Significance
The primary cilium is an organelle typically found on postmitotic vertebrate cells. Cilia serve as antennae to receive signals from extracellular space and thus play important roles in both development and disease. Understanding the mechanisms controlling their formation (ciliogenesis) is of great importance. Ciliogenesis is known to depend on basal bodies, but although major steps have been described at a morphological level, the underlying mechanism and its regulation remain poorly understood. In our study, we characterized Cep164, a key component of basal bodies that is crucial for ciliogenesis. We show that one major function of Cep164 is to recruit a protein kinase, TTBK2, to basal bodies. Once localized correctly, TTBK2 then functions in distal appendage assembly and primary cilia formation.
Keywords: primary cilium, centrosome
Abstract
Primary cilia play critical roles in development and disease. Their assembly is triggered by mature centrioles (basal bodies) and requires centrosomal protein 164kDa (Cep164), a component of distal appendages. Here we show that loss of Cep164 leads to early defects in ciliogenesis, reminiscent of the phenotypic consequences of mutations in TTBK2 (Tau tubulin kinase 2). We identify Cep164 as a likely physiological substrate of TTBK2 and demonstrate that Cep164 and TTBK2 form a complex. We map the interaction domains and demonstrate that complex formation is crucial for the recruitment of TTBK2 to basal bodies. Remarkably, ciliogenesis can be restored in Cep164-depleted cells by expression of chimeric proteins in which TTBK2 is fused to the C-terminal centriole-targeting domain of Cep164. These findings indicate that one of the major functions of Cep164 in ciliogenesis is to recruit active TTBK2 to centrioles. Once positioned, TTBK2 then triggers key events required for ciliogenesis, including removal of CP110 and recruitment of intraflagellar transport proteins. In addition, our data suggest that TTBK2 also acts upstream of Cep164, contributing to the assembly of distal appendages.
The primary cilium (PC) functions as an antenna-like signaling organelle typically found on postmitotic cells (1–3). It consists of a microtubule-based axoneme enclosed within a ciliary membrane, and its assembly is triggered at the basal body (4, 5). The basal body, in turn, is derived from one of the two centrioles that make up the centrosome, specifically the mature (or “mother”) centriole (M centriole) (6–9). Long erroneously considered a vestigial organelle, the PC has emerged as a key structure for sensing extracellular stimuli and hence plays crucial roles in cellular responses to both mechanical and chemical cues. In vertebrates, PC function has been linked to the regulation of many important aspects of embryonic development as well as tissue homeostasis in adulthood (2, 10); moreover, defects in ciliary assembly or function have been linked to a large number of human diseases known as ciliopathies (3, 6, 11–13).
PC structure and formation have long been studied at a morphological level (14, 15), but a molecular understanding of the regulation of PC assembly and resorption is only beginning to emerge (5, 7, 8, 16, 17). In cultured cells, PC formation generally occurs when cells exit the cell cycle to enter quiescence (Go), and, conversely, PCs are often resorbed when cells reenter the cell cycle. Thus, PC formation can readily be triggered by serum starvation of some cultured cells, including telomerase-immortalized retinal pigment epithelial (RPE-1) cells. Early steps in PC formation include the docking of membrane vesicles to centrioles (14, 18, 19), the removal of the capping protein CP110 from the distal part of the M centriole (20, 21), the recruitment of intraflagellar transport (IFT) protein complexes (22–24), the formation of a transition zone at the membrane (25, 26), and, finally, the outgrowth of the ciliary axoneme (27, 28).
An important role in PC formation resides with specific components of the M centriole, termed distal appendages (14, 15). These appendages are considered critical for the early docking of Golgi-derived membrane vesicles and the subsequent anchorage of the basal body underneath the plasma membrane. Following the discovery of centrosomal protein 164kDa (Cep164), the first marker for distal appendages (29), several additional distal appendage proteins (DAPs) have recently been identified and functionally linked to ciliogenesis. These include centrosomal protein 83kDa (Cep83)/CCDC41, centrosomal protein 89kDa (Cep89)/CCDC123, SCLT1, and FBF1 (18, 19, 30, 31). The DAP Cep164 was discovered in a screen for components that are critical for PC formation (29). Subsequently, mutations in Cep164 were linked to ciliopathies, providing direct proof for the importance of this protein in human pathophysiology (32). At a mechanistic level, Cep164 was shown to be required at an early stage of PC formation, notably for the docking of membrane vesicles to the basal body (18). Moreover, two components of the vesicle transport machinery, the small GTPase Rab8 and its guanine–nucleotide exchange factor Rabin8, were identified as interaction partners of Cep164 (18). Despite this progress, the precise molecular functions of Cep164 remain to be fully understood.
Importantly, protein kinases have also been implicated in ciliogenesis and in cilia-related diseases. These include Nek1 and Nek8, two members of the family of NIMA-related kinases (33, 34), and Tau Tubulin Kinase 2 (TTBK2), a member of the casein kinase 1 family (35–37).
Here, we report that Cep164 and TTBK2 form a complex and that formation of this complex at M centrioles is essential for ciliogenesis. We show that the noncatalytic C-terminal domain of TTBK2 interacts with Cep164 and that formation of the complex critically depends on the WW domain within the N-terminal domain of Cep164. We also provide evidence that Cep164 is a likely physiological substrate of TTBK2. Use of chimeric TTBK2–Cep164 constructs in siRNA-rescue experiments leads us to conclude that a main function of Cep164 consists of the recruitment of TTBK2 to M centrioles. Once localized correctly, TTBK2 is then in a position to trigger PC formation through phosphorylation of appropriate substrates. Interestingly, our data also reveal a role for TTBK2 in the assembly of distal appendages. Overexpression of active kinase in fact enhances not only the association of DAPs with existing appendages but also triggers their occasional recruitment to daughter centrioles (D centrioles).
Results
Loss of Cep164 Leads to Early Defects in Cilium Assembly.
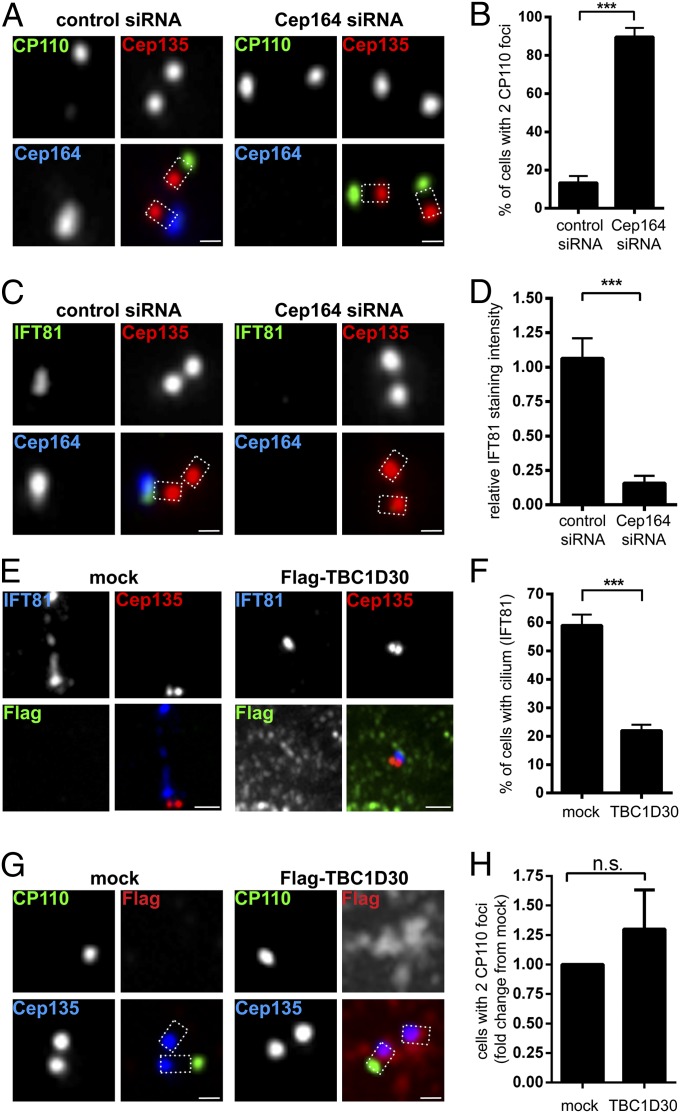
Cep164 is critical for ciliogenesis in human cells (18, 19, 29). This requirement can readily be demonstrated by siRNA-mediated depletion of Cep164 from RPE-1 cells, which results in strong suppression of PC formation (Fig. S1 A and B). To better understand the function of Cep164 in ciliogenesis, we first monitored the fate of two proteins previously implicated in the process, CP110 and IFT81. Whereas CP110 suppresses ciliogenesis and must be removed from M centrioles (19–21), the IFT complex component IFT81 is essential for ciliogenesis and must be recruited (38, 39). In serum-starved control cells, CP110 was virtually undetectable at M centrioles, but the protein persisted at all centrioles in most Cep164-depleted cells (Fig. 1 A and B). Conversely, IFT81 was recruited to M centrioles in controls, but not in Cep164-depleted cells (Fig. 1 C and D).
Fig. 1.
Depletion of Cep164 impairs early events in ciliogenesis independently of Rab8. RPE-1 cells were transfected with the indicated siRNA oligonucleotides (A–D) or plasmids (E–H) and serum starved. After fixation and permeabilization, cells were stained with the indicated antibodies and examined by indirect immunofluorescence microscopy. Cep135 staining was used to label proximal ends of centrioles. Dashed lines in merged images outline the centrioles. [Scale bars: 0.5 μm (A, C, and G) and 1 μm (E).] (A) Depletion of Cep164 from RPE-1 cells prevents removal of distal centriolar protein CP110 from the M centriole. (B) Quantification of the effects of Cep164 depletion on CP110 removal. Graph represents the summary of three experiments. (C) Loss of Cep164 impairs the recruitment of IFT81 to the M centriole. (D) Quantification of the effects of Cep164 depletion on IFT81 recruitment. Graph represents the summary of two experiments. (E) Transient expression of TBC1D30 (Rab8 GAP) impairs ciliogenesis, but not IFT81 localization to M centriole. (F) Quantification of the effects of TBC1D30 expression on ciliogenesis. Graph represents the summary of three experiments. (G) Transient expression of TBC1D30 does not prevent removal of CP110 from M centrioles. (H) Quantification of the effects of TBC1D30 expression on the CP110 removal. Graph represents the summary of four experiments. ***P < 0.001; n.s., not significant.
It has recently been demonstrated that Cep164 plays a role in vesicular docking, most likely through Rab8a activation (18). To determine whether the above phenotypes could be attributed to impaired Rab8a activity, we transiently expressed Flag–TBC1D30, a Rab8a GTPase-activating protein (GAP) known to interfere with Rab8a activity (40). Expression of the Rab8a GAP strongly suppressed PC formation (Fig. 1F), attesting to its functionality; however, IFT81 was still recruited to M centrioles (Fig. 1E), and CP110 was still removed (Fig. 1 G and H). We conclude that attenuation of Rab8a activity by expression of its GAP, TBC1D30, failed to phenocopy the defects observed after Cep164 siRNA depletion, indicating that initial IFT81 recruitment and CP110 removal occur independently of the Cep164/Rab8a pathway.
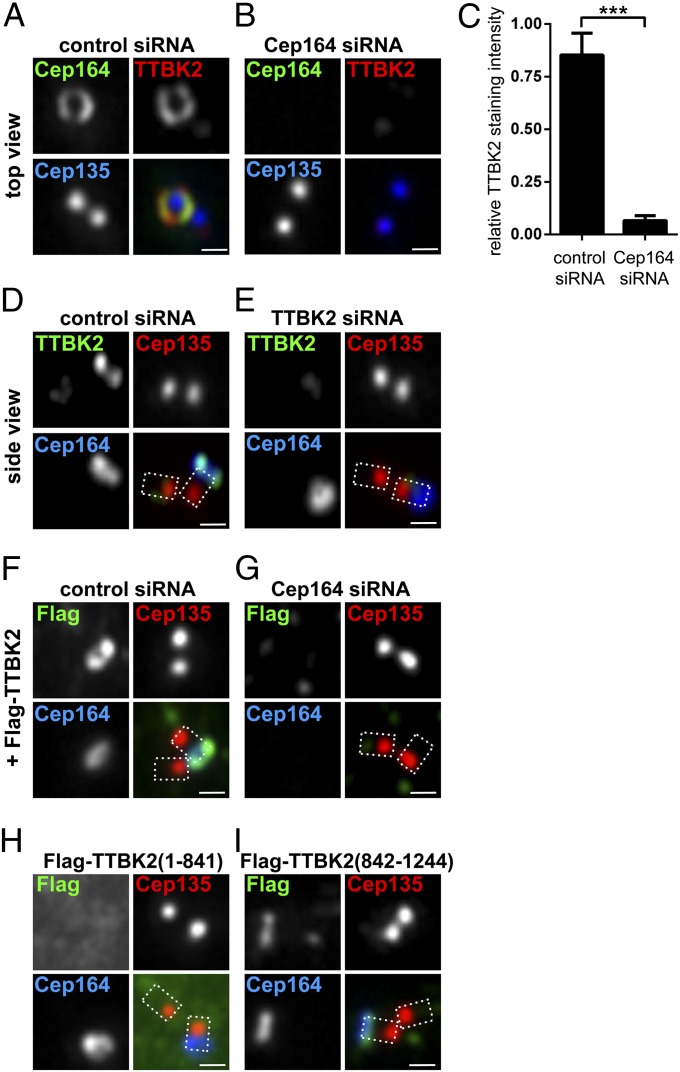
When considering alternative modes of action of Cep164, we were intrigued by a recent study implicating murine TTBK2 in ciliogenesis (35). Moreover, TTBK2 was found to bind Cep164 in a yeast two-hybrid screen (32). Thus, we decided to explore a possible functional connection between Cep164 and TTBK2. Reminiscent of Cep164 depletion, TTBK2-depleted RPE-1 cells failed to form PCs upon serum starvation (Fig. S1 C and D), and CP110 was not removed from M centrioles (Fig. S1E). Endogenous TTBK2 localized at M centrioles (Fig. 2A), confirming and extending previous results (35). Interestingly, in top views, TTBK2 showed a ring-like staining coincident with Cep164, consistent with appendage association (Fig. 2A). Localization of TTBK2 was dependent on the presence of Cep164 (Fig. 2 B and C) but not vice versa (Fig. 2 D and E). Exogenous Flag-tagged TTBK2 also colocalized with Cep164 (Fig. 2F), and, again, this staining was lost upon Cep164 depletion (Fig. 2G).
Fig. 2.
Cep164 is required for TTBK2 localization to the M centriole. Transfection and immunofluorescence experiments were carried out as described in the Fig. 1 legend. Cep135 staining was used to label proximal ends of centrioles. (Scale bars: 0.5 μm.) (A) Staining for endogenous Cep164 and TTBK2 reveals colocalization in a ring-like pattern at the M centriole, consistent with colocalization at distal appendages. (B) Depletion of Cep164 prevents recruitment of TTBK2 to the M centriole. (C) Quantification of the effects of Cep164 depletion on TTBK2 recruitment. Graph represents the summary of two experiments. ***P < 0.001. (D and E) Cep164 does not require TTBK2 for its localization to distal appendages of the M centriole. (F and G) Depletion of Cep164 prevents the recruitment of Flag-TTBK2 to the M centriole. (H) Flag–TTBK2 (1–841), lacking the Cep164 binding region (Fig. 3E), fails to localize to the M centriole. (I) Flag–TTBK2 (842–1244), containing the Cep164 binding region, localizes to the M centriole.
TTBK2–Cep164 Complex Formation and Cep164 Phosphorylation.
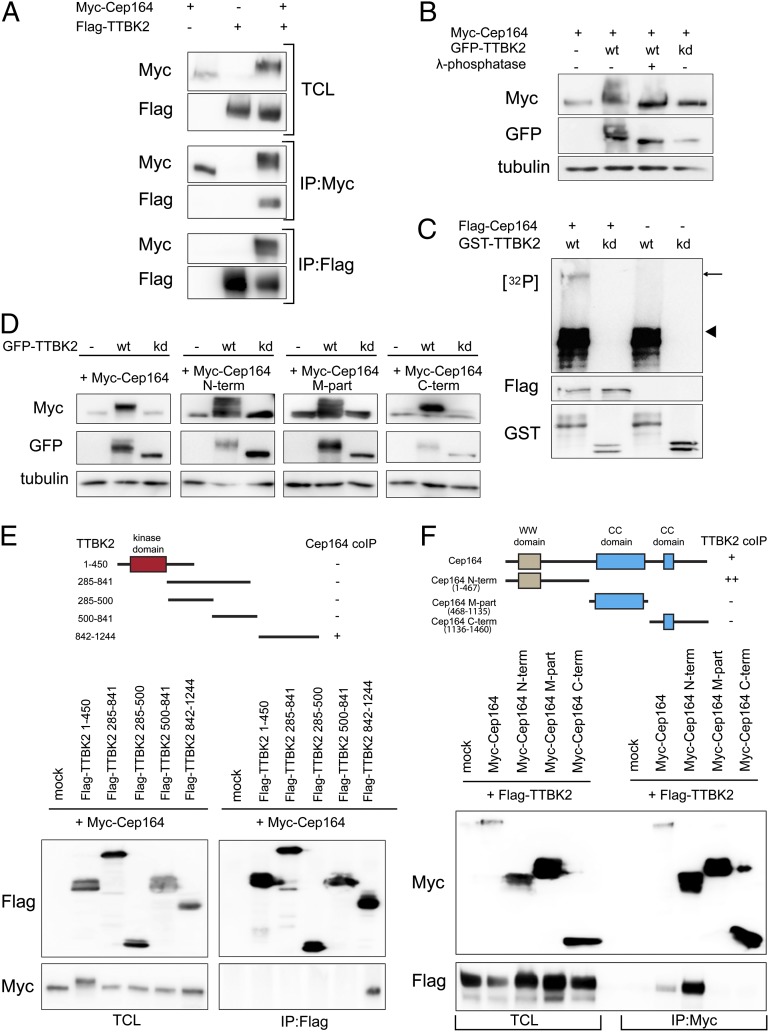
Given the dependency of TTBK2 localization on Cep164 and a previously suggested interaction between the two proteins (32), we asked whether Cep164 and TTBK2 might form a complex. Indeed, Myc–Cep164 and Flag–TTBK2 could readily be coimmunoprecipitated from HEK293T cells (Fig. 3A). Furthermore, the electrophoretic mobility of Myc–Cep164 was retarded in the presence of Flag–TTBK2, suggesting that Cep164 undergoes TTBK2-mediated phosphorylation. In support of this conclusion, the TTBK2-induced mobility upshift of Myc–Cep164 was dependent on TTBK2 activity and sensitive to lambda phosphatase (Fig. 3B). It is noteworthy that TTBK2 itself also migrated more slowly than a kinase dead (kd) mutant [D163A (41)], and this retarded migration was abolished by phosphatase treatment (Fig. 3B), indicating that TTBK2 undergoes autophosphorylation. Depletion of TTBK2 from RPE-1 cells caused a loss of higher migrating forms of endogenous Cep164 (Fig. S2A), supporting the conclusion that Cep164 is phosphorylated in a TTBK2-dependent manner in vivo.
Fig. 3.
Cep164 is a binding partner and substrate of TTBK2. (A) HEK293T cells were transfected with the indicated constructs and subjected to immunoprecipitation (IP)–Western experiments. Upper shows total cell lysate (TCL). Western blots were performed by using the anti-tag antibodies listed to the left, and IPs were performed by using either anti-Myc (Myc–Cep164; Middle) or anti-Flag antibodies (Flag–TTBK2; Bottom). (B) Protein extracts, prepared from HEK293T cells transfected with Myc–Cep164 and GFP–TTBK [wild-type (WT) or kinase dead (kd) mutant], were treated with or without λ-phosphatase and analyzed by Western blotting. Note that Cep164 undergoes TTBK2-mediated phosphorylation, as indicated by a mobility shift that is sensitive to λ-phosphatase treatment. (C) In vitro-translated Flag–Cep164 was immunoprecipitated by using anti-Flag antibody and subjected to a kinase assay in the presence of [γ-32P]ATP and recombinant GST–TTBK2 (1-450) WT or kd protein. Arrow and arrowhead point to phosphorylated Flag–Cep164 and autophosphorylated GST–TTBK2 (1–450), respectively. (D) Protein extracts of transfected HEK293T cells were analyzed by Western blotting. Note that expression of GFP–TTBK2, but not kd mutant, led to prominently retarded electrophoretic mobility of Myc-tagged Cep164, Cep164 N term, Cep164 M part, and Cep164 C term. Also, note concomitant increases in levels of Cep164 and Cep164 C term. (E and F) Transfected HEK293T cells were subjected to IP–Western experiments. (Upper) Schematic representations of the constructs used are shown, and the symbols +, ++, or − summarize the efficacy of coimmunoprecipitation for each combination. (E) Domain mapping of TTBK2 identifies the C-terminal part (842–1244) as sufficient for Cep164 binding. (F) Domain mapping of Cep164 identifies the N-terminal part (1–467) as sufficient for interaction with TTBK2.
To explore a possible direct kinase–substrate relationship, we asked whether TTBK2 is able to phosphorylate Cep164 in vitro. Recombinant GST–TTBK2 (1–450; catalytic domain), but not the corresponding kd mutant construct (D163A), was indeed able to phosphorylate in vitro-translated Flag–Cep164 (Fig. 3C). In vivo, TTBK2 triggered a pronounced Cep164 upshift, not only when coexpressed with full-length protein, but also with Cep164 N-term, M-part, or C-term fragments, demonstrating the existence of multiple phosphorylation sites distributed throughout Cep164 (Fig. 3D). Together these results identify Cep164 as a likely physiological substrate of TTBK2. Analysis of the C-term fragment of Cep164 by mass spectrometry (MS) identified seven distinct phosphorylation sites (Fig. S2B); additional sites undoubtedly remain to be identified, both within the C-term fragment and the two other Cep164 moieties. Although the physiological consequences of Cep164 phosphorylation remain to be determined, we note that TTBK2 caused a striking accumulation of Cep164 full-length protein as well as C-term fragment (Fig. 3D). This finding raises the possibility that TTBK2-dependent phosphorylation triggers Cep164 stabilization (see also below).
To map the interaction domains between TTBK2 and Cep164, we performed coimmunoprecipitation experiments using deletion mutants of Cep164 and TTBK2, respectively. We identified the C-terminal noncatalytic region of TTBK2 (842–1244) and the N-terminal part of Cep164 (1–467) as being important for complex formation (Fig. 3 E and F). Although phosphatase treatment clearly reduced phosphorylation of the interaction partners, it did not detectably affect the stability of the complex (Fig. S2C), arguing against a major regulatory role of phosphorylation in TTBK2–Cep164 complex formation.
Importantly, the C-terminal part of TTBK2 (842–1244) was sufficient for centriole localization, but the Flag–TTBK2 (1–841) mutant failed to localize (Fig. 2 H and I), confirming and extending data obtained for murine TTBK2 (35). These results support our conclusion that the association of TTBK2 with M centrioles is mediated by direct binding to Cep164.
Cep164 N Terminus Suppresses Ciliogenesis Through Sequestration of TTBK2.
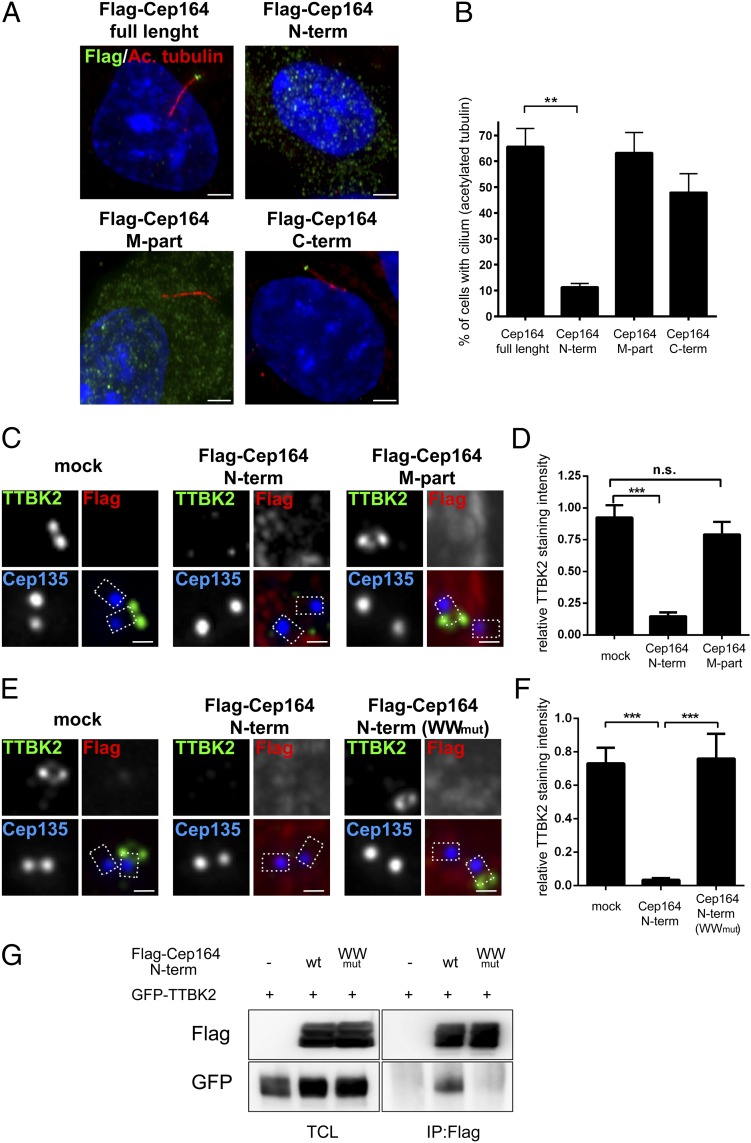
Having established a physical interaction between Cep164 and TTBK2, we next asked to what extent this interaction might contribute to explaining the role of these two proteins in ciliogenesis. First, we examined the consequences of overexpression of various Cep164 constructs, either full-length or truncated. Overexpression of full-length Cep164, M part or C term did not detectably interfere with the formation of PCs, as monitored by staining for acetylated tubulin; in stark contrast, overexpression of Cep164 N term almost completely suppressed ciliogenesis (Fig. 4 A and B). Virtually identical results were obtained when using alternative methods for assessing PC formation (Fig. S3A), demonstrating that these effects cannot be explained by an inhibitory influence of Cep164 on tubulin acetylation. We conclude that a strong correlation exists between the ability of Cep164 fragments to bind TTBK2 (Fig. 3F) and their ability to exert a dominant-negative effect on PC formation (Fig. 4B).
Fig. 4.
Excess of Cep164 N-terminal fragment impairs cilia formation by WW domain-mediated sequestration of TTBK2. Transfections and immunofluorescence experiments were carried out as outlined in the Fig. 1 legend. Cep135 staining was used to label proximal ends of centrioles. [Scale bars: 2.5 μm (A) and 0.5 μm (C and E).] (A) Localizations and impact on ciliogenesis of Cep164 full-length and deletion mutants (N term, 1–467; M part, 468–1135; C term, 1136–1460). PCs were visualized by staining for acetylated tubulin; note the dominant-negative effect of Cep164 N term. (B) Quantification of the data shown in A. Graph represents the summary of three experiments. (C) Expression of Cep164 N term, but not Cep164 M part, prevents TTBK2 localization to M centrioles. (D) Quantification of the data shown in C. Graph represents the summary of two experiments. (E) Expression of the WWmut mutant version of Cep164 N term (1–467) fails to impair M centriole localization of TTBK2. (F) Quantification of the data shown in E. Graph represents the summary of two experiments. (G) HEK293T cells were transfected with the indicated constructs and subjected to IP–Western experiments. Note that Cep164 N term (WWmut) fails to coimmunoprecipitate TTBK2. **P < 0.01; ***P < 0.001; n.s., not significant.
To determine whether the observed correlation might indicate causality, we next examined the consequences of expressing the above Cep164 constructs on TTBK2 localization in RPE-1 cells. Whereas endogenous TTBK2 localized to distal appendages of M centrioles in mock or Flag–Cep164 M-part transfected cells, the centriole association of this kinase was strongly diminished in cells transfected with Flag–Cep164 N term (Fig. 4 C and D). Because Cep164 N term itself localizes diffusely throughout the cytoplasm (Fig. 4A; see also ref. 18); this result points to sequestration of TTBK2 away from centrioles, a conclusion confirmed by using Flag–TTBK2 (Fig. S3B). Interestingly, the N-terminal part of Cep164 contains a predicted WW domain (56–89) that has been suggested to play a role in ciliogenesis (18). Furthermore, the WW domains of other proteins have previously been implicated in protein–protein interactions (42, 43). Thus, we tested a possible involvement of the Cep164 WW domain in TTBK2 binding. A mutant version of Cep164 N term (WWmut), in which two amino acid substitutions disrupt the structural integrity of the WW domain [(Y74AY75A) (18, 42)], failed not only to sequester TTBK2 away from centrioles (Fig. 4 E and F), but also to coimmunoprecipitate GFP–TTBK2 (Fig. 4G). This result demonstrates that the WW domain of Cep164 is functionally important and required for efficient binding to TTBK2.
The above results suggested that the recruitment of TTBK2 to M centrioles might represent a key function of Cep164. If so, one would predict that the displacement of TTBK2 should phenocopy the loss of Cep164. Although the overexpression of Flag–Cep164 N term in RPE-1 cells did not detectably affect either the levels or localization of endogenous Cep164 (Fig. S3 C and D), cells expressing Flag–Cep164 N term failed to trigger either removal of CP110 (Fig. S3E) or centriole recruitment of IFT81 (Fig. S3F). This result demonstrates that centriole-associated Cep164 cannot trigger these early steps of ciliogenesis when TTBK2 is mislocalized.
Functional Consequences of TTBK2–Cep164 Complex Formation.
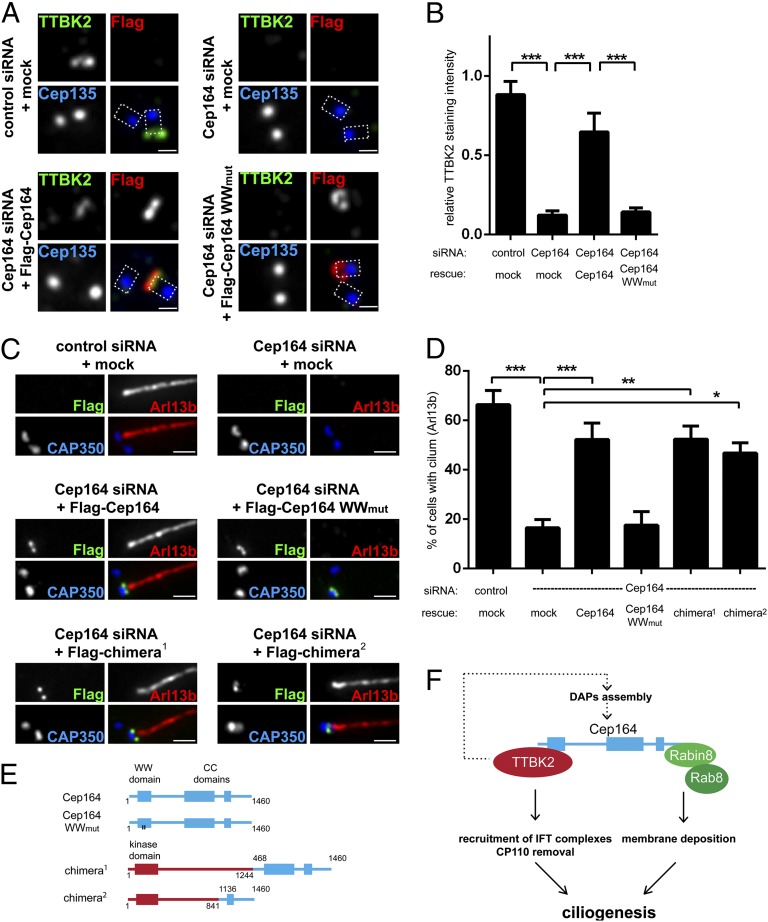
On the basis of the results described above, we hypothesized that one of the key functions of Cep164 consists of the recruitment of TTBK2 to the appendages of M centrioles. This recruitment, mediated by the WW domain within the N terminus of Cep164, appears then to be sufficient to trigger both CP110 removal and IFT81 recruitment in preparation for ciliogenesis. To rigorously test this hypothesis, we carried out a series of siRNA-depletion and rescue experiments. The extent of Cep164 depletion achieved in these experiments is illustrated in Fig. S4A. Expression of a siRNA-resistant version of Cep164 (Cep164 RNAi-res) in Cep164-depleted cells restored TTBK2 levels at M centrioles to ∼70% of controls, and, importantly, this rescue required an intact WW domain within Cep164 (Fig. 5 A and B). In parallel experiments, PC formation could be restored to Cep164-depleted cells by expression of Cep164 RNAi-res wild type (WT), but not the corresponding WWmut construct (Fig. 5 C and D).
Fig. 5.
Tethering of TTBK2 to M centrioles rescues ciliogenesis in the absence of full-length Cep164. (A–D) Transfections for siRNA-depletion/rescue experiments were carried out in RPE-1 cells, using the indicated siRNA oligonucleotides and rescue constructs; cells were then processed as outlined in the legend to Fig. 1. Cep135 staining visualizes proximal site of centrioles, and dashed lines in the merged images outline the centrioles. [Scale bars: 0.5 μm (A) and 1 μm (C).] (A) Expression of Cep164 full-length protein, but not the WWmut mutant, restores TTBK2 levels at M centrioles in Cep164-depleted cells. (B) Quantification of the rescue data shown in A. Graph represents the summary of three experiments. (C) Representative images illustrating the ability of the indicated constructs to rescue ciliogenesis. Staining for CAP350 and Arl13b was used to visualize centrioles and PC formation, respectively. (D) Quantification of the rescue of the rescue data shown in C. Graph represents the summary of at least four experiments. (E) Schematic representations of chimeric constructs used for siRNA-depletion/rescue experiments. Cep164 moieties are depicted in blue, and TTBK2 moieties are in red. (F) Model summarizing the crucial functions of TTBK2 and Cep164 domains in PC formation (see also Discussion). *P < 0.05; **P < 0.01; ***P < 0.001.
To confirm the critical role of the N-terminal domain of Cep164 for TTBK2 recruitment and ciliogenesis, we examined the functionality of TTBK2–Cep164 fusion proteins. The structures of these chimeric proteins are illustrated in Fig. 5E. Remarkably, PC formation could be restored in Cep164-depleted cells by reexpression of two different fusion proteins in which TTBK2 is directly linked to C-terminal (appendage-targeting) domains of Cep164 (Fig. 5 C and D and Fig. S4B). These results demonstrate that enforced M centriole targeting of TTBK2 via fusion to C-terminal fragments of Cep164 is sufficient to allow PC formation, even in the absence of full-length Cep164. To explore the possibility of a complete bypass of Cep164, we also targeted TTBK2 to M centrioles via fusion to the DAP Cep89 (19, 31). However, although three different TTBK2–Cep89 chimeras (Fig. S4C) all conferred M centriole localization to TTBK2, none were able to restore ciliogenesis in Cep164-depleted cells (Fig. S4D). At face value, these negative results indicate that the C terminus of Cep164 is not dispensable, in line with a recent study identifying an essential role of this domain in the binding of Rab8a/Rabin and the recruitment of ciliary vesicles (18). We thus conclude that Cep164 comprises at least two functionally distinct domains: Whereas the C-terminal domain is crucial for centriole association and the recruitment of membrane vesicles (18), the N-terminal WW domain functions primarily to recruit TTBK2 to the M centriole; this kinase is then proposed to trigger at least two early steps in ciliogenesis, CP110 removal and IFT81 recruitment (Fig. 5F).
Evidence for an Additional Role of TTBK2 in the Assembly of Distal Appendages.
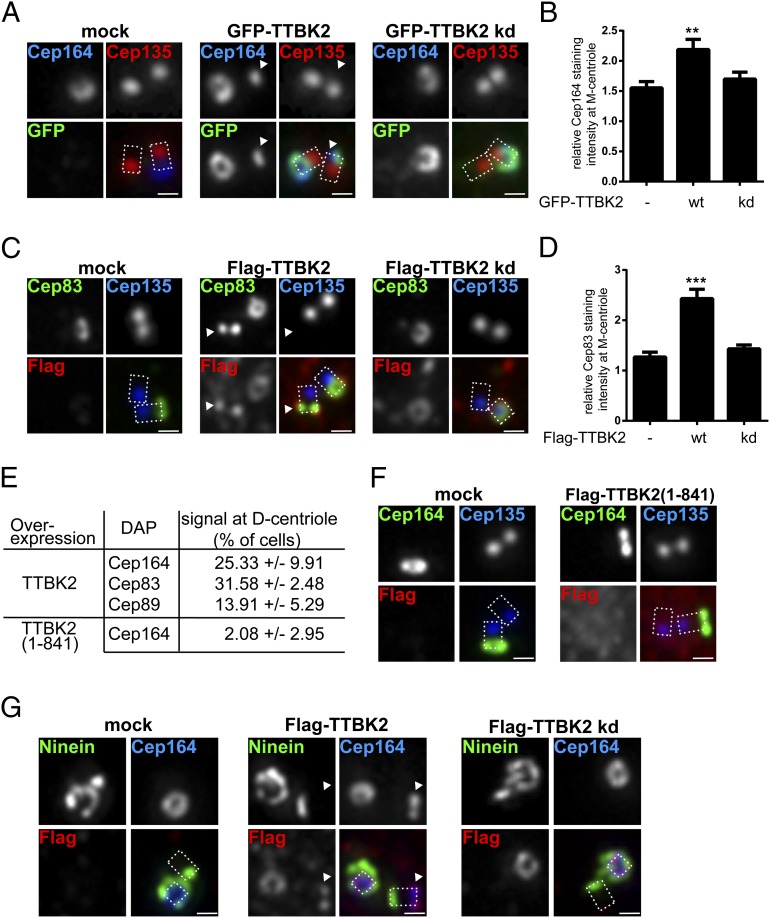
The above model for the role of TTBK2 in ciliogenesis emphasizes kinase activities downstream of Cep164. However, some of our results hint at additional activities upstream of Cep164. In particular, we had observed that coexpression of active TTBK2 with Cep164 resulted in the accumulation of Cep164, possibly reflecting stabilization (Fig. 3D). To follow up on this observation, we used immunofluorescence microscopy to monitor the localization of Cep164 and other DAPs in response to expression of exogenous TTBK2 in RPE-1 cells. Overexpression of TTBK2 WT, but not a kd mutant, markedly increased Cep164 levels at the M centriole (Fig. 6 A and B), and similar effects were seen for the DAPs Cep83 (Fig. 6 C and D) and Cep89 (Fig. S5 A and B). Moreover, and most remarkably, we also observed the accumulation of GFP- or Flag-tagged TTBK2 at the distal ends of D centrioles (Fig. 6 A and B). As quantified in Fig. 6E, this unexpected localization was accompanied by the concomitant recruitment of the DAPs Cep164 (Fig. 6A), Cep83 (Fig. 6C), and Cep89 (Fig. S5A). This finding strongly indicates that excess TTBK2 activity is able to trigger aspects of centriole maturation, notably the recruitment of DAPs to immature centrioles. Interestingly, a TTBK2 mutant (1–841) defective in Cep164 binding was impaired in its ability to induce ectopic assembly of DAPs at D centrioles, indicating that the C-terminal domain of TTBK2 is important for this function (Fig. 6 E and F).
Fig. 6.
TTBK2 promotes assembly of DAPs at both M and D centrioles. Transfections of RPE-1 cells and immunofluorescence experiments were carried out as outlined in the Fig. 1 legend. Cep135 staining was used to label proximal ends of centrioles, and dashed lines in merged images outline the centrioles. (Scale bars: 0.5 μm.) Arrowheads point to distal parts of D centrioles. (A) Transient overexpression of GFP–TTBK2 promotes Cep164 localization to both M and D centrioles. (B) Quantification of the effects of TTBK2 overexpression on Cep164 levels at M centrioles. In cells where both centrioles showed Cep164 staining, the centriole showing more prominent Cep164 signal was considered as the M centriole. Graph represents the summary of two experiments. (C) Transient overexpression of Flag–TTBK2 promotes Cep83 localization to both M and D centrioles. (D) Quantification of the effects of TTBK2 overexpression on Cep83 levels at M centrioles. Graph represents the summary of two experiments. (E) Frequency of D centriole localizations of Cep164, Cep83, and Cep89 observed in transfected cells. Results were collected from at least two experiments. (F) Lack of influence of Flag–TTBK2 (1–841) overexpression on Cep164 localization. (G) Flag–TTBK2 promotes distal D centriole localization of Cep164 but not Ninein. Flag–TTBK2 kd is shown for control. **P < 0.01; ***P < 0.001.
To exclude the possibility that the unexpected phenotype produced by excess TTBK2 activity might reflect generic effects on centriole structure and/or cell-cycle progression, we also examined the effects of TTBK2 overexpression on the localization of Ninein and Cep170, two proteins known to associate with both subdistal appendages (29, 44) and the proximal ends of centrioles (45, 46). Although Flag–TTBK2 promoted localization of Cep164 to the distal end of D centrioles, no recruitment to these ectopic sites was seen for either Ninein (Fig. 6G) or Cep170 (Fig. S5C). Likewise, the asymmetry in Ninein localization between M and D centrioles was maintained in cells in which Flag–TTBK2 localized to D centrioles (Fig. S5D). These experiments demonstrate that TTBK2 specifically promotes the assembly of distal appendage components but not subdistal appendage proteins. We conclude that the range of activities of TTBK2 is not limited to events that occur downstream of Cep164. Instead, our data suggest that TTBK2 also contributes to the assembly of distal appendages, hinting at the existence of a positive feedback loop centered on the Cep164–TTBK2 complex (Fig. 5F).
Discussion
The appendage protein Cep164 plays a key role in the formation of PCs (18, 29). Here we have explored the mechanistic basis for this requirement. We demonstrate that a major role of Cep164 consists of the recruitment of a casein kinase I family member, TTBK2, to appendages of M centrioles. We show that Cep164 interacts via its N-terminal WW domain with the C-terminal domain of TTBK2 and that this interaction is required for both the recruitment of TTBK2 to appendages and for PC formation. The impairment of ciliogenesis in Cep164-depleted cells can be rescued by reexpression of Cep164 WT, but not a WW domain mutant that is defective in TTBK2 binding. Furthermore, ciliogenesis can also be restored by reexpression of chimeric proteins in which TTBK2 is targeted to distal appendages via fusion to the C-terminal domain of Cep164. Once associated with appendages, TTBK2 is then in the appropriate place to trigger ciliogenesis by regulating both the recruitment of IFT components and the release of CP110 from M centrioles (see also ref. 35).
In a recent study, the C-terminal moiety of Cep164 was implicated in the binding of a Rab8a/Rabin complex, which in turn controls the docking of Golgi-derived vesicles to M centrioles (18). A role for Rab8a in ciliogenesis is well supported by data showing that GTP loading of Rab8a is important for both centrosome localization and vesicle trafficking (40, 47–49). By overexpression of TBC1D30, a Rab8a GAP, we could readily confirm that interference with Rab8a activity strongly suppresses PC formation. However, excess TBC1D30 did not detectably influence either the recruitment of IFT81 or the removal of CP110 from centrioles, arguing that these two key events are regulated independently of the Rab8a/Rabin interaction with Cep164.
As reported previously (18) and confirmed here, a N-terminal fragment of Cep164 exerts a dominant-negative effect on PC formation. Our finding that the N-terminal Cep164 fragment localizes to the cytoplasm and thus sequesters TTBK2 away from M centrioles provides an attractive explanation for the observed phenotype. Our siRNA-depletion/rescue experiments identified the WW domain within the N terminus of Cep164 as critical for TTBK2 recruitment and PC formation, clearly demonstrating the importance of the Cep164–TTBK2 interaction for ciliogenesis. This conclusion is strengthened further by our demonstration that PC formation could be restored to Cep164-depleted cells by chimeric proteins in which TTBK2 was fused to the C-terminal domain of Cep164. This finding strongly suggests that centriole recruitment of TTBK2 represents the key function of the N-terminal domain of Cep164, although it would be premature to exclude additional, perhaps more subtle, functions for this domain. The C-terminal moiety of Cep164, conversely, is both necessary and sufficient for correct localization to centriolar appendages. In addition, this domain has been implicated in the docking of Golgi-derived vesicles during early stages of PC formation (Fig. 5F) (18). This result provides a plausible explanation for the failure of our attempts at bypassing Cep164 through expression of TTBK2–Cep89 chimeras in Cep164-depleted cells.
Recently, information has begun to emerge on proteins that are likely to function upstream of Cep164 and TTBK2. These include Cep83/CCDC41 (19, 30) and C2CD3 (50–52). Although a strict requirement for Cep83/CCDC41 in Cep164 localization has been questioned (30), our data clearly demonstrate that Cep164 is both necessary and sufficient for M centriole association of TTBK2, in line with a model in which Cep83/CCDC41 acts upstream of Cep164 (19). C2CD3, a C2 domain protein required for ciliogenesis in mammals (50–52), has also been shown to be important for the recruitment of TTBK2 to basal bodies (52). This protein was reported to localize to centriolar satellites (52) as well as the distal ends of both M and D centrioles (51, 52). Furthermore, C2CD3 is required not only for ciliogenesis but also for centriole biogenesis (51), indicating that it functions hierarchically upstream in the assembly of appendages.
Our current understanding of TTBK2 function emphasizes events in ciliogenesis that are presumed to occur downstream of Cep164-mediated recruitment to distal appendages. However, in the course of our studies, we obtained evidence for an additional role of this kinase in the assembly of DAP complexes. In particular, we discovered that overexpression of TTBK2 led to the accumulation of Cep164 and the concomitant enrichment of Cep164 as well as other DAPs at the distal ends of M centrioles. In addition, and most surprisingly, excess TTBK2 was also able to trigger the recruitment of DAPs to the distal ends of D centrioles. Given that Cep164 is required to position TTBK2 for the downstream functions of this kinase, as monitored by CP110 removal and IFT81 recruitment, the ability of overexpressed TTBK2 to enhance the recruitment of Cep164 and other DAPs to distal appendages may appear puzzling. If TTBK2 were strictly required for distal appendage formation, one might in fact have expected to see an impairment of Cep164 assembly in response to depletion or sequestration of TTBK2. However, this impairment was not observed in either this study (Fig. 2 D and E and Fig. S3 C and D) or elsewhere (19, 35). One possible explanation for this conundrum is that TTBK2 functions redundantly to other kinases, particularly members of the casein kinase 1 family, in DAP assembly. In addition, we consider it attractive to postulate the existence of a positive feedback loop centered on the TTBK2–Cep164 complex (Fig. 5F). A self-promoting recruitment mechanism may enhance the local concentration of this complex, leading to higher phosphorylation of substrates that are critical for DAP assembly. In support of this hypothesis, we emphasize that the induction of DAP recruitment by overexpressed TTBK2 required not only an intact catalytic domain but also the C-terminal domain that binds Cep164 (Fig. 6 E and F). In light of a previous report on the regulation of TTBK2 activity (41), one could also argue that the C-terminal domain of TTBK2 may be required for regulation of kinase activity. Although we cannot rigorously exclude this possibility, we note that our rescue experiments with TTBK2–Cep164 chimeras demonstrate that constructs lacking the C-terminal part of TTBK2 exhibited sufficient kinase activity to restore ciliogenesis in Cep164-depleted cells (Fig. 5 C and D and Fig. S4B).
Mutations in both Cep164 and TTBK2 are causally related to human disease. In particular, mutations in Cep164 have been implicated in the etiology of nephronophthisis-related ciliopathies, and a link to DNA-damage response signaling has been proposed (32). In line with this notion, the N terminus of Cep164 was proposed to interact with components of DNA damage pathways (53). Conversely, truncating mutations in human TTBK2 were identified as causative for the neurodegenerative disorder spinocerebellar ataxia type 11 (36, 54). Our domain-mapping experiments predict that the longest ataxia-associated TTBK2 truncation described, 450X (36), is unable to form a stable complex with Cep164 (Figs. 2H and 3E). Thus, the inability of ataxia-associated forms of TTBK2 to bind Cep164 provides a plausible explanation for the localization defects of these truncation mutants in murine cells (35). In consideration of the existence of the TTBK2–Cep164 complex characterized in this study, it is intriguing that mutations in the two proteins have been linked to different disease spectra, suggesting cell-type-specific functions and/or redundancies. Also, it might be rewarding to explore whether TTBK2 plays a role in nephronophthisis-related ciliopathies, and, conversely, whether Cep164 mutations can be found in ataxia patients.
TTBK2 was originally named for its ability to phosphorylate tau protein on two serine residues and thus regulate tau oligomerization (55). Tau is unlikely to be the only target, but little is presently known about the physiological substrates of TTBK2. It is interesting, therefore, that TTBK2 readily phosphorylates Cep164 in vitro and, most likely, in vivo. Given the colocalization of the two proteins and their intimately linked roles in ciliogenesis, Cep164 appears to be an attractive candidate substrate of TTBK2. Our preliminary analyses of Cep164 phosphorylation by TTBK2 point to a multitude of phosphorylation sites distributed throughout the protein, suggesting complex regulation. This finding notwithstanding, it will be interesting to explore the functional consequences of Cep164 phosphorylation in future studies. Furthermore, considering that TTBK2 controls the recruitment of IFT components as well as the dissociation of CP110 from M centrioles (ref. 35 and this study), these proteins, as well as their potential binding partners (20, 56, 57), represent attractive candidates for TTBK2 substrates. Understanding the functional consequences of substrate phosphorylation by TTBK2 as well as the interaction of TTBK2 with other kinases implicated in ciliogenesis (33, 34, 37) will undoubtedly be critical for a mechanistic understanding of ciliogenesis.
Materials and Methods
Cell Culture and Transfections.
RPE-1 and HEK293T cells were propagated as described (21) and transfected with Lipofectamine LTX (Invitrogen) and TransIT-LT1 (Mirus), respectively. siRNA experiments were performed by using Oligofectamine (Invitrogen) and described control (GL2) and Cep164-specific oligonucleotides at concentrations of 50 nM (29). To knock down TTBK2, Silencer siRNA (Life Technologies; s44813 and s44814) was used at a concentration of 5 nM. Efficient knockdown was routinely verified by immunofluorescence staining and/or Western blotting. Following transfections of RPE-1 cells, ciliogenesis was promoted either by serum starvation for 24 h or, in the case of the experiments described in Fig. 4 A and B and Fig. S3A, the use of 0.5 μM Cytochalasin D (Sigma).
Cloning.
Insert-containing entry vectors for use in the GATEWAY system (Invitrogen) were generated by PCR, using Pfu Ultra II Fusion DNA polymerase (Agilent). Constructs were verified by sequencing and subsequently cloned into pDEST-Myc (58), pDEST-Flag (58), pDEST-GST (Invitrogen), or pg-LAP1 (Addgene plasmid 19702) GATEWAY destination vectors. The human cDNAs used here have been described: Cep164 (29), TTBK2 (35), TBC1D30 (40), and Cep89 (31). Inserts for TTBK2–Cep164/Cep89 fusions, containing a 3xGly linker, were constructed by overlapping PCR. Mutants were prepared by site-directed mutagenesis (Agilent). All primers are listed in Table S1.
Cell Lysis and Immunoprecipitation.
At 24 h posttransfection, cells were washed in PBS and lysed in Lysis buffer [20 mM Tris⋅Cl, pH 7.4, 150 mM NaCl, 25mM β-glycerol phosphate, 0.5% Triton-X-100, 0.5% Igepal CA630 (all from Sigma), and 1× Complete proteasome inhibitors (Roche)]. Following centrifugation (15,000 × g for 10 min at +4 °C), cleared extracts were incubated (6 h at +4 °C in an orbital shaker) with anti-Myc (clone 9E10) or anti-Flag (clone M2) G protein Sepharose (GE Healthcare)-coupled antibodies. Immune complexes were pelleted, washed, and subsequently analyzed by SDS/PAGE and Western blotting. Where indicated, samples were treated with λ-phosphase (New England BioLabs) (30 °C for 20–90 min). Luminescence was detected by using a SuperSignal Femto Maximum Sensitivity Substrate (Thermo Scientific) in the LAS3000 system (GE Healthcare). Where appropriate, contrast and/or brightness of images were adjusted by using Photoshop CS5 (Adobe). The following antibodies were used: rabbit anti-Cep164 (29), mouse anti-FLAG (clone M2; Sigma), mouse anti-Myc (clone 9E10), goat anti-GST (45-001-369; GE Healthcare), rabbit anti-GFP (ab290; Roche), rabbit anti-TTBK2 (HPA018113; Sigma), horseradish peroxidase (HRP)-conjugated goat anti-mouse and anti-rabbit secondary antibodies (BioRad), and HRP-conjugated donkey anti-goat secondary antibody (Santa Cruz Biotechnology).
Recombinant Protein Purification.
BL21-RIL bacteria, transformed with appropriate plasmids, were grown overnight (37 °C) in LB medium (100 μg/mL Ampicillin) and expanded the following day for another 3–5 h. Expression of recombinant proteins was induced by 0.2 mM IPTG (20 °C, 12 h). After centrifugation, bacteria were resuspended in 3 mL of Lysis buffer (20 mM Tris⋅Cl, pH 7.4, 150 mM NaCl, 1% Triton-X-100, 1 mM DTT, 1× Complete proteasome inhibitors) and repeatedly sonicated (2×, 20 s) and freeze-thawed. Following clearing of the extract (15,000 × g for 10 min at +4 °C), GST-tagged recombinant proteins were purified (2 h at +4 °C in an orbital shaker) by using Glutathione Sepharose 4B (GE Healthcare), washed (20 mM Tris⋅Cl, pH 7.4, 300 mM NaCl, 1% Triton X-100, 1 mM DTT, 1× Complete proteasome inhibitors), eluted [50 mM Tris⋅Cl, pH 8, 50 mM NaCl, 10 mM Glutathione, 1 mM DTT, 5% (vol/vol) glycerol], and stored at −80 °C. Chemicals were purchased from Sigma.
In Vitro Translation and Kinase Assay.
Flag–Cep164 was obtained by using the in vitro translation TNT Quick system (Promega), mixed with Lysis buffer (see above; cell lysis), and subjected to anti-Flag immunoprecipitation (3 h at +4 °C in an orbital shaker) and washing. In vitro kinase assays were performed in 50 mM Tris, pH 7.4, 10 mM MgCl2, and 0.1 mM EGTA in the presence of 10 μM ATP, 2 μCi of [γ-32P]ATP and recombinant kinase (30 °C for 30 min). To terminate reactions, samples were mixed with 2× sample buffer, boiled for 5 min, and subjected to SDS/PAGE and PVDF membrane transfer. 32P incorporation was detected by using Kodak BioMAX films (Sigma).
Immunofluorescence Microscopy.
Methanol fixation, blocking, incubation with primary and secondary antibodies, and washing were performed as described (45). The antibodies used were as follows: mouse antiacetylated tubulin (6-11B-1; Sigma), mouse antiglutamylated tubulin (GT335), rabbit anti-Arl13b (17711; Proteintech), mouse anti-Flag (M2; Sigma), rabbit anti-Cep83 (HPA038161; Sigma), rabbit Cep89 (31), rabbit anti-TTBK2 (HPA018113; Sigma; Alexa 488 or 647-labeled), goat anti-CAP350 (59) (Alexa 647-labeled), rabbit anti-Cep135 (60) (Alexa 555- or 647-labeled), rabbit anti-CP110 (60) (Alexa 488-labeled), rabbit anti-Cep164 (29) (Alexa 488- or 647-labeled), rat anti-IFT81 (38), Alexa 488 anti-rabbit, Alexa 488 anti-rat, Alexa 555 anti-mouse, and Alexa 555 anti-rabbit (all from Invitrogen). Direct labeling of primary antibodies was performed by using an Alexa-antibody labeling kit (Invitrogen). Imaging was performed on a DeltaVision system (Applied Precision) with a 60×/1.2 or 100×/1.4 Apo plan oil immersion objective. Image stacks were taken with a z distance of 0.2 μm, deconvolved (conservative ratio, three to five cycles), and projected as maximal intensity images by using SoftWoRX (Applied Precision). Where appropriate, contrast and/or brightness were adjusted by using Photoshop CS5 (Adobe). For cell counts, 100 (siRNA experiments), 25–50 (transgene expression experiments), or 15–25 (siRNA rescue experiments) cells per condition and experiment were analyzed. Where indicated, a densitometry analysis within selected regions of interest was performed in 16-bit TIFF images by using ImageJ, analyzing 15–20 cells per experiment and condition. Data are presented as relative staining intensity (staining intensity of a protein of interest normalized to the staining intensity of the centriolar marker Cep135).
Rescue Experiments.
At 4–5 h after transfection of RPE-1 cells, medium was changed to prevent cell death. At 26–30 h later, cells were transfected with appropriate siRNA oligonucleotides. At 72 h after the initial plasmid transfection, medium was changed (1% FBS) to promote the formation of PC for 24 h. Control experiments were carried out in parallel to verify efficient knockdown of Cep164 protein levels by Western blotting.
Phosphorylation Site Mapping by MS.
Myc–Cep164 C-term protein complexes were immunoprecipitated from HEK293T cell extracts 24 h after transfection. Proteins were reduced, alkylated, digested with trypsin overnight, and then purified with C18 Microspin columns (Harvard Apparatus) according to manufacturer's instructions. Eluted peptides were separated by using an Easy-Nano-LC system, and liquid chromatography–tandem MS analysis was performed on a hybrid LTQ-Orbitrap mass spectrometer (both from Thermo Scientific). Obtained spectra were searched by Mascot against the human proteome database (UniProt). Carbamidomethylation was set as fixed modification. Oxidation and phosphorylation (Ser/Thr/Tyr) were considered as variable modification. Results were visualized and analyzed by using the Scaffold package (Proteome Software).
Statistical Analyses.
Statistical analyses (Student t test and one-way ANOVA with multiple comparison tests) were performed by using Prism (Version 6; GraphPad Software). P < 0.05, P < 0.01, and P < 0.001 were considered as statistically significant differences. Results are presented as mean plus SEM.
Supplementary Material
Acknowledgments
We thank Peter Jackson, Francis Barr, James Sillibourne, Kathryn Anderson, and Sarah Goetz for reagents; Elena Nigg, the staff of the Biozentrum Imaging facility (notably Alexia Ferrand) and the Proteomics core facility (Timo Glatter) for assistance; and all members of the E.A.N. laboratory for helpful discussions. This work was supported by Swiss National Science Foundation Grants 31003A_132428 and 310030B_149641 and the University of Basel. L.C. was supported by a long-term fellowship from the Federation of European Biochemical Societies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401777111/-/DCSupplemental.
References
- 1.Singla V, Reiter JF. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science. 2006;313(5787):629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: A signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137(1):32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123(Pt 4):499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishikawa H, Marshall WF. Ciliogenesis: Building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12(4):222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 6.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139(4):663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Seeley ES, Nachury MV. The perennial organelle: Assembly and disassembly of the primary cilium. J Cell Sci. 2010;123(Pt 4):511–518. doi: 10.1242/jcs.061093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193(3):435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michaud EJ, Yoder BK. The primary cilium in cell signaling and cancer. Cancer Res. 2006;66(13):6463–6467. doi: 10.1158/0008-5472.CAN-06-0462. [DOI] [PubMed] [Google Scholar]
- 11.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27(8):307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma N, Berbari NF, Yoder BK. Ciliary dysfunction in developmental abnormalities and diseases. Curr Top Dev Biol. 2008;85:371–427. doi: 10.1016/S0070-2153(08)00813-2. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3(2):207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 16.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol. 2009;19(17):1498–1502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464(7291):1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt KN, et al. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol. 2012;199(7):1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanos BE, et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27(2):163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130(4):678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19(12):1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Pazour GJ, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151(3):709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164(2):255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3(11):813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43(8):776–784. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192(6):1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 28.Reiter JF, Blacque OE, Leroux MR. The base of the cilium: Roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13(7):608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179(2):321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joo K, et al. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci USA. 2013;110(15):5987–5992. doi: 10.1073/pnas.1220927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sillibourne JE, et al. Primary ciliogenesis requires the distal appendage component Cep123. Biol Open. 2013;2(6):535–545. doi: 10.1242/bio.20134457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaki M, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150(3):533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Upadhya P, Birkenmeier EH, Birkenmeier CS, Barker JE. Mutations in a NIMA-related kinase gene, Nek1, cause pleiotropic effects including a progressive polycystic kidney disease in mice. Proc Natl Acad Sci USA. 2000;97(1):217–221. doi: 10.1073/pnas.97.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, et al. A defect in a novel Nek-family kinase causes cystic kidney disease in the mouse and in zebrafish. Development. 2002;129(24):5839–5846. doi: 10.1242/dev.00173. [DOI] [PubMed] [Google Scholar]
- 35.Goetz SC, Liem KF, Jr, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151(4):847–858. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houlden H, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39(12):1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- 37.Kuhns S, et al. The microtubule affinity regulating kinase MARK4 promotes axoneme extension during early ciliogenesis. J Cell Biol. 2013;200(4):505–522. doi: 10.1083/jcb.201206013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhogaraju S, et al. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341(6149):1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83(2):S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178(3):363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouskila M, et al. TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, activity, localization and development. Biochem J. 2011;437(1):157–167. doi: 10.1042/BJ20110276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513(1):30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 43.Ingham RJ, et al. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25(16):7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guarguaglini G, et al. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16(3):1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1(10):965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krauss SW, et al. Downregulation of protein 4.1R, a mature centriole protein, disrupts centrosomes, alters cell cycle progression, and perturbs mitotic spindles and anaphase. Mol Cell Biol. 2008;28(7):2283–2294. doi: 10.1128/MCB.02021-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 48.Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22(20):1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci USA. 2011;108(7):2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoover AN, et al. C2cd3 is required for cilia formation and Hedgehog signaling in mouse. Development. 2008;135(24):4049–4058. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balestra FR, Strnad P, Flückiger I, Gönczy P. Discovering regulators of centriole biogenesis through siRNA-based functional genomics in human cells. Dev Cell. 2013;25(6):555–571. doi: 10.1016/j.devcel.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 52.Ye X, Zeng H, Ning G, Reiter JF, Liu A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci USA. 2014;111(6):2164–2169. doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008;22(5):587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauer P, et al. Spinocerebellar ataxia type 11 (SCA11) is an uncommon cause of dominant ataxia among French and German kindreds. J Neurol Neurosurg Psychiatry. 2010;81(11):1229–1232. doi: 10.1136/jnnp.2009.202150. [DOI] [PubMed] [Google Scholar]
- 55.Tomizawa K, Omori A, Ohtake A, Sato K, Takahashi M. Tau-tubulin kinase phosphorylates tau at Ser-208 and Ser-210, sites found in paired helical filament-tau. FEBS Lett. 2001;492(3):221–227. doi: 10.1016/s0014-5793(01)02256-6. [DOI] [PubMed] [Google Scholar]
- 56.Tsang WY, Dynlacht BD. CP110 and its network of partners coordinately regulate cilia assembly. Cilia. 2013;2(1):9. doi: 10.1186/2046-2530-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145(6):914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 58.Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125(Pt 5):1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- 59.Yan X, Habedanck R, Nigg EA. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol Biol Cell. 2006;17(2):634–644. doi: 10.1091/mbc.E05-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.