Significance
Among mammals, only primates can accumulate high levels of xanthophyll carotenoids in their retinas. We discovered that, unlike most other mammals, the major xanthophyll cleavage enzyme β,β-carotene-9′,10′-dioxygenase (BCO2) is inactive in humans, explaining the unique accumulation of lutein, zeaxanthin, and meso-zeaxanthin in primate macula. We confirmed this discovery by demonstrating that BCO2 knockout mice accumulate zeaxanthin in their retinas.
Abstract
The macula of the primate retina uniquely concentrates high amounts of the xanthophyll carotenoids lutein, zeaxanthin, and meso-zeaxanthin, but the underlying biochemical mechanisms for this spatial- and species-specific localization have not been fully elucidated. For example, despite abundant retinal levels in mice and primates of a binding protein for zeaxanthin and meso-zeaxanthin, the pi isoform of glutathione S-transferase (GSTP1), only human and monkey retinas naturally contain detectable levels of these carotenoids. We therefore investigated whether or not differences in expression, localization, and activity between mouse and primate carotenoid metabolic enzymes could account for this species-specific difference in retinal accumulation. We focused on β,β-carotene-9′,10′-dioxygenase (BCO2, also known as BCDO2), the only known mammalian xanthophyll cleavage enzyme. RT-PCR, Western blot analysis, and immunohistochemistry (IHC) confirmed that BCO2 is expressed in both mouse and primate retinas. Cotransfection of expression plasmids of human or mouse BCO2 into Escherichia coli strains engineered to produce zeaxanthin demonstrated that only mouse BCO2 is an active zeaxanthin cleavage enzyme. Surface plasmon resonance (SPR) binding studies showed that the binding affinities between human BCO2 and lutein, zeaxanthin, and meso-zeaxanthin are 10- to 40-fold weaker than those for mouse BCO2, implying that ineffective capture of carotenoids by human BCO2 prevents cleavage of xanthophyll carotenoids. Moreover, BCO2 knockout mice, unlike WT mice, accumulate zeaxanthin in their retinas. Our results provide a novel explanation for how primates uniquely concentrate xanthophyll carotenoids at high levels in retinal tissue.
The macular pigment of the human retina consists of the xanthophyll carotenoids, lutein, zeaxanthin, and their metabolite, meso-zeaxanthin (1–3). Macular pigments are highly concentrated in the foveal area of the human retina. Their highest concentration can reach 1 mM, making the macula a visible yellow spot at the center of the human retina. These xanthophylls are known to play antioxidant and light-screening roles in nature (4–6). Clinical studies have shown that lutein and zeaxanthin supplementation can reduce the risk of age-related macular degeneration (AMD), a leading cause of blindness in the developed world (4, 7, 8). Recently, the Age-Related Eye Disease Study 2 (AREDS2) showed that daily oral supplementation with 10 mg lutein and 2 mg zeaxanthin was associated with 10–18% reduced progression to advanced AMD over the 5-y study relative to original Age-Related Eye Disease Study (AREDS) formulations without lutein and zeaxanthin, and the authors recommended adding lutein and zeaxanthin to replace β-carotene in the formulation for safety and efficacy reasons (9, 10).
The spatially and chemically specific accumulation of high concentrations of xanthophylls in the retina is a unique feature of the primate retina relative to other mammals, but the biochemical basis for this high degree of specificity is unclear. High-affinity binding proteins might drive this process. Mice and humans express a zeaxanthin-binding protein, the pi isoform of glutathione S-transferase (GSTP1), abundantly in the retina, yet only the human retina naturally contains zeaxanthin and meso-zeaxanthin (11, 12). Thus, other biochemical mechanisms underlying accumulation of the human macular pigment need to be considered.
β,β-Carotene-9′,10′-dioxygenase (BCO2, also known as BCDO2) is a xanthophyll carotenoid cleavage enzyme which can cleave carotenoids in many animals eccentrically at the 9–10 or 9′-10′ carbon–carbon double bonds into C13- and C27-apocarotenoids, or further into C14-dialdehyde (13, 14). Its physiological function is supported by genetic evidence from several species. A nonsense mutation in the sheep BCO2 gene that introduces a stop codon at amino acid 66 of the 575-aa full-length protein is strongly associated with a yellow fat phenotype due to the high amount of lutein that deposits in tissue (15). Cis-acting and tissue-specific regulatory mutations that inhibit the expression of BCO2 in domestic chickens can cause them to have yellow skin secondary to carotenoid accumulation (16). Genetic variations in the BCO2 gene have been reported to increase carotenoid levels in bovine adipose tissue and milk (17). In vitro biochemical studies have also demonstrated that ferret BCO2 can cleave lutein and zeaxanthin (13).
BCO2, β,β-carotene-15,15′-monooxygenase (BCO1), and retinal pigment epithelium-specific 65 kDa protein (RPE65) are members of the polyene oxygenase gene family. BCO1, which is also known as BCMO1, cleaves β-carotene symmetrically at the 15–15′ position to yield two molecules of retinal, the key pigment in the visual cycle, and it is inactive with xanthophyll (oxygenated) carotenoids such as lutein and zeaxanthin (18, 19). RPE65, an essential enzyme in the visual cycle, participates in the conversion of all-trans-retinyl palmitate to 11-cis-retinol, and various mutations are associated with Leber congenital amaurosis type 2 and retinitis pigmentosa (18, 20–22). BCO2 may also play important roles in ocular tissues, but a physiological function has not been established. Based on the fact that high amounts of lutein and zeaxanthin exist in the human macula, we hypothesized that either human BCO2 is not expressed in human retina or it is an inactive carotenoid cleavage enzyme. In this study, we tested these proposed hypotheses by investigating the expression, distribution, and activity of BCO2 in human, monkey, and mouse retina.
Results
Expression and Localization of BCO2 in Mammalian Retina.
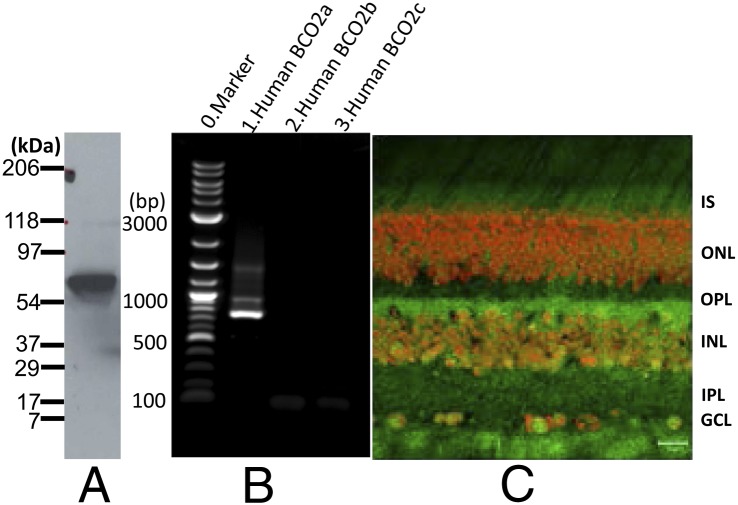
To determine if BCO2 is expressed in the human retina, Western blots were performed using an anti-BCO2 antibody. A ∼63-kDa polypeptide was detected in the total protein preparation of human retina (Fig. 1A). The human BCO2 gene expresses five protein splice variants: a, b, c, d and e, which are composed of 579, 545, 539, 506, and 474 aa, respectively [National Center for Biotechnology Information (NCBI) database] that differ only at the N terminus, and all of which, except for isoform d, preserve the enzymes’ active sites. According to the mobility of BCO2 revealed on Western blot, isoforms a, b, and c are the most likely candidates. RT-PCR detected the mRNA of isoform a but not b or c (Fig. 1B). By immunohistochemistry (IHC), BCO2 is localized throughout the monkey retina, especially in the inner segments, inner retina, and ganglion cells (Fig. 1C). We also examined BCO2’s expression in mouse retina and identified a ∼63-kDa polypeptide (Fig. 2A) on a Western blot with mouse retina lysates. A single ∼640-bp amplicon (Fig. 2B) suggested that mouse retina expresses only one BCO2 isoform. As observed in monkey retina, IHC results showed that mouse BCO2 is expressed throughout the retina and retinal pigment epithelium (RPE) (Fig. 2C).
Fig. 1.
Expression of BCO2 in the human retina and localization in monkey retina. (A) Western blot of human retina lysates probed with anti-BCO2. The BCO2 polypeptide has a mobility of ∼65 kDa [calculated molecular weight (MW) = 65,674 for the BCO2a isoform]. (B) RT-PCR with human retina cDNA and isoform-specific oligonucleotide primers. Human BCO2 isoform a (1) but not isoforms b (2) or c (3) could be identified. (C) IHC of perfusion-fixed monkey retina cryosection probed with antibody directed against BCO2 (green). Propidium iodide (red) permits visualization of nuclear layers. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; IS, inner segment layer; ONL, outer nuclear layer; OPL, outer plexiform layer. Scale bar: 20 µm.
Fig. 2.
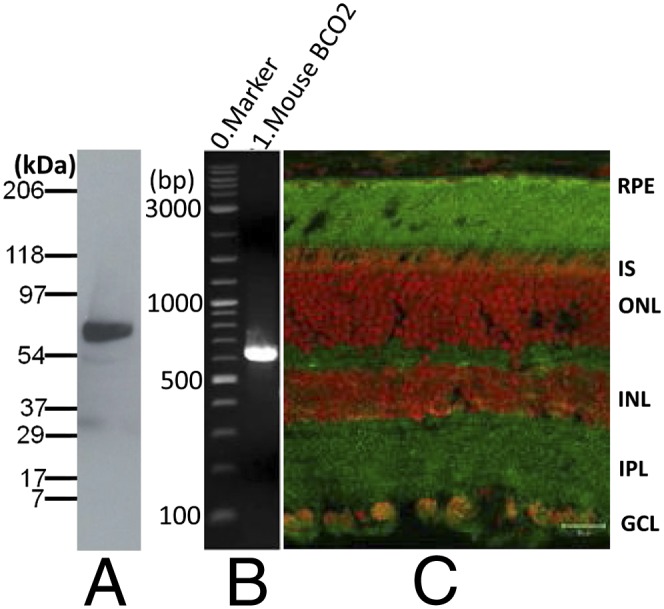
Expression and localization of BCO2 in the mouse retina. (A) Western blot of mouse retina lysate with anti-mouse BCO2 antibody. The mobility of the mouse BCO2 polypeptides is ∼60 kDa (calculated MW = 60,142). (B) RT-PCR with mouse BCO2-specific primers and mouse retina cDNA. (C) IHC of albino mouse retina cryosection probed with polyclonal anti-BCO2 antibody (green) and contrasted with propidium iodide (red) which binds nucleic acids. Labeling of retina and RPE is specific, as it was absent in the negative control (omission of primary antibody). Scale bar: 20 µm.
Carotenoid Cleavage Assays.
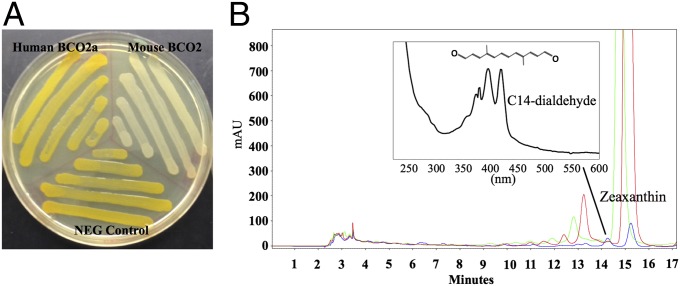
Next, we investigated the enzyme activity of human and mouse BCO2 using a cell-based carotenoid cleavage assay. These assays have been used to test the activity of many carotenoid oxygenases with zeaxanthin, β-carotene, and lycopene (23–28). In our first experiments, a plasmid of various BCO2 constructs was transfected into a bacterium that had been engineered to carry a zeaxanthin synthesis plasmid. If following transfection, the bacterium remains yellow in color, it indicates that the enzyme is inactive; if the carotenoid coloration disappears, it suggests that the enzyme is a functional enzyme. The yellow color of zeaxanthin was faded in the bacteria transfected with mouse BCO2 plasmids (Fig. 3A), demonstrating that mouse BCO2 can cleave zeaxanthin. Human BCO2 isoform a (human BCO2a) failed to cleave zeaxanthin, retaining the yellow color. The cleavage products of each assay were then analyzed by HPLC (Fig. 3B). Chromatograms of the human BCO2 experiments were identical to those of the control experiments, consistent with absence of cleavage activity. In the chromatogram of the mouse BCO2 experiment, the elution peak of zeaxanthin almost disappeared, whereas a new peak appeared at 14.22 min whose diode-array spectrum was consistent with C14-dialdehyde (also known as rosafluene dialdehyde; Fig. 3B, Inset), a previously reported major cleavage product of zeaxanthin by BCO2 (14). These experiments demonstrate that human retinal BCO2 is an inactive enzyme, whereas mouse BCO2 is an active carotenoid cleavage enzyme.
Fig. 3.
Enzymatic activity assays of human and mouse BCO2. (A) Expression of human, mouse BCO2 cDNA, or empty expression vector in E. coli strains engineered for zeaxanthin production. Yellow color indicates absence of enzymatic cleavage activity. White color demonstrates cleavage of zeaxanthin. (B) HPLC analysis on a cyano column of extracts from E. coli engineered to synthesize zeaxanthin which had been cotransformed with BCO2 or control vectors (blue trace, mouse BCO2; red trace, human BCO2a; green trace, control; Inset, absorption spectra of a cleavage product). The identity of zeaxanthin was confirmed by comparison with PDA spectra and by coelution with authentic standard. The PDA spectrum of the peak labeled as C14-dialdehyde (rosafluene dialdehyde) was consistent with a previous report of a major cleavage product of zeaxanthin by mouse BCO2 (14). Retention times of the late eluting peaks are slightly offset due to variations in HPLC chromatographic conditions.
Protein Sequence and Structure Analysis.
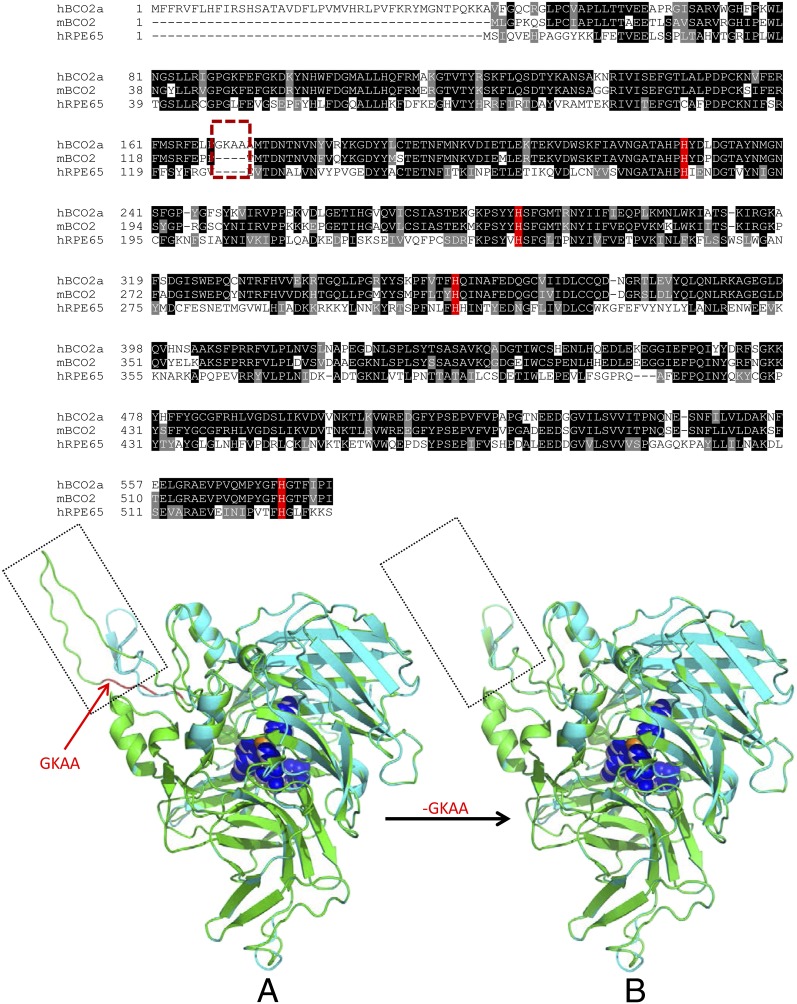
To explain the functional difference between human BCO2 and mouse BCO2, we analyzed the amino acid sequences of human BCO2a and mouse BCO2. Sequence alignments (Fig. 4, Upper) show that the major difference between human BCO2a enzyme and mouse BCO2 is the presence of 4 aa residues, GKAA, in human BCO2 (Fig. 4), suggesting the loss of an alternate splice site in the human gene. An NCBI protein database search revealed that this GKAA insertion is apparently unique to primates and is absent in cows, sheep, rats, mice, and ferrets. GKAA is also absent in human and mouse RPE65 that can actively cleave all-trans-retinyl esters. Inspecting the human and mouse BCO2 genes reveals that GKAA represents an extension of human BCO2 exon 3 caused by use of an alternate donor splice site (Fig. S1). Deletion of GKAA between P125 and M127 and replacement of A126 by T are expected to induce a substantial change in structure in this area, but no significant difference is found at their enzymatic cleavage domains, as all four key histidine residues are conserved (shaded red in Fig. 4, Upper). We also compared the 3D homology structure of human BCO2a with that of mouse BCO2 using RPE65 crystal structure (PDB 3fsn.1.a) as a template, and we confirmed that there are no significant differences between human BCO2a and mouse BCO2 cleavage domains (Fig. 4, Lower). As expected, the GKAA deletion in the N-terminal half of human BCO2 caused a structural rearrangement (boxes in Fig. 4, Lower). Next, the activity of human BCO2 enzyme with its GKAA sequence deleted was tested using the cell-based carotenoid cleavage assay to determine if enzymatic activity could be restored. The modified expression vector of human BCO2 in which the nucleotides encoding GKAA were deleted was transfected into Escherichia coli carrying a zeaxanthin synthesis plasmid. These bacteria retained their yellow color, indicating that simple deletion of GKAA cannot convert human BCO2 into an active carotenoid cleavage enzyme. Replacing the N-terminal 197 aa of human BCO2a with the N-terminal 150 aa of mouse BCO2 or substitution of a wide range of amino acid residues of human BCO2 with the corresponding residues of mouse BCO2 could not restore human BCO2’s cleavage activity (Fig. S2 and Table S1). Conversely, insertion of GKAA into mouse BCO2 inactivated the enzyme, demonstrating that the GKAA insertion alone was sufficient to inactivate mouse BCO2. Further studies in which we combined deletion of GKAA from human BCO2 with several key amino acid substitutions selected from the mouse BCO2 sequence still could not rescue human BCO2’s function, implying that there has been considerable genetic drift since primate BCO2 first acquired the GKAA insertion and lost its xanthophyll cleavage function.
Fig. 4.
Sequence and structure alignment of human and mouse BCO2. (Upper) Alignment of human BCO2 isoform a, mouse BCO2, and human RPE65 amino acid sequences. Histidine residues that coordinate the iron required for activity in RPE65 are shaded red. The hatched box denotes the deletion of GKAA in mouse BCO2. (Lower) Comparison of 3D structures of (A) human BCO2a (green) and mouse BCO2 (cyan), and (B) chimeric human BCO2 (green) and mouse BCO2 (cyan). The boxes identify the structural differences between human BCO2a and mouse BCO2. Histidine residues (blue in human, white in mouse BCO2) coordinating iron (orange) in human and mouse BCO2s are shown as spheres in the center of BCO2.
Carotenoid-Binding Affinity Measurements by Surface Plasmon Resonance.
The 3D protein structure of an apocarotenoid cleavage oxygenase from the cyanobacterium Synechocystis sp. PCC6803 demonstrates that a substrate-binding tunnel is present near the cleavage domain (29). Without access to the substrate-binding tunnel, carotenoids cannot be cleaved. This led us to investigate whether or not there is a difference in carotenoid-binding affinities between human and mouse BCO2 by surface plasmon resonance (SPR). Fig. S3 shows the recombinant proteins used for SPR experiments. Table 1 and Fig. S4 show that human BCO2a has much weaker binding affinity for xanthophylls than mouse BCO2. For example, the KD of meso-zeaxanthin to human BCO2a is around 40-fold higher than that of mouse BCO2 and even human serum albumin (HSA). These data demonstrate that the binding affinities between xanthophylls and mouse BCO2 are stronger than those associated with human BCO2a, implying that weak binding of carotenoids may be the reason why human BCO2 has lost its cleavage function.
Table 1.
Equilibrium dissociation constant (KD) determined by SPR in micromolars
| Carortenoids | Human BCO2a | Mouse BCO2 | HSA |
| Lutein | 25.00 ± 2.00 | 1.17 ± 0.02 | 2.40 ± 0.20 |
| Zeaxanthin | 19.00 ± 2.00 | 1.39 ± 0.03 | 2.20 ± 0.20 |
| Meso-zeaxanthin | 46.00 ± 4.00 | 1.10 ± 0.02 | 4.50 ± 0.40 |
Detection of Zeaxanthin in the Retina of BCO2 Knockout Mice.
Finally, we tested if induced inactivity of BCO2 can cause the retinal accumulation of xanthophyll carotenoids by feeding zeaxanthin to BCO2 knockout (BCO2−/−) mice. Carotenoid contents were extracted from the retinas, RPE/choroids, and lenses of BCO2−/− mice, respectively, and zeaxanthin was identified according to zeaxanthin standard’s retention time and UV–visible spectrum from HPLC-photodiode array (PDA) detectors on two different chromatographic column systems (cyano and chiral) and by mass spectrometry. Fig. 5 shows that 0.66 ng zeaxanthin were detected per BCO2−/− mouse’s retinas, whereas zeaxanthin cannot be detected by HPLC in the retina of WT mice, demonstrating that BCO2 is a key enzyme for preventing xanthophyll accumulation in most mammalian retinas. Zeaxanthin levels in RPE/choroid of BCO2−/− mice are around three times higher than that of the WT mice. No zeaxanthin was detected in the lenses of BCO2−/− mice or WT mice.
Fig. 5.
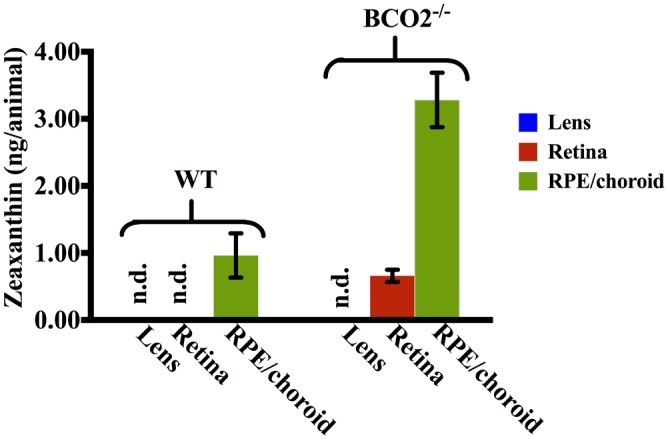
Zeaxanthin levels in the ocular tissues of BCO2−/− and WT mice. Zeaxanthin contents in lens (blue), retina (red), and RPE/choroid (green) were measured by normal-phase HPLC. n.d., not detectable. Each experiment was repeated five times on batches of five mice. Error bars represent SDs.
Discussion
The yellow pigment of the human retina’s macula lutea (yellow spot) was first noted by anatomists over 200 y ago (30), and the pigment was chemically identified to be lutein, zeaxanthin, and meso-zeaxanthin about 30 y ago (1). Many clinical studies have already indicated that lutein and zeaxanthin supplementation may reduce the risk of AMD (4, 7–10); however, the protective mechanisms and physiological functions of the macular carotenoid pigments have generally been studied only in vitro because no other nonprimate mammals accumulate carotenoids at high levels in their retinas (31). Birds, amphibians, and reptiles accumulate their ocular carotenoids through a distinct mechanism involving xanthophyll esterification to fatty acids. In the past, our laboratory focused on the identification and characterization of specific binding proteins for lutein, zeaxanthin, and meso-zeaxanthin. Although these proteins may be required for the uptake and stabilization of these compounds in the human retina, they are not sufficient to explain why xanthophylls are not found in other mammalian retinas, because these binding proteins are often found in these animals’ retinas, too.
Recent investigations on various mammalian BCO2 enzymes have shown that BCO2 can generally cleave xanthophylls such as lutein and zeaxanthin, and BCO2 mutations can cause the tissues of chickens, cows, and sheep to develop a deep yellow color due to carotenoid deposition (13–17). To explain the unique deposition of xanthophyll carotenoids in the primate retina, we hypothesized that either human BCO2 is not expressed in human retina, or it is an inactive carotenoid cleavage enzyme. We found that a ∼63-kDa polypeptide was strongly labeled by anti-BCO2 antibodies when probed against human, monkey, or mouse retinal total protein preparations and tissue sections, and RT-PCR also detected mRNA of human and mouse BCO2, demonstrating that BCO2 is expressed in both the human and mouse retina. Subsequently, we used a cell-based carotenoid cleavage assay, which showed that human BCO2 is an inactive carotenoid cleavage enzyme, whereas mouse BCO2 can clearly cleave zeaxanthin. Our finding that human BCO2 is an inactive carotenoid cleavage enzyme is consistent with other published reports. In an investigation on carotenoid uptake mechanisms, human ARPE-19 cells specifically took up high amounts of xanthophyll carotenoids, but no xanthophyll cleavage products were identified in these cells (32). Moreover, no eccentric cleavage products were detected in human subjects given labeled β-carotene supplements (33, 34). Recent genetic analyses have reported no association between SNPs of human BCO2 and macular pigment optical density, also implying that human BCO2 cannot cleave macular pigments (35). Furthermore, our mouse feeding studies showed that BCO2−/− mice accumulate zeaxanthin in their retinas, but WT mice do not (Fig. 5), and our SPR results revealed that the binding affinities between a variety of carotenoids and mouse BCO2 are around 10- to 40-times stronger than those between the same carotenoids and human BCO2, demonstrating that the inability to bind potential substrate carotenoids in the active site may be the reason that human BCO2 is an inactive xanthophyll cleavage enzyme. Persistent expression of BCO2 in the human retina despite lack of carotenoid cleavage activity implies that it may serve an as yet unknown enzymatic and/or regulatory function akin to the retinoid isomerohydrolase activity acquired by its relative in the RPE, RPE65.
To explore the reasons underlying the difference of enzymatic activities between human and mouse BCO2, we examined their primary structures. Sequence alignment (Fig. 4) showed the four key histidine residues responsible for enzyme activity and carotenoid cleavage are conserved. The 3D homology structures of human and mouse BCO2 show that the insertion of GKAA in human BCO2 unfolds a short β-stranded loop present in mouse BCO2 into a much larger unstructured loop (boxes in Fig. 4, Lower). As this is the only major structural change between human and mouse BCO2, disruption of this loop may be key in preventing human BCO2 from cleaving zeaxanthin by disrupting access to the substrate binding tunnel, and, indeed, insertion of GKAA into mouse BCO2 inactivated the carotenoid cleavage enzyme. On the other hand, deletion of GKAA from human BCO2, replacement of the N-terminal domain of human BCO2 with mouse BCO2, or various substitutions of mouse BCO2 amino acids into corresponding residues of human BCO2 did not restore cleavage function to human BCO2 alone or in combination with the GKAA deletion. All these data suggest that there has been considerable genetic drift since primate BCO2 became inactive which complicates genetically engineered reconstitution of human BCO2’s active site. Further attempts to restore human BCO2 activity should be guided by crystallography, but unfortunately, the 3D structures of BCO2s have not been solved, and the 3D structure of the RPE65’s binding site for all-trans-retinyl palmitate is not well characterized (36, 37). Solving the crystal structures of human BCO2 and mouse BCO2 will facilitate our understanding of these two enzymes’ interactions with xanthophylls.
This work reveals that the inactivity of BCO2 with xanthophylls such as zeaxanthin causes primates to be the only mammals that highly accumulate xanthophyll carotenoids in their retinas. It appears that many, if not all, nonprimate mammals break down xanthophylls in ocular and nonocular tissues, but this alone would not account for the selective uptake and the specific localization of just lutein, zeaxanthin, and meso-zeaxanthin in the primate retina when there are around 50 carotenoids in the human diet and 15–20 in the serum. Several transport proteins, such as scavenger receptor class B, member 1 (SR-B1), ATP-binding cassette, subfamily A, member 1 (ABCA1), and HDL, are known to be involved in the process of macular pigment uptake (32, 38–40), and the effects of their regulators, PPAR and ISX, also need to be considered in the carotenoid absorption process (41, 42). During et al. reported that SR-B1 is involved in xanthophyll uptake into cultured retinal cells (32). Carotenoid supplementation did not elevate the carotenoid levels in the retina of the WHAM chicken whose ABCA1 transport protein is defective, and many studies have shown a significant positive association between the concentration of serum HDL and that of serum lutein and zeaxanthin (38–40). The other known carotenoid cleavage enzyme, BCO1, also could be considered a contributor to this selective uptake process. The expression of human BCO1 was detected in the cytoplasm of most human tissues (18, 21, 24, 43), and enzymatic assays show that BCO1 prefers to cleave the major carotenoids in human serum, such as β-carotene, lycopene, and β-cryptoxanthin but not lutein or zeaxanthin (18, 19). Thus, BCO1 can also be considered a participant in the selective targeting of just lutein and zeaxanthin to the human retina. Within the retina itself, the specific binding proteins, StARD3 (lutein) and GSTP1 (zeaxanthin and meso-zeaxanthin), appear to drive the spatial localization to the foveal region because the human retinal distribution of these binding proteins matches well with that of the macular carotenoid pigments (12, 44).
In summary, human BCO2 is present in the human retina, but it showed no carotenoid cleavage activity, whereas mouse BCO2 is clearly an active carotenoid cleavage enzyme. Losing the ability to capture lutein and zeaxanthin efficiently appears to cause human BCO2 to become an inactive xanthophyll carotenoid cleavage enzyme whose other physiological functions in the human retina remain to be elucidated. Thus, inactivity of BCO2 facilitates xanthophyll deposition in the macula of the human and monkey retina.
Materials and Methods
Tissue Procurement and Preparation.
Human donor eyes were procured through the Utah Lions Eye Bank in compliance with tenets of the Declaration of Helsinki. Dissection of the globes was performed within 24 h postmortem on ice and under dim light. Mouse retinas were also harvested under dim light. Human and mouse tissues were washed twice with PBS and homogenized in 10 mM Tris⋅HCl buffer (pH 7.4) containing 0.2 mM PMSF and 10 μg/mL aprotinin to prepare total protein extracts. All animal procedures and experiments were approved by the University of Utah Institutional Animal Care and Use Committees.
Western Blots.
Proteins were separated and transferred to membranes as described before (44). The membranes were washed and incubated overnight with the primary anti-BCO2 antibody (Proteintech Group, Inc.) at a dilution of 1:5,000. The membranes were incubated with horseradish peroxidase-conjugated secondary antibody [1:1,000 polyclonal goat anti-rabbit IgG (H+L)-HRP] for 2 h at room temperature. Membranes were developed using ECL Plus Western blot detection reagents (Amersham Biosciences).
RT-PCR.
Total RNA (2 μg) was prepared from human retina and mouse retina. cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen). PCR amplification was performed using a 1-μL reaction mixture as a template. PCRs were performed using the following primer pairs: human BCO2a forward, 5′-AGG AGT CAT TCT GCC ACT GC-3′; and human BCO2a reverse, 5′-TCT CTG GAG GAA CCC GAA TAA CC -3′; human BCO2b forward, 5′-GAC ATG GCA ACT GGA TCT TGA AGG -3′; and human BCO2b reverse, 5′-TCT CTG GAG GAA CCC GAA TAA CC -3′; human BCO2c forward, 5′-ATG GAA CGA TCT GGT GCT CTC ATG-3′; and human BCO2c reverse, 5′-TTC TCT CCA AAC CAC ATC AAC C -3′; mouse BCO2 forward, 5′- TGA CCC ATG CAA GAG CAT CTT TGA ACG -3′; and mouse BCO2 reverse, 5′-AGC CCT GGT CCT CAA AGG CAT TGA TTT G -3′. Products were evaluated by electrophoresis on an ethidium bromide-stained 1% agarose gel in 1× TBE buffer. The amplicons were cloned and sequenced for verification.
IHC.
Confocal microscope images were derived from a Macaca mulatta monkey or from mouse eyes as described earlier (44). Monkey retinas were chosen because of the availability of high quality perfusion-fixed tissue and the 96% homology of human and monkey BCO2. Shortly after fixation, cryosections (12-μm thick) were cut, rinsed in 0.1 M phosphate buffer containing 0.1% Triton X-100 (PBT), and blocked for 1 h using 10% (vol/vol) normal donkey serum in PBT. Antibodies directed against human BCO2 (1:2,000 dilution) or mouse BCO2 (1:2,000 dilution) were applied overnight at 4 °C. After the samples had been rinsed in PBT (3× 10 min), FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were applied for 2 h at room temperature. Immunolocalization was imaged using a Zeiss LSM 510 confocal microscope set to an optical slice of <0.9 μm. Control sections, which were not incubated in primary antibodies, were processed in parallel and found to be negative.
Carotenoid Cleavage Assays.
The expression vectors, EX-Z2826-B05 (human BCO2a) and EX-Mm13746-B05 (mouse BCO2), were purchased from GeneCopoeia. The cDNAs were cloned into pReceiver-B05 vector with a Tac promoter, N-GST tag, and TEV protease site. EX-NEG-B05 (Empty pReceiver-B05) was used as a negative control. All three vectors were transformed into E. coli BL21 carrying an expression plasmid for zeaxanthin biosynthesis (45, 46). The transformed E. coli were grown overnight at 37 °C on LB solid medium containing 100 µg/mL ampicillin and 35 µg/mL chloramphenicol. Selected colonies were picked into 5 mL LB medium containing 100 µg/mL ampicillin and 35 µg/mL chloramphenicol and grown at 37 °C with shaking until the OD600 was 0.5–0.7. Then, protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM. After growing the cultures for an additional 3–5 h, they were then streaked on the same LB solid medium and incubated at 25 °C for 2–3 d in darkness. In this system, carotenoid cleavage activity is visualized qualitatively by the lack of accumulating carotenoids which results in the absence of the yellow color characteristic of the control cells containing carotenoid synthesis enzymes but no cleavage enzymes. The bacteria on the plates were collected for HPLC analysis to confirm the presence of typical carotenoid cleavage products such as C14-dialdehyde. The extracted carotenoid contents were dried and redissolved in the HPLC mobile phase. HPLC separation was carried out on a C30 column (YMC Europe GmbH; 25 cm length and 4.6 mm i.d.) as before (47).
To try to restore the carotenoid cleavage activity of human BCO2, substitution of amino acid residues of human BCO2 with the corresponding residues of mouse BCO2 was performed according to Fig. S2 and Table S1. All of the chimera constructs were generated using human BCO2a, human BCO2b, or mouse BCO2 expression vectors as templates, respectively. All of the primers as shown in Table S1 were designed using the QuikChange Primer Design Program (Agilent Technologies). The mutagenesis experiments were carried out with a QuikChange II XL Site-Directed Mutagenesis Kit or QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies). After confirmation by DNA sequencing, all of the constructs were cotransfected with the zeaxanthin synthesis plasmid into BL21 to assay for carotenoid cleavage activity as described above.
SPR Binding Studies.
SPR analyses were conducted as before (48, 49). Briefly, the proteins (human BCO2a, mouse BCO2, and HSA) at 10–50 μg/mL in 10 mM sodium acetate (pH 5.0) were immobilized on the sensor chip surface using standard amine coupling to obtain a density of 10–12 kilo-response units. Each of the three carotenoids was dissolved in DMSO to achieve a high concentration, and 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes pH 7.4) with 0.01% Tween-20 and 3% DMSO were used as the running buffer. Typically, the analyte concentration series spanned 0.01–50 µM. Five of these blanks were analyzed at the beginning of the analysis, and the remaining blanks were interspersed throughout the analysis for double-referencing purposes. Each of the analytes was run in triplicate. The analyte concentration series was injected as twofold dilutions (seven dilutions) in running buffer using the FastStep (SensiQ Technologies, Inc.) gradient injection mode.
Three-Dimensional Structure of BCO2s.
Using RPE65 (PDB 3fsn.1.a) as their templates, the protein homology structures of human and mouse BCO2 were prepared through PyMol (Schrödinger, LLC).
Mouse Feeding Studies.
Twenty-five 3-mo-old BCO2−/− mice (bred at the University of Utah vivarium using founders from Case Western Reserve University), and twenty-five 3-mo-old WT C57/BL6 mice were fed with DSM ActiLease zeaxanthin beadlets mixed with their chow (∼2.6 mg per mouse per day) for 4 wk after first receiving a vitamin A-deficient chow (AIN-93) for 4 wk to help promote carotenoid uptake. Retinas, RPE/choroid, and lenses of mice from each feeding group were pooled together in batches of five pairs of tissue, respectively. Zeaxanthin levels in mouse ocular tissues were analyzed by HPLC on a cyano column (Microsorb 250 mm × 4.6 mm; Rainin Instrument Co.) as before (47), with confirmation of identity by retention time and UV–visible spectrum, and mass spectrum versus authentic zeaxanthin standard. Chiral HPLC analysis confirmed the presence of 3R,3′R-zeaxanthin and absence of 3R,3′S-meso-zeaxanthin.
Supplementary Material
Acknowledgments
We thank Dr. Francis Cunningham (University of Maryland) for the kind gift of plasmids to produce carotenoid-accumulating E. coli stains. We also thank Dr. Johannes von Lintig (Case Western Reserve University) for kindly providing the BCO2−/−mice and a critical reading of the manuscript. This work was supported by National Eye Institute Grants EY-11600 and EY-14800, Kemin Health, the Lowy Medical Research Institute, and by unrestricted grants to the Department of Ophthalmology (University of Utah) from Research to Prevent Blindness. W.B. is the recipient of a Research to Prevent Blindness Senior Investigator Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402526111/-/DCSupplemental.
References
- 1.Bone RA, Landrum JT, Tarsis SL. Preliminary identification of the human macular pigment. Vision Res. 1985;25(11):1531–1535. doi: 10.1016/0042-6989(85)90123-3. [DOI] [PubMed] [Google Scholar]
- 2.Bone RA, Landrum JT. Macular pigment in Henle fiber membranes: A model for Haidinger’s brushes. Vision Res. 1984;24(2):103–108. doi: 10.1016/0042-6989(84)90094-4. [DOI] [PubMed] [Google Scholar]
- 3.Khachik F, Bernstein PS, Garland DL. Identification of lutein and zeaxanthin oxidation products in human and monkey retinas. Invest Ophthalmol Vis Sci. 1997;38(9):1802–1811. [PubMed] [Google Scholar]
- 4.Beatty S, Boulton M, Henson D, Koh HH, Murray IJ. Macular pigment and age related macular degeneration. Br J Ophthalmol. 1999;83(7):867–877. doi: 10.1136/bjo.83.7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krinsky NI, Landrum JT, Bone RA. Biologic mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr. 2003;23:171–201. doi: 10.1146/annurev.nutr.23.011702.073307. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead AJ, Mares JA, Danis RP. Macular pigment: A review of current knowledge. Arch Ophthalmol. 2006;124(7):1038–1045. doi: 10.1001/archopht.124.7.1038. [DOI] [PubMed] [Google Scholar]
- 7.Bone RA, et al. Macular pigment in donor eyes with and without AMD: A case-control study. Invest Ophthalmol Vis Sci. 2001;42(1):235–240. [PubMed] [Google Scholar]
- 8.Landrum JT, et al. A one year study of the macular pigment: The effect of 140 days of a lutein supplement. Exp Eye Res. 1997;65(1):57–62. doi: 10.1006/exer.1997.0309. [DOI] [PubMed] [Google Scholar]
- 9.Age-Related Eye Disease Study 2 (AREDS2) Research Group Secondary analyses of the effects of lutein/zeaxanthin on age-related macular degeneration progression: AREDS2 Report No. 3. JAMA Ophthalmol. 2014;132(2):142–149. doi: 10.1001/jamaophthalmol.2013.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Age-Related Eye Disease Study 2 (AREDS2) Research Group Lutein/zeaxanthin for the treatment of age-related cataract: AREDS2 randomized trial report no. 4. JAMA Ophthalmol. 2013;131(7):843–850. doi: 10.1001/jamaophthalmol.2013.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, et al. Dietary wolfberry upregulates carotenoid metabolic genes and enhances mitochondrial biogenesis in the retina of db/db diabetic mice. Mol Nutr Food Res. 2013;57(7):1158–1169. doi: 10.1002/mnfr.201200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhosale P, et al. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J Biol Chem. 2004;279(47):49447–49454. doi: 10.1074/jbc.M405334200. [DOI] [PubMed] [Google Scholar]
- 13.Mein JR, Dolnikowski GG, Ernst H, Russell RM, Wang XD. Enzymatic formation of apo-carotenoids from the xanthophyll carotenoids lutein, zeaxanthin and β-cryptoxanthin by ferret carotene-9′,10′-monooxygenase. Arch Biochem Biophys. 2011;506(1):109–121. doi: 10.1016/j.abb.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amengual J, et al. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25(3):948–959. doi: 10.1096/fj.10-173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Våge DI, Boman IA. A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries) BMC Genet. 2010;11:10. doi: 10.1186/1471-2156-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson J, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4(2):e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian R, Pitchford WS, Morris CA, Cullen NG, Bottema CD. Genetic variation in the beta, beta-carotene-9′,10′-dioxygenase gene and association with fat colour in bovine adipose tissue and milk. Anim Genet. 2010;41(3):253–259. doi: 10.1111/j.1365-2052.2009.01990.x. [DOI] [PubMed] [Google Scholar]
- 18.Lindqvist A, Andersson S. Biochemical properties of purified recombinant human beta-carotene 15,15′-monooxygenase. J Biol Chem. 2002;277(26):23942–23948. doi: 10.1074/jbc.M202756200. [DOI] [PubMed] [Google Scholar]
- 19.dela Seña C, et al. Substrate specificity of purified recombinant human β-carotene 15,15′-oxygenase (BCO1) J Biol Chem. 2013;288(52):37094–37103. doi: 10.1074/jbc.M113.507160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fierce Y, et al. In vitro and in vivo characterization of retinoid synthesis from beta-carotene. Arch Biochem Biophys. 2008;472(2):126–138. doi: 10.1016/j.abb.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hessel S, et al. CMO1 deficiency abolishes vitamin A production from beta-carotene and alters lipid metabolism in mice. J Biol Chem. 2007;282(46):33553–33561. doi: 10.1074/jbc.M706763200. [DOI] [PubMed] [Google Scholar]
- 22.Boulanger A, et al. Identification of beta-carotene 15,15′-monooxygenase as a peroxisome proliferator-activated receptor target gene. FASEB J. 2003;17(10):1304–1306. doi: 10.1096/fj.02-0690fje. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer C, et al. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem. 2001;276(17):14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- 24.Redmond TM, et al. Identification, expression, and substrate specificity of a mammalian beta-carotene 15,15′-dioxygenase. J Biol Chem. 2001;276(9):6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz SH, Qin X, Zeevaart JA. Characterization of a novel carotenoid cleavage dioxygenase from plants. J Biol Chem. 2001;276(27):25208–25211. doi: 10.1074/jbc.M102146200. [DOI] [PubMed] [Google Scholar]
- 26.Sun Z, et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta. 2008;228(5):789–801. doi: 10.1007/s00425-008-0781-6. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz SH, Qin X, Loewen MC. The biochemical characterization of two carotenoid cleavage enzymes from Arabidopsis indicates that a carotenoid-derived compound inhibits lateral branching. J Biol Chem. 2004;279(45):46940–46945. doi: 10.1074/jbc.M409004200. [DOI] [PubMed] [Google Scholar]
- 28.García-Limones C, et al. Functional characterization of FaCCD1: A carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J Agric Food Chem. 2008;56(19):9277–9285. doi: 10.1021/jf801096t. [DOI] [PubMed] [Google Scholar]
- 29.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci. 2006;63(19-20):2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Home E. An account of the orifice in the retina of the human eye, discovered by Professor Soemmering: To which are added proofs of this appearance being extended to the eyes of other animals. Philos Trans R Soc Lond. 1798;88:332–345. [Google Scholar]
- 31.Li B, Ahmed F, Bernstein PS. Studies on the singlet oxygen scavenging mechanism of human macular pigment. Arch Biochem Biophys. 2010;504(1):56–60. doi: 10.1016/j.abb.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.During A, Doraiswamy S, Harrison EH. Xanthophylls are preferentially taken up compared with beta-carotene by retinal cells via a SRBI-dependent mechanism. J Lipid Res. 2008;49(8):1715–1724. doi: 10.1194/jlr.M700580-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krinsky NI, Wang XD, Tang G, Russell RM. Mechanism of carotenoid cleavage to retinoids. Ann N Y Acad Sci. 1993;691:167–176. doi: 10.1111/j.1749-6632.1993.tb26168.x. [DOI] [PubMed] [Google Scholar]
- 34.Yeum KJ, Russell RM. Carotenoid bioavailability and bioconversion. Annu Rev Nutr. 2002;22:483–504. doi: 10.1146/annurev.nutr.22.010402.102834. [DOI] [PubMed] [Google Scholar]
- 35.Meyers KJ, et al. Genetic determinants of macular pigments in women of the Carotenoids in Age-Related Eye Disease Study. Invest Ophthalmol Vis Sci. 2013;54(3):2333–2345. doi: 10.1167/iovs.12-10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiser PD, Golczak M, Lodowski DT, Chance MR, Palczewski K. Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc Natl Acad Sci USA. 2009;106(41):17325–17330. doi: 10.1073/pnas.0906600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chander P, Gentleman S, Poliakov E, Redmond TM. Aromatic residues in the substrate cleft of RPE65 protein govern retinol isomerization and modulate its progression. J Biol Chem. 2012;287(36):30552–30559. doi: 10.1074/jbc.M112.364596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connor WE, Duell PB, Kean R, Wang Y. The prime role of HDL to transport lutein into the retina: Evidence from HDL-deficient WHAM chicks having a mutant ABCA1 transporter. Invest Ophthalmol Vis Sci. 2007;48(9):4226–4231. doi: 10.1167/iovs.06-1275. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y, et al. Association of variants in the LIPC and ABCA1 genes with intermediate and large drusen and advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52(7):4663–4670. doi: 10.1167/iovs.10-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loane E, et al. Transport and retinal capture of lutein and zeaxanthin with reference to age-related macular degeneration. Surv Ophthalmol. 2008;53(1):68–81. doi: 10.1016/j.survophthal.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Lobo GP, et al. Genetics and diet regulate vitamin A production via the homeobox transcription factor ISX. J Biol Chem. 2013;288(13):9017–9027. doi: 10.1074/jbc.M112.444240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr Rev. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 43.Bhatti RA, et al. Expression of beta-carotene 15,15′ monooxygenase in retina and RPE-choroid. Invest Ophthalmol Vis Sci. 2003;44(1):44–49. doi: 10.1167/iovs.02-0167. [DOI] [PubMed] [Google Scholar]
- 44.Li B, Vachali P, Frederick JM, Bernstein PS. Identification of StARD3 as a lutein-binding protein in the macula of the primate retina. Biochemistry. 2011;50(13):2541–2549. doi: 10.1021/bi101906y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunningham FX, Jr, Sun Z, Chamovitz D, Hirschberg J, Gantt E. Molecular structure and enzymatic function of lycopene cyclase from the cyanobacterium Synechococcus sp strain PCC7942. Plant Cell. 1994;6(8):1107–1121. doi: 10.1105/tpc.6.8.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham FX, Jr, et al. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell. 1996;8(9):1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhosale P, Serban B, Zhao Y, Bernstein PS. Identification and metabolic transformations of carotenoids in ocular tissues of the Japanese quail Coturnix japonica. Biochemistry. 2007;46(31):9050–9057. doi: 10.1021/bi700558f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rich RL, Quinn JG, Morton T, Stepp JD, Myszka DG. Biosensor-based fragment screening using FastStep injections. Anal Biochem. 2010;407(2):270–277. doi: 10.1016/j.ab.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vachali P, Li B, Nelson K, Bernstein PS. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch Biochem Biophys. 2012;519(1):32–37. doi: 10.1016/j.abb.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.