Significance
Cellular signals evoke rapid and broad changes in gene regulatory networks. To uncover these network dynamics, we developed an approach able to monitor primary targets of a transcription factor (TF) based solely on gene regulation, in the absence of detectable binding. This enabled us to follow the transient propagation of a nitrogen (N) nutrient signal as a direct impact of the master TF Basic Leucine Zipper 1 (bZIP1). Unexpectedly, the largest class of primary targets that exhibit transient associations with bZIP1 is uniquely relevant to the rapid and dynamic propagation of the N signal. Our ability to uncover this transient network architecture has revealed the “dark matter” of dynamic N nutrient signaling in plants that has previously eluded detection.
Keywords: transcription regulation, nitrogen signaling, systems biology, gene networks
Abstract
The dynamic nature of gene regulatory networks allows cells to rapidly respond to environmental change. However, the underlying temporal connections are missed, even in kinetic studies, as transcription factor (TF) binding within at least one time point is required to identify primary targets. The TF-regulated but unbound genes are dismissed as secondary targets. Instead, we report that these genes comprise transient TF–target interactions most relevant to rapid signal transduction. We temporally perturbed a master TF (Basic Leucine Zipper 1, bZIP1) and the nitrogen (N) signal it transduces and integrated TF regulation and binding data from the same cell samples. Our enabling approach could identify primary TF targets based solely on gene regulation, in the absence of TF binding. We uncovered three classes of primary TF targets: (i) poised (TF-bound but not TF-regulated), (ii) stable (TF-bound and TF-regulated), and (iii) transient (TF-regulated but not TF-bound), the largest class. Unexpectedly, the transient bZIP1 targets are uniquely relevant to rapid N signaling in planta, enriched in dynamic N-responsive genes, and regulated by TF and N signal interactions. These transient targets include early N responders nitrate transporter 2.1 and NIN-like protein 3, bound by bZIP1 at 1–5 min, but not at later time points following TF perturbation. Moreover, promoters of these transient targets are uniquely enriched with cis-regulatory motifs coinherited with bZIP1 binding sites, suggesting a recruitment role for bZIP1. This transient mode of TF action supports a classic, but forgotten, “hit-and-run” transcription model, which enables a “catalyst TF” to activate a large set of targets within minutes of signal perturbation.
Signal propagation through gene regulatory networks (GRNs) enables organisms to rapidly respond to changes in environmental signals. For example, dynamic GRN studies in plants have uncovered genome-wide responses that occur within as little as 3 min following a nitrogen (N) nutrient signal perturbation (1). However, many of the underlying rapid and temporal network connections between transcription factors (TFs) and their targets elude detection even in fine-scale time-course studies (2, 3), as current methods, such as chromatin immunoprecipitation (ChIP), require stable TF binding in at least one time point to identify primary targets (4–6). However, recent models suggest that GRNs built solely on TF binding data are insufficient to recapture transcriptional regulation (7–9). Compounding this dilemma, TFs have been found to stably bind to only a small percentage (5–32%) of the TF-regulated genes across eukaryotes (4–6, 10–13). Because TF binding is required to define the primary targets in current GRN studies, the large set of TF-regulated but not TF-bound genes must be categorically dismissed as indirect or secondary targets (4, 5, 11–13). We report an alternative—and more intriguing—conclusion that these typically dismissed targets comprise the “dark matter” of rapid and transient signal transduction that has previously eluded detection across eukaryotes.
To capture these rapid and dynamic network connections that elude detection by biochemical TF binding assays, we developed an approach that can identify primary targets based on a functional readout—TF-induced gene regulation—even in the absence of detectable TF binding. Our case study focuses on the master TF Basic Leucine Zipper 1 (bZIP1), a central integrator of metabolic signaling including sugar (14–16) and N nutrient signals (17, 18). To uncover the underlying dynamic GRNs, we temporally perturbed both bZIP1 and the N signal it transduces in a cell-based system designed for temporal TF perturbation. This cell-based system, named TARGET (transient assay reporting genome-wide effects of transcription factors), which involves inducible TF nuclear localization, is able to identify primary TF targets based solely on TF-induced gene regulation, as shown for a well-studied TF involved in plant hormone signaling—abscisic acid insensitive 3 (ABI3) (19). In our current study, by adapting a micro-ChIP protocol (20) to the cell-based TARGET system, we were able to monitor primary targets based on either TF-induced gene regulation or TF binding quantified in the same cell samples, enabling a direct comparison. The use of isolated cells allowed us to capture rapid and transient regulatory events including the formation of TF–DNA complexes within 1–5 min from the onset of TF translocation to the nucleus. Such a short-lived interaction would likely be missed in planta, as effective protein–DNA cross-linking in intact plant tissues requires prolonged (for a minimum of 15 min) infiltration under vacuum. Unexpectedly, the primary TF targets that are regulated by but not stably bound to bZIP1—termed “transient”—were the most biologically relevant to rapid transduction of the N signal. These transient TF targets include first-responder genes, induced as early as 3–6 min after N signal perturbation in planta (1). This discovery suggests that the current “gold standard” of GRNs built solely on the intersection of TF binding and TF regulation data misses a large and important class of transient TF targets, which are at the heart of dynamic networks. Moreover, the shared features of these transient bZIP1 targets and their role in rapid N signaling provides genome-wide support for a classic but largely forgotten model of “hit-and-run” transcription (21). This transient mode-of-action can enable a master TF to catalytically and rapidly activate a large set of genes in response to a signal.
Results
Temporal Perturbation of Both bZIP1 and the N Signal It Transduces.
To identify how bZIP1 mediates the rapid propagation of an N signal in a GRN, we temporally perturbed both bZIP1 and the N signal it transduces in the cell-based TARGET system (SI Appendix, Fig. S1 A and B) (19). bZIP1, which is ubiquitously expressed across all root cell types (22), was transiently overexpressed in root protoplasts as a glucocorticoid receptor::bZIP1 (GR::bZIP1) fusion protein, enabling temporal induction of nuclear localization by dexamethasone (DEX) (SI Appendix, Fig. S1A) (19). Transfected root cells expressing the GR::bZIP1 fusion protein were sequentially treated with (i) inorganic N (+/−N), (ii) cycloheximide (+/− CHX), and (iii) DEX (+/−DEX) (SI Appendix, Fig. S1C). The N treatment can induce posttranslational modifications of bZIP1 (14) or influence bZIP1 partners by transcriptional or posttranscriptional mechanisms (SI Appendix, Fig. S1B). DEX treatment induces TF nuclear import (SI Appendix, Fig. S1A) (19). Further, genes regulated by DEX-induced TF import are deemed primary targets, as a CHX pretreatment blocks translation of downstream regulators, as previously shown in the TARGET system (19) and in planta (23) (SI Appendix, Fig. S1A). Importantly, to eliminate any side effects caused by CHX pretreatment, we only considered genes in our analysis whose transcriptome response to DEX-induced TF nuclear import is the same in either the presence or absence of CHX (Materials and Methods and SI Appendix, Supplemental Methods). Such bZIP1 primary targets identified based on gene regulation following DEX-induced TF import were identified using Affymetrix ATH1 microarrays. In parallel, primary targets identified by TF binding were identified in a micro–ChIP-Seq assay (20) using anti-GR antibodies. Both transcriptome and ChIP-Seq data were obtained 5 h after the DEX-induced nuclear import of bZIP1, from the same cell samples, enabling a direct comparison (SI Appendix, Fig. S1 C and D). Regarding the N signal, we identified 328 N-responsive genes in our cell-based experiments (SI Appendix, Fig. S2, Table S1, and Dataset S1). These N-responsive genes significantly overlap with the N-responsive genes identified in whole seedlings exposed to a similar N treatment (NH4NO3) (17) and from roots treated with nitrate (24, 25), including a dynamic study (1) (121/328, P < 0.001) (SI Appendix, Fig. S3, Table S2, and Dataset S1). The N-responsive genes in our cell-based experiments are enriched with genes that respond to N treatment across all root cell types in planta (P = 8.8E-13, hypergeometric distribution) (26).
Primary Targets of bZIP1 Can Be Identified by Either TF Regulation or TF Binding.
We first identified bZIP1 primary targets based solely on TF-induced gene regulation. A total of 901 genes were identified as primary bZIP1 targets based on significant regulation in response to DEX-induced TF nuclear import, compared with minus DEX controls [ANOVA analysis; false discovery rate (FDR) adjusted P value < 0.05] (SI Appendix, Figs. S1D and S4A,Tables S3–S5, and Dataset S1). These DEX-responsive genes are deemed to be primary targets of bZIP1, as pretreatment of the samples with CHX (before DEX-induced TF nuclear import) blocks translation of mRNAs of primary bZIP1 targets, thus preventing changes in the mRNA levels of secondary targets in the GRN. To control for the potential side effects of CHX, this list of bZIP1 primary targets excluded genes whose DEX-induced mRNA response was altered by CHX treatment (Materials and Methods and SI Appendix, Supplemental Methods). With regard to the N signal, 28 out of the 901 bZIP1 primary targets were regulated in response to a significant N Treatment × TF interaction (P < 0.01) (SI Appendix, Fig. S5, Table S6, and Dataset S1). This could reflect a posttranslational modification of bZIP1 by the N signal or the N-induced modification of bZIP1 partners at the transcriptional and/or posttranslational level (SI Appendix, Fig. S1B). We next identified bZIP1 primary targets based solely on TF–DNA binding. Genes bound by bZIP1 were identified as genic regions enriched in the ChIP DNA, compared with the background (input DNA), using the QuEST peak-calling algorithm (SI Appendix, Fig. S4C) (27). This identified 850 genes with significant bZIP1 binding (FDR < 0.05) (SI Appendix, Fig. S1D, Table S7, and Dataset S1), which included validated bZIP1 targets identified by single gene studies (e.g., ASN1 and ProDH) (16). We note that ChIP-Seq can potentially detect genes directly bound to bZIP1, as well as genes indirectly bound by bZIP1 through bridging interactors. Thus, to independently assess whether primary targets identified either by TF binding or TF regulation were due to direct binding of bZIP1, we performed cis-element analysis (SI Appendix, Fig. S4 B and D). The bZIP1-bound genes and the bZIP1-regulated genes are each highly significantly enriched in known bZIP1 binding sites, based on analysis of de novo cis-motifs using MEME (28) or known cis-motif enrichment using Elefinder (29) (SI Appendix, Fig. S4 B and D).
Integration of TF Regulation and TF Binding Data Identifies Three Modes-of-Action for bZIP1 and Its Primary Targets: Poised, Stable, and Transient.
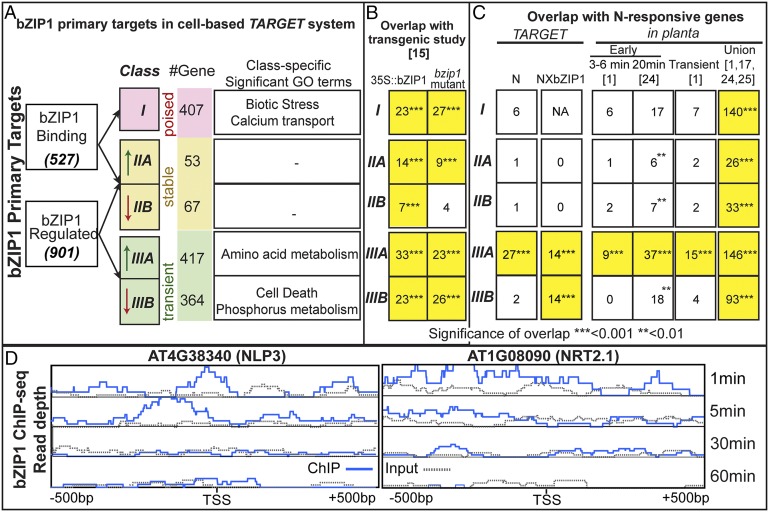
To understand the underlying mechanisms by which bZIP1 propagates N signals through a GRN, we integrated primary targets identified by either TF-induced gene regulation or TF binding. To enable a direct comparison of transcriptome and TF binding data, of the 850 genes bound to bZIP1, we omitted 187 genes not represented on the ATH1 microarray. We also omitted 136 genes that did not pass the stringent filters for effects of protoplasting or DEX and CHX treatment (Materials and Methods and SI Appendix, Supplemental Methods). This resulted in a filtered total of 527 bZIP1-bound genes (Fig. 1A). The resulting list of 1,308 high-confidence primary targets of bZIP1 identified by either TF-mediated gene regulation (901 genes) or TF binding (527 genes) were integrated and analyzed for biological relevance to the N signal (Fig. 1). The intersection of the TF regulation and TF binding data identified three classes of primary targets, representing distinct modes-of-action for bZIP1 in N signal propagation (Fig. 1A and SI Appendix, Table S8, and Dataset S1). Class I targets (407 genes) were deemed “poised,” as they are bound to bZIP1 but show no significant TF-induced gene regulation. Class II targets (120 genes) are deemed “stable,” as they are both bound and regulated by bZIP1. Unexpectedly, class III targets (781 genes)—the largest class of bZIP1 primary target genes—were deemed transient, as they are regulated by bZIP1 perturbation but not detectably bound to it. We note that these are not indirect TF targets, as ChIP-Seq is able to detect direct or indirect binding by bZIP1—that is, as part of a protein complex. They also cannot be dismissed as secondary targets of bZIP1, as they are regulated in response to DEX-induced bZIP1 perturbation performed in the presence of CHX, which blocks the regulation of secondary targets.
Fig. 1.
Class III transient targets of bZIP1 are uniquely associated with rapid N signaling. (A) Primary bZIP1 targets identified by either bZIP1-induced regulation or bZIP1 binding assayed in the same root protoplast samples. Intersection of these datasets revealed three distinct classes of primary targets: (class I) poised, TF-bound but not TF-regulated; (class II) stable, TF-bound and TF-regulated; and (class III) transient, TF-regulated but no detectable binding. Classes II and III are subdivided into activated or repressed, with their associated overrepresented GO terms (FDR < 0.01) listed. bZIP1 primary targets detected in protoplasts (A) were compared with bZIP1-regulated genes in planta (B) (15), and N-regulated genes in plants (1, 17, 24, 25) (C) [size of overlap is listed, and significance is indicated by asterisks (yellow, P < 0.001)]. Class III transient targets are uniquely enriched in genes related to rapid N signaling (1, 24). (C) Class IIIA target genes (NLP3 and NRT2.1) show transient bZIP1 binding at 1 and 5 min after nuclear import of bZIP1 but not at later time points (30 and 60 min) (D).
To next explore the biological relevance of the three distinct classes of primary bZIP1 targets, we examined the following features: (i) enrichment of cis-regulatory elements (Fig. 2), (ii) comparison with bZIP1-regulated genes in planta (Fig. 1B), and (iii) biological relevance to N signal transduction in isolated cells (Fig. 1 A and C) and in planta (Fig. 1C). This comparative analysis uncovered features common to all three classes of bZIP1 targets as well as specific features of class III transient targets that are uniquely relevant to rapid N signal propagation. The features shared by all three classes of bZIP1 primary targets are (i) bZIP1 binding sites—all three classes of genes deemed to be bZIP1 primary targets share enrichment of known bZIP1 binding sites in their promoters (E < 0.01, Fig. 2); (ii) in planta relevance to bZIP1—all three classes of bZIP1 primary targets identified in the cell-based TARGET system were validated by their significant overlap with bZIP1-regulated genes identified in transgenic plants, either by comparison with a 35S::bZIP1 overexpression line (100/449 genes; 22% overlap; P < 0.001) or a transfer-DNA (T-DNA) insertion mutant in bZIP1 (89/488 genes; 18.2% overlap; P < 0.001) (15) (Fig. 1B); (iii) N regulation in planta—bZIP1 was predicted to be a master regulator in N response (17, 18), and in support of this, all three classes of bZIP1 primary targets in protoplasts are significantly enriched with N-responsive genes in planta (1, 17, 24, 25) (438/1,308 genes, P < 0.001) (Fig. 1C); and (iv) known bZIP1 functions—all three classes of targets show enrichment of gene ontology (GO) terms associated with other known bZIP1 functions (e.g., stimulus/stress) (SI Appendix, Fig. S6). Specifically, bZIP1 is reported as a master regulator in response to darkness and sugar starvation (14, 15). Consistent with this, all three classes of bZIP1 primary targets share a significant overlap (P < 0.001) with genes induced by sugar starvation and extended darkness (30).
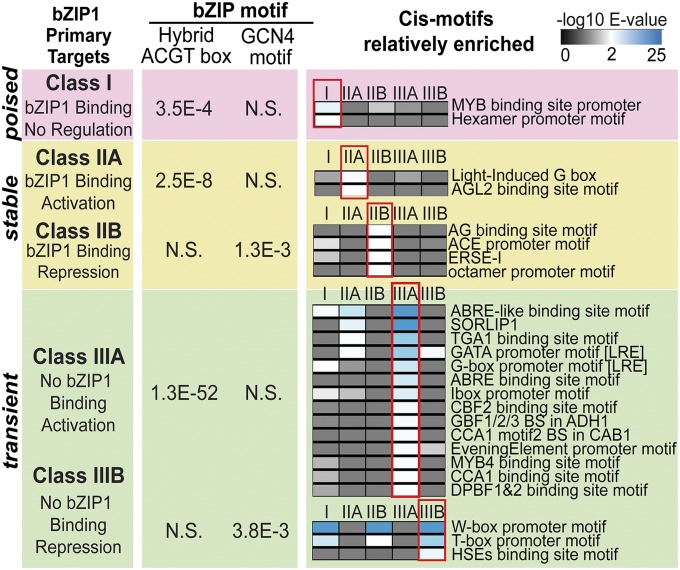
Fig. 2.
Class III bZIP1 transient targets are specifically enriched in coinherited cis-motif elements. The significance of the overrepresentation of the known bZIP binding motif hybrid ACGT box (15) and GCN4 binding motif (31) are listed for each class of bZIP1 primary targets. In addition to these bZIP binding sites, the significance of enrichment of coinherited cis-regulatory motifs is shown as a heat-map specific to each subclass.
In addition to these common features consistent with the role of bZIP1 in planta (14, 15, 17), we uncovered distinctive features for the class III transient bZIP1 primary targets specifically relevant to rapid N signaling. These class-specific features are outlined below.
Class I Poised Targets (TF Binding Only).
Class I bZIP1 primary targets (407 genes) that are bound but not regulated by bZIP1 are significantly enriched in genes involved in response to biotic/abiotic stimuli and transport of divalent ions (FDR < 0.01) (Fig. 1A and SI Appendix, Fig. S6). They are also significantly enriched in the known bZIP1 binding site “hybrid ACGT box” (E = 3.5e-4), supporting the notion that they are valid primary targets of bZIP1 (Fig. 2). This suggests that bZIP1 is bound to and poised to activate these target genes, possibly in response to a signal or a TF partner not present in our experimental conditions.
Class II Stable Targets (TF Binding and Regulation).
Class II targets (120 genes) are regulated and bound by bZIP1. This 23% overlap (P < 0.001) between transcriptome and ChIP-Seq data (Fig. 1A) is comparable to the relatively low overlap observed for other TF perturbation studies performed in planta [23% ABI3 (10), 5% ASR5 (11), 20–30% KNOTTED1 (12)] and in other eukaryotes [8% BRCA1 (4), 32% LRH-1 (13)]. Thus, our class II stable bZIP1 targets correspond to the gold standard set typically identified in TF studies across eukaryotes (4, 5, 10–13). Further, our cis-element analysis suggests that bZIP1 functions to activate or repress target gene expression via two distinct binding sites (Fig. 2). The targets activated by bZIP1 (class IIA) are significantly enriched with the hybrid ACGT box bZIP1 binding site (E = 2.5e-8) (Fig. 2). By contrast, genes repressed by bZIP1 (class IIB) are enriched with the bZIP binding site GCN4 (E = 1.3e-3) (Fig. 2). Interestingly, the GCN4 motif was reported to mediate N and amino acid starvation sensing in yeast (31), suggesting a conserved link between bZIPs and nutrient sensing across eukaryotes. Finally, class II targets share the “stimulus/stress” GO terms with other classes, but surprisingly, no significant biological terms unique to class II targets were identified (Fig. 1A and SI Appendix, Fig. S6).
Class III Transient Targets (TF Regulation but No Detectable TF Binding).
Unexpectedly, the largest group of bZIP1 primary targets (781 genes) is represented by the class III transient targets—that is, primary targets regulated by bZIP1 perturbation but not detectably bound by it (Fig. 1A). Paradoxically, class IIIA transient targets that are activated by bZIP1 are the most significantly enriched in the known bZIP1 binding site (E = 1.3e-52) (Fig. 2), despite their lack of detectable bZIP1 binding. Class IIIB targets repressed by bZIP1 are significantly enriched in a distinct bZIP binding site, GCN4 (E = 3.8e-3) (Fig. 2). Intriguingly, both of these known bZIP1 binding sites in the class III transient genes are also observed in the class II stable target genes (TF-bound and TF-regulated) (Fig. 2). The lack of detectable TF binding for class III targets likely represents a transient or weak interaction of bZIP1 and these primary targets, rather than an indirect interaction, as the ChIP-Seq protocol can also detect indirect binding (e.g., via interacting TF partners). The trivial explanation that the mRNAs for class IIIA genes are stabilized by CHX or bZIP1 is not supported by our data, as the CHX effect was accounted for by filtering out genes whose response to DEX-induced nuclear localization of bZIP1 is altered by CHX treatment (Materials and Methods and SI Appendix, Supplemental Methods). Instead, the class III primary targets likely represent a transient interaction between bZIP1 and its targets. Indeed, 41 genes from class III transient targets have detectable bZIP1 binding at one or more of the earlier time points (1, 5, 30, and 60 min) measured by ChIP-Seq, following DEX-induced TF nuclear import (Fig. 1D and SI Appendix, Table S12, and Dataset S1). These class III transient genes are uniquely relevant to rapid N signaling, as described below.
The Class III Transient bZIP1 Primary Targets Comprise “First Responders” in Rapid N signaling.
In line with its role as a master regulator in an N response gene network, all three classes of bZIP1 primary targets uncovered in our cell-based study are significantly enriched with N-responsive genes observed in whole plants (1, 17, 24, 25) (Fig. 1C, overlap with the “union” of N-responsive genes in planta). Unexpectedly, the transient class III bZIP1 targets—regulated by but not stably bound to bZIP1—are uniquely relevant to rapid and dynamic N signaling in planta (Fig. 1C). This conclusion is based on the following evidence: (i) The class IIIA transient bZIP1 targets have the largest and most significant overlap (P < 0.001; Fig. 1C) with the 147 genes induced by N signals in our cell-based TARGET study (SI Appendix, Table S1, and Dataset S1). (ii) Only class III transient bZIP1 targets have a significant enrichment in genes involved in N-related biological processes (enrichment of GO terms; P < 0.01), including amino acid metabolism (Fig. 1A and SI Appendix, Fig. S8, Table S10, and Dataset S1), a role also supported by in planta studies of bZIP1 (16). (iii) The class III transient genes comprise the bulk of the bZIP1 targets in the N assimilation pathway (SI Appendix, Fig. S7, Table S9, and Dataset S1), including the “early N responders,” such as the high-affinity nitrate transporter 2.1 (NRT2.1), induced rapidly (<12 min) and transiently following N signal perturbation in planta (1). (iv) The class III transient targets exclusively comprise all of the genes regulated by a N Treatment × bZIP1 interaction (28 genes) (Fig. 1C and SI Appendix, Fig. S5). These include well-known early mediators of N signaling induced at 6–12 min after N provision (1), including the NIN-like transcription factor 3 (NLP3; At4g38340) (32) and the LOB domain-containing protein 39 (LBD39) TF (At4g37540) (33). NLP3 belongs to the NIN-like TF family, which plays an essential role in nitrate signaling (32). In our study, NLP3 is a transient bZIP1 target whose up-regulation by bZIP1 is dependent on the N signal (SI Appendix, Fig. S5, Table S6, and Dataset S1). LBD39, which has been reported to fine-tune the magnitude of the N response in planta (33), is a transient bZIP1 target that is only induced by bZIP1 in the presence of the N signal in our cell-based study (SI Appendix, Fig. S5, Table S6, and Dataset S1). This N Treatment × bZIP1 interaction could be a posttranslational modification of bZIP1, reminiscent of its posttranslational modification in response to other abiotic signals (e.g., sugar and stress signals) (16). The N Treatment × bZIP1 interaction could also involve translational/transcriptional effects of the N signal on its interacting TF partners, as depicted in SI Appendix, Fig. S1B.
(v) Most importantly, class III transient target genes are uniquely enriched in genes that respond early and transiently to the N signal in planta (Fig. 1C). Although all three classes of bZIP1 target genes have significant intersections with N-regulated genes in planta (P < 0.001) (1, 17, 24, 25) (Fig. 1C, union of N response genes in planta), only class IIIA transient targets have a significant overlap with genes induced transiently or early in response to an N signal (within 3–6 min) (P < 0.001), based on fine-scale kinetic studies of N treatments performed in planta (1) (Fig. 1C and SI Appendix, Table S11, and Dataset S1). These transient bZIP1 targets include known early N responders, such as the TFs LBD38 (At3g49940) and LBD39 (At4g37540), which respond to N signals in as early as 3–6 min (1), and are involved in regulating N uptake and assimilation genes in planta (33). Additionally, class IIIA transient targets are uniquely enriched in rapid N responders (Fig. 1C and SI Appendix, Table S11, and Dataset S1), identified as genes induced within 20 min after a supply of 250 μM nitrate to roots (24), including the nitrate transporters NRT3.1 and NRT2.1. This result further supports the notion that the class IIIA transient bZIP1 targets are specifically relevant to a rapid N signaling response in planta.
A Transient Mode of bZIP1 Action Invokes a Hit-and-Run Model for N Signaling.
The significant enrichment of N-relevant genes in class III targets links the transient mode-of-action of bZIP1 with early and transient aspects of N nutrient signaling (Fig. 1 C and D). We posit that this transient mode-of-action could allow a small number of bZIP1 molecules to initiate and catalyze a large response to an N signal in the GRN within minutes, without having to wait for a significant buildup of the bZIP1 protein. Two unique properties of class III transient targets support this hypothesis. First, pioneer TFs have been shown to facilitate and/or initiate gene expression (2, 34). Accordingly, we should be able to detect bZIP1 binding to the promoter of class III transient targets at very early time points after DEX-induced nuclear localization of the GR–bZIP1 fusion protein (e.g., within minutes). Second, cis-motif analysis of target genes of a pioneer TF in Drosophila highlighted the specific enrichment of other TF binding motifs in close proximity to the pioneer TF motif (35), suggesting either active recruitment or passive enabling of binding by additional TF partners. By this model, the promoters of class III transient bZIP1 targets should show specific enrichment for binding sites of other TFs in addition to bZIP1. Indeed, we find bZIP1 shares both of these properties, as detailed below.
To experimentally determine if any of the class III transient targets are bound by bZIP1 at very early time points, we performed ChIP-Seq analysis on four additional time points after the DEX-induced nuclear import of bZIP1. This revealed 41 genes from class III transient targets that have detectable bZIP1 binding at one or more of the earlier time points (1, 5, 30, and 60 min) (Fig. 1D and SI Appendix, Table S12, and Dataset S1) but are not bound by bZIP1 at the 5 h time point of our original study (Fig. 1A). Crucially, these 41 transiently bound bZIP1 targets are significantly enriched in GO terms related to the N signal (e.g., amino acid metabolism, P < 0.05). The validated bZIP1 binding site (hybrid ACGT motif) (14–16) is enriched in the promoters of these 41 genes (E = 2.7e-3) as well as in the remaining class III transient targets (E = 1e-26). These transiently bound bZIP1 targets include NLP3, a key early regulator of nitrate signaling in plants (32). In our study, NLP3 is bound by bZIP1 at very early time points (1 and 5 min) but not at the later points (30 and 60 min) following TF perturbation (Fig. 1D). Similarly, the promoter of an early response gene encoding the high-affinity nitrate transporter NRT2.1 (1) is bound by bZIP1 as early as 1 and 5 min after the DEX-induced nuclear import of bZIP1, but binding is weakened at 30 min and disappears at 60 min (Fig. 1D). In summary, this time-course analysis provides physical evidence that some class III targets are indeed transiently bound to bZIP1, only at very early time points after bZIP1 nuclear import (1–5 min). We note that such transient TF binding is difficult to capture, unless multiple early time points are designed for ChIP-Seq study. However, our cell-based TARGET system can identify primary targets based on the outcome of TF binding (e.g., TF-induced gene regulation), even if TF binding is highly transient (e.g., within seconds) or is never bound stably enough to be detected at any time point.
Finally, the hypothesis that bZIP1 acts as a “pioneer/catalyst” TF in N signal propagation through a GRN is further supported by cis-motif analysis. Specifically, the promoters of class III transient bZIP1 target genes contained the largest number and most significant enrichment of cis-regulatory motifs, in addition to bZIP1 binding sites (Fig. 2). In particular, the class IIIA transient activated genes contain the most significant enrichment of the known bZIP1 binding site (E = 1.3e-52) and are specifically enriched in coinherited cis-elements that belong to the bZIP, MYB, and GATA families (36) (Fig. 2 and SI Appendix, Supplemental Results). These results support the hypothesis that bZIP1 is a pioneer/catalyst TF that interacts and/or recruits other TFs, including other bZIPs and/or MYB/GATA binding factors, to temporally coregulate target genes in response to an N signal (Fig. 3). Indeed, bZIP1 has been reported to interact with other TFs in vitro (37) (SI Appendix, Table S13, and Dataset S1) and in vivo (14, 15, 37). This list of bZIP1 interactors includes bZIP25, a gene in the class III transient bZIP1 primary targets. In support of a collaborative relationship between bZIP1 and the GATA family TFs in mediating the N response, one GATA TF was reported to be nitrate-inducible and involved in regulating energy metabolism, thus serving as a functional analog to bZIP1 (38). Taken together, the transient binding of bZIP1 and enrichment of coinherited binding sites for additional TFs specifically in class III transient bZIP1 targets support a role for bZIP1 as a TF pioneer/catalyst (35) and a model for hit-and-run transcription (21), as depicted in Fig. 3 and discussed below.
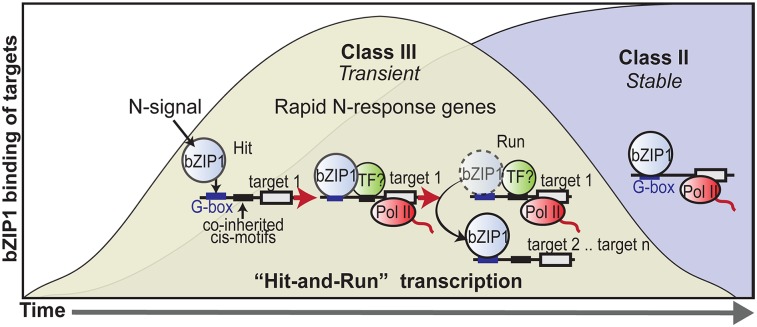
Fig. 3.
A hit-and-run transcription model enables bZIP1 to rapidly and catalytically activate genes in response to an N signal. The transient mode-of-action for class III bZIP1 targets follows a classic model for hit-and-run transcription (21). In this model, transient interactions of bZIP1 with class III targets (the “hit”) lead to recruitment of the transcription machinery and possibly other TFs. Next, the transient nature of the bZIP1–target interaction (the “run”) enables bZIP1 to catalytically activate a large set of rapidly induced genes (e.g., target 2 … target n) biologically relevant to rapid transduction of the N signal.
Discussion
Our discovery of a large and typically overlooked class of transient primary targets of the master TF bZIP1 introduces a novel perspective in the general field of dynamic GRNs. Dynamic TF–target binding studies across eukaryotes have captured many transient TF targets (2, 3). However, even those fine-scale time-series ChIP studies likely miss highly temporal connections, as they require biochemically detectable TF binding in at least one time point to identify primary TF targets. Key to our discovery of the transient targets of bZIP1 involved in rapid N signaling is our ability to identify primary targets based on TF-induced changes in mRNA that can occur even in the absence of detectable TF binding. The cell-based system also enabled us to detect rapid and transient binding within 1 min of TF nuclear import, owing to rapid fixation of protein–DNA complexes in plant cells lacking a cell wall. Importantly, the in planta relevance of our cell-based TARGET studies (Fig. 1A) confirms and complements data from bZIP1 T-DNA mutants and transgenic plants (15) (Fig. 1B), which are unable to distinguish primary from secondary targets or capture transient TF–target interactions. Therefore, the transient interactions between bZIP1 and its targets uncovered in our cell-based TARGET system help to refine our understanding of the in planta mechanism of bZIP1.
Our discovery of these transient TF targets adds a new perspective to the field of dynamic GRNs. Recent time-series studies in yeast by Lickwar et al. reported transitive TF–target binding described as a “tread-milling” mechanism, in which a TF exhibits weak and transitive binding to some of its targets, resulting in a lower level of gene activation (9). The transient bZIP1targets detected in our study do not fit this tread-milling model, as there is no significant difference between the expression fold-change distributions for class III transient targets versus class II stable targets. Instead, we conceptualize the transient TF–target interactions we have uncovered to a classic, but largely forgotten, hit-and-run model of transcription proposed in the 1980s (21) (Fig. 3). This hit-and-run model posits that a TF can act as a trigger to organize a stable transcriptional complex, after which transcription by RNA polymerase II can continue without the TF being bound to the DNA (21).
In support of this hit-and-run transcription model, class III transient targets include genes that are rapidly and transiently bound by bZIP1 at very early time points (1–5 min) after TF nuclear import and whose level of expression is maintained at a higher level, despite being no longer bound by bZIP1 at later time points. Continued regulation of the bZIP1 targets (after bZIP1 is no longer bound) might be mediated by other TF partners recruited by the catalyst TF (Fig. 3). This model is supported by the enrichment of cis-motifs coinherited with the known bZIP1 binding motif (14–16) in the class III transient targets (Fig. 2). This finding also supports other explanatory models for “continuous” TF networks (7, 8), which converge on the idea that TF binding data alone is insufficient to fully characterize regulatory networks and that other factors (including chromatin and other TFs) may influence the action of a master TF. In this transient mode-of-action, bZIP1 can activate genes in response to an N signal (“the hit”), whereas the transient nature of the TF–target association (“the run”) enables bZIP1 to act as a TF “catalyst” to rapidly induce a large set of genes needed for the N response. In support of this “catalytic” TF model, the global targets of bZIP1 N signaling are broad, covering 32% of the directly regulated targets of NLP7 related to the N signal, a well-studied master regulator of the N response (6). Importantly, the class III transient bZIP1 targets play a unique role in mediating a rapid, early, and biologically relevant response to the N signal in planta. This hit-and-run model, supported by our results for bZIP1, could represent a general mechanism for the deployment of an acute response to nutrient sensing, as well as other signals.
Importantly, our results have significance beyond bZIP1 and N signaling and indeed transcend plants. Across eukaryotes, TFs are found to bind only to a small percentage of their regulated targets, as shown in plants (10–12), yeast (5), and animals (4, 13). The large number of TF-regulated but unbound genes, including the false negatives of ChIP-Seq (39), must be dismissed as putative secondary targets in approaches that can only identify primary targets based on TF–DNA binding. Instead, we show that these typically dismissed targets, which can be identified as primary TF targets by a functional readout in our cell-based TARGET approach (e.g., TF-induced regulation), are crucial for rapid and dynamic signal propagation, thus uncovering the dark matter of signal transduction that has been missed. More broadly, our approach is applicable across eukaryotes and can also be adapted to studying cell-specific GRNs, by using GFP-marked cell lines in the assay (22). Moreover, this approach can identify primary targets even in cases where TF binding can never be physically detected. The transient targets thus uncovered will reveal the elusive temporal interactions that mediate rapid and dynamic responses of GRNs to external signals.
Materials and Methods
Protoplast Preparation and Cell Sorting.
Root protoplasts were prepared, transfected, and sorted as in ref. 19. We used 40 µg of pBeaconRFP_35S::GR::bZIP1 plasmid DNA per 1 × 106 cells (19). Protoplast suspensions were treated sequentially with either 20 mM KNO3 and 20 mM NH4NO3 (N) or 20 mM KCl (control) for 2 h, CHX (35 µM in DMSO), or solvent alone as mock for 20 min, and then with either DEX (10 µM in EtOH) or solvent alone as mock for 5 h at room temperature. Treated protoplast suspensions were FACS cell-sorted as in ref. 19 to collect ∼10,000 RFP-positive cells expressing the RFP reporter on the vector.
Analysis of Microarray Data.
Microarray intensities were normalized using the GC-robust multiarray averaging (GCRMA) package and analyzed using a three-way ANOVA. The significant gene list was filtered to account for the effect of protoplasting, CHX, and DEX (see SI Appendix, Supplemental Methods for details). Genes significantly regulated by the N signal and/or by DEX-induced bZIP1 nuclear localization were selected with an FDR cutoff of 5%. Genes significantly regulated by the interaction of the N treatment and bZIP1 (N Treatment × bZIP1) were selected with a P (ANOVA) cutoff of 0.01. The above analysis was initially performed on the DEX-responsive transcriptome using samples exposed to +CHX, and this gene list was then filtered to remove genes whose DEX response is altered in the minus (–CHX) sample, as described in SI Appendix, Supplemental Methods.
ChIP-Seq Data Analysis.
Illumina reads were filtered and aligned to the Arabidopsis thaliana genome (TAIR10) and clonal reads were removed. TF binding peaks were called using the QuEST package (27) and assigned to genes within 500 bp downstream of the peak.
Supplementary Material
Acknowledgments
We thank D. Shasha, M. Katari, and D. Tranchina for support in bioinformatics and statistical analysis. This work was supported by National Institutes of Health (NIH) Grants R01-GM032877 (to G.M.C.) and R01-GM078279 (to K.D.B.), National Science Foundation Grants MCB-0929338 (to G.M.C., Y.L., and K.D.B.) and DBI-0923128 (to W.R.M.), NIH National Research Service Award Grant GM095273 (to A.M.-C.), and Agence Nationale de la Recherche (ANR) (NitroNet) Grant ANR 11 PDOC 020 01 and the Centre National de la Recherche Scientifique (Projets Exploratoires Pluridisciplinaires Bio Math Info 2012–2013: SuperRegNet) (to G.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data from all microarray assays reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54049). The raw sequencing data from ChIP-Seq assays have been deposited in the Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/sra (accession no. SRX425878).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404657111/-/DCSupplemental.
References
- 1.Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11(12):R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ni L, et al. Dynamic and complex transcription factor binding during an inducible response in yeast. Genes Dev. 2009;23(11):1351–1363. doi: 10.1101/gad.1781909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang KN, et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife. 2013;2:e00675. doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorski JJ, et al. Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Res. 2011;39(22):9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes TR, de Boer CG. Mapping yeast transcriptional networks. Genetics. 2013;195(1):9–36. doi: 10.1534/genetics.113.153262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 7.Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21(4):611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Walhout AJM. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011;12(4):109. doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484(7393):251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mönke G, et al. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 2012;40(17):8240–8254. doi: 10.1093/nar/gks594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arenhart RA, et al. New insights into aluminum tolerance in rice: The ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol Plant. 2014;7(4):709–721. doi: 10.1093/mp/sst160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolduc N, et al. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012;26(15):1685–1690. doi: 10.1101/gad.193433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianco S, Brunelle M, Jangal M, Magnani L, Gévry N. LRH-1 governs vital transcriptional programs in endocrine-sensitive and -resistant breast cancer cells. Cancer Res. 2014;74(7):2015–2025. doi: 10.1158/0008-5472.CAN-13-2351. [DOI] [PubMed] [Google Scholar]
- 14.Baena-González E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448(7156):938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 15.Kang SG, Price J, Lin PC, Hong JC, Jang JC. The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol Plant. 2010;3(2):361–373. doi: 10.1093/mp/ssp115. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich K, et al. Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell. 2011;23(1):381–395. doi: 10.1105/tpc.110.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutiérrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105(12):4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM. Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Syst Biol. 2010;4(1):111. doi: 10.1186/1752-0509-4-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bargmann BO, et al. TARGET: A transient transformation system for genome-wide transcription factor target discovery. Mol Plant. 2013;6(3):978–980. doi: 10.1093/mp/sst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahl JA, Collas P. MicroChIP—A rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36(3):e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaffner W. Gene regulation. A hit-and-run mechanism for transcriptional activation? Nature. 1988;336(6198):427–428. doi: 10.1038/336427a0. [DOI] [PubMed] [Google Scholar]
- 22.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302(5652):1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 23.Eklund DM, et al. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell. 2010;22(2):349–363. doi: 10.1105/tpc.108.064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132(2):556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136(1):2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA. 2008;105(2):803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valouev A, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5(9):829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Swaminathan K, Hudson ME. Rapid, organ-specific transcriptional responses to light regulate photomorphogenic development in dicot seedlings. Plant Physiol. 2011;156(4):2124–2140. doi: 10.1104/pp.111.179416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krouk G, et al. A systems approach uncovers restrictions for signal interactions regulating genome-wide responses to nutritional cues in Arabidopsis. PLOS Comput Biol. 2009;5(3):e1000326. doi: 10.1371/journal.pcbi.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill DE, Hope IA, Macke JP, Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: Requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 32.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 33.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21(11):3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magnani L, Eeckhoute J, Lupien M. Pioneer factors: Directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27(11):465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22(4):656–665. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yilmaz A, et al. AGRIS: The Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 2011;39(Database issue):D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehlert A, et al. Two-hybrid protein-protein interaction analysis in Arabidopsis protoplasts: Establishment of a heterodimerization map of group C and group S bZIP transcription factors. Plant J. 2006;46(5):890–900. doi: 10.1111/j.1365-313X.2006.02731.x. [DOI] [PubMed] [Google Scholar]
- 38.Bi YM, et al. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005;44(4):680–692. doi: 10.1111/j.1365-313X.2005.02568.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods. 2012;9(6):609–614. doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.