Significance
Microbial photoreceptors, bacteriophytochromes, absorb near-infrared light, which penetrates deep into animal tissues and is harmless. Bacteriophytochromes delivered as genes can be used to control biological activities in live animals via external light sources. However, the lack of understanding of light-induced conformational changes has hindered development of bacteriophytochrome-based optogenetic tools. Here, we offer a proof of principle that homodimeric bacteriophytochromes can be engineered to activate heterologous output domains that require homodimerization. We constructed an adenylate cyclase, which can control cAMP-dependent processes in live animals in response to light in the near-infrared spectral window. When expressed in cholinergic neurons of a roundworm Caenorhabditis elegans, near-infrared light-activated adenylate cyclase affected worm behavior in a light-dependent manner.
Keywords: phytochrome, protein engineering, adenylyl cyclase, cAMP, locomotion
Abstract
Bacteriophytochromes sense light in the near-infrared window, the spectral region where absorption by mammalian tissues is minimal, and their chromophore, biliverdin IXα, is naturally present in animal cells. These properties make bacteriophytochromes particularly attractive for optogenetic applications. However, the lack of understanding of how light-induced conformational changes control output activities has hindered engineering of bacteriophytochrome-based optogenetic tools. Many bacteriophytochromes function as homodimeric enzymes, in which light-induced conformational changes are transferred via α-helical linkers to the rigid output domains. We hypothesized that heterologous output domains requiring homodimerization can be fused to the photosensory modules of bacteriophytochromes to generate light-activated fusions. Here, we tested this hypothesis by engineering adenylate cyclases regulated by light in the near-infrared spectral window using the photosensory module of the Rhodobacter sphaeroides bacteriophytochrome BphG1 and the adenylate cyclase domain from Nostoc sp. CyaB1. We engineered several light-activated fusion proteins that differed from each other by approximately one or two α-helical turns, suggesting that positioning of the output domains in the same phase of the helix is important for light-dependent activity. Extensive mutagenesis of one of these fusions resulted in an adenylate cyclase with a sixfold photodynamic range. Additional mutagenesis produced an enzyme with a more stable photoactivated state. When expressed in cholinergic neurons in Caenorhabditis elegans, the engineered adenylate cyclase affected worm behavior in a light-dependent manner. The insights derived from this study can be applied to the engineering of other homodimeric bacteriophytochromes, which will further expand the optogenetic toolset.
Light has advantages over chemical means of regulating biological processes because it acts noninvasively and provides superior spatial and temporal resolution (1). Optogenetic approaches that rely on algal and archael channelrhodopsins controlling specific animal neurons opened up a new era in neurobiology (2, 3). Photoreceptors of several other types have been engineered to regulate biological processes and used in cell cultures and transparent animals (4, 5). However, application of optogenetic tools in heme-rich animal tissues has been hindered by high scattering and poor penetration of visible light. Light in the near-infrared window (NIRW), which encompasses the spectral region of ∼680–880 nm, penetrates animal tissues much better than light outside the NIRW (6). A significant fraction of NIRW light can pass through several centimeters of human tissues (7–9), which makes NIRW light a promising means of controlling biological processes in animals. Absence of photoreceptors of NIRW light in most animal tissues is an additional advantage that makes NIRW light harmless (10). This is in contrast to blue light, which is absorbed by flavins and porphyrins, and therefore promotes photooxidative damage (11).
Phytochromes are photoreceptors that absorb light in the NIRW of the spectrum (12–14). The photosensory modules of these photoreceptors covalently bind bilin chromophores. Plant and cyanobacterial phytochromes bind phycocyanobilins or phycoerythrobilins, whereas bacteriophytochromes bind biliverdin IXα. As the first product of heme turnover, biliverdin IXα is naturally present in animal cells, which makes bacteriophytochromes preferred over plant and cyanobacterial phytochromes, whose chromophore synthesis requires dedicated enzymes. Further, absorption wavelength maxima of bacteriophytochromes are red shifted compared with the absorption maxima of plant and cyanobacterial phytochromes. This results in a 2- to 10-fold gain in the penetration depth of light through mammalian tissues (7, 10). Up to now, bacteriophytochrome engineering for optogenetic applications has lagged behind the engineering of photoreceptors of other types (4, 5), including engineering of plant phytochromes (15, 16). The major obstacle to bacteriophytochrome engineering has been the lack of understanding of the mechanisms through which light-induced conformational changes are transduced to regulate output activities (12–14, 17).
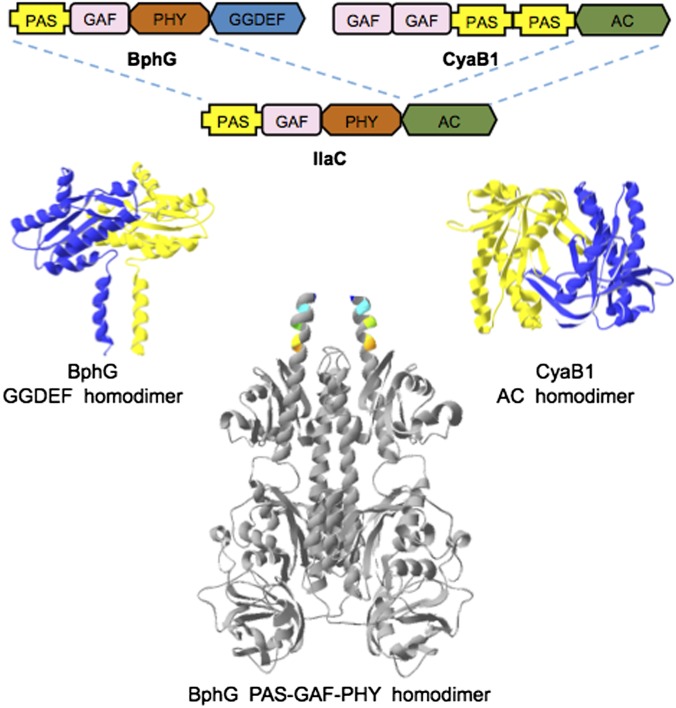
Most or all bacteriophytochromes function as homodimeric enzymes, usually histidine kinases and, more rarely, diguanylate cyclases (DGCs). Enzymatic activities of both histidine kinases and DGCs require precise alignment of two monomers in a homodimer. In the case of DGCs, their product, cyclic dimeric GMP (c-di-GMP), is synthesized from two GTP molecules at the interface between two Gly-Gly-Asp-Glu-Phe (GGDEF) domains (Pfam database) (18) responsible for DGC activity. Each GGDEF domain brings a substrate molecule to the catalytic site (19, 20). In the inhibited state, the photosensory modules apparently prevent enzymatic domains from forming a properly aligned homodimer, whereas light-induced conformational changes restore enzymatically productive domain alignment. Although the exact nature of conformational changes is unknown, it is likely that they are mediated by the α-helical linkers that connect the photosensory modules to the output domains (21–23) (Fig. 1). Further, in all DGCs whose regulation has been studied at the structural level, enzyme activation has been shown or predicted to occur via alignment of the rigid GGDEF domains rather than via intradomain conformational changes (19, 20). Based on these considerations, we reasoned that bacteriophytochromes can regulate diverse output activities that depend on proper alignment of the two output domains (24).
Fig. 1.
Domain architectures, and 3D models of the components used in IlaC engineering. R. sphaeroides BphG is bacteriophytochrome DGC; Nostoc sp. CyaB1 is homodimeric AC. Details on 3D models are provided in SI Materials and Methods.
Here we demonstrate that bacteriophytochrome photosensory modules can indeed regulate heterologous output domains. We engineered a series of photoactivated adenylate cyclases (ACs) designated NIRW light-activated AC (IlaC), by fusing a photosensory module from the Rhodobacter sphaeroides bacteriophytochrome DGC, BphG1 (25, 26), to an AC domain from the Nostoc sp. CyaB1 protein (27) (Fig. 1), which, like all bacterial type III ACs, works as a homodimer (28, 29). We chose to make IlaCs because cAMP is an important second messenger involved in regulation of diverse biological processes (28, 29). The ability to manipulate cAMP levels in specific cells or tissues in live animals is of significant interest in biomedical research. Earlier, we and others described naturally occurring blue-light–activated ACs (30–32) whose utility in optogenetic applications in cell cultures and small animal models has been demonstrated (32–34). However, undesirable effects of blue light and its poor penetration through nontransparent tissues present major obstacles for the use of blue-light–activated ACs, which can be solved by IlaCs. Further, the insights emerged from IlaC engineering are expected to facilitate engineering of new NIRW light-activated optogenetic tools.
Results
Components of the NIRW Light-Activated AC.
For the photosensory module of the NIRW light-activated AC, we chose the PAS–GAF–PHY module from BphG1 from R. sphaeroides, where PAS, GAF, and PHY (phytochrome) are protein domain names (Pfam database). The truncated derivative of BphG1, BphG, where the PAS–GAF–PHY module is linked to a GGDEF domain, functions as a light-activated DGC (25) (Fig. 1). BphG was particularly attractive because (i) absorption maxima of its dark [red-absorbing (Pr)] and lit [far-red–absorbing (Pfr)] forms, 712 and 756 nm, respectively, lie within the NIRW, and (ii) its DGC activity is activated by light by ∼11-fold, the largest photodynamic range (fold activation) among bacteriophytochromes for which such a ratio has been quantified (26).
For the output AC domain, we looked for a protein (i) whose AC activity is confined to the AC domains, i.e., where regulatory domains are not required for basal activity, and (ii) whose AC activity in the dark can be detected in a bacterial screening system. CyaB1 from Nostoc sp. fit these requirements (27). The native CyaB1 protein has the following domain architecture, GAF–GAF–PAS–PAS–AC (where AC domain is responsible for AC activity (Fig. 1). The C-terminal AC domain of CyaB1 untangled from the regulatory domains possesses some AC activity (27).
To monitor cAMP synthesis, we used Escherichia coli BL21[DE3] cya, which lacks the endogenous AC, Cya. In this strain, expression of the chromosomal lacZ gene encoding β-galactosidase is low because of the absence of activation by the cAMP-responsive protein (CRP) also known as catabolite activator protein (35). BL21[DE3] cya produces white colonies on agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (31). The C-terminal AC domain of CyaB1 (amino acids 585–857) restores lacZ expression, thus generating blue colonies. To endow this bacterial system with the ability to synthesize biliverdin IXα, we introduced the cyanobacterial heme oxygenase gene ho1 (36), whose product converts heme synthesized by E. coli into biliverdin IXα.
Engineering a NIRW Light-Activated AC.
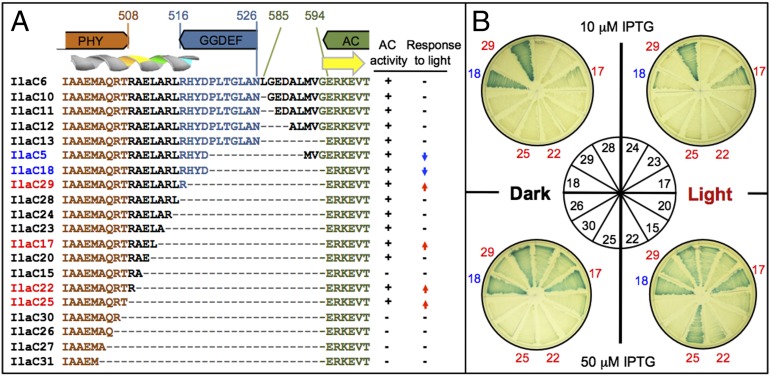
We synthesized the DNA fragment encoding the AC domain of CyaB1 (amino acids 585–857) and fused it to amino acid 526 in the unstructured (loop) region of the GGDEF domain of BphG. Leu585 of CyaB1 is also in the loop region, 8 aa upstream of the first structural element, β-strand, of the AC domain (Fig. 1 and Fig. S1). The unstructured linkers were meant to prevent potential steric interference between the fusion partners. In accord with this intent, the chimeric protein, IlaC6, possessed AC activity, yet this activity was nonresponsive to light (Fig. 2A). Next, we determined the minimal AC domain size that retained enzymatic activity in the BphG–CyaB1 fusions. We fixed the fusion point in BphG at amino acid 526 and progressively shortened the AC domain, from Leu585 to Glu594 of CyaB1, where Glu594 is in the predicted β-strand of the AC domain (Fig. 2A). All fusions in this series, Ilac10–13, proved to be enzymatically active and nonresponsive to light (Fig. 2A). In the next round of engineering, the AC domain border was fixed at Glu594 but the unstructured region of the GGDEF domain and the α-helical linker extending from the PHY domain were subject to shortening. The fusion to Arg507 of BphG (IlaC30) and fusions containing shorter BphG fragments (IlaC26, -27, and -31) had no AC activity (Fig. 2 A and B) likely because of the steric hindrance between the fusion components, thus the engineering space was limited to amino acids 508–526 of BphG.
Fig. 2.
IlaC engineering strategy. (A) A subset of BphG–CyaB1 protein fusion sequences, their AC activities, and responsiveness to light. The amino acid colors correspond to the colors of protein domains shown above protein sequences. Brown, PHY domain; blue, GGDEF domain; green, AC domain; and black, interdomain linkers. Predicted secondary structure elements, α-helix, and β-strand, are shown above sequences. AC activity: +, active; −, inactive, according to the lacZ plate assays (see B). Response to light: −, no response; ↑, activation; and ↓, inactivation. (B) Images of the lacZ plate assays of AC activity. Selected E. coli BL21[DE3] (pETilaC# and pT7-ho1-1) strains from A were grown on LB agar containing X-Gal and IPTG, either in the dark (Left of center) or in the red light (Right). Blue colony color indicates cAMP–CRP-induced lacZ expression. Each strain expressing an IlaC# was plated in a sector of each of the four plates. The plating guide is in the Center. Dark-blue numbers, light-inactivated IlaC protein; red numbers, light-activated IlaC proteins. IlaC expression from pETilaC# was induced at two IPTG levels: 10 μM (Upper plates) and 50 μM (Lower plates).
One of the fusions in this region, IlaC18, produced a protein whose AC activity was higher in the dark than in the light, i.e., light-inactivated AC (Fig. 2 A and B). This result showed that light induces conformational changes that are sufficient to misalign the enzymatically productive AC homodimer. Notably, light-inactivated fusions were also obtained at other fusion points, e.g., IlaC5 (Fig. 2A). Four fusions, IlaC29 (Arg516), IlaC17 (Leu512), IlaC22 (Arg509), and IlaC25 (Thr508) produced the desired, photoactivated enzymes. The AC activity of these fusions differed from each other as judged by colony color on X-Gal agar at two different isopropyl-1-thio-β-d-galactopyranoside (IPTG) concentrations used to regulate IlaC expression levels (Fig. 2B).
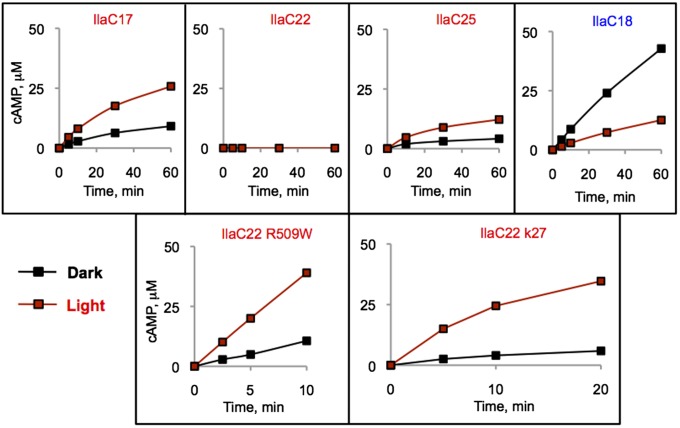
Three photoactivated cyclases, IlaC17, IlaC22, and IlaC25, as well as one photoinactivated cyclase, IlaC18, were overexpressed as a C-terminal His6 fusion and purified. The AC activity of IlaC18 was decreased by threefold upon irradiation with 700-nm light (Fig. 3), whereas the activities of IlaC17 and IlaC25 were increased by light by approximately twofold (Fig. 3). In agreement with the colony phenotypes, the basal AC activity of IlaC17 was higher than the activities of IlaC25 or IlaC22. The AC activity of IlaC22 in vitro was below detection (Fig. 3).
Fig. 3.
Kinetics of cAMP accumulation by the purified IlaC proteins in the dark and light. (Upper row) First-generation light-responsive proteins: IlaC18, light-inactivated AC; IlaC17, 22 and 25, light-activated AC. (Lower row) IlaC22 mutants with improved photodynamic ranges: IlaC22 R509W and IlaC k27. AC activity was measured at room temperature. cAMP was quantified by high-performance liquid chromatography (HPLC). Black traces, dark (dim green light); red traces, irradiation with 700-nm light.
Improving Photodynamic Range of the Photoactivated ACs.
Because the photodynamic range of the first-generation NIRW light-activated ACs were significantly lower than the photodynamic range of BphG, we intended to improve this parameter by random PCR-based mutagenesis. Here, we focused on IlaC22 as a template because of its low dark activity. In the light, IlaC22 produced blue colonies only when expressed at high, 50 μM, IPTG concentration. We therefore screened the library of the PCR-mutagenized IlaC22 gene for blue colony appearance at low, 10 μM, IPTG concentration. After screening ∼105 mutant clones, we found mutants with significantly increased AC activity. The best one had an R509W (BphG numbering) substitution, right at the junction between the PHY and AC domains (Fig. 2A).
The photodynamic range of AC activity of the purified IlaC22 R509W was approximately fourfold (Fig. 3). To further improve the photodynamic range, we mutagenized the ilaC22 R509W gene at high mutation frequency. Our primary goal was to decrease the dark AC activity of IlaC22 R509W. We therefore searched for white colonies at high, 100 μM, IPTG, in the dark. The strategy used in this screen is illustrated in Fig. S2. After screening of >105 mutant clones, we identified a derivative, IlaC22 k27, that had significantly lower activity in the dark but only slightly lower activity in the light, compared with IlaC22 R509W (Fig. 3), which resulted in the photodynamic range of sixfold. IlaC22 k27 was found to contain three new mutations, compared with IlaC22 R509W, two of which were in the BphG photosensory module (L164M and Q371H) and one in the AC domain (Q670R of CyaB1) (Fig. S3). Because the photodynamic range of IlaC22 k27 was within twofold of the photodynamic range of the BphG, we did not attempt to increase it further. Instead, we focused on modifying another important parameter, i.e., stability of the photoactivated state.
Extending Lifetime of the Light-Activated State of an AC.
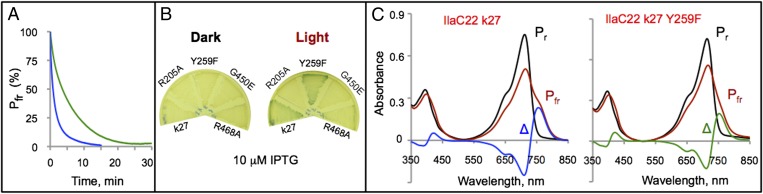
Following photoactivation, bacteriophytochromes in the lit (Pfr) state spontaneously return to the ground, dark (Pr), state via thermal reversion (12). The half-life of IlaC22 k27 in the Pfr state is 46 ± 3 s (Fig. 4A). A relatively short half-life is desirable for optogenetic applications that require short pulses of cAMP, whereas applications involving sustained light-induced increases in cAMP levels, will benefit from enzymes with more stable lit state.
Fig. 4.
Photochemical characterization of IlaC variants. (A) Kinetics of the dark recovery of IlaC derivatives from the lit (Pfr) state. Plotted are changes in absorbance at 755 nm over time following 5-min excitation with 700-nm light. Blue trace, IlaC22 k27; green trace, IlaC22 k27 Y259F. Half of the Pfr form of IlaC22 k27 decayed after 46 ± 3 s and half of the Pfr form of the IlaC22 k27 Y259F mutant decayed after 197 ± 9 s. (B) Plate assays of the IlaC k22 dark recovery mutants. (C) Light-induced spectral changes in IlaC22 derivatives. Black (Pr), before irradiation; red (Pfr), after irradiation with 700-nm light; Δ, difference (Pfr − Pr) spectra. IlaC22 k27 (Left); IlaC22 k27 Y259F (Right).
To increase lifetime of the lit state of IlaC22 k27, we relied on success in extending this parameter in the Arabidopsis thaliana phytochrome PhyB (37, 38). Four residues in the proximity of the chromophore involved in controlling dark recovery of PhyB are conserved in BphG (Fig. S4). We introduced in IlaC22 k27 the same mutations as those that prolonged the half-life of PhyB. Three mutants (R205A, G450E, and R468A) resulted in the loss of AC activity and were discarded (Fig. 4B). The fourth mutant, IlaC22 k27 Y259F, was purified and characterized in vitro. Its absorption maxima were slightly shifted compared with IlaC22 k27, i.e., Pr (713 nm) and Pfr (755 nm) (Fig. 4C), and the half-life of thermal reversion was significantly (by 4.3-fold), increased to 197 ± 9 s, compared with IlaC22 k27 (Fig. 4A).
The higher stability of the lit state of IlaC22 k27 Y259F was expected to allow its use in the pulsed light regiments, which may decrease such negative effects of constant irradiation as tissue heating. To test this possibility, we compared the effect of IlaC22 k27 and IlaC22 k27 Y259F on the cAMP–CRP-dependent lacZ gene expression in E. coli (Fig. S5). Whereas both ACs showed similar increases in lacZ expression in constant light, compared with the dark, when pulsed light (30 s light, 90 s dark) was used, the IlaC22 k27 Y259F mutant outperformed IlaC22 k27 (Fig. S5), as expected.
NIRW Light-Activated Control of Caenorhabditis elegans Behavior.
To test performance of a NIRW light-activated AC in an animal model, we expressed IlaC k27 in cholinergic neurons of the roundworm C. elegans. Previously, a blue-light–activated AC, PACα, has been used in C. elegans as a tool for optogenetic manipulation of behavior (33). Expression of PACα in the cholinergic neurons, using the promoter for the vesicular acetylcholine transporter, unc-17, followed by stimulation with blue light, resulted in increased locomotory activity (33). However, blue light activates a C. elegans avoidance response and is toxic upon prolonged irradiation (33, 39), thus confounding the interpretation of the behavioral response to the optogenetic manipulation. Therefore, IlaC could be advantageous for behavioral analyses in C. elegans.
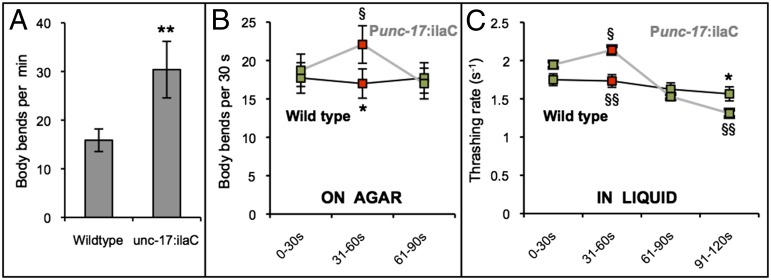
We generated transgenic animals expressing IlaC22 k27 in cholinergic neurons using the unc-17 promoter. IlaC22 k27 transgenic animals cultivated on an agar surface and exposed to daily light from the environment were more active than wild-type animals, as evident by the higher frequency of their body bends (Fig. 5A). Hyperactivity is characteristic of animals with increased activity of cAMP-dependent protein kinase A (PKA), such as mutants for the gene kin-2, which encodes for the regulatory subunit of PKA (40). To test the effect of red light, we grew wild-type and Punc-17:ilaC animals in the dark for a single generation. Individual animals were transferred to an agar surface that did not contain food. Animals were monitored under monochromatic light-emitting diode (LED) irradiation for 90 s. During this time, body bends were counted during the following light regiment: green light 0–30 s, red 31–60 s, and green 61–90 s. Whereas control animals did not alter their locomotory activity in response to red light (Fig. 5B), Punc-17:ilaC animals performed more body bends in the presence of red light (Fig. 5B and Movie S1).
Fig. 5.
Red-light stimulated locomotion of animals expressing IlaC22 k27 in C. elegans cholinergic neurons (Punc-17:ilaC). (A) When exposed to environmental light on an agar surface, transgenic Punc-17:ilaC animals have higher frequency of anterior body bends relative to wild-type animals (two-tailed Student t test, **P < 0.001, n ≥ 12). (B) Red (650 nm) light increases the number of body bends performed on an agar surface by Punc-17:ilaC animals (two-tailed Wilcoxon rank-sum test, §P < 0.05, n = 11). (C) Red light increases the thrashing rate in liquid of Punc-17:ilaC animals (two-tailed Wilcoxon rank-sum test, §P < 0.05). After reexposure to green light (532 nm; 61- to 120-s interval), Punc-17:ilaC animals thrash slower compared with their first green light exposure (two-tailed Wilcoxon rank-sum test, §§P < 0.005) and compared with wild-type controls (two-tailed Student t test, *P < 0.05, n ≥ 20). Two transgenic strains were analyzed, both showing similar results. Analysis of body bends on an agar surface was performed on the strain NQ721 and analysis of the thrashing rate in liquid was performed on the strain NQ719.
We also monitored the effects of AC activation in cholinergic neurons on the frequency of thrashing movements in liquid medium. Swimming animals, which were reared in the dark, were video monitored for a total of 2 min during the following light regiment: green 0–30 s, red 31–60 s, green 61–120 s. Whereas control animals did not alter their thrashing rate in response to red light, Punc-17:ilaC animals increased their thrashing rates when exposed to red light (Fig. 5C). Interestingly, their thrashing rates significantly decreased during the second exposure to green light (Fig. 5C), suggesting poststimulatory fatigue. Thus, our results indicate that the NIRW light-activated AC can be used as a tool for optogenetic manipulation of cAMP levels in animals.
Discussion
Unique photochemical properties of bacteriophytochromes, i.e., (i) light absorbance in the NIRW, optimal for use in red-blooded animals, (ii) naturally available in animal cells chromophore (biliverdin IXα), and (iii) innocuous nature of NIRW light, position them as superior photoreceptors for optogenetic applications in animals (10). However, bacteriophytochrome engineering for optogenetic applications has been hindered by the lack of understanding of mechanisms through which light-induced conformational changes induce changes in output domains. The two most common bacteriophytochrome types, histidine kinases and DGCs, operate as homodimeric enzymes, where proper alignment of two monomeric kinase or GGDEF domains is essential for enzymatic activity. We postulated that α-helices extending from the PAS–GAF–PHY photosensory modules are primarily responsible for alignment of the output domains (Fig. 1) and hypothesized that the light-induced movements mediated by the α-helices can regulate alignment of unrelated, heterologous domains whose activity requires properly aligned homodimers. The NIRW light-activated ACs, IlaCs, constructed in this study, are consistent with our assumption. The recently solved structures of the photosensory module of the Deinoccoccus bacteriophytochrome showed that the distance between the tips of the signaling α-helices changes by as much as ∼30 Å in response to light (41).
What lessons can we draw for future homodimeric bacteriophytochrome engineering? First, the choice of the output domains and screening scheme are important. For example, the ability of the AC domains of CyaB1 to spontaneously homodimerize helped us to distinguish between enzymatically active and inactive fusions (Fig. 2 A and B). The high sensitivity of the screening system used here was useful for detecting relatively low (twofold) photodynamic ranges (Fig. 2B). An engineering strategy that involves (i) construction of permissive fusions (involving unstructured linkers) as the first step, (ii) subsequent minimization of the output domain size, and finally, (iii) shortening (or possibly, extending) signaling α-helices, may also be universally applicable. Another lesson learned is that photoactivation ratios of the first-generation light-responsive fusions can be significantly improved by extensive mutagenesis.
Are there limitations of homodimeric bacteriophytochrome engineering? First, it is imperative that energy associated with the light-induced conformational changes is sufficient to manipulate the output domain alignment. For overly tight homodimers, monomer interactions may have to be weakened. Another important issue is reasonable correspondence in distances between fusion points in the signaling and output components. We estimated that the distances between the α-helical residues of BphG that resulted in photoactivated fusions were in the range of 11–22 Å, whereas the distances between the N-terminal β-strands of the catalytically inactive CyaB1 AC domain homodimer were ∼40 Å (Fig. S1). These distances turned out to fit nicely in the 10–40 Å spread between the α-helices of the Deinococcus BphP in the dark and lit states (41). Whether proteins whose homodimers are separated by larger distances can be made light-responsive is unclear.
We found not one but several photoactivated IlaC fusions. Interestingly, they differed from each other by either 3 or 4 residues (IlaC29 = IlaC17 + 4 aa, IlaC25 = IlaC17 − 4 aa, and IlaC22 = IlaC17 − 3 aa) (Fig. 2A), which places their AC domains roughly in the same helical phase but separated by one or two α-helical turns (1 turn = 3.6 aa) (42) (Fig. S1). This result is consistent with the model where α-helices act as rods involved in aligning rigid output domains located at their tips. It is therefore unsurprising that the 1 or 2-aa shifts out of helical phase destroy proper domain alignment and result in the loss of light responsiveness (Fig. 2A). The involvement of α-helices in mediating intramolecular signal transduction is not unique to bacteriophytochromes, and other researchers have used such helices for heterologous domain replacements (43–49).
IlaC expressed in neurons of the roundworm C. elegans affected behavior in response to light. In the case of C. elegans, the main advantages of IlaCs over blue-light activated ACs are the absence of the photoavoidance response and the lack of phototoxicity associated with prolonged exposure to blue light (33, 39). However, we anticipate that IlaCs will prove most useful in applications in deep mammalian tissues inaccessible by blue light.
Our demonstration that homodimeric bacteriophytochromes are amenable to protein engineering (24) and recent progress in structural understanding of dark-to-light conformational changes (41) should encourage design of new NIRW light-activated proteins. Because activities of many signal transduction components depend on homodimerization, including membrane receptors, cyclic nucleotide phosphodiesterases, certain protein phosphatases, proteases, nucleases, and transcription factors, we expect significant expansion of the optogenetic toolset involving NIRW light.
Materials and Methods
Microbiological Methods.
E. coli BL21[DE3] cya lacking endogenous adenylate cylase CyaA (26) and containing two plasmids, pT7-ho1-1 (25) that expresses the Synechocystis sp. heme oxygenase ho1 (36) and pETilaC# (expressing IlaC proteins) were used for IlaC screening. Strains were grown at 30 °C in LB supplemented with X-Gal (40 μg/mL), ampicillin (50 μg/mL), and kanamycin (25 μg/mL). IPTG was added for induction of IlaC proteins. For light-sensitive experiments, Petri dishes were placed onto All-Red (660 nm) LED grow light panels 225 (30.5 × 30.5 cm; LED Wholesalers). Light irradiance was ∼0.2 mW cm−2. For growth in the dark, Petri dishes were wrapped in aluminum foil.
Recombinant DNA Techniques.
The DNA fragment of bphG1 gene (RSP4191) encoding the photosensory PAS–GAF–PHY module was amplified by PCR from the R. sphaeroides 2.4.1 genome. The DNA fragment of cyaB1 from Nostoc (formerly Anabaena) sp. PCC 7120 was synthesized by BioBasics with the codon use optimized for R. sphaeroides. Two fragments were joined by fusion PCR using GoTaq (Promega) and subsequently cloned into the XbaI and HindIII sites of pET23a(+) (Invitrogen) to yield a series of plasmids, pETilaC#. Each of these plasmids encodes a unique BphG-CyaB1::His6 fusion protein. Site-directed mutagenesis was performed using a QuikChange kit (Stratagene). Error-prone PCR mutagenesis was carried out using GeneMorph II Random Mutagenesis kit (Agilent Technologies).
The Punc-17:ilaC plasmid, pNQ149, was constructed using the MultiSite Gateway Three-Fragment Vector Construction kit (Invitrogen). pNQ149 combines the unc-17 promoter (obtained from the Promoeome library in the pdonrP4-P1r entry vector), the ilaC22 k27 DNA, and the unc-54 3′ UTR in the pdonrP2R-P3 entry vector.
Protein Purification and AC Assays.
The IlaC proteins were purified as C-terminal His6-tagged fusions using Ni-affinity chromatography (Novagen). AC assays were performed using freshly purified proteins at room temperature, essentially as described earlier (50). Detailed protocols are available in SI Materials and Methods.
Spectroscopy.
Electronic absorption spectra were recorded with a UV-1601 spectrophotometer (Shimadzu) at room temperature. Protein solution (100 μL) in a 10-mm light path quartz cuvette was irradiated by 1-W (700 nm) LED directly in the spectrophotometer from the top of the cuvette.
C. elegans Cultivation and Transgenesis.
Animals were cultivated on nematode growth medium (NGM) agar and were fed E. coli DA837 derived from strain OP50 (51). All experiments were performed on hermaphrodites. The following strains were used in this study: N2 (Bristol), NQ719 qnEx386[Punc-17:iLac; Pmyo-2:mCherry], and NQ721 qnEx388[Punc-17:iLac; Pmyo-2:mCherry]. Transgenic animals were created by microinjection (52) using a Leica DMIRB inverted differential interference contrast microscope equipped with an Eppendorf Femtojet microinjection system. N2 animals were injected with 25 ng/μL of pNQ149 in combination with 5 ng/μL of pCFJ90 (Pmyo-2:mCherry) and 120 ng/μL 1-kb molecular weight ladder (New England Biolabs).
C. elegans Behavioral Assays.
Animals were grown in the dark for one generation on NGM agar seeded with DA837 bacteria. For the assays performed on agar, L4 larvae were transferred to NGM plates seeded with DA837 and grown to early adulthood overnight. Individual adult animals were transferred in the presence of green light onto an NGM plate without bacteria and left unperturbed for 15 min. Animals were observed using a Leica MZ16 stereomicroscope. During this time, the animals were exposed to green light for 30 s, red light for 30 s, and green light for 30 s. Body bends were counted by the observer. Irradiation was provided by a LED Color Changing kit IP66 (LED Wholesalers). No biliverdin IXα was added to the agar.
For the swimming assays, we followed with minor modifications the protocols described by Weissenberger et al. (33). L4 larvae were transferred to NGM plates seeded with DA837 bacteria supplemented with 1 mM biliverdin hydrochloride (Sigma) and grown to early adulthood for 1 d. Individual early adult animals were transferred in the presence of green light into a 10-μL drop of NGM and M9 in a 1:1 ratio supplemented with 1 mM biliverdin hydrochloride. Animals were video monitored for 2 min using a USB 2.0 monochrome camera (ImagingSource, model DMK 72AUC02), mounted on a Leica MZ16 stereomicroscope. During the 2-min recording, the animals were exposed to green light for 30 s, red light for 30 s, and green light again for 60 s. Body bend frequency was counted by observing the video recordings.
Supplementary Material
Acknowledgments
We thank Katrina Forest for stimulating discussions, J. Clark Lagarias for the source of heme oxygenase ho1, and Ryan Griesbach and Steve Schoeber [recipients of National Science Foundation Wyoming Experimental Program to Stimulate Competitive Research (EPSCoR) undergraduate research fellowships] and Benjamin Freedman (recipient of the University of Pennsylvania Ramesh fellowship for undergraduates) for technical help. This study was supported by National Institutes of Health (NIH) Grants 5R21CA167862 (to M.G.), 5P20RR016474 (a subproject to M.G.), and R01NS064030 (to D.M.R.). J.S.-L. was supported by NIH 5P20RR016474 and M.D.N. was supported by T32HL07713. T.M.J. was a recipient of undergraduate research fellowships from the Wyoming National Aeronautics and Space Administration Space Grant Consortium and the National Science Foundation Wyoming EPSCoR.
Footnotes
Conflict of interest statement: M.-H.R. and M.G. are coauthors on the patent application “Near-infrared light activated proteins” US Patent 20,130,030,041; WO 2013016693 filed by the University of Wyoming.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324301111/-/DCSupplemental.
References
- 1.Toettcher JE, Voigt CA, Weiner OD, Lim WA. The promise of optogenetics in cell biology: Interrogating molecular circuits in space and time. Nat Methods. 2011;8(1):35–38. doi: 10.1038/nmeth.f.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deisseroth K. Optogenetics. Nat Methods. 2011;8(1):26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams SC, Deisseroth K. Optogenetics. Proc Natl Acad Sci USA. 2013;110(41):16287. doi: 10.1073/pnas.1317033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pathak GP, Vrana JD, Tucker CL. Optogenetic control of cell function using engineered photoreceptors. Biol Cell. 2013;105(2):59–72. doi: 10.1111/boc.201200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller K, Weber W. Optogenetic tools for mammalian systems. Mol Biosyst. 2013;9(4):596–608. doi: 10.1039/c3mb25590e. [DOI] [PubMed] [Google Scholar]
- 6.Weissleder R. A clearer vision for in vivo imaging. Nat Biotechnol. 2001;19(4):316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 7.Wan S, Parrish JA, Anderson RR, Madden M. Transmittance of nonionizing radiation in human tissues. Photochem Photobiol. 1981;34(6):679–681. doi: 10.1111/j.1751-1097.1981.tb09063.x. [DOI] [PubMed] [Google Scholar]
- 8.Cubeddu R, Pifferi A, Taroni P, Torricelli A, Valentini G. Noninvasive absorption and scattering spectroscopy of bulk diffusive media: An application to the optical characterization of human breast. Appl Phys Lett. 1999;74:874–876. [Google Scholar]
- 9.Byrnes KR, et al. Light promotes regeneration and functional recovery and alters the immune response after spinal cord injury. Lasers Surg Med. 2005;36(3):171–185. doi: 10.1002/lsm.20143. [DOI] [PubMed] [Google Scholar]
- 10.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev. 2013;42(8):3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hockberger PE, et al. Activation of flavin-containing oxidases underlies light-induced production of H2O2 in mammalian cells. Proc Natl Acad Sci USA. 1999;96(11):6255–6260. doi: 10.1073/pnas.96.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auldridge ME, Forest KT. Bacterial phytochromes: More than meets the light. Crit Rev Biochem Mol Biol. 2011;46(1):67–88. doi: 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- 14.Ulijasz AT, Vierstra RD. Phytochrome structure and photochemistry: Recent advances toward a complete molecular picture. Curr Opin Plant Biol. 2011;14(5):498–506. doi: 10.1016/j.pbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 15.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461(7266):997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller K, et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41(7):e77. doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Möglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem Photobiol Sci. 2010;9(10):1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- 18.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7(10):724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 20.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc Natl Acad Sci USA. 2008;105(38):14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Kuk J, Moffat K. Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc Natl Acad Sci USA. 2009;106(37):15639–15644. doi: 10.1073/pnas.0902178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Ren Z, Kuk J, Moffat K. Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature. 2011;479(7373):428–432. doi: 10.1038/nature10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomelsky M, Ryu MH. Near-infrared light-activated proteins. US Patent 20,130,030,041; WO 2013016693 (pending). Publication date Jan 31, 2013.
- 25.Tarutina M, Ryjenkov DA, Gomelsky M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J Biol Chem. 2006;281(46):34751–34758. doi: 10.1074/jbc.M604819200. [DOI] [PubMed] [Google Scholar]
- 26.Ryu MH, Gomelsky M. 2014. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol, 10.1021/sb400182x.
- 27.Kanacher T, Schultz A, Linder JU, Schultz JE. A GAF-domain-regulated adenylyl cyclase from Anabaena is a self-activating cAMP switch. EMBO J. 2002;21(14):3672–3680. doi: 10.1093/emboj/cdf375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha SC, Sprang SR. Structures, mechanism, regulation and evolution of class III nucleotidyl cyclases. Rev Physiol Biochem Pharmacol. 2006;157:105–140. doi: 10.1007/112_0603. [DOI] [PubMed] [Google Scholar]
- 29.Linder JU. Class III adenylyl cyclases: Molecular mechanisms of catalysis and regulation. Cell Mol Life Sci. 2006;63(15):1736–1751. doi: 10.1007/s00018-006-6072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schröder-Lang S, et al. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4(1):39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- 31.Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285(53):41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stierl M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286(2):1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissenberger S, et al. PACα—an optogenetic tool for in vivo manipulation of cellular cAMP levels, neurotransmitter release, and behavior in Caenorhabditis elegans. J Neurochem. 2011;116(4):616–625. doi: 10.1111/j.1471-4159.2010.07148.x. [DOI] [PubMed] [Google Scholar]
- 34.Efetova M, et al. Separate roles of PKA and EPAC in renal function unraveled by the optogenetic control of cAMP levels in vivo. J Cell Sci. 2013;126(Pt 3):778–788. doi: 10.1242/jcs.114140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293(2):199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 36.Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98(19):10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ádám É, et al. Altered dark- and photoconversion of phytochrome B mediate extreme light sensitivity and loss of photoreversibility of the phyB-401 mutant. PLoS ONE. 2011;6(11):e27250. doi: 10.1371/journal.pone.0027250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Stankey RJ, Vierstra RD. Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 2013;161(3):1445–1457. doi: 10.1104/pp.112.208892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edwards SL, et al. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 2008;6(8):e198. doi: 10.1371/journal.pbio.0060198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schade MA, Reynolds NK, Dollins CM, Miller KG. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Genetics. 2005;169(2):631–649. doi: 10.1534/genetics.104.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takala H, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509(7499):245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. 2nd Ed. NY: Worth Publishers; 2000. [Google Scholar]
- 43.Anantharaman V, Balaji S, Aravind L. The signaling helix: A common functional theme in diverse signaling proteins. Biol Direct. 2006;1:25. doi: 10.1186/1745-6150-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126(5):929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 45.Möglich A, Ayers RA, Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385(5):1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanchan K, et al. Transmembrane signaling in chimeras of the Escherichia coli aspartate and serine chemotaxis receptors and bacterial class III adenylyl cyclases. J Biol Chem. 2010;285(3):2090–2099. doi: 10.1074/jbc.M109.051698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diensthuber RP, Bommer M, Gleichmann T, Möglich A. Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure. 2013;21(7):1127–1136. doi: 10.1016/j.str.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 48.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105(31):10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler K, Schultz A, Schultz JE. The S-helix determines the signal in a Tsr receptor/adenylyl cyclase reporter. J Biol Chem. 2012;287(19):15479–15488. doi: 10.1074/jbc.M112.348409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J Bacteriol. 2005;187(5):1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis MW, et al. Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function. J Neurosci. 1995;15(12):8408–8418. doi: 10.1523/JNEUROSCI.15-12-08408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5(12):3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.