Adaptive nucleic acid-based immune systems of bacteria and archaea provided new tools for genome editing applications and transcription silencing. Despite the rapid success in developing targetable nucleases based on the clustered regularly interspersed short palindromic repeats (CRISPR)-associated (Cas) enzymes, our understanding of how these ribonucleoprotein complexes recognize and process their DNA targets remains rudimentary. A report in PNAS (1) delves into the mechanism of R-loop formation by two very different CRISPR/Cas systems, Cas9 and Cascade (CRISPR-associated complex for antiviral defense).
Our ability to tailor the genomes of various organisms has been revolutionized by the development of targetable nucleases with flexible specificity (2). These powerful reagents generate double-strand breaks (DSBs) in the chromosomal DNA with exquisite specificity. Once generated, the DSB is repaired through nonhomologous end-joining or through homologous recombination. The former often leads to gene inactivation, whereas the latter incorporates the desired genetic alterations by using the information from an exogenously supplied template to introduce, correct, or disrupt specific genes.
Three types of targetable nucleases are currently in use: zinc-finger nucleases, transcription activator-like effector nucleases (TALENs), and the RNA-guided nucleases. Zinc-finger nucleases consist of multiple Zn2+-finger DNA-binding modules selected or engineered to confer specificity for a particular triplet of consecutive base pairs. DNA recognition modules of TALENs are derived from transcription factors found in plant-pathogenic bacteria. Each DNA-binding module of a TALEN specifically interacts with a single base pair, yielding a straightforward recognition code. Two sets of zinc-finger arrays or transcription activator-like effector arrays are engineered to bind the two adjacent DNA sites in a head-to-head orientation, and are fused to the nuclease domains of FokI restriction enzyme. Dimerization activates FokI, allowing it to generate the desired DSB. Requirement for FokI dimerization is an important contributor to cutting specificity, as DSB can only occur when both DNA sequence recognition arrays are bound at a specified distance from one another.
RNA-guided nucleases are based on the CRISPR/Cas enzymes. CRISPR/Cas-based targetable nucleases are the newest addition to the genome engineering toolbox (3). These reagents represent an ingenious adaptation of the sophisticated and diverse nucleic acid-based adaptive immune system of bacteria and archaea. In the CRISPR/Cas system, the ribonucleoprotein complex delivers its nuclease to the desired genome locus by recognizing its target sequence via Watson–Crick base-pairing between the RNA component of the complex and the DNA locus. There are three types of CRISPR/Cas immune systems classified as such based on the distinct operon organizations (4). Among these, type II CRISPR/Cas systems gained the highest popularity in genome engineering applications because of their robustness and relative simplicity: the type II ribonucleoprotein complex is made by a single-subunit 180-kDa Cas9 protein and a dual-guide RNA consisting of crRNA and transactivating crRNA (trcrRNA) that can be substituted with a chimeric single-guide RNA. The Cas9–RNA complex executes both R-loop formation—whereby it forms a hybrid duplex with one of the strands in the protospacer—and DNA cleavage. Efficient target DNA recognition requires a 20-nt-long guide sequence complementary to the target DNA (referred to as spacer and protospacer, respectively) followed by the protospacer-adjacent motif (PAM), which, in the host bacteria, distinguishes foreign and own DNA.
Despite current excitement about harnessing biochemical properties of Cas9 for genome editing applications and transcription control, Cas9-based RNA-guided nucleases are far from being a universal solution for all genome editing needs. In fact, a robust, highly specific, universally applicable nuclease has not been developed thus far. The range of cleavage efficiencies of each of the three major nuclease classes varies greatly among the targets, with no obvious correlation with affinity or even with a sequence context. There is also an issue of an off-target cleavage, which is common for Cas9-based reagents that cut some secondary targets containing mismatches with guide RNA at similar efficiencies as they cut the intended target. Such promiscuity has an obvious biological explanation: a 20-bp complementary region within the R-loop is excessive if the goal is to protect bacteria with a 107-bp genome from the infection by the virus whose genome is typically less than 106 bp. By accommodating mismatches, the host’s immune system may respond to the agility of the ever-mutating phage genomes, which is good for bacteria but may cause problems for the genome-editing applications for which they were not evolved.
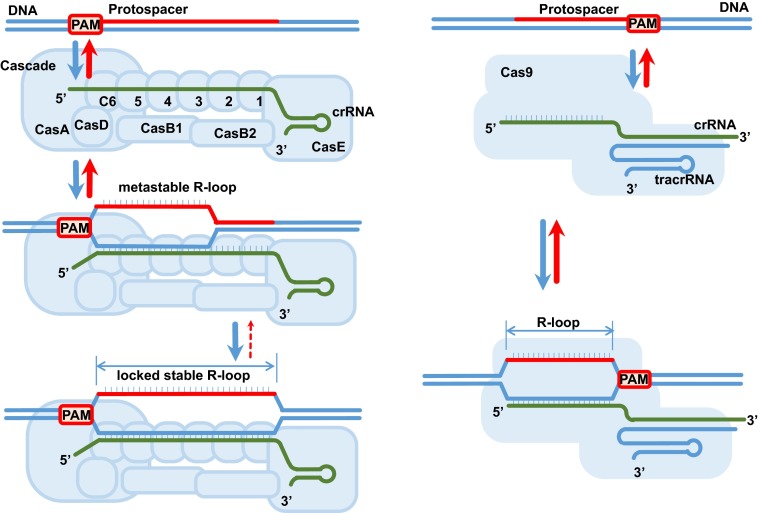
Reengineering the Cas9 protein to be less tolerant of the RNA/DNA mismatches would reduce off-target effects and improve targeting specificity. To achieve this end, one needs to have a detailed understanding of the mechanism underlying Cas9 specificity or promiscuity, as well as the mechanism of R-loop formation. Recent structural studies defined the detailed molecular architecture of the Cas9 ribonucleoprotein (5, 6). In PNAS, Szczelkun et al. describe a single-molecule study that revealed a detailed mechanism of the R-loop formation by Cas9 and compared it to a more complex type IE Cascade system, both from Streptococcus thermophiles (1). Cascade comprises 11 protein subunits (CasA1B2C6D1E1): crRNA is capped at the 5′ and 3′ termini by CasA and CasE subunits, respectively; six CasC subunits form a right-handed helical filament on extended crRNA; a CasB dimer is positioned along the CasC-RNA filament connecting CasA and CasE; and CasD functions as a hinge that connects CasC6 to CasA (7). Cascade binding to the complementary protospacer generates a 33-nt R-loop, which is longer and more stable than the 20-nt R-loop generated by Cas9. Target DNA cleavage is carried out by the Cas3 helicase/nuclease. Similar to Cas9, Cascade uses PAM to distinguish own and invading DNA. In contrast to Cas9, which requires 4-nt PAM (GGNG), Cascade’s PAM is promiscuous and short, with AA supporting optimal cleavage.
Szczelkun et al. (1) used magnetic tweezers, a single-molecule technique that allows stretching and twisting individual dsDNA molecules anchored between the surface of a fluidic cell and a magnetic bead. Bead rotation untwists or overtwists the dsDNAmolecule, resulting in negative or positive supercoils, respectively. Formation of the
Szczelkun et al. describe a single-molecule study that revealed a detailed mechanism of the R-loop formation by Cas9 and compared it to a more complex type IE Cascade system.
R-loop locally separates the DNA duplex and changes the torque profile, making magnetic tweezers an ideal approach to monitor enzyme-mediated R-loop formation and measure its stability. When the DNA molecules contained appropriate protospacers and PAM sequences, the authors could directly monitor and quantify torque-dependence of the R-loop formation and dissolution in the two systems. Several important observations were made. Most notably, this study has unambiguously established the directionality of the R-loop formation process in both Cas9 and Cascade systems, whereby Cas9 and Cascade initiate the R-loop formation at the PAM, which exerts the kinetic control over R-loop formation but has no effect on the R-loop stability. Thus, both Cas9 and Cascade verify the presence of PAM and the target by a kinetic proofreading mechanism. Although a unified model for the formation and dissociation of the R-loop in the two structurally different systems is derived, Cas9 and Cascade were also found to differ from one another in several important aspects. The rate-limiting steps in the R-loop formation by Cascade appears to be the initial loop formation followed by its conversion into a stable R-loop. In contrast, association of Cas9 with DNA was protein concentration-dependent, suggesting that it is inferior to Cascade with respect to association: productive Cas9-mediated R-loop formation requires higher protein concentrations in vitro, which likely translates into slower, less-efficient targeting in the cell.
Another peculiar feature of the Cascade mechanism is the presence of the “lock” revealed by analysis of the truncated protospacers. Initial R-loop expansion results in a metastable R-loop that readily dissociates when negative supercoiling is removed. If given sufficient time, the R-loop becomes “locked” in a stable configuration consistent with the stable complex poised for Cas3 recruitment and subsequent DNA degradation (Fig. 1). A high-resolution structure of Cascade that would provide an atomistic depiction of the locking mechanism remains to be solved, but the electron microscopy reconstructions hint at a significant conformational change within the CasC nucleoprotein filament upon target binding (7), not dissimilar to that observed in the filaments of RecA recombinase and DnaA-replication origin melting protein (8). This result is not surprising considering that all systems that identify homology within the DNA duplex face common physical challenges. In contrast to RecA, which requires ATP and can initiate pairing and strand exchange at either end of the homologous locus (9), Cascade is cofactor independent and directional.
Fig. 1.
R-loop formation by Cascade and Cas9. Overall architectures of Cascade (Left) and Ca9 (Right) ribonucleoproteins and their respective R-loops. Although both complexes initiate R-loop formation by recognizing their respective PAM sequences flanking the protospacer, the Cascade reaction proceeds through a metastable intermediate, which is then locked to form a stable R-loop.
The detailed mechanisms of the Cas9- and Cascade-mediated R-loop formation revealed in this study will feed into the ongoing structural analyses of the two systems. Engineering a locking mechanism into Cas9 may improve both its fidelity and affinity for the target, making it a more efficient cutting agent. Combining structural data with the precise molecular mechanism will also be instrumental in improving the Cas9 fidelity and reducing the off-target cutting, fulfilling the dream of a perfect reagent for genome editing.
Supplementary Material
Footnotes
The author declares no conflict of interest.
See companion article on page 9798 of issue 27 in volume 111.
References
- 1.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111(27):9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 3.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duderstadt KE, Berger JM. A structural framework for replication origin opening by AAA+ initiation factors. Curr Opin Struct Biol. 2013;23(1):144–153. doi: 10.1016/j.sbi.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ragunathan K, Joo C, Ha T. Real-time observation of strand exchange reaction with high spatiotemporal resolution. Structure. 2011;19(8):1064–1073. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]