 8.9 in DDAH [27] apoenzymes, yet ADI and DDAH remain active down to pH 5 and 6.1, respectively.
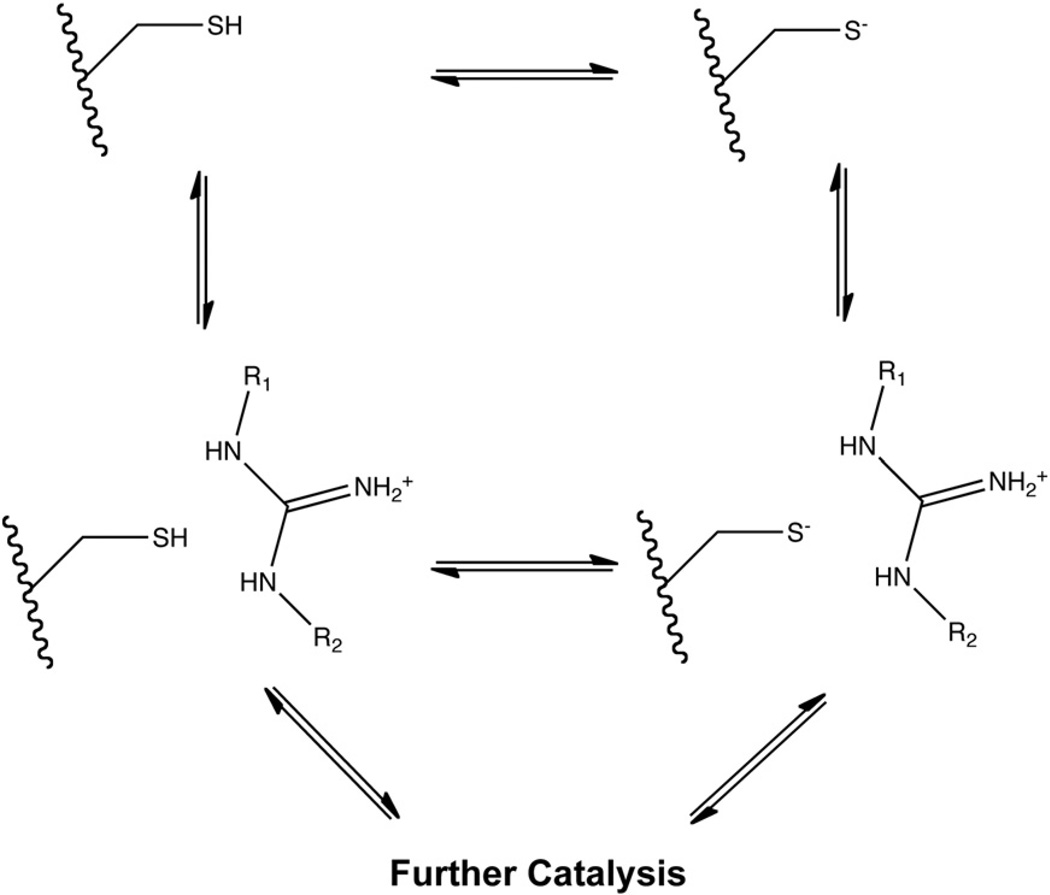
Three protonation state models for the core Cys are proposed to explain these results. In the first model, coined the “substrate-assisted” mechanism, the core Cys is predominantly protonated in free enzyme, but binding of protonated substrate lowers the pKa of the core Cys. The resulting deprotonation of the core Cys is supported by UV difference spectroscopy of DDAH with positively charged ligands [27]. In this case, binding of the positively charged guanidinium appears to shield the core Cys from the electrostatic effects of the nearby Asp side chains and induce deprotonation of the Cys into an active thiolate nucleophile, shifting its pKa to 6.9 in ADI and 6.1 in DDAH [27,35].
In the second model, only the minor fraction of enzyme with the core Cys in the thiolate form at physiological pH is active, while the major fraction in the thiol form is inactive. This is termed a reverse protonation mechanism. pKa measurements of the core Cys in PADs 1, 3 and 4 by inactivation with the neutral iodoacetamide and positively charged 2-chloroacetamidine suggest that positive charge does not significantly affect the pKa of the core Cys, altering it from 8.3 with iodoacetamide to 7.9 with 2-chloroacetamidine in PAD4, and from 8.8 to 9.5 in PAD1 [28,37]. In addition, a large inverse deuterium solvent isotope effect has been observed with PAD4 during both normal catalysis and haloacetamidine inactivation, consistent with existence of a thiolate-imidazolium ion pair [28,34]. These results suggest that at least some substrate binds to apoenzyme which contains a core Cys thiolate anion forming a salt bridge with the core His in its resting state, stabilizing these residues in their reactive forms. Interestingly, the overlap of the titration curves for these residues imply that only about 15% of the enzyme would exist in this active state at the pH optimum.
In the third model, the protonated thiol form of the core Cys is reactive. In this model, deprotonation of the core Cys occurs in concert with nucleophilic attack on the guanidine carbon. Computational studies with PAD4 have suggested that a protonated Cys is energetically more favorable than a Cys thiolate in the Michaelis complex [38,39]. In addition, the core Cys residue in AGAT is also proposed to be predominantly protonated in the resting state, possibly activated by Asp305 for attack [33]. These three proposals for Cys protonation state can be summarized and linked by a thermodynamic box (Fig. 6) [28], although the kinetically preferred pathway for the reaction of substrate with the core Cys has not yet been firmly established in any pentein.
Fig. 6.
Proposed protonation states of pentein core Cys.
4.2. Hydrolase mechanism
In the hydrolases, the Nω–Cζ bond of arginine substrates is cleaved to free the Nω substituent and form the S-alkylthiouronium adduct. The adduct is retained by the covalent link and positioned through bidentate interaction with the conserved lateral Asp residue. However, at this step in catalysis, the anterior carboxylate is generally proposed to make only a single interaction with the Nω of the substrate. In ADI, PAD and AgD, the switch of the anterior Asp from bidentate to monodentate interactions allows the second oxygen in the Asp’s carboxylate side chain to form a hydrogen bond to the hydrolytic water molecule. DDAH, which uses an anterior Glu residue to form a monodentate interaction with the substrate, maintains this interaction with the thiouronium adduct. A nearby Thr in Pseudomo-nas aeruginosa DDAH or Ser in human DDAH-1 instead uses its hydroxyl group to form a hydrogen bond with the nucleophilic water [16–18].
The core His, generally proposed to be in its neutral form following formation of the S-alkyl thiouronium adduct, activates the bound nucleophilic water by deprotonation. The resulting hydroxide ion attacks the thiouronium carbon to form a second tetrahedral adduct. This intermediate collapses, and the core Cys leaves to free an uriedo product and regenerate apoenzyme.
The essential role of the core His in activating water for hydrolysis of the covalent thiouronium has been demonstrated by mutagenesis. In ADI, mutation of the core His to Ala and incubation with l-arginine results in buildup of the thiouronium adduct, suggesting it is essential for nucleophilic attack by water [20]. A similar study with a core His mutant of DDAH and a modified substrate also showed buildup of the thiouronium adduct [9,23].
4.3. Amidinotransferase mechanism
Unlike the hydrolases which cleave the Nω—Cζ bond, the amidinotranferases cleave the Nδ—Cζ bond of arginine substrates to free the Nδ substituent and form an alternate S-alkylthiouronium adduct. This thiouronium contains only one carbon, the arginine substrate’s Cζ carbon. The amidinotransferases provide an example of how similar catalytic machinery can select for cleavage of a different N—C bond by reorienting the substrate.
The amidine donor substrate leaves as a free amino compound (e.g. l-ornithine in AGAT) following donation of the amidino group to the core Cys to form the thiouronium adduct. An amidine acceptor substrate (e.g. glycine in AGAT) then enters and binds to the active site, where its amino group is positioned for attack on the thiouronium. The amino group of the acceptor substrate is then deprotonated by the core His and attacks the thiouronium carbon in a manner analogous to the nucleophilic attack by water in the hydrolases. This presumably results in formation of a second tetrahedral adduct, which collapses to release the guanidine product and regenerate free enzyme.
A direct acceptor-to-donor amidine transfer mechanism has been ruled out in favor of a covalent intermediate due to ping pong kinetics [40,41], ligand-bound structures showing overlap between the arginine and glycine substrates [42,43], and intermediate trapping experiments [44,45].
It is notable that the thiouronium adduct formed during amidinotransferase catalysis appears to be significantly more stable against hydrolysis than the analogous intermediate formed during hydrolase catalysis. Despite the presence of the same core residues, the amidinotransferases can somehow stabilize the adduct until a suitable amidine acceptor substrate can be bound. One possible explanation is a subtle reorganization of the active site; in an overlay of the arginine-bound structures of ADI and AGAT, a slightly different orientation of the core residues relative to one another can be observed. Such reorganization is feasible because the active sites of both AGAT and AIAT are flexible, supported by the induced-fit mechanism those enzymes use for substrate binding [13,42]. However, the basis for the stability of the thiouronium adduct in the amidinotransferases has not yet been studied in detail.
4.4. Dihydrolase mechanism
In the dihydrolase reaction, the S-alkyl thiouronium adduct formation is followed by two hydrolysis reactions, which cleave all three C–N bonds in the guanidine and free the guanidine carbon as carbon dioxide or bicarbonate. The dihydrolases are not as well characterized as the hydrolases or amidinotranferases, and consequently only limited mechanistic information is available. Work with Pseudomonas cepacia and P. aeruginosa lysates supports the proposed role of SADH as a dihydrolase. These crude lysate studies demonstrated both consumption of Nα-succinylarginine and production of Nα-succinylornithine and two equivalents of ammonia [11,46]. This activity was abolished by disruption of the SADH gene [47], implying that SADH is responsible for the observed activity. Furthermore, purified SADH has been shown to produce ammonia from synthetic Nα-succinylarginine, and cocrystallization experiments with Nα-succinylarginine substrate have resulted in electron density matching the expected Nα-succinylornithine product [12]. SADH does not appear to recognize urea or Nα-succinylcitrulline as substrates in lysates [11]. No urea appears to be produced, but low levels of succinylcitrulline were identified in crude cell extracts, suggesting that it could be a reaction intermediate or byproduct [46].
Although SADH has not been mechanisticallycharacterizedindepth, it seems likely that SADH operates with a mode of catalysis similar to that of the other catalytic penteins, based on the presence of the core catalytic machinery in the crystal structure of SADH [12]. The active site of SADH contains a lateral Asn residue, in contrast to the lateral Asp residues that appear in the hydrolases and amidinotransferases. It has been proposed that this change alters the electrostatic environment of the Michaelis complex and any reaction intermediates, which may facilitate formation of a second covalent enzyme intermediate and attack by a second water molecule [12]. The presence of an Asn residue in the lateral position has been sufficient for some successful functional annotations of dihydrolase activity [7]. However, it may be oversimplistic to suggest that this single substitution is responsible for so large a change in catalytic mechanism without more detailed experimental evidence, as there are likely other differences that account for the mechanistic divergence of the dihydrolases.
5. Discussion
Apparent divergent evolution of enzyme superfamilies from a common promiscuous ancestor has been hypothesized in other mechanistically diverse superfamilies, especially those in which a portion of the catalytic mechanism (here, the formation of a thiouronium adduct) is conserved [48–53]. This hypothesis suggests that enzymes with promiscuous but not necessarily optimized activity serve as the ancestors of enzymes optimized for a single function. At least one member of the pentein superfamily, AIAT, displays a significant amount of substrate promiscuity [13]. In addition, catalytic promiscuity has also been observed in the pentein superfamily. For some examples, small amounts of hydrolysis have been observed in hog kidney AGAT in the absence of the acceptor substrate glycine. However, this hydrolysis activity is much slower (0.01-fold) than the corresponding amidinotransfer reaction [54]. In DDAH, the “activated” substrate S-methylthiocitrulline has been used in combination with a core His mutation to study the S-alkylthiouronium adduct [23] and its partitioning into amidinotransfer and hydrolysis reactions. Unlike the primary amino leaving group in the natural substrate Nω-dimethyl-l-arginine, the methanethiol leaving group of S-methylthiocitrulline does not require protonation by the core His in the first half reaction leading up to formation of the adduct. This adduct is chemically analogous to the one proposed for the penteins. Using DDAH as a catalyst, the adduct reacts with exogenous amines in a manner similar to the amidinotransferases, underscoring the catalytic similarity that exists among the penteins.
Covalent inactivation of Bacillus cereus ADI by l-canavanine results in buildup of an irreversible oxythiocarbamate adduct, the result of one additional hydrolysis of the oxythiouronium [55]. This oxythiocarbamate could be analogous to a potential thiocarbamate intermediate in the dihydrolase mechanism (not shown).
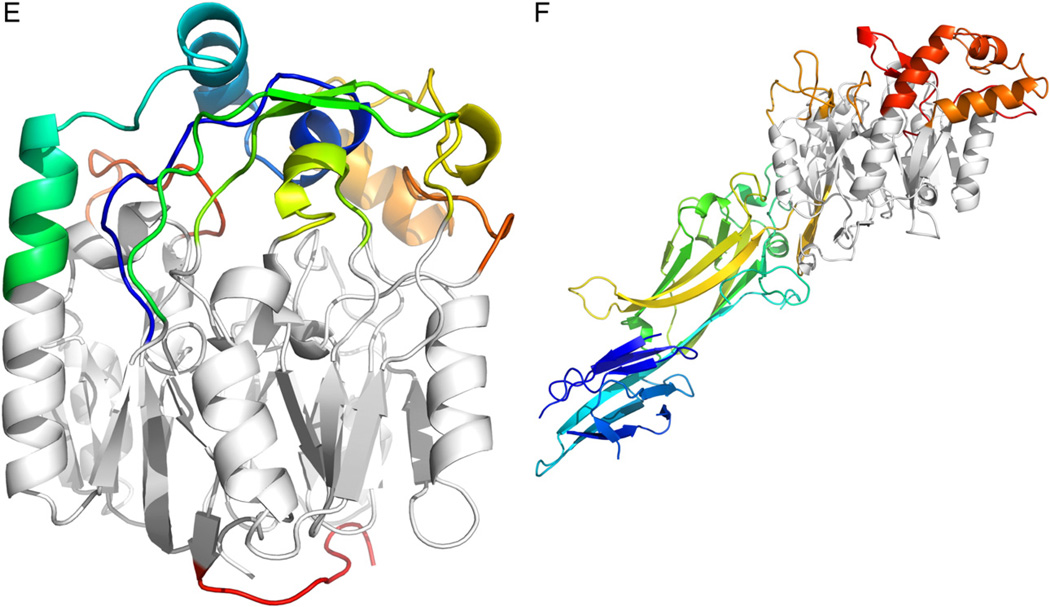
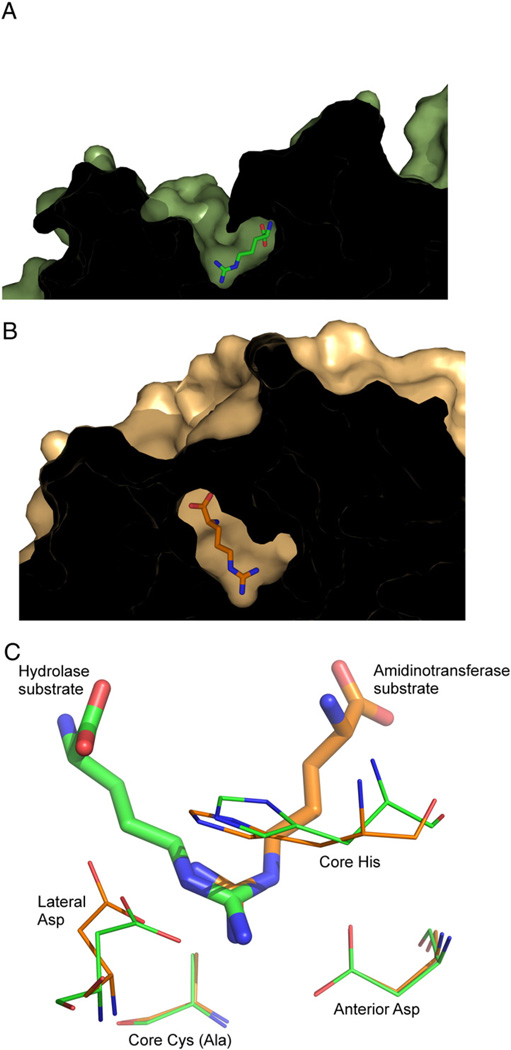
Finally, it is evident from the crystal structures that many penteins contain two channels leading to the active site (Fig. 7). The donor substrate for the amidinotransferases binds in a channel that is also present in the hydrolases but is believed in hydrolases to allow water access to the active site and provide an exit for the ammonia or methylamine leaving groups. It is possible that this channel in the hydrolases is a vestigial substrate binding channel for the amidinotransfer substrates that is retained from a common promiscuous ancestor.
Fig. 7.
Structures of A) hydrolase (Green) and B) amidinotransferase (Orange) active sites, and C) an overlay of the two active sites. The hydrolase is ADI from P. aeruginosa (PDB ID: 2a9g) and the amidinotransferase is human AGAT (PDBID: 4jdw). Substrate is positioned at different relative angles, presumable to facilitate cleavage of a different C–N bond. In both structures, the core Cys is mutated to Ala to enable formation of a stable substrate complex.
Together, the slow hydrolytic activity found in an amidinotransferase, the recapitulation of amidinotransfer activity in DDAH [23], and the two hydrolysis steps involved in canavanine’s inactivation of B. cereus ADI provide examples of catalytic promiscuity. This evidence is consistent with the hypothesis that an ancestral enzyme may have once fulfilled both hydrolase and amidinotransferase roles and evolved separately after a gene duplication event. In this scenario, the ancestor enzyme would have used the superimposable pentein catalytic core and conserved mechanistic features to form the thiouronium adduct upon reaction with guanidines, but accepted both amines and water as the second nucleophile. The low sequence similarity and various oligomerization states found in the superfamily suggest that the enzymes diverged far in the past [53], and the similar core machinery suggests a gene duplication event [48]. Following this proposed ancient gene duplication event, the promiscuous activity would have reduced as each enzyme evolved higher specificity for hydrolase, amidinotransferase or dihydrolase activity.
6. Conclusion
The pentein superfamily is a mechanistically diverse superfamily containing enzymes that modify guanidines. They display hydrolase, dihydrolase and amidinotransferase activities, but share common structural and catalytic features. They possess the same β/α propeller structure and although they use different residues to recognize a wide variety of substrates, they contain a similar set of conserved catalytic core residues. All known catalytic penteins begin the reaction with nucleophilic attack by the core Cys residue, assisted by the core His as a general acid catalyst, to form a thiouronium adduct, which is followed in the different catalytic families by breakage of one or more of the C—N guanidine bonds. Although the amidinotransferases and hydrolases cleave different C—N bonds, they are able to reuse the same catalytic machinery by reorienting the substrate by approximately 120°. The mechanistic information that has been gathered on the penteins is consistent with proposing their divergent evolution from a catalytically promiscuous ancestor [48]. A further understanding of how the various activities present in the catalytic penteins have evolved will not only be useful for learning about the history of the superfamily, but will also provide new insight for drug discovery and directed enzyme evolution of novel enzyme activities.
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (GM69754), the Robert A. Welch Foundation (F-1572), a seed grant from the Texas Institute for Drug and Diagnostic Development (Welch Foundation Grant No. H-F-0032), and fellow-shipsto Thomas Linsky from the Welch Foundation and the University of Texas Graduate School.
Abbreviations
- ADI
arginine deiminase
- DDAH
dimethylarginine dimethylaminohydrolase
- AgD
arginine deiminase
- PAD
peptidylarginine deiminase
- AGAT
arginine:glycine amidinotransferase
- AIAT
arginine:inosamine-phosphate amidinotransferase
- SADH
Nα-succinylarginine dihydrolase
References
- 1.Groft CM, Beckmann R, Sali A, Burley SK. Crystal structures of ribosome anti-association factor IF6. Nature Structural & Molecular Biology. 2000 Dec;7:1156–1164. doi: 10.1038/82017. [DOI] [PubMed] [Google Scholar]
- 2.Paoli M. An elusive propeller-like fold. Nature Structural Biology. 2001 Sep;8:744–745. doi: 10.1038/nsb0901-744. [DOI] [PubMed] [Google Scholar]
- 3.Teichmann SA, Murzin AG, Chothia C, Determination of protein function. evolution and interactions by structural genomics. Current Opinion in Structural Biology. 2001 Jun;11:354–363. doi: 10.1016/s0959-440x(00)00215-3. [DOI] [PubMed] [Google Scholar]
- 4.Hartzoulakis B, Rossiter S, Gill H, O’Hara B, Steinke E, Gane PJ, Hurtado-Guerrero R, Leiper JM, Vallance P, Rust JM, Selwood DL. Discovery of inhibitors of the pentein superfamily protein dimethylarginine dimethylaminohydrolase (DDAH) by virtual screening and hit analysis. Bioorganic & Medicinal Chemistry Letters. 2007 Jul;17:3953–3956. doi: 10.1016/j.bmcl.2007.04.095. [DOI] [PubMed] [Google Scholar]
- 5.Groft CM, Beckmann R, Sali A, Burley SK. Crystal structures of ribosome anti-association factor IF6. Nature Structural Biology. 2000 Dec;7:1156–1164. doi: 10.1038/82017. [DOI] [PubMed] [Google Scholar]
- 6.Gartmann M, Blau M, Armache J, Mielke T, Topf M, Beckmann R. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. The Journal of Biological Chemistry. 2010 May;285:14848–14851. doi: 10.1074/jbc.C109.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirai H, Mokrab Y, Mizuguchi K. The guanidino-group modifying enzymes: structural basis for their diversity and commonality. Proteins. 2006 Sep;64:1010–1023. doi: 10.1002/prot.20863. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, Li L, Wu R, Feng X, Li Z, Yang H, Wang C, Guo H, Galkin A, Herzberg O, Mariano PS, Martin BM, Dunaway-Mariano D. Kinetic analysis of Pseudomonas aeruginosa arginine deiminase mutants and alternate substrates provides insight into structural determinants of function. Biochemistry. 2006 Jan;45:1162–1172. doi: 10.1021/bi051591e. [DOI] [PubMed] [Google Scholar]
- 9.Stone EM, Person MD, Costello NJ, Fast W. Characterization of a transient covalent adduct formed during dimethylarginine dimethylaminohydrolase catalysis. Biochemistry. 2005 May;44:7069–7078. doi: 10.1021/bi047407r. [DOI] [PubMed] [Google Scholar]
- 10.Hong L, Fast W. Inhibition of human dimethylarginine dimethylaminohydrolase-1 by S-nitroso-L-homocysteine hydrogen Analysis peroxide, quantification and implications for hyperhomocysteinemia. The Journal of Biological Chemistry. 2007 Nov;282:34684–34692. doi: 10.1074/jbc.M707231200. [DOI] [PubMed] [Google Scholar]
- 11.Jann A, Stalon V, Wauven CV, Leisinger T, Haas D. N-Succinylated intermediates in an arginine catabolic pathway of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 1986 Jul;83:4937–4941. doi: 10.1073/pnas.83.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tocilj A, Schrag JD, Li Y, Schneider BL, Reitzer L, Matte A, Cygler M. Crystal structure of N-succinylarginine dihydrolase AstB bound to substrate, product an enzyme from the arginine catabolic pathway of Escherichia coli. The Journal of Biological Chemistry. 2005 Apr;280:15800–15808. doi: 10.1074/jbc.M413833200. [DOI] [PubMed] [Google Scholar]
- 13.Fritsche E, Bergner A, Humm A, Piepersberg W, Huber R. Crystal structure of L-arginine:inosamine-phosphate amidinotransferase StrB1 from Streptomyces griseus: an enzyme involved in streptomycin biosynthesis. Biochemistry. 1998 Dec;37:17664–17672. doi: 10.1021/bi981949p. [DOI] [PubMed] [Google Scholar]
- 14.Jones JE, Causey CP, Lovelace L, Knuckley B, Flick H, Lebioda L, Thompson PR. Characterization and inactivation of an agmatine deiminase from Helicobacter pylori. Bioorganic Chemistry. 2010 Apr;38:62–73. doi: 10.1016/j.bioorg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arita K, Hashimoto H, Shimizu T, Nakashima K, Yamada M, Sato M. Structural basis for Ca(2+)-induced activation of human PAD4. Nature Structural & Molecular Biology. 2004 Aug;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 16.Leiper J, Nandi M, Torondel B, Murray-Rust J, Malaki M, O’Hara B, Rossiter S, Anthony S, Madhani M, Selwood D, Smith C, Wojciak-Stothard B, Rudiger A, Stidwill R, McDonald NQ, Vallance P. Disruption of methylarginine metabolism impairs vascular homeostasis. Nature Medicine. 2007 Feb;13:198–203. doi: 10.1038/nm1543. [DOI] [PubMed] [Google Scholar]
- 17.Frey D, Braun O, Briand C, Vasák M, Grütter MG. Structure. Vol. 14. London England: 1993: May, 2006. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors; pp. 901–911. [DOI] [PubMed] [Google Scholar]
- 18.Murray-Rust J, Leiper J, McAlister M, Phelan J, Tilley S, Santa Maria J, Vallance P, McDonald N. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase. Nature Structural Biology. 2001 Aug;8:679–683. doi: 10.1038/90387. [DOI] [PubMed] [Google Scholar]
- 19.Das K, Butler GH, Kwiatkowski V, Clark AD, Yadav P, Arnold E. Structure. Vol. 12. London England: 1993: Apr, 2004. Crystal structures of arginine deiminase with covalent reaction intermediates; implications for catalytic mechanism; pp. 657–667. [DOI] [PubMed] [Google Scholar]
- 20.Galkin A, Lu X, Dunaway-Mariano D, Herzberg O. Crystal structures representing the Michaelis complex and the thiouronium reaction intermediate of Pseudomonas aeruginosa arginine deiminase. The Journal of Biological Chemistry. 2005 Oct;280:34080–34087. doi: 10.1074/jbc.M505471200. [DOI] [PubMed] [Google Scholar]
- 21.Llácer JL, Polo LM, Tavárez S, Alarcón B, Hilario R, Rubio V. The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine transcarbamylase and agmatine deiminase and a snapshot of agmatine deiminase catalyzing its reaction. Journal of Bacteriology. 2007 Feb;189:1254–1265. doi: 10.1128/JB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Arita K, Bhatia M, Knuckley B, Lee Y, Stallcup MR, Sato M, Thompson PR. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006 Oct;45:11727–11736. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linsky TW, Monzingo AF, Stone EM, Robertus JD, Fast W. Promiscuous partitioning of a covalent intermediate common in the pentein superfamily. Chemistry & Biology. 2008 May;15:467–475. doi: 10.1016/j.chembiol.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Monzingo AF, Hu S, Schaller TH, Robertus JD, Fast W. Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide. Biochemistry. 2009 Sep;48:8624–8635. doi: 10.1021/bi9007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu X, Galkin A, Herzberg O, Dunaway-Mariano D. Arginine deiminase uses an active-site cysteine in nucleophilic catalysis of L-arginine hydrolysis. Journal of the American Chemical Society. 2004 May;126:5374–5375. doi: 10.1021/ja049543p. [DOI] [PubMed] [Google Scholar]
- 26.Galkin A, Kulakova L, Sarikaya E, Lim K, Howard A, Herzberg O, Structural insight into arginine degradation by arginine deiminase. an antibacterial and parasite drug target. The Journal of Biological Chemistry. 2004 Apr;279:14001–14008. doi: 10.1074/jbc.M313410200. [DOI] [PubMed] [Google Scholar]
- 27.Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of dimethylargininase (DDAH) from Pseudo-monas aeruginosa. Biochemistry. 2006 May;45:5618–5630. doi: 10.1021/bi052595m. [DOI] [PubMed] [Google Scholar]
- 28.Knuckley B, Bhatia M, Thompson PR. Protein arginine deiminase 4: evidence for a reverse protonation mechanism. Biochemistry. 2007 Jun;46:6578–6587. doi: 10.1021/bi700095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005 Aug;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 30.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Haloacetamidine-based inactivators of protein arginine deiminase 4 (PAD4): evidence that general acid catalysis promotes efficient inactivation. Chembiochem: A European Journal of Chemical Biology. 2010 Jan;11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llácer JL, Polo LM, Tavárez S, Alarcón B, Hilario R, Rubio V. The gene cluster for agmatine catabolism of Enterococcus faecalis: study of recombinant putrescine transcarbamylase and agmatine deiminase and a snapshot of agmatine deiminase catalyzing its reaction. Journal of Bacteriology. 2007 Feb;189:1254–1265. doi: 10.1128/JB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janowitz T, Kneifel H, Piotrowski M. Identification characterization of plant agmatine iminohydrolase the last missing link in polyamine biosynthesis of plants. FEBS Letters. 2003 Jun;544:258–261. doi: 10.1016/s0014-5793(03)00515-5. [DOI] [PubMed] [Google Scholar]
- 33.Fritsche E, Humm A, Huber R. Substrate binding and catalysis by L-arginine: glycine amidinotransferase—a mutagenesis and crystallographic study. European Journal of Biochemistry / FEBS. 1997 Jul;247:483–490. doi: 10.1111/j.1432-1033.1997.00483.x. [DOI] [PubMed] [Google Scholar]
- 34.Knuckley B, Causey CP, Pellechia PJ, Cook PF, Thompson PR. Haloacetamidine-based inactivators of protein arginine deiminase 4 (PAD4): evidence that general acid catalysis promotes efficient inactivation. Chembiochem: A European Journal of Chemical Biology. 2010 Jan;11:161–165. doi: 10.1002/cbic.200900698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Li Z, Wang C, Xu D, Mariano PS, Guo H, Dunaway-Mariano D. The electrostatic driving force for nucleophilic catalysis in L-arginine deiminase: a combined experimental and theoretical study. Biochemistry. 2008 Apr;47:4721–4732. doi: 10.1021/bi7023496. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Kulakova L, Li L, Galkin A, Zhao Z, Nash TE, Mariano PS, Herzberg O, Dunaway-Mariano D. Mechanisms of catalysis and inhibition operative in the arginine deiminase from the human pathogen Giardia lamblia. Bioorganic Chemistry. 2009 Oct;37:149–161. doi: 10.1016/j.bioorg.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity kinetic studies of PADs 1, 3 and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010 Jun;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke Z, Wang S, Xie D, Zhang Y. Born-Oppenheimer ab initio QM/MM molecular dynamics simulations of the hydrolysis reaction catalyzed by protein arginine deiminase 4. The Journal of Physical Chemistry. B. 2009 Dec;113:16705–16710. doi: 10.1021/jp9080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ke Z, Zhou Y, Hu P, Wang S, Xie D, Zhang Y. Active site cysteine is protonated in the PAD4 Michaelis complex: evidence from Born–Oppenheimer ab initio QM/ MM molecular dynamics simulations. The Journal of Physical Chemistry. 2009;B 113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGuire DM, Tormanen CD, Segal IS, Van Pilsum JF. The effect of growth hormone and thyroxine on the amount of L-arginine:glycine amidinotransferase in kidneys of hypophysectomized rats. Purification and some properties of rat kidney transamidinase. The Journal of Biological Chemistry. 1980 Feb;255:1152–1159. [PubMed] [Google Scholar]
- 41.Ronca G, Vigi V, Grazi E. Transamidinase of hog kidney: V. Kinetic studies. The Journal of Biological Chemistry. 1966 Jun;241:2589–2595. [PubMed] [Google Scholar]
- 42.Humm A, Fritsche E, Steinbacher S, Huber R. Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. The EMBO Journal. 1997 Jun;16:3373–3385. doi: 10.1093/emboj/16.12.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humm A, Fritsche E, Steinbacher S. Structure and reaction mechanism of L-arginine:glycine amidinotransferase. Biological Chemistry. 1997 Apr;378:193–197. [PubMed] [Google Scholar]
- 44.Walker JB. Arginine-ornithine transamidination in kidney. The Journal of Biological Chemistry. 1956 Aug;221:771–776. [PubMed] [Google Scholar]
- 45.Grazi E, Rossi N. Transamidinase of hog kidney: VII. Cysteine at the amidine-binding site. The Journal of Biological Chemistry. 1968 Feb;243:538–542. [PubMed] [Google Scholar]
- 46.Vander Wauven C, Stalon V. Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. Journal of Bacteriology. 1985 Nov;164:882–886. doi: 10.1128/jb.164.2.882-886.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider BL, Kiupakis AK, Reitzer LJ. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. Journal of Bacteriology. 1998 Aug;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chemistry & Biology. 1999 Apr;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- 49.Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Archives of Biochemistry and Biophysics. 2005 Jan;433:59–70. doi: 10.1016/j.abb.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 50.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annual Review of Biochemistry. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 51.Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Current Opinion in Chemical Biology. 2006 Oct;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Current Opinion in Chemical Biology. 2006 Oct;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Poelarends GJ, Veetil VP, Whitman CP. The chemical versatility of the beta-alpha-beta fold: catalytic promiscuity and divergent evolution in the tautomerase superfamily. Cellular and Molecular Life Sciences: CMLS. 2008 Nov;65:3606–3618. doi: 10.1007/s00018-008-8285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratner S, Rochovansky O. Biosynthesis of guanidinoacetic acid: I. Purification and properties of transamidinase. Archives of Biochemistry and Biophysics. 1956;63:277–295. doi: 10.1016/0003-9861(56)90044-3. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Li Z, Chen D, Lu X, Feng X, Wright EC, Solberg NO, Dunaway-Mariano D, Mariano PS, Galkin A, Kulakova L, Herzberg O, Green-Church KB, Zhang L. Inactivation of microbial arginine deiminases by L-canavanine. Journal of the American Chemical Society. 2008 Feb;130:1918–1931. doi: 10.1021/ja0760877. [DOI] [PubMed] [Google Scholar]