Significance
Mechanisms of protein–ligand binding have long generated interest in wide circles, including enzyme catalysis, receptor function, and drug design. This study reconciles some of the conflicting views. Using simulations of a model protein–ligand system, we demonstrate that neither conformational selection (CS) nor induced fit (IF) is the sole mechanism governing protein–ligand binding. Instead, both intrinsic and extrinsic factors can shift the binding mechanism between CS and IF. CS dominates when both protein conformational transition rate and ligand concentration are low. With the increase of either factor, the binding mechanism shifts to IF. The rigorous theoretical framework established here can guide experiments and simulations in determining the fraction of binding events achieved via either CS or IF.
Keywords: protein–ligand complex, conformational dynamics, diffusion-influenced bimolecular reaction, induced-fit fraction
Abstract
This study aimed to shed light on the long debate over whether conformational selection (CS) or induced fit (IF) is the governing mechanism for protein–ligand binding. The main difference between the two scenarios is whether the conformational transition of the protein from the unbound form to the bound form occurs before or after encountering the ligand. Here we introduce the IF fraction (i.e., the fraction of binding events achieved via IF), to quantify the binding mechanism. Using simulations of a model protein–ligand system, we demonstrate that both the rate of the conformational transition and the concentration of ligand molecules can affect the IF fraction. CS dominates at slow conformational transition and low ligand concentration. An increase in either quantity results in a higher IF fraction. Despite the many-body nature of the system and the involvement of multiple, disparate types of dynamics (i.e., ligand diffusion, protein conformational transition, and binding reaction), the overall binding kinetics over wide ranges of parameters can be fit to a single exponential, with the apparent rate constant exhibiting a linear dependence on ligand concentration. The present study may guide future kinetics experiments and dynamics simulations in determining the IF fraction.
The binding of proteins to small molecules (i.e., ligands) is central to many essential biological functions, including enzyme catalysis, receptor activation, and drug action. Generally, significant differences in protein conformation exist between the unbound and bound states, as exemplified by hemoglobin upon binding oxygen (1–4) and HIV-1 protease upon binding a substrate or a drug molecule (5). In the latter as well as some other cases (6–10), loops and other groups collapse around the bound ligand, leading to a closed binding pocket. The conformational redistribution and dynamics of the protein molecule exhibited during the binding process can potentially play a critical role in determining the magnitude of the rate constant as well as the mechanism of ligand binding (11, 12). Two mechanistic models have emerged as archetypes. In the induced-fit (IF) model, one assumes that, owing to interactions with the incoming ligand, the protein transitions from an “inactive” conformation to an “active” conformation (13). In the conformational-selection (CS) model, one assumes that the protein can preexist in the active conformation with a low probability, and it is when the protein is in this conformation that the ligand comes into contact, leading to productive binding (14). Both models have garnered defenders and detractors (15–19). This study aimed to shed light on the long debate over whether CS or IF is the governing mechanism for protein–ligand binding.
It has been suggested that observation of the active conformation without the ligand, akin to constitutive activity of receptors, is direct evidence of CS (17, 19). However, detractors of CS have noted that, at least for cases with a closed binding pocket in the active conformation, direct binding to the latter conformation cannot proceed (9, 15). In some cases, a partially closed conformation has been observed by a sensitive probe such as paramagnetic relaxation enhancement (20) or in molecular dynamics simulations. Accordingly, a revised model known as extended CS has been put forward (21–28), whereby the ligand binds to the partially closed conformation and then the protein–ligand system evolves to the bound state with the closed binding pocket. Although the divide between CS and IF is somewhat blurred by extended CS, strictly speaking the latter is an IF model, in the sense that the ligand binds to an inactive conformation (i.e., the partially closed conformation) before the protein adopts the final active conformation with the closed binding pocket. Indeed, a strict CS mechanism is not possible for a protein whose active conformation features a closed binding pocket. In any event, mere observation of the active conformation in the unbound state cannot be taken as proof of the CS mechanism. According to the Boltzmann distribution, every conformation, including the active conformation, has a certain equilibrium probability. Whether the active conformation can be observed depends on the magnitude of its equilibrium probability as well as the sensitivity of the probe. The binding mechanism should not change just because the probe has become more sensitive.
It thus seems that neither CS nor IF should be the sole dominant mechanism governing protein–ligand binding. What, then, are the determinants of binding mechanism? Hammes et al. (29) and Daniels et al. (30) have suggested that an increase in ligand concentration can shift the binding mechanism from CS to IF, because a higher ligand concentration would make binding more likely. The assumption is that that would increase the chance for the binding to occur before the conformational transition, but one cannot be certain without additional information about the dynamics and interactions of the protein and ligand molecules. Others have suggested that the timescale of the protein conformational transition, relative to the timescale of the ligand diffusional approach to the binding pocket, controls the binding mechanism (12), but the effect of ligand concentration was not studied.
To unequivocally determine the binding mechanism, one has to follow the protein–ligand relative translation and the protein internal motion, from the unbound state until two reactant molecules form the bound product. This process involves disparate types of dynamics, including ligand diffusion, protein conformational transition, and the final binding reaction. As the simplest model, protein conformational transition has been treated as gating, that is, the transitions between two conformational states are approximated as rate processes (31–33). The transition rates were initially assumed to be unaffected by protein–ligand interactions. More recently it was recognized that protein–ligand interactions necessarily influence the conformational transition rates and such influence is an essential ingredient of molecular recognition (12, 34, 35). Accordingly, the transition rates were assigned different values depending on whether the ligand is inside or outside the binding pocket, resulting in the dual-transition-rates model.
Here we studied the binding mechanism and kinetics of a system consisting of a concentration of ligand molecules surrounding a protein molecule whose conformational dynamics follows the dual-transition-rates model (Fig. 1A). From dynamics simulations, we calculate the IF fraction (i.e., the fraction of binding events achieved via IF) and show that the binding mechanism is shifted by both the rate of protein conformational transition and the concentration of ligand molecules. CS dominates at slow conformational transition and low ligand concentration. With the increase of either quantity, the binding mechanism shifts from CS to IF. The overall binding kinetics over wide ranges of parameters can be fit to a single exponential, with the apparent binding rate constant exhibiting a linear dependence on ligand concentration. The concentration dependence of the binding kinetics thus yields little information on the binding mechanism, but kinetics experiments and dynamics simulations can be designed to determine the IF fraction.
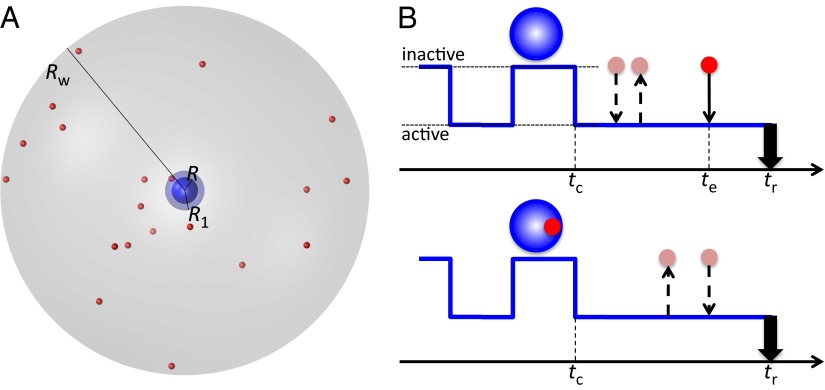
Fig. 1.
The model protein–ligand system and its binding mechanism. (A) A spherical protein is surrounded by point-like ligand molecules inside a spherical container (with radius Rw). The protein can transition between an inactive conformation and active conformation, and the transition rates depend on whether a ligand molecule is in the binding pocket (with inner and outer radii R and R1, respectively). (B) A binding event achieved through either the conformational selection (Upper) or the induced fit (Lower) mechanism. The crucial difference is whether the last inactive-to-active transition (at time tc) before the binding reaction (at tr) occurs with or without a loosely bound ligand molecule.
Results and Discussion
The Protein–Ligand System.
Our model system, illustrated in Fig. 1A, has the essential ingredients of ligand-binding proteins that undergo conformational transitions. Its simplicity allowed us to comprehensively explore the determinants of binding mechanism. The protein is modeled as an immobile sphere (radius R) with a thin-shell binding pocket (inner and outer radii R and R1 = 1.1R) that fluctuates between an inactive and active state. The ligand molecules, totaling N and each treated as a point-like particle, are confined between the protein surface (at radial distance r = R) and an outer wall (at r = Rw = 11R), mimicking the typical experimental condition with ligand excess. The resulting ligand concentration is . The basic setup of the system is similar to one in a previous study (33), but with the key difference that the transition rates between the inactive and active conformations depend on the positions of the ligand molecules, as further described below.
Each ligand molecule undergoes diffusion (with diffusion constant D) and interacts with the protein, giving rise to an interaction energy , where s = a or i for the active or inactive state and r denotes the position of the ligand molecule. is assumed to be uniformly 0, whereas is assumed to switch smoothly but sharply from 0 when r > R1 to U0 when R < r < R1:
The parameter L (set to 0.005R) measures the sharpness of the switch. We will refer to a ligand molecule with R < r < R1 as loosely bound. The ligand molecules do not have direct interactions with each other. Hereafter length and time will be reported in units of R and R2/D, respectively.
As already pointed out, we model the transitions of the protein between the inactive and active conformations as rate processes, with transition rates ω+ and ω–:
We use ω∞± to denote the values of the transition rates when all of the ligand molecules are far away from the protein. In that situation, the equilibrium constant between the active and inactive conformations is ω∞+/ω∞–. When the ligand molecules are at positions , the new equilibrium constant, given by the ratio of the transition rates, should satisfy the following detailed-balance relation:
where β is the inverse of the product of the Boltzmann constant and the absolute temperature. This generalizes a similar relation when only a single ligand molecule is present (12, 34). For simplicity, we set . Correspondingly, the inactive-to-active transition rate carries all of the influence of the protein–ligand interactions. We chose ω∞+/ω∞– = 0.1 and exp(–βU0) = 100, such that the equilibrium constant favors the inactive conformation when all of the ligands are far away but favors the active conformation when a ligand molecule is loosely bound. Such a shift in the more stable conformation from the inactive to the active is typical of ligand-binding proteins.
When the protein molecule is in the active conformation, a loosely bound ligand molecule has a chance to react with the protein to form the final complex. We model this last step also as a rate process (with rate constant γ), as done previously (33, 36, 37). We chose γ = 10; for a single protein–ligand pair with such a parameter value the binding process can be described as largely diffusion-controlled (as opposed to reaction-controlled) (34).
Overall the binding process involves three different types of dynamics: ligand diffusion, protein conformational transition, and the final binding reaction. Even though the ligand molecules do not directly interact with each other, they experience any conformational transition of the protein at the same time. The latter event introduces coupling between the ligand molecules (33), and as a result our protein–ligand system is many-body in nature. We carried out dynamics simulations of the system for ω– from 2 × 10−3 to 2 × 102 and for N from 5 to 100. In real units, the corresponding half-lives of the active conformation would be roughly 0.01 ms to 0.1 ns, and the corresponding ligand concentrations would be 0.2–4 mM. Each simulation was followed until either a binding reaction or a cutoff time. In the former case the time of its occurrence and whether it occurred via CS or IF were recorded.
Both Protein Dynamics and Ligand Concentration Can Shift the Binding Mechanism.
We use a strict definition of CS in this study (Fig. 1B, Upper). That is, a binding event is classified as CS only if no ligand molecule was inside the binding pocket when the protein transitioned to the active conformation for the last time (at tc) before reaction (at tr). The binding reaction would have to be between the protein and a ligand molecule that entered the binding pocket at some time (te) between tc and tr. Before tc, the protein could have made multiple transitions between the inactive and active conformations; between tc and te, one or more ligand molecules could have entered and left the binding pocket.
The alternative is that a ligand molecule first entered the binding pocket and the protein then made the last transition to the active conformation (Fig. 1B, Lower). The subsequent binding event is classified as IF. Between tc and tr, the loosely bound ligand molecule could have left and reentered the binding pocket, or could even be replaced by another ligand molecule. Irrespective of these nuances, the fact remains that the final conformational transition was caused by a loosely bound ligand molecule (thereby preparing the protein for the binding reaction), and hence an IF classification is appropriate.
From simulations for the transition rate ω– from 2 × 10−3 to 2 × 102 and for the number N of ligand molecules from 5 to 100, we determined the fraction, ΦIF, of binding reactions classified as IF. The results are shown as symbols in Fig. 2A. At a given N (or, equivalently, ligand concentration), ΦIF increases with increasing . For example, at N = 5, ΦIF increases from 0.10 at ω– = 2 × 10−3 to 0.99 at ω– = 2 × 102. As N increases, the IF fraction increases rapidly at the lowest ω– and less so at intermediate ω–, and shows little variation at ω– ≥ 1. Correspondingly, the shift from CS to IF upon increasing ω– becomes more and more gradual.
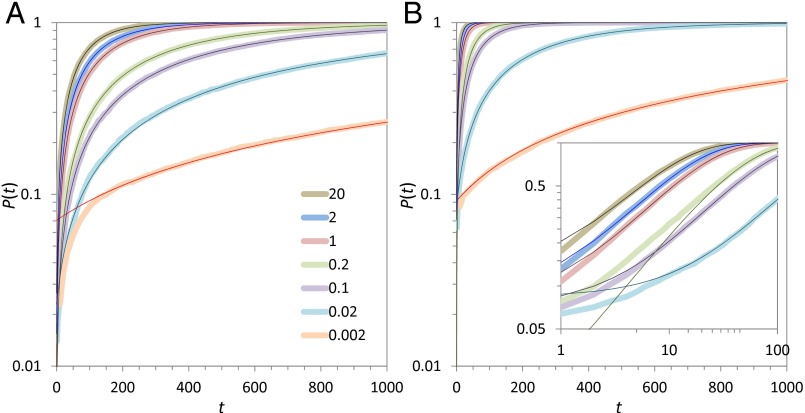
Fig. 2.
Dependence of the IF fraction (ΦIF) on conformational transition rate (ω–) and ligand concentration. (A) ΦIF values from the simulations at N = 5, 10, 20, 40, and 100, displayed as symbols from bottom to top. Error bars represent SDs of ΦIF values obtained from five batches of 1,000 simulations each. The curves show fits to Eq. 1. (Inset) Dependences of the fitting parameters ωtr, m, and ν on N; on the log-log scale shown, power-law dependences appear as linear. (B) Two-dimensional surface showing the simultaneous dependence of ΦIF on ω– and N.
We were able to fit the dependence of ΦIF on ω– to the following function:
| [1] |
where ωtr would be the midpoint of the shift from CS to IF if ν = 1, m directly controls the sharpness of the shift, and a ν that is less than 1 serves to raise ΦIF at ω– << ωtr and thus make the transition to IF less gradual (Fig. 2A, curves). All of the three parameters have strong dependences on N and can be fit to a power law (Fig. 2A, Inset). Applying the latter fits in Eq. 1, we can plot the dependence of ΦIF on ω– and N as a two-dimensional surface (Fig. 2B). This surface shows that CS dominates only when both the transition rate and the ligand concentration are small. The IF fraction increases when either the transition rate or the ligand concentration increases.
The effects of transition rate and ligand concentration on the shifting binding mechanism can be easily understood from our model. As described above, for a binding reaction to be CS, two requirements have to be satisfied. The protein must first transition to the active conformation while free of any ligand molecule and then stay in the active conformation until a ligand molecule enters the binding pocket and completes the binding reaction. At a low ligand concentration, the protein is free of any ligand molecule for most of the time, including some occasions when the protein transitions to the active conformation, thus allowing the first requirement of CS to be satisfied. If after such an occasion a ligand molecule enters the binding pocket and conformational transition is slow, then a binding reaction will occur before the protein has a chance to make another transition, thus satisfying the second requirement of CS.
An increase in ligand concentration makes it more likely to violate the first requirement of CS, whereas an increase in transition rate makes it more likely to violate the second requirement. Either way, the binding mechanism will shift toward IF, as further explained next. At a high ligand concentration, a ligand molecule will be inside the binding pocket most of the time, including occasions when the protein makes an inactive-to-active transition. After such an apparently ligand-induced transition, a low transition rate will likely keep the protein in the active conformation until the binding reaction. However, regardless of the level of ligand concentration, after a ligand molecule enters the binding pocket, a high transition rate will produce many transitions between the inactive and active conformations before the ligand molecule escapes. If one such inactive-to-active transition is followed by a binding reaction, then IF is again the binding mechanism.
Binding Kinetics Can Be Fit to a Single Exponential.
The ligand binding kinetics can be monitored by the probability, P(t), of product formation, which equals the fraction of reacted simulations at time t and was calculated from the recorded lifetimes. For each and each N in the aforementioned ranges, P(t) can be well fit to a single exponential:
| [2] |
where P0 is the nominal initial value of P(t) and kapp is the apparent rate constant for protein–ligand binding. In Fig. 3 we display the P(t) data and their fits for a range of ω– and N = 5 and 40. The correct initial value of P(t) for our system is 0. The fitting values of P0 are overall very close to 0 but rise to ∼0.1 at the lowest transition rate (i.e., ω– = 2 × 10−3) or the highest concentrations (N = 40 and 100). The deviation of from 0 indicates a small-amplitude fast phase, which would be within the dead time of typical experimental measurements.
Fig. 3.
The time dependence of the probability of product formation, fit to a single exponential. Simulation data and fits are displayed as thick and thin curves, respectively. (A) N = 5. The ω– values are shown as legend. (B) N = 40. (Inset) Short-time portions of the curves (except for the lowest ω– value) displayed on the log-log scale.
The kapp values are displayed in Fig. 4A as functions of N for different ω–. The dependence on N is close to being linear, as if the binding process obeys ordinary bimolecular reaction kinetics. This is despite the disparate types of dynamics involved and the many-body nature of the system.
Fig. 4.
The apparent rate constant for protein–ligand binding. (A) Nearly linear dependence of kapp on N. From bottom to top, the ω– values are 0.002, 0.02, 0.1, 0.2, 1, 2, 20, and 200. (Inset) Results for the six lowest ω– values displayed on the log-log scale. (B) Dependence of kapp on ω–, along with fits (dashed curves) to Eq. 3, at N = 5, 10, 20, 40, and 100 (from bottom to top). Error bars represent SDs of kapp values obtained from five batches of 1,000 simulations each; in many cases they are smaller than the corresponding symbols and hence are invisible.
At fixed N, kapp increases with increasing ω– (Fig. 4B). Therefore, speeding up conformational transition always accelerates ligand binding. The dependence of kapp on ω– can be fit to the following function:
| [3] |
where k∞ is the maximal kapp achieved when conformational transition is infinitely fast, ωhf is the value of ω– at which kapp is half maximal, and n measures the sharpness of the rise of kapp as ω– increases. In line with the linear dependence of kapp on N (Fig. 4A), k∞ exhibits a nearly linear dependence on N, whereas ωhf and n are nearly constant, around 0.9 and 0.8, respectively. With the conformational transition rate spanning five orders of magnitude (ω– from 2 × 10−3 to 2 × 102), only two orders of magnitude variation is found for kapp (Fig. 4B). The much narrower range of kapp accentuates the fact that conformational transition is but one of the three types of dynamics involved in the ligand-binding process.
It is interesting to note that the conditions under which IF prevails (i.e., high ligand concentration or high transition rate) are also the ones under which , the apparent rate constant for protein–ligand binding, is higher.
Comparison with a Four-State Model.
In many studies (15, 18, 19, 29, 38), a kinetic scheme involving four states has been used to model the binding kinetics:
where we have assigned ω∞± to the rate constants for the conformational transitions of the unbound protein and ω± to the counterparts when a ligand molecule is loosely bound. We further equate both the binding rate constants ki+ and ka+ to the Smoluchowski rate constant 4πDR1 and obtain the unbinding rate constants using the equilibrium constants and . In our model, the product is formed from with rate constant γ, but for the parameters studied this step can be approximated as very fast. To go from to , there are two pathways. The CS pathway has as the intermediate, whereas the IF pathway has as the intermediate.
By making the steady-state approximation for and , we obtain the apparent rate constant for protein–ligand binding:
| [4] |
It can be easily seen that the first term comes from the reactive flux along the CS pathway, whereas the second term is the counterpart along the IF pathway. The IF fraction can thus be obtained as
In Fig. 5 we compare the kapp and ΦIF predictions of the kinetic scheme with our simulation results. kapp is well predicted, except for the highest ligand concentration along with the lowest transition rate.* However, ΦIF is only semiquantitatively predicted. The plateauing at both the low and high ω– ends seems to be overly abrupt.
Fig. 5.
Predictions of kapp and ΦIF by the four-state kinetic scheme. (A) Comparison of predicted kapp (solid curves) against the simulation data, shown by the same dashed curves as in Fig. 4B. (B) Comparison of predicted ΦIF (solid curves) against the simulation data, shown by the same dashed curves as in Fig. 2A.
Concluding Remarks
Mechanisms of protein–ligand binding have long generated interest in wide circles, including enzyme catalysis, receptor function, and drug design. We have presented a comprehensive study on the binding mechanism and kinetics of a model protein–ligand system. Our results reconcile some of the conflicting views. In particular, the rate of protein conformational transition has been emphasized by some (12) as an intrinsic factor for dictating the binding mechanism, whereas the concentration of ligand molecules has been emphasized by others (29, 30) as an extrinsic factor. We show here that both factors can shift the binding mechanism between CS and IF and propose to use the IF fraction (i.e., the fraction of binding events achieved via IF) to quantify the binding mechanism. CS dominates (i.e., IF fraction is low) only when both conformational transition is slow and ligand concentration is low. With the increase of either conformational transition rate or ligand concentration the binding mechanism shifts to IF (i.e., toward high IF fraction).
Whereas it is very easy to change ligand concentrations, it may also be possible to perturb conformational transition rates by mutations. If conformations are separated by an energy barrier, mutations targeting the barrier are expected to directly affect the transition rates (while perhaps minimally affecting the conformational equilibrium). Examples include mutations in the hinge region of a protein that undergoes domain opening/closure (38–40). It will be interesting to test whether such mutations can produce a shift in binding mechanism.
Despite the many-body nature of the system and the involvement of multiple types of dynamics, and in contrast to the shifting binding mechanism, the binding kinetics is surprisingly simple. The time-dependent probability of product formation over wide ranges of model parameters can be fit to a single-exponential, and the apparent rate constant for protein–ligand binding has a linear dependence on ligand concentration. The concentration dependence of the binding kinetics thus provides little information on the binding mechanism of our system, even though such data have been suggested as a means for mechanism determination (19, 38).
However, we have found an apparent correlation between kapp and ΦIF (i.e., higher kapp corresponds to higher ΦIF, and vice versa). The correlation comes about because the same factors control both kapp and ΦIF. The increase (decrease) of either conformational transition rate or ligand concentration leads to the increase (decrease) of both kapp and ΦIF.
Our study does not support the idea (17, 19, 41) that sampling of the active conformation in the unbound state is evidence for CS. This finding in part reflects the strict definition of CS adopted here, which involves only the final inactive-to-active transition before the binding reaction (Fig. 1B, Upper). The mechanism is CS if this transition occurs when no ligand molecule is inside the binding pocket. Sampling of the active conformation in the unbound state is a necessary condition for CS but does not guarantee it. In fact, if the protein rapidly transitions between the inactive and active conformations (when the energy barrier separating them is low) in the unbound state, it may do so also while a ligand molecule is loosely bound. The latter situation, as our study indicates, will push the mechanism toward IF. Fundamentally, binding mechanism is dictated by the interplay of the various types of dynamics involved in the binding process, rather than by equilibrium properties alone.
The kind of dynamics simulations with a concentration of ligand molecules will be difficult to perform beyond the model system studied here. It is thus very helpful that the often-invoked four-state kinetic scheme is not bad for modeling the binding process. It makes good predictions for the apparent rate constant for protein–ligand binding and at least semiquantitative predictions even for the IF fraction.† The problem is then reduced to the determination of six elemental rate constants. Experimentally, single-molecule fluorescence resonance energy transfer measurements are now making such determination possible (38), and NMR spectroscopy is capable of determining at least the four conformational transition rates (10). It is also possible to compute these intra- and intermolecular rate constants by dynamics simulations. The resulting IF fraction will provide a full picture of the binding mechanism.
Computational Methods
The Simulations.
Three different events were simulated: ligand diffusion, protein conformational transition, and binding reaction. Ligand positions were updated according to the Ermak–McCammon algorithm (42):
where r0 is the old position, r is the new position after the timestep Δt (set to ), F0 is the force calculated from the interaction potential , and is a Gaussian random vector with zero mean and unit variance. Both the protein surface and the outer wall were treated as reflecting, with any new attempted position across a boundary placed back to the old position (36).
Both the transitions between conformations and the binding reaction were modeled as rate processes and implemented in similar ways. If the rate constant of a rate process has values ω0 and ω, respectively, before and after the ligand position update, then the chance for the reaction to occur in the present timestep is . The latter quantity was compared with a random number uniformly distributed between 0 and 1 to determine whether the reaction indeed took place. Any loosely bound ligand molecule could react with the protein. In the unlikely event (for the low ligand concentrations studied here) that more than one ligand molecule was loosely bound, they each could independently react with the protein.
Determination of Binding Mechanism.
To determine whether a binding reaction event was preceded by the CS or IF mechanism, we tracked the trajectories of the protein conformation and the ligand positions. For each set of model parameters, the simulations were repeated 5,000 times with different random number seeds. Initially the protein was assigned to the inactive and active conformations according to their equilibrium probabilities [ and , respectively] in the unbound state, and the ligand molecules were uniformly distributed outside the binding pocket.
For a binding event to be counted as CS, the protein would have to make an inactive-to-active transition while all of the ligand molecules were outside the binding pocket and stay in the active conformation until a ligand molecule loosely binds to and react with the protein. Any binding event not counted as CS was counted as IF. Each simulation was followed until either a binding reaction occurred or the time reached a preset cutoff time (tcut), which was 103 for all but the lowest ω– (i.e., 2 × 10−3) and 104 in the latter case. The binding reaction was assumed to be irreversible. Among the simulations that were terminated by binding reactions, the IF fraction was calculated. Alternatively, the growth of the IF count over time was fit to a single exponential similar to Eq. 2, and the extrapolated value at infinite time divided by the total number of simulations was taken as the final ΦIF. These methods yielded results that differ by less than 2%.
Monitoring of Binding Kinetics.
To monitor the binding kinetics of the protein–ligand system, the lifetimes of the simulations were recorded. From these lifetimes, the probability of product formation (i.e., the fraction of reacted simulations) at times up to tcut was calculated.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM58187.
Footnotes
The authors declare no conflict of interest.
*With a high ligand concentration and a low transition rate, there is some chance for more than one ligand molecule to be loosely bound. By allowing for at most one ligand molecule to be loosely bound (via moving the reflecting boundary from R to R1 when one ligand molecule is loosely bound), kapp is slightly reduced and is in better agreement with the prediction of the kinetic scheme. The restriction to single occupancy also slightly lowers the IF fraction (8% at N = 100 and ω– = 0.1).
†A simple application of the kinetic scheme is to predict how the strength of the interaction energy (i.e., |U0|) affects kapp and ΦIF. An increase in exp(–βU0) will lead to an increase in ω+ and correspondingly an increase in the reactive flux along the IF pathway (without affecting the CS pathway; see Eq. 4). As a result, both kapp and ΦIF would increase. This prediction was confirmed by simulations.
This article is a PNAS Direct Submission.
References
- 1.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12(1):88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 2.Perutz MF. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970;228(5273):726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 3.Szabo A. Kinetics of hemoglobin and transition state theory. Proc Natl Acad Sci USA. 1978;75(5):2108–2111. doi: 10.1073/pnas.75.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton WA, Henry ER, Hofrichter J. Application of linear free energy relations to protein conformational changes: The quaternary structural change of hemoglobin. Proc Natl Acad Sci USA. 1991;88(10):4472–4475. doi: 10.1073/pnas.88.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller M, et al. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989;246(4934):1149–1152. doi: 10.1126/science.2686029. [DOI] [PubMed] [Google Scholar]
- 6.Spurlino JC, Lu GY, Quiocho FA. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266(8):5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 7.Müller CW, Schulz GE. Structure of the complex between adenylate kinase from Escherichia coli and the inhibitor Ap5A refined at 1.9 A resolution. A model for a catalytic transition state. J Mol Biol. 1992;224(1):159–177. doi: 10.1016/0022-2836(92)90582-5. [DOI] [PubMed] [Google Scholar]
- 8.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 9.Rivas-Pardo JA, et al. Crystal structure, SAXS and kinetic mechanism of hyperthermophilic ADP-dependent glucokinase from Thermococcus litoralis reveal a conserved mechanism for catalysis. PLoS ONE. 2013;8(6):e66687. doi: 10.1371/journal.pone.0066687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341(6148):899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgen AS, Roberts GC, Feeney J. Binding of flexible ligands to macromolecules. Nature. 1975;253(5494):753–755. doi: 10.1038/253753a0. [DOI] [PubMed] [Google Scholar]
- 12.Zhou H-X. From induced fit to conformational selection: A continuum of binding mechanism controlled by the timescale of conformational transitions. Biophys J. 2010;98(6):L15–L17. doi: 10.1016/j.bpj.2009.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44(2):98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgen AS. Conformational changes and drug action. Fed Proc. 1981;40(13):2723–2728. [PubMed] [Google Scholar]
- 15.Sullivan SM, Holyoak T. Enzymes with lid-gated active sites must operate by an induced fit mechanism instead of conformational selection. Proc Natl Acad Sci USA. 2008;105(37):13829–13834. doi: 10.1073/pnas.0805364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Changeux JP, Edelstein S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol Rep. 2011;3:19. doi: 10.3410/B3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiefhaber T, Bachmann A, Jensen KS. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol. 2012;22(1):21–29. doi: 10.1016/j.sbi.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Vogt AD, Di Cera E. Conformational selection is a dominant mechanism of ligand binding. Biochemistry. 2013;52(34):5723–5729. doi: 10.1021/bi400929b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449(7165):1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 21.Arora K, Brooks CL., 3rd Large-scale allosteric conformational transitions of adenylate kinase appear to involve a population-shift mechanism. Proc Natl Acad Sci USA. 2007;104(47):18496–18501. doi: 10.1073/pnas.0706443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35(10):539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daily MD, Phillips GN, Jr, Cui Q. Many local motions cooperate to produce the adenylate kinase conformational transition. J Mol Biol. 2010;400(3):618–631. doi: 10.1016/j.jmb.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant BJ, McCammon JA, Gorfe AA. Conformational selection in G-proteins: Lessons from Ras and Rho. Biophys J. 2010;99(11):L87–L89. doi: 10.1016/j.bpj.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bucher D, Grant BJ, McCammon JA. Induced fit or conformational selection? The role of the semi-closed state in the maltose binding protein. Biochemistry. 2011;50(48):10530–10539. doi: 10.1021/bi201481a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva D-A, Bowman GR, Sosa-Peinado A, Huang X. A role for both conformational selection and induced fit in ligand binding by the LAO protein. PLOS Comput Biol. 2011;7(5):e1002054. doi: 10.1371/journal.pcbi.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nussinov R, Ma B. Protein dynamics and conformational selection in bidirectional signal transduction. BMC Biol. 2012;10(1):2. doi: 10.1186/1741-7007-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clore GM. Interplay between conformational selection and induced fit in multidomain protein-ligand binding probed by paramagnetic relaxation enhancement. Biophys Chem. 2014;186:3–12. doi: 10.1016/j.bpc.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci USA. 2009;106(33):13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniels KG, et al. Ligand concentration regulates the pathways of coupled protein folding and binding. J Am Chem Soc. 2014;136(3):822–825. doi: 10.1021/ja4086726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCammon JA, Northrup SH. Gated binding of ligands to proteins. Nature. 1981;293(5830):316–317. doi: 10.1038/293316a0. [DOI] [PubMed] [Google Scholar]
- 32.Szabo A, Shoup D, Northrup SH, McCammon JA. Stochastically gated diffusion-influenced reactions. J Chem Phys. 1982;77(9):4484–4493. [Google Scholar]
- 33.Zhou H-X, Szabo A. Theory and simulation of stochastically-gated diffusion-influenced reactions. J Phys Chem. 1996;100(7):2597–2604. [Google Scholar]
- 34.Cai L, Zhou H-X. Theory and simulation on the kinetics of protein-ligand binding coupled to conformational change. J Chem Phys. 2011;134(10):105101. doi: 10.1063/1.3561694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc Natl Acad Sci USA. 2011;108(21):8651–8656. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou HX. Kinetics of diffusion-influenced reactions studied by Brownian dynamics. J Phys Chem. 1990;94(25):8794–8800. [Google Scholar]
- 37.Zhou HX, Szabo A. Theory and simulation of the time-dependent rate coefficients of diffusion-influenced reactions. Biophys J. 1996;71(5):2440–2457. doi: 10.1016/S0006-3495(96)79437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim E, et al. A single-molecule dissection of ligand binding to a protein with intrinsic dynamics. Nat Chem Biol. 2013;9(5):313–318. doi: 10.1038/nchembio.1213. [DOI] [PubMed] [Google Scholar]
- 39.Marvin JS, Hellinga HW. Manipulation of ligand binding affinity by exploitation of conformational coupling. Nat Struct Biol. 2001;8(9):795–798. doi: 10.1038/nsb0901-795. [DOI] [PubMed] [Google Scholar]
- 40.Millet O, Hudson RP, Kay LE. The energetic cost of domain reorientation in maltose-binding protein as studied by NMR and fluorescence spectroscopy. Proc Natl Acad Sci USA. 2003;100(22):12700–12705. doi: 10.1073/pnas.2134311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Dima RI, Thirumalai D. Allosteric communication in dihydrofolate reductase: signaling network and pathways for closed to occluded transition and back. J Mol Biol. 2007;374(1):250–266. doi: 10.1016/j.jmb.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 42.Ermak DL, McCammon JA. Brownian dynamics with hydrodynamic interactions. J Chem Phys. 1978;69(4):1352–1360. [Google Scholar]