Significance
Excessive activation of poly(ADP-ribose) (PAR) polymerase (PARP) is intimately linked to cell death in a variety of organ systems. It has long been thought that the cell death caused by excessive activation of PARP occurs through the catalytic consumption of NAD+ followed by reduction of ATP and bioenergetic collapse. This study shows that the bioenergetic collapse is caused not by the consumption of NAD+ but by PAR-dependent inhibition of hexokinase activity leading to defects in glycolysis. These results are consistent with the notion that cell death induced by excessive PARP activity (parthanatos) is an active cell-death program that is initiated by PAR signaling.
Abstract
Excessive poly(ADP-ribose) (PAR) polymerase-1 (PARP-1) activation kills cells via a cell-death process designated “parthanatos” in which PAR induces the mitochondrial release and nuclear translocation of apoptosis-inducing factor to initiate chromatinolysis and cell death. Accompanying the formation of PAR are the reduction of cellular NAD+ and energetic collapse, which have been thought to be caused by the consumption of cellular NAD+ by PARP-1. Here we show that the bioenergetic collapse following PARP-1 activation is not dependent on NAD+ depletion. Instead PARP-1 activation initiates glycolytic defects via PAR-dependent inhibition of hexokinase, which precedes the NAD+ depletion in N-methyl-N-nitroso-N-nitroguanidine (MNNG)-treated cortical neurons. Mitochondrial defects are observed shortly after PARP-1 activation and are mediated largely through defective glycolysis, because supplementation of the mitochondrial substrates pyruvate and glutamine reverse the PARP-1–mediated mitochondrial dysfunction. Depleting neurons of NAD+ with FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, does not alter glycolysis or mitochondrial function. Hexokinase, the first regulatory enzyme to initiate glycolysis by converting glucose to glucose-6-phosphate, contains a strong PAR-binding motif. PAR binds to hexokinase and inhibits hexokinase activity in MNNG-treated cortical neurons. Preventing PAR formation with PAR glycohydrolase prevents the PAR-dependent inhibition of hexokinase. These results indicate that bioenergetic collapse induced by overactivation of PARP-1 is caused by PAR-dependent inhibition of glycolysis through inhibition of hexokinase.
Pharmacological inhibition or genetic deletion of poly(ADP-ribose) polymerase-1 (PARP-1) is dramatically protective against a variety of toxic insults including ischemia reperfusion injury in the heart, brain, and other organs (1, 2). PARP-1 activation also may play a role in several neurologic disorders such as Parkinson disease (PD), Alzheimer's disease (AD), autoimmune encephalomyelitis, and multiple sclerosis (3–5). PARP-1 activation plays a prominent role in necrotic cell death; the necrotic cell-death program initiated by PARP-1 activation has been designated “parthanatos” to distinguish it from other forms of cell death (6, 7). Cell death via PARP-1 activation is thought to occur through the formation of poly(ADP-ribose) (PAR), which acts as a death signal to cause the release of apoptosis-inducing factor (AIF) from the mitochondria (8–10). AIF then translocates to the nucleus, causing nuclear condensation, genomic DNA fragmentation, and cell death (9, 10).
One of key features of parthanatos is consumption of NAD+ caused by parylation, the addition of PAR on PARP-1 itself and ribosylation of PARP-1 substrates (6, 11). Accompanying the NAD+-dependent parylation is a drop in cellular ATP levels and metabolic collapse that have been ascribed to the consumption of NAD+ and subsequent restoration of NAD+ levels, requiring four ATP molecules for each NAD+ molecule (12). Early studies suggested that the decrements in NAD+ and ATP led to cell death through energy collapse (12–14). However, many studies have challenged this notion, because cell death has been shown to be independent of the loss of cellular energy stores (11, 15, 16) but dependent on PAR signaling (8–11). In cells lacking the PAR-degradative enzyme PAR glycohydrolase (PARG), activation of PARP-1 leads to cell death that does not require NAD+ depletion. Instead, cell death occurs through PAR activation of parthanatos (17, 18). In addition, reductions in ATP levels have been shown to precede the reduction in NAD+, thus challenging the idea that the reduction in ATP is caused by the consumption of NAD+ (15, 16). Thus the initiator of the mechanism underlying the decrements in ATP after PARP-1 activation is not known. Here we show that that the bioenergetic collapse after PARP-1 activation is not dependent on NAD+ depletion but instead is caused by the PAR-dependent inhibition of glycolysis that occurs through the inhibition of hexokinase (HK). This study suggests that pharmacological interventions independent of NAD+ are potential targets to prevent bioenergetic defects and cell death in diseases that involve PARP-1 activation.
Results
PARP Activation Induces Defects in Glycolysis.
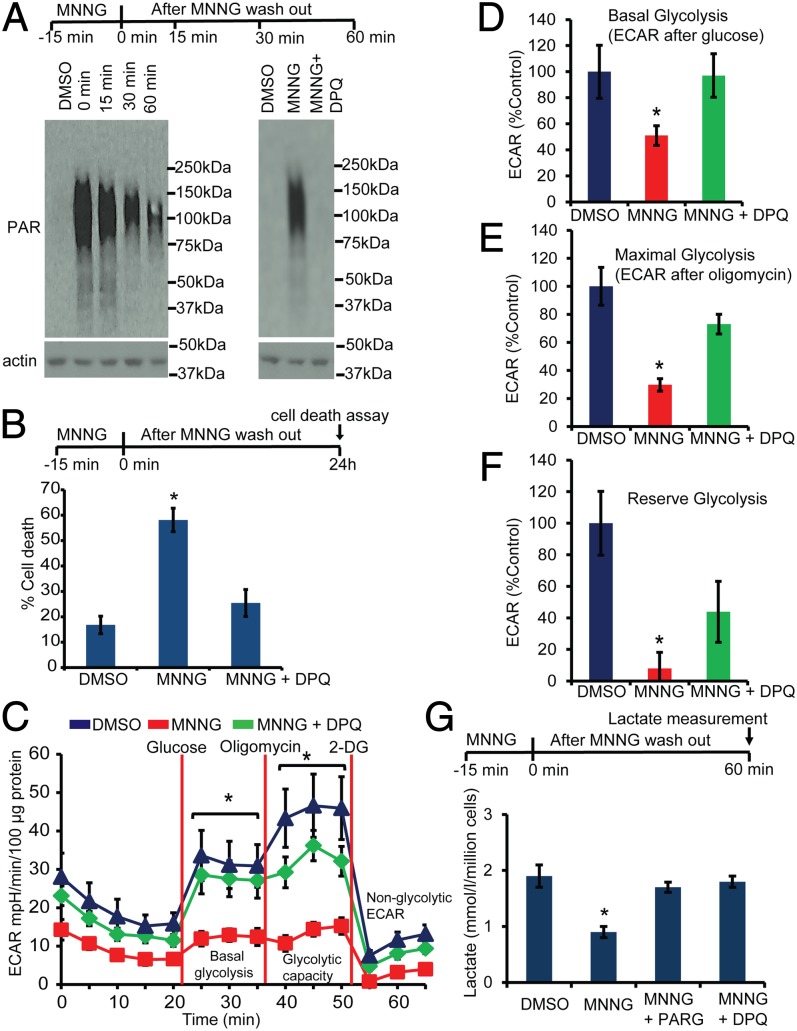
To analyze the effects of PARP-1 activation on glycolysis, a comprehensive real-time analysis of glycolytic flux was assessed via an XF24 Seahorse Flux Analyzer (Seahorse Bioscience) in N-methyl-N-nitroso-N′-nitroguanidine (MNNG)–treated mouse cortical neurons in the presence and absence of the potent PARP inhibitor 3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ) (Fig. 1). MNNG is a potent, widely used activator of PARP (11, 19). Treatment of cortical neurons with 50 μM MNNG for 15 min induces PARP activation as determined by PAR formation via an antibody to PAR (Fig. 1A). PAR is detected immediately after the 15-min treatment of cortical neurons with MNNG and is present for at least 60 min after MNNG administration. DPQ completely prevents the formation of PAR, as previously described (8) (Fig. 1A). Cell death monitored by Hoechst and propidium iodide staining 24 h after MNNG treatment is dependent on PARP-1, because DPQ prevents cell death, as previously described (8) (Fig. 1B). Under these conditions, glycolytic flux (basal glycolysis, glycolytic capacity, and glycolytic reserve) as assessed by the extracellular acidification rate (ECAR) was analyzed by the sequential addition of glucose, oligomycin, and 2-deoxyglucose in glucose-free Seahorse assay medium (Fig. 1C). Basal glycolysis is inhibited significantly 1 h after the 15-min MNNG treatment (Fig. 1D), whereas the maximal glycolytic capacity (Fig. 1E) and reserve glycolysis (Fig. 1F) are abolished almost completely by MNNG. DPQ completely preserved the glycolytic function in MNNG-treated neurons (Fig. 1C). To determine whether these defects in glycolysis are caused by PAR formation, cortical neurons were transduced with an adenovirus expressing the PAR-degrading enzyme PARG (8, 10). Glycolysis was assessed via lactate production (20, 21) in the MNNG-treated mouse cortical neurons. MNNG induces a 58% reduction in lactate levels that is prevented by PARG and DPQ (Fig. 1G). Taken together, these results suggest that PARP-1 activation causes severe defects in glycolysis that are mediated by PAR.
Fig. 1.
PARP activation induces defects in glycolysis. (A) Western blots showing PAR formation in cortical neurons after treatment for 15 min with MNNG (50 μM). DPQ (30 μM) inhibits MNNG-induced PAR formation. (B) Cell-death assessment in mouse cortical neurons treated with MNNG ± DPQ. Cell death was assessed using propidium iodide and Hoechst staining. n = 6; *P < 0.01 vs. control. (C) ECAR analysis in MNNG-treated cortical neurons. Cortical neurons were treated with MNNG for 15 min; then medium was replaced with glucose-free Seahorse medium for ECAR analysis. n = 6; *P < 0.01 vs. control. (D) Basal ECAR was calculated relative to the DMSO control. After the nonglycolytic ECAR was subtracted, the basal ECAR was 20.97 ± 4.2, 10.15 ± 1.5, and 20.34 ± 3.4 milli-pH (mpH)⋅min−1⋅100 µg protein−1 for DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. n = 6; *P < 0.01 vs. control. (E) Maximal glycolysis after the addition of oligomycin was calculated relative to DMSO control. After the nonglycolytic ECAR was subtracted, the maximal ECAR was 34.36 ± 4.6, 10.2 ± 1.5, and 25.10 ± 2.4 mpH⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. n = 6; *P < 0.01 vs. control. (F) Glycolytic reserve capacity in cortical neurons treated with MNNG ± DPQ. After the nonglycolytic ECAR was subtracted, the reserve ECAR was 13.39 ± 2.7, 1.05 ± 1.3, and 5.87 ± 2.5 mpH⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. n = 6; *P < 0.05 vs. control. (G) Lactate production in cortical neurons treated with MNNG ± DPQ. PARG cultures were transduced with adenovirus expressing PARG on DIV 10 and were treated with MNNG on DIV 12. Lactate was measured in the medium 1 h after MNNG treatment. n = 4; *P < 0.05 vs. control. Data represent mean ± SEM.
PARP Activation Causes Mitochondrial Dysfunction.
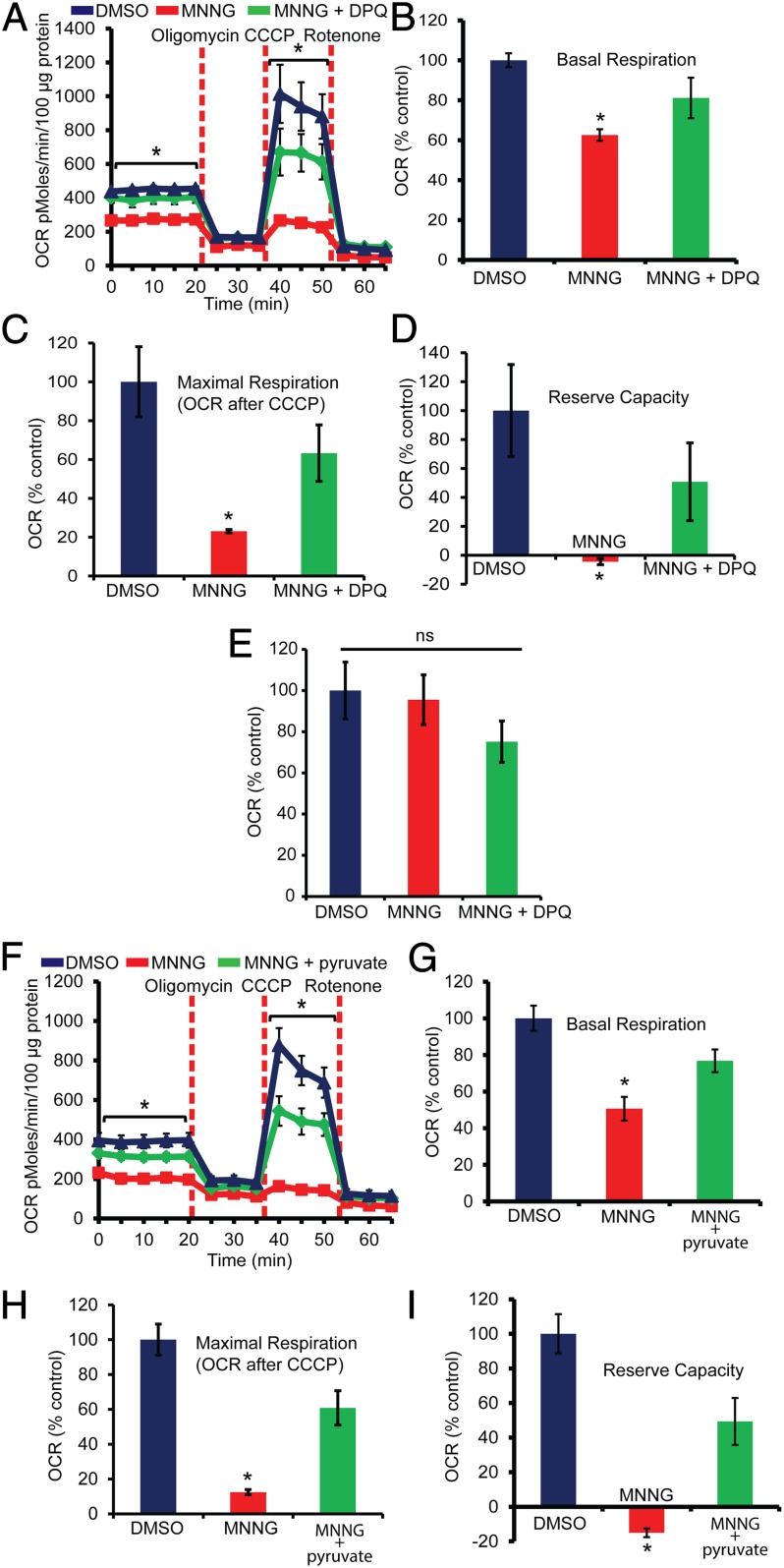
To determine whether PARP-1 activation causes mitochondrial dysfunction, we monitored the mitochondrial oxygen consumption rate (OCR) in mouse cortical neurons treated with MNNG (Fig. 2A). Basal mitochondrial OCR is decreased significantly in MNNG-treated cortical neurons, and DPQ completely rescues the reduction in OCR (Fig. 2B). MNNG induces a significant reduction in maximal OCR (Fig. 2C) and completely abolishes the reserve respiratory capacity (Fig. 2D). DPQ substantially protects against these MNNG-induced defects in mitochondrial respiration. Interestingly, the mitochondrial electron transport chain appears to be intact, because there are no apparent proton leaks 1 h after MNNG treatment (Fig. 2E). These results are in agreement with previously published studies in which Krebs cycle intermediates rescue neurons from PARP-1–dependent cell death (22). Moreover, these observations suggest that the changes in mitochondrial OCR may be caused primarily by the reduced availability of substrates for mitochondrial oxidative phosphorylation and not by defective mitochondrial function. Mitochondrial function is directly dependent on glycolytically derived pyruvate (23), which is a substrate for oxidative phosphorylation (23, 24). To assess whether the impaired OCR is caused by the reduced availability of pyruvate, we supplemented pyruvate to bypass the intermediate steps in glycolysis in the assay medium containing glutamine and assessed OCR (Fig. 2F). The addition of pyruvate to the assay medium completely preserved the basal mitochondrial OCR in MNNG-treated neurons (Fig. 2G). Similarly, the maximal OCR and reserve capacity were preserved significantly by pyruvate in MNNG-treated neurons (Fig. 2 H and I). These data show that PAR can cause a reduction in mitochondrial function because of the reduced availability of glycolytically derived pyruvate.
Fig. 2.
PARP activation causes mitochondrial dysfunction. (A) Analysis of mitochondrial OCR in MNNG-treated cortical neurons. Cortical neurons were treated with MNNG for 15 min; then medium was replaced with Seahorse medium containing 10 mM glucose. n = 5; *P < 0.01 vs. control. (B) The basal OCR was calculated relative to the DMSO control. After the nonmitochondrial OCR was subtracted, the basal OCR was 347.34 ± 12.05 pmol⋅min−1⋅100 µg protein−1 in DMSO-treated cultures vs. 217.19 ± 10.1 in MNNG-treated cultures. n = 5; *P < 0.05 vs. control. (C) The maximal OCR after the addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP) was calculated relative to DMSO control. After the nonmitochondrial OCR was subtracted, the maximal OCR was 843.44 ± 151, 195.15 ± 7, and 533.79 ± 122.23 pmol⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. n = 5; *P < 0.01 vs. control. (D) Relative reserve respiratory capacity in cortical neurons treated with MNNG ± DPQ. n = 5; *P < 0.01 vs. control. After the nonmitochondrial OCR was subtracted, the reserve OCR was 496.39 ± 158, −2.03 ± 10.5, and 252.04 ± 133.7 pmol⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. (E) Proton leaks were calculated by subtracting the rotenone OCR values from the oligomycin OCR values. The data show no significant difference between MNNG and DMSO cultures. (F) The OCR was recorded in cultures treated with MNNG in the presence or absence of 2 mM pyruvate and 2 mM glutamine. n = 5–7. (G) Relative basal OCR in cultures treated with MNNG ± pyruvate. After correction for nonmitochondrial OCR, the basal OCR was 273.18 ± 18.74, 138.32 ± 17.8, and 209.7 ± 16.98 pmol⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + pyruvate-treated cultures, respectively. (H) The maximal OCR after the addition of CCCP was calculated relative to the DMSO control. After the correction for nonmitochondrial OCR, the maximal OCR was 652.75 ± 58, 81.24 ± 10, and 397.7 ± 64 pmol⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + pyruvate-treated cultures, respectively. (I) Respiratory reserve capacity in cultures treated with MNNG ± pyruvate. After correction for nonmitochondrial OCR, the reserve OCR was 379.57 ± 43, -57.08 ± 9, and 187.3 ± 51 pmol⋅min−1⋅100 µg protein−1 in DMSO-, MNNG-, and MNNG + pyruvate-treated cultures, respectively. Data represent mean ± SEM; *P < 0.01.
Glycolytic Defects Caused by PARP Activation Do Not Require NAD+ Depletion.
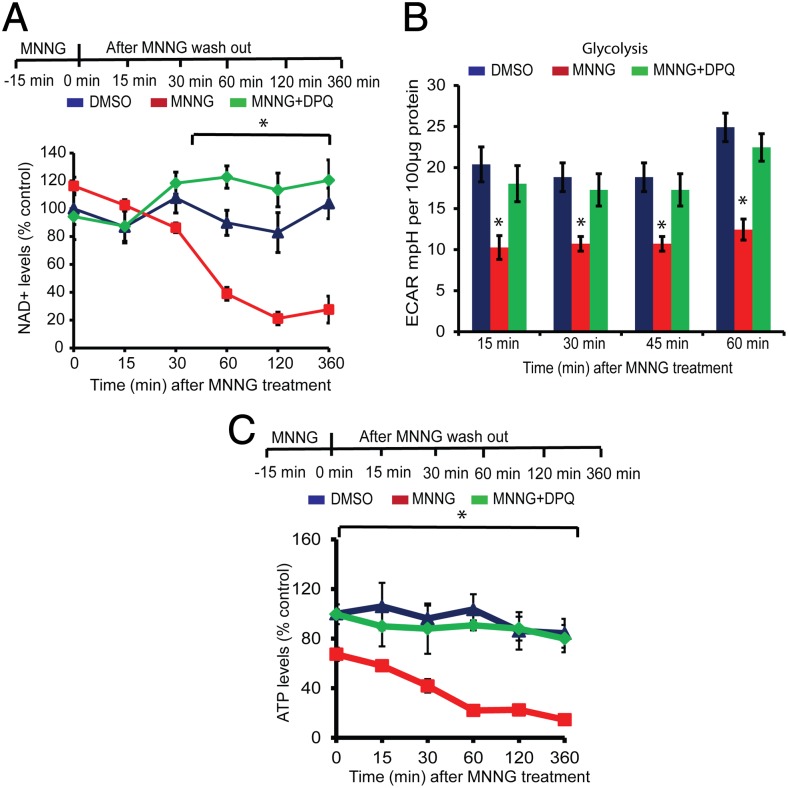
NAD+ levels were monitored after treatment of cortical neurons with 50 μM MNNG for 15 min. There was no appreciable change in NAD+ levels between 0 and 30 min after MNNG treatment (Fig. 3A), when there was robust evidence of PAR formation (Fig. 1A), but NAD+ levels were decreased significantly at 1 h (Fig. 3A) and remained depressed up to 6 h after the MNNG treatment (Fig. 3A). Glycolytic function was assessed 15, 30, and 45 min after MNNG treatment, time points at which there is no appreciable loss of NAD+ levels, and at 60 min after MNNG treatment (Fig. 3B). Glycolytic function was reduced significantly at 15 min and was reduced further at 30 and 45 min after MNNG treatment, with no further reduction at 60 min (Fig. 3B). ATP levels were reduced following MNNG treatment, paralleling the reduction in glycolytic function (Fig. 3C). Of note, the decrease in ATP levels occurs before a significant loss of NAD+. DPQ completely prevents the decrease in glycolytic function and ATP levels at all time points.
Fig. 3.
Glycolytic defects precede NAD+ depletion. (A) NAD+ measurements in mouse cortical neurons treated with 50 μM MNNG for 15 min. NAD+ was quantified at the indicated time points after MNNG treatment. NAD+ levels in cortical neurons at 360 min after DMSO or MNNG treatment were 142.66 ± 15.08, 37.95 ± 13.18, and 165.49 ± 20.14 pmol per million cells in DMSO-, MNNG-, and MNNG + DPQ-treated cultures, respectively. n = 4; *P < 0.01 vs. control (B) Glycolytic flux in the presence 10 mM glucose in mouse cortical neurons treated with MNNG ± DPQ was measured directly after MNNG washout in an XF 24 analyzer. n = 5, *P < 0.01 vs. control. (C) ATP measurements in MNNG-treated cortical neurons at the indicated time points. At 360 min after MNNG treatment, the ATP concentration was 0.0604 ± 0.0144 nM/μg protein, compared with 0.3499 ± 0.0481 in DMSO-treated and 0.3320 ± 0.0456 in MNNG + DPQ-treated cultures. n = 4; *P < 0.01 vs. control. Data represent mean ± SEM.
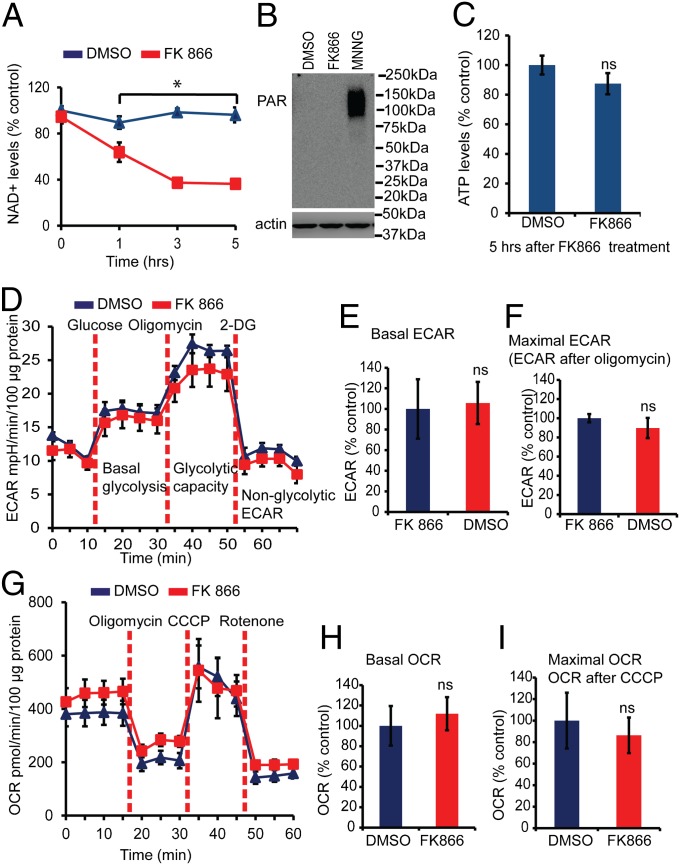
To confirm that the defects in glycolysis and mitochondrial respiration are not caused by a reduction in NAD+ levels, FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, was used to decrease NAD+ levels independent of PARP activation (25). Treatment of cortical neurons with FK866 leads to a decrease in NAD+ levels (Fig. 4A) equivalent to those induced by MNNG treatment. FK866 fails to activate PARP under these conditions, as determined by PAR immunoblot analysis (Fig. 4B), and it fails to reduce ATP levels (Fig. 4C). In contrast, MNNG treatment leads to PAR activation (Fig. 1A), decreases NAD+ levels (Fig. 3A), and reduces ATP levels (Fig. 3C). Glycolytic flux (basal glycolysis, glycolytic capacity, and glycolytic reserve) as assessed by ECAR was analyzed by the sequential addition of glucose, oligomycin, and 2-deoxyglucose in glucose-free Seahorse assay medium 5 h after FK866 treatment, when NAD+ levels are significantly decreased (Fig. 4 A and D). Under these conditions FK866 did not cause any changes in basal glycolysis (Fig. 4 D and E), maximal glycolytic capacity, or reserve glycolysis (Fig. 4 D and F). Mitochondrial OCR also was monitored in mouse cortical neurons treated with FK866 (Fig. 4G). FK866 did not cause any changes in basal or maximal mitochondrial OCR (Fig. 4 H and I). Thus, NAD+ depletion induced by FK866 does not cause glycolytic or mitochondrial defects as assessed by ECAR and OCR analysis.
Fig. 4.
Glycolytic defects do not require NAD+ depletion. (A) Quantification at the indicated time points of NAD+ in mouse cortical neurons treated with 10 μM FK866. At 5 h of treatment the NAD+ level was 48.36 ± 1.50 pmol per million cells in FK866-treated neurons and 128.10 ± 8.65 in DMSO-treated neurons. n = 3; *P < 0.01 vs. control. (B) PAR Western blots showing no PAR formation in cortical neurons treated with 10 μM FK866 or DMSO for 5 h. Treatment with 50 μM MNNG for 15 min induces PAR formation in mouse cortical neurons. The experiment was repeated two times with similar results. (C) ATP levels were determined in mouse cortical neurons treated with 10 μM FK866 or DMSO for 5 h. There are no significant differences in ATP levels in the FK866- and DMSO-treated cultures. At 5 h of treatment the ATP level was 0.3001 ± 0.028 nM/μg protein in FK866-treated neurons and 0.337 ± 0.019 in DMSO-treated neurons. (D) ECAR measurements in cortical neurons treated with 10 μM FK866 or DMSO for 5 h. n = 9. (E) After the nonglycolytic ECAR was subtracted, the basal ECAR was calculated relative to the DMSO. The ECAR was 6.32 ± 1.8 mpH⋅min⋅−1⋅100 µg protein−1 in DMSO-treated cultures and 6.69 ± 1.2 in FK866-treated cultures. n = 9. (F) The maximal ECAR after the addition of oligomycin was calculated relative to control. After the nonglycolytic ECAR was subtracted, the ECAR was 14.75 ± 0.6 and 13.2 ± 1.5 mpH⋅min⋅−1⋅100 µg protein−1 in DMSO- and FK866-treated cultures, respectively. n = 9. (G) OCR was measured in an XF24 Seahorse analyzer in mouse cortical neurons. Treatment with 10 μM FK866 or DMSO for 5 h does not affect the mitochondrial function in mouse cortical neurons. n = 7. (H) Basal OCR was calculated relative to DMSO control. After the nonmitochondrial OCR was subtracted, the basal OCR was 234.08 ± 45.5 and 261.83 ± 37.9 mpH⋅min⋅−1⋅100 µg protein−1 in DMSO- and FK866-treated cultures, respectively. n = 7. (I) Maximal OCR after the addition of CCCP was calculated relative to the control. After the nonmitochondrial OCR was subtracted, the OCR was 763.41 ± 197 and 659.15 ± 126 mpH⋅min⋅−1⋅100 µg protein−1 in DMSO- and FK866-treated cultures, respectively. n = 7. FK866 does not alter the mitochondrial function in mouse cortical neurons. Data represent mean ± SEM.
PARP Activation Inhibits HK.
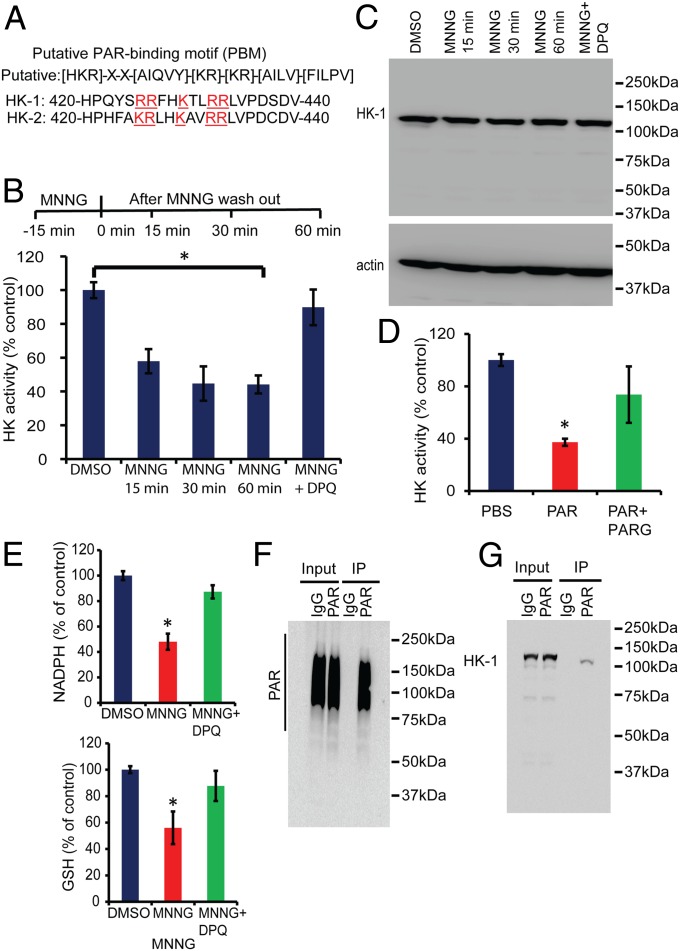
To identify how PARP activation inhibits glycolysis, glycolytic proteins were examined for PAR-binding domains including the PAR-binding motif (PBM), the PAR-binding zinc finger (PBZ) domain, the macro domain, and the WWE domain, a domain with conservative multiple sequence alignment of two tryptophan (W) and a glutamate (E) residues (26). HK-1 and HK-2 contain a strong PBM, as previously described (27) (Fig. 5A). Because HK catalyzes the first regulatory step in glycolysis (28), HK activity was assessed in MNNG-treated cortical neurons. MNNG treatment of cortical neurons leads to a significant reduction in HK activity 15 min after the MNNG treatment (Fig. 5B), when defects in glycolysis are observed (Fig. 3B), but NAD+ levels are not reduced (Fig. 3A). HK activity continues to decrease after MNNG treatment (Fig. 5B), similar to the reduction in glycolysis (Fig. 3B). The reduction in HK activity is reversed by pretreatment with DPQ (Fig. 5B). In addition, MNNG does not reduce the total HK levels, as determined by Western blot (Fig. 5C). To determine whether PAR inhibits HK activity, an in vitro HK enzymatic assay was performed in neuronal cell lysates in the presence of PAR or predigested PAR with PARG as a control. PAR directly inhibits HK activity, but PARG-digested PAR is not able to inhibit HK activity (Fig. 5D). Besides serving as an essential glycolytic enzyme, HK activity also is required to maintain redox homeostasis via the pentose phosphate pathway (20, 21, 29). Pentose phosphate pathway-dependent redox homeostasis is a critical cell survival pathway required to maintain adequate levels of the antioxidant glutathione (GSH) (20, 21, 29). Accordingly, NADPH and GSH levels were monitored as additional indices of HK activity in mouse cortical neurons 1 h after MNNG treatment. Both NADPH and GSH levels were decreased significantly by MNNG treatment, and DPQ prevented the decrements in NADPH and GSH (Fig. 5E). To determine whether PAR binds to HK, cortical neurons were treated with MNNG and immunoprecipitated with a PAR antibody (Fig. 5F). PAR coimmunoprecipitated HK-1 from cortical neurons following MNNG treatment (Fig. 5G). Taken together, these results suggest that the reduction in HK activity is PARP dependent and potentially occurs in a PAR-dependent manner.
Fig. 5.
PARP activation inhibits HK. (A) HK-1 and HK-2 contain strong PAR PBM between amino acids 420–440 (mouse HK1: Q6GQU1 and mouse HK2: O08528). The putative consensus PBM consists of ∼20 amino acids characterized by the presence of hydrophobic amino acids (H: ACGVILMFYW) spaced by basic amino acids (B: HKR) and the accumulation of basic amino acids at the N-terminal site of the motif (K/R). (B) HK activity in mouse cortical neurons treated with MNNG ± DPQ. Samples were collected 15, 30, and 60 min after MNNG treatment. The HK activity was measured at 0.67 ± 0.03 units per milligram of protein in DMSO-treated neurons, whereas in MNNG-treated neurons the activity was measured at 0.39 ± 0.04, 0.30 ± 0.06, and 0.29 ± 0.035 units per milligram of protein at 15, 30, and 60 min, respectively, after MNNG treatment. n = 4; *P < 0.01 vs. control. (C) Western blots show that there is no change in HK-1 expression after MNNG treatment in mouse cortical cultures. (D) HK activity in cell lysates from neuronal cells in the presence of exogenous PAR polymer or PARG-digested PAR polymer. PAR incubation in the lysates inhibits HK activity. The HK activity in PBS-incubated lysates was measured at 0.38 ± 0.016 units per milligram of protein; the HK activity in PAR-treated lysates was 0.16 ± 0.01 units per milligram of protein. n = 3; *P < 0.01 vs. control. (E) Quantification of NADPH 1 h after MNNG treatment. DPQ recovers NADPH levels after MNNG treatment. The NADPH level was 12.01 ± 0.42 pmol per well containing 1 million cells in DMSO-treated cultures and 5.77 ± 0.76 in MNNG-treated cultures. NADPH was estimated using an NADP/NADPH assay kit (catalog no. ab65349; Abcam). Reduced GSH was quantified by HPLC using a C18 column. Samples for GSH estimation were collected 1 h after MNNG treatment. The GSH level was 321.40 ± 8.62 ng/mg protein in DMSO-treated cultures, 180.08 ± 40.07 ng/mg protein in MNNG-treated cultures, and 281 ± 36.8 ng/mg protein in MNNG + DPQ-treated cultures. n = 3; *P < 0.05 vs. control. (F) Immunoprecipitation of PAR using a custom-made PAR monoclonal antibody (clone #21) in mouse cortical neurons. PAR formation was stimulated by MNNG treatment for 15 min, and samples were collected 15 min after MNNG washout. PAR blots were developed using PAR antibody (clone #19). (G) Immunoblots show coimmunoprecipitation of HK-1 with PAR. Neuronal cultures were treated with MNNG for 15 min, and samples were collected 15 min after MNNG washout. PAR antibody (clone #21) was used to immunoprecipitate PAR, and the blots were developed with HK-1 antibody after SDS/PAGE and Western blotting on a nitrocellulose membrane.
Discussion
The major finding of this article is that PARP activation leads to inhibition of glycolysis that is not dependent on NAD+ depletion. PARP activation causes significant decrements in glycolytic flux as determined by a significant reduction in basal glycolysis, glycolytic capacity, glycolytic reserve, and lactate production. Moreover, there are PARP-dependent reductions in mitochondrial function as determined by mitochondrial OCR and reserve respiratory capacity. These reductions in glycolytic and mitochondrial function occur before the reduction in NAD+, suggesting that the reduction in NAD+ levels is not responsible for the bioenergetics failure that occurs after PARP activation. Consistent with this notion are the observations that reducing NAD+ levels, via inhibition of nicotinamide phosphoribosyltransferase with FK866, equivalent to the reduction of NAD+ levels following PARP activation, does not cause any defects in glycolysis and mitochondrial function. Prior studies postulated that NAD+ depletion was responsible for the glycolytic and bioenergetics failure in PARP-1 activation (22, 30). Our data, on the other hand, suggest that NAD+ depletion does not play a major role in the glycolytic and bioenergetics failure. Moreover, the mitochondrial dysfunction is not caused by direct damage to mitochondria by PARP action, because the mitochondrial electron transport chain is intact because there are no proton leaks. These results are in agreement with previously published studies showing that Krebs cycle intermediates rescue neurons from PARP-1–dependent cell death (22). Because pyruvate and glutamine completely restore the defects in mitochondrial function, the changes are likely to be caused by the defects in glycolysis and by the decreased availability of substrates for mitochondrial oxidative phosphorylation. We cannot exclude the possibility that the NAD+ depletion contributes to and maintains the glycolytic and bioenergetics failure after the defects are set in motion by PAR via PARP activation.
Our results indicate that PARP activation leads to a decrement in HK activity. Because studies suggest that PARP activation plays an important role in many pathophysiological states, including stroke, PD, and AD (3–5), it is likely that PARP-dependent HK inhibition plays a role in the pathophysiology of these disorders. In support of this notion, redistribution and a decrease in HK activity has been shown to play a critical role in oxidative stress and neurodegeneration in AD (31). Similarly, overexpression of HK protects against neurodegeneration in mouse models of PD (32). Future studies are needed to explore whether PAR-induced HK inhibition occurs in other pathophysiological states associated with PARP activation, but past studies with MNNG suggest that our findings should extend to many if not all pathophysiological states associated with PARP activation.
The decrement in HK activity observed with PARP activation likely accounts for the decreased glycolytic flux. HK is the first regulatory enzyme in glycolysis that converts glucose into glucose 6-phosphate (23). Glucose 6-phosphate is required for glycolysis, and it is also an important substrate for maintaining NADPH and GSH levels via the pentose phosphate pathway. Importantly HK is inhibited by PARP activation, and the time course of inhibition parallels the decrements in glycolysis and mitochondrial function that are observed after PARP activation. HK contains a PBM, and after PARP activation it coimmunoprecipitates with PAR, indicating that it is a PAR-binding protein. PAR directly inhibits HK, as indicated by the in vitro HK enzymatic assay. Because of HK’s critical role in glycolysis and maintaining bioenergetics, it is highly probable that direct binding of PAR to HK after PARP activation leads to its inhibition and the subsequent reductions in glycolysis and bioenergetics. It is likely that inhibition of PAR binding to HK via blocking peptides or small molecular inhibitors could prevent the energy deficits induced by PARP activation.
Previously it was shown that PARP activation leads to cell death via parthanatos, a process involving the PAR-dependent release of AIF from the mitochondria. Because AIF and HK interact (33, 34), it is conceivable that the PAR-induced release of AIF could contribute to reductions in HK activity and subsequent glycolytic and bioenergetics defects. Future studies are required to sort out the importance of the AIF, HK, and PAR interactions in parthanatos.
In summary, we report that PARP activation leads to NAD+-independent glycolytic and bioenergetic failure that likely is caused by the inhibition of HK by PAR. These observations provide important insights into cell death mechanisms that are induced by PARP activation.
Materials and Methods
More detailed information on the materials and methods is provided in SI Materials and Methods.
Primary Neuronal Culture Preparation.
Primary cortical cell cultures were prepared as previously described (35).
Measurements of OCR.
Mitochondrial OCR in mouse cortical neurons was measured in an XF24 Extracellular Flux Analyzer (Seahorse Bioscience), as described previously (36–38).
Measurement of Glycolysis as ECAR.
Glycolytic flux (basal glycolysis, glycolytic capacity, and glycolytic reserve) as assessed by ECAR were analyzed by the sequential addition of glucose, oligomycin, and 2-deoxyglucose in an XF24 flux analyzer (Seahorse Biosciences) as previously described (37).
Immunoblot Analysis.
Immunoblots were performed on neuronal culture lysates after exposure to MNNG for 15 min and thereafter collected at 0, 15, 30 and 60 min in lysis buffer.
Immunoprecipitation.
Fifteen minutes after MNNG treatment, neuronal cultures from a six-well plate (Nunc Thermo Scientific) were collected in 0.5 mL lysis buffer (PBS containing 1 mM EDTA, 1 mM EGTA, 0.5% SDS, 1% Nonidet P-40, 0.25 mM sodium orthovanadate, 0.25 mM PMSF, 2.5 µg/mL leupeptin, 2.5 µg/mL aprotinin) and incubated for 30 min at 4 °C with constant agitation. After centrifugation (10,000 × g, 4 °C for 10 min), the resulting supernatants were subjected to immunoprecipitation by incubation at 4 °C overnight with a preformed complex of Pure ProteomeTM Kappa IgG binder magnetic beads (Millipore) and an anti-PAR antibody (clone #21, custom designed at Bio-Rad, AbD Serotec GmbH). After overnight incubation at 4 °C, the samples were washed three times with Tris-buffered saline containing 0.1% Tween 20, and the samples were eluted with Laemmli sample loading buffer (Bio-Rad Laboratories) and subsequent heating at 100 °C for 5 min. The eluted samples were run on an SDS/PAGE, and Western blots were developed using anti-PAR (clone #19, custom designed at Bio-Rad, AbD Serotec GmbH), anti–HK-1 (Cell Signaling) primary antibodies, and HRP goat anti-human IgG (Fab')2 (Abcam) and HRP donkey anti-rabbit IgG (GE Healthcare) secondary antibodies. The immunoblots were visualized with enhanced chemiluminescence (Pierce) in an ImageQuant LAS 4000 mini imaging analyzer (GE Healthcare).
NAD+ and ATP Measurements.
NAD+ was measured using a NAD/NADH quantification KIT (Sigma) after treatment with 50 μM MNNG for 15 min in the presence or absence of DPQ or 10 μM FK866 or DMSO.
HK Activity Assay.
HK activity was measured according to the Worthington protocol (Worthington Biochemical Corp.) with slight modifications.
Generation of PAR Antibodies from the HuCAL Platinum Library.
Antibodies against PAR polymer were generated using the Human Combinatorial Antibody Library (HuCAL) platform (Bio-Rad AbD Serotec, Germany). HuCAL consists of billions of recombinant human Fab antibodies to generate highly specific monoclonal antibodies. Purified PAR was purchased from Trevigen Inc., and highly specific antibodies were generated using HuCAL technology at AbD Serotec. The panning in HuCAL technology involves phage display selection that consists of antigen (PAR) immobilization on magnetic beads, followed by HuCAL library incubation and removal of nonspecific antibodies. In this case, all antibodies that detect free ADP ribose were removed. Phages encoding antibodies specific for PAR were amplified, and highly specific human monoclonal recombinant antibodies were generated (AbD Serotec). Multiple highly specific clones detecting PAR were screened in both human and mouse cells. Here we used clones #21 and #19.
Statistical Analysis.
One-way ANOVA was used followed by Tukey’s post hoc test for multiple comparisons or a t test for paired comparisons. Data represent mean ± SEM.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH)/National Institute on Drug Abuse Grant DA000266, NIH/National Institute of Neurological Disorders and Stroke Grants NS067525 and NS38377, and American Heart Association Grant 12SDG9310031. G.G.P. holds a Tier I Canada Research Chair in Proteomics. G.G.P.’s and J.-P.G.’s research was supported by the Canadian Institutes of Health Research. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases at The Johns Hopkins University School of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405158111/-/DCSupplemental.
References
- 1.Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173(1):2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szabó C, Dawson VL. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19(7):287–298. doi: 10.1016/s0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16(10):1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strosznajder JB, Czapski GA, Adamczyk A, Strosznajder RP. Poly(ADP-ribose) polymerase-1 in amyloid beta toxicity and Alzheimer’s disease. Mol Neurobiol. 2012;46(1):78–84. doi: 10.1007/s12035-012-8258-9. [DOI] [PubMed] [Google Scholar]
- 5.Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54(3):375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 6.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci (Landmark Ed) 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, et al. Molecular definitions of cell death subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19(1):107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrabi SA, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103(48):18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu SW, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA. 2006;103(48):18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatokun AA, Dawson VL, Dawson TM. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol. 2014;171(8):2000–2016. doi: 10.1111/bph.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pieper AA, Verma A, Zhang J, Snyder SH. Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol Sci. 1999;20(4):171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- 13.Berger NA, Berger SJ. Metabolic consequences of DNA damage: The role of poly (ADP-ribose) polymerase as mediator of the suicide response. Basic Life Sci. 1986;38:357–363. doi: 10.1007/978-1-4615-9462-8_39. [DOI] [PubMed] [Google Scholar]
- 14.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci USA. 1999;96(24):13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto S, et al. Poly(ADP-ribose) polymerase impairs early and long-term experimental stroke recovery. Stroke. 2002;33(4):1101–1106. doi: 10.1161/01.str.0000014203.65693.1e. [DOI] [PubMed] [Google Scholar]
- 16.Paschen W, Oláh L, Mies G. Effect of transient focal ischemia of mouse brain on energy state and NAD levels: No evidence that NAD depletion plays a major role in secondary disturbances of energy metabolism. J Neurochem. 2000;75(4):1675–1680. doi: 10.1046/j.1471-4159.2000.0751675.x. [DOI] [PubMed] [Google Scholar]
- 17.Koh DW, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci USA. 2004;101(51):17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Feng X, Koh DW. Activation of cell death mediated by apoptosis-inducing factor due to the absence of poly(ADP-ribose) glycohydrolase. Biochemistry. 2011;50(14):2850–2859. doi: 10.1021/bi101829r. [DOI] [PubMed] [Google Scholar]
- 19.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 20.Almeida A, Bolaños JP, Moncada S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc Natl Acad Sci USA. 2010;107(2):738–741. doi: 10.1073/pnas.0913668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrero-Mendez A, et al. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11(6):747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- 22.Ying W, Chen Y, Alano CC, Swanson RA. Tricarboxylic acid cycle substrates prevent PARP-mediated death of neurons and astrocytes. J Cereb Blood Flow Metab. 2002;22(7):774–779. doi: 10.1097/00004647-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 23.Lunt SY, Vander Heiden MG. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 24.Bélanger M, Allaman I, Magistretti PJ. Brain energy metabolism: Focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14(6):724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Pittelli M, et al. Inhibition of nicotinamide phosphoribosyltransferase: Cellular bioenergetics reveals a mitochondrial insensitive NAD pool. J Biol Chem. 2010;285(44):34106–34114. doi: 10.1074/jbc.M110.136739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krietsch J, et al. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med. 2013;34(6):1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gagné JP, et al. Quantitative proteomics profiling of the poly(ADP-ribose)-related response to genotoxic stress. Nucleic Acids Res. 2012;40(16):7788–7805. doi: 10.1093/nar/gks486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith TA. Mammalian hexokinases and their abnormal expression in cancer. Br J Biomed Sci. 2000;57(2):170–178. [PubMed] [Google Scholar]
- 29.Rodriguez-Rodriguez P, Fernandez E, Almeida A, Bolaños JP. Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell Death Differ. 2012;19(10):1582–1589. doi: 10.1038/cdd.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alano CC, et al. NAD+ depletion is necessary and sufficient for poly(ADP-ribose) polymerase-1-mediated neuronal death. J Neurosci. 2010;30(8):2967–2978. doi: 10.1523/JNEUROSCI.5552-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saraiva LM, et al. Amyloid-β triggers the release of neuronal hexokinase 1 from mitochondria. PLoS ONE. 2010;5(12):e15230. doi: 10.1371/journal.pone.0015230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corona JC, Gimenez-Cassina A, Lim F, Díaz-Nido J. Hexokinase II gene transfer protects against neurodegeneration in the rotenone and MPTP mouse models of Parkinson’s disease. J Neurosci Res. 2010;88(9):1943–1950. doi: 10.1002/jnr.22357. [DOI] [PubMed] [Google Scholar]
- 33.Jeong NY, Yoo YH. Cerulenin-induced apoptosis is mediated by disrupting the interaction between AIF and hexokinase II. Int J Oncol. 2012;40(6):1949–1956. doi: 10.3892/ijo.2012.1401. [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Zhang H, Lu W, Huang P. Role of mitochondria-associated hexokinase II in cancer cell death induced by 3-bromopyruvate. Biochim Biophys Acta. 2009;1787(5):553–560. doi: 10.1016/j.bbabio.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrabi SA, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17(6):692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper O, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci Transl Med. 2012;4(141):141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abe Y, Sakairi T, Beeson C, Kopp JB. TGF-β1 stimulates mitochondrial oxidative phosphorylation and generation of reactive oxygen species in cultured mouse podocytes, mediated in part by the mTOR pathway. Am J Physiol Renal Physiol. 2013;305(10):F1477–F1490. doi: 10.1152/ajprenal.00182.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill BG, et al. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393(12):1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.