Significance
DNA cut-and-paste transposons are discrete DNA segments that move from place to place within genomes via excision from a donor site by double-strand DNA breaks and insertion into a target site. These events are mediated by nucleoprotein complexes whose assembly regulates and coordinates breakage and joining. Multiple protein–protein and protein–DNA interactions are involved in assembly of these nucleoprotein complexes. The nucleoprotein complexes that mediate the movement of the bacterial transposon Tn7 are particularly elaborate, requiring four Tn7-encoded proteins. Here we define specific protein–protein interactions between the central regulator of Tn7 transposition, TnsC, and both the transposase that carries out the chemical steps of transposition and the target-selecting protein.
Keywords: photocrosslinking, protein–protein interaction, transpososome, transposable element
Abstract
The excision of transposon Tn7 from a donor site and its insertion into its preferred target site, attachment site attTn7, is mediated by four Tn7-encoded transposition proteins: TnsA, TnsB, TnsC, and TnsD. Transposition requires the assembly of a nucleoprotein complex containing all four Tns proteins and the DNA substrates, the donor site containing Tn7, and the preferred target site attTn7. TnsA and TnsB together form the heteromeric Tn7 transposase, and TnsD is a target-selecting protein that binds specifically to attTn7. TnsC is the key regulator of transposition, interacting with both the TnsAB transposase and TnsD-attTn7. We show here that TnsC interacts directly with TnsB, and identify the specific region of TnsC involved in the TnsB–TnsC interaction during transposition. We also show that a TnsC mutant defective in interaction with TnsB is defective for Tn7 transposition both in vitro and in vivo. Tn7 displays cis-acting target immunity, which blocks Tn7 insertion into a target DNA that already contains Tn7. We provide evidence that the direct TnsB–TnsC interaction that we have identified also mediates cis-acting Tn7 target immunity. We also show that TnsC interacts directly with the target selector protein TnsD.
In DNA cut-and-paste transposition, the transposon is excised from the donor site and then integrated into a new insertion site. These reactions are mediated by nucleoprotein complexes called transpososomes (1, 2). Understanding the mechanism and regulation of transposition requires identification of the protein–DNA and protein–protein interactions that underlie transpososome assembly and activity.
The transpososomes that promote transposition of the bacterial transposon Tn7 are particularly elaborate. Tn7 encodes five transposition proteins—TnsA, TnsB, TnsC, TnsD, and TnsE—that mediate transposition to two classes of target sites (3–5). TnsABC+D promote target site-specific insertion of Tn7 into its preferred chromosomal target site attTn7 (3), whereas TnsABC+E promote Tn7 insertion into non-attTn7 sites on conjugal plasmids (6, 7). Thus, TnsABC form the core of the transposition machinery. This viewpoint is reinforced by the finding that although WT TnsABC alone do not promote transposition, transposition with TnsABC can occur when TnsC is activated by gain-of-function mutations that allow TnsC target binding and transposase activation in the absence of TnsD or TnsE (8, 9). In contrast, other characterized transposition systems involve only one or two transposition proteins (10).
Tn7 transposition also requires ATP (11), given that TnsC is an ATP-dependent DNA-binding protein (12) and an ATPase (9). It should be noted that ATP is not required for the chemical steps of transposition (13); rather, it regulates assembly of the Tn7 transpososomes (14). TnsC contains AAA+ ATPase motifs (15). Bacteriophage Mu, whose breakage and joining reactions are performed by the transposase MuA, also uses the ATP-dependent target-binding protein MuB to regulate transposition (16, 17).
Our work is focused on the TnsABC+D system. All of these Tns proteins, as well as the DNA substrates for transposition (i.e., the Tn7 ends and the attTn7 target DNA), must assemble into the elaborate transpososome in which transposition actually occurs (11, 18). The targeting protein TnsD binds to a specific sequence in attTn7 (11, 19). Subsequent TnsC recruitment involves both TnsC–DNA interactions that depend on TnsD-induced distortions in attTn7 (20, 21) and likely TnsC–TnsD interactions that we previously detected using yeast two-hybrid assays (19). Although we have shown that a TnsCD-attTn7 complex can form (20), TnsC also may be recruited to attTn7 as part of a TnsA-TnsC complex, because TnsA and TnsC can copurify as a TnsA2C2 heterotetramer (22, 23). The stoichiometery of the Tns proteins in the target complex remains to be directly established, but we have observed an TnsACD-attTn7 complex (23). Analysis of the stoichiometery of the Tns proteins in a posttransposition complex has revealed the likely presence of multiple TnsA2C2s in this complex (23).
TnsA and TnsB together form the Tn7 transposase (13). TnsB is a sequence-specific DNA-binding protein that binds to multiple sites at both transposon ends (24, 25). TnsB also mediates DNA breakage and joining at the 3′ ends of the transposon and is a member of the RNase H transposase-retroviral integrase superfamily (26). TnsA is a nuclease that mediates cleavage at the 5′ ends of the transposon (27, 28). Although TnsA lacks specific DNA-binding activity, it is positioned at the transposon ends by interaction with TnsB (29). Thus, TnsA and TnsB collaborate to excise Tn7 from the donor site and insert it into a target site. Notably, TnsA and TnsB are interdependent, with breakage and joining occurring only in the presence of both proteins, even when their catalytic activity has been abolished by mutations in their active sites (26, 27).
TnsA and TnsB do not promote breakage and joining in the absence of TnsC, however. How does TnsC activate the TnsAB transposase? As noted above, TnsA and TnsC can interact directly, forming a TnsA2C2 heterotetramer. The structure of a cocrystal of TnsA/TnsC504–555 has been solved (22), locating the TnsC region that interacts with TnsA to the very C-terminal region of TnsC. The resulting model for TnsA–TnsC504–555 interaction also led to the proposal that the TnsC495–501 region, which is lysine-rich, may play a significant role in interacting with the donor DNA near the transposon end. Such an interaction could be the basis for the stabilization of transpososomes by the presence of TnsA and in its role in stabilizing the TnsACD-target complex (18, 23, 30).
We have previously suggested that the transposition regulator TnsC and the transposase subunit TnsB interact (31), but a direct TnsB–TnsC interaction has not yet been demonstrated. Support for the view that TnsB and TnsC interact is that TnsB bound to a Tn7 end-containing DNA and a TnsD- and TnsE-independent gain-of-function TnsC mutant, TnsCA225V, bound to a target DNA can form a donor-target complex in the presence of the crosslinker glutaraldehyde (18).
Other observations suggesting a TnsB–TnsC interaction come from the study of Tn7 cis-acting target immunity, i.e., the process that inhibits the insertion of Tn7 into a target DNA that already contains Tn7 ends (31, 32). A target DNA already containing Tn7 ends is immune to Tn7 insertion because TnsB can bind to the Tn7 ends, leading to an increase in the local concentration of TnsB. This increase in TnsB concentration on the target DNA results in ATP hydrolysis by TnsC that attempts to bind to the target DNA, thereby clearing TnsC from that potential target DNA (30, 31). Thus, the key step in Tn7 target immunity is TnsB-induced inhibition of the binding of TnsC to the Tn7-containing target DNA, blocking the formation of a stable TnsC-target complex (30, 31). We have visualized this TnsB-induced dissociation of TnsC from attTn7 by analysis of Tns-attTn7 complexes by EMSAs (30). Such target immunity established by transposase-induced ATP-dependent dissociation of the target-binding protein from a target DNA containing the Mu ends occurs in the Mu system as well (33).
Thus, several lines of evidence support the view that TnsB interacts with TnsC; however, direct demonstration of this point has not yet been reported.
What regions of TnsB and TnsC might interact? We previously suggested that the region of TnsB that interacts with TnsC lies at the C terminus of TnsB. We isolated mutants of TnsB that have reduced effectiveness of transposition immunity (30). These “immunity bypass” mutants are located in the C-terminal region of TnsB at TnsBP686S, TnsBV689M, and TnsBP690L, TnsB being 702-aa long. We demonstrated that these TnsB mutants have reduced ability to promote dissociation of the TnsCD-attTn7 target complex in vitro, consistent with the view that the region of TnsB in which the immunity bypass mutations are located interacts with TnsC (30). Moreover, whereas short C-terminal TnsB WT peptides can promote TnsCD-attTn7 complex dissociation, such C-terminal TnsB peptides from immunity bypass mutants do not promote target complex dissociation (30). Here we identify a region of TnsC that is critical to TnsB–TnsC interaction using a photocrosslinking assay and analysis of the effects of TnsC mutants on TnsB-dependent transposition activities.
TnsC also plays a key role in target site selection, being recruited to the TnsD-attTn7 complex to form a TnsCD-attTn7 complex (11, 20). TnsD binds specifically to attTn7, thereby identifying this preferred target site (11, 19). A key step in TnsC recruitment is the interaction of TnsC with TnsD-induced distortions in attTn7 (20, 21). We previously detected TnsC–TnsD interactions using a yeast two-hybrid assay (19). In the present work, we used affinity chromatography to show that TnsC also interacts directly with the attTn7-binding protein TnsD. Thus, TnsC is central to transposition and participates in multiple protein–protein and protein–DNA interactions.
Results
TnsC Interacts Directly with TnsB.
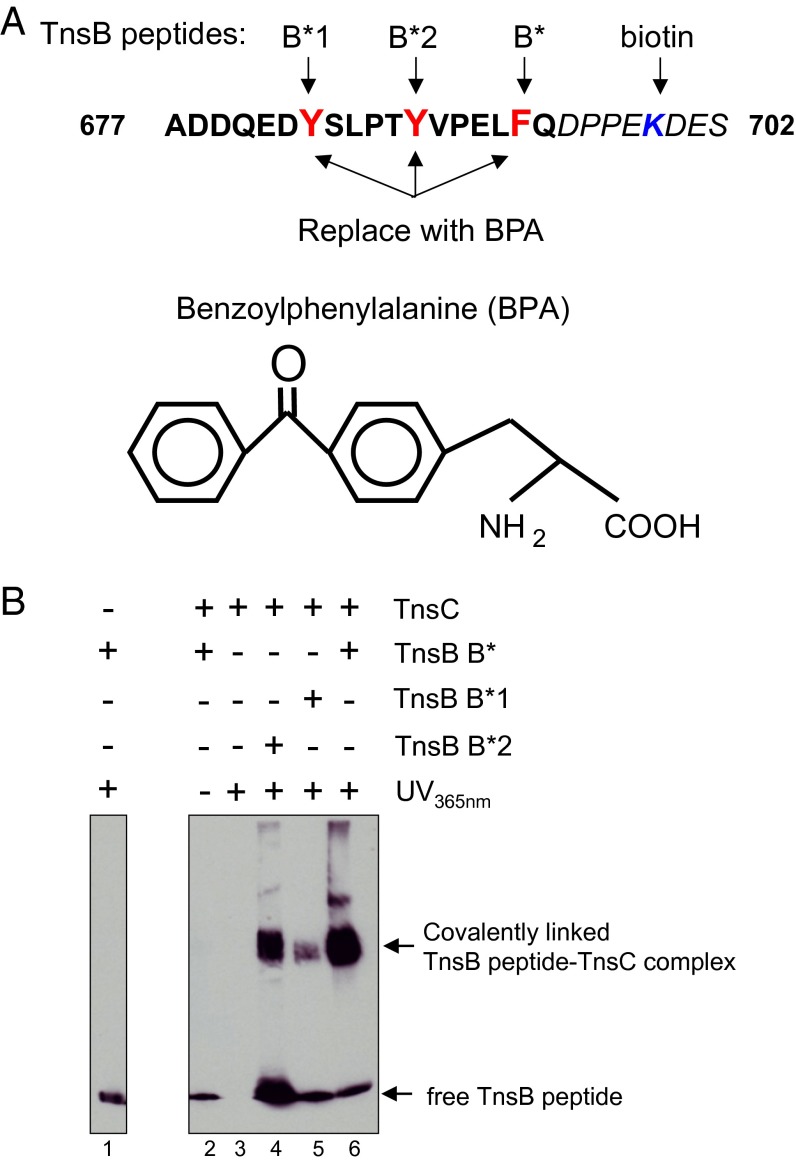
Previous work has suggested that TnsB can interact with TnsC (31), but this interaction had not been directly demonstrated until now. To show that TnsB and TnsC interact directly, we developed a photocrosslinking assay. We exploited the fact that TnsB can promote the TnsC-mediated dissociation of the target complex TnsCD-attTn7, a reaction that we have suggested depends on the interaction of TnsB with TnsC (30), and that this TnsB-dependent dissociation of a TnsC-containing target complex underlies Tn7 target immunity (31). We also previously demonstrated that a TnsB peptide, the carboxyl-terminal peptide TnsB677–694, is sufficient to promote dissociation of a TnsC target complex (30). Thus, for the TnsB partner in the crosslinking reaction, we used the C-terminal peptide TnsB677–702 containing the photoactivatable crosslinker benzoylphenylalanine (BPA) (34, 35) (Fig. 1A).
Fig. 1.
UV crosslinking of TnsC with BPA-substituted TnsB peptides. (A) BPA-substituted TnsB peptides. The structure of BPA is shown. Various TnsB peptides were synthesized in which K699 (blue) was derivatized with biotin and BPA replaced the indicated aromatic acids (red). (B) BPA-containing TnsB peptides were incubated with TnsC and exposed to UV light as indicated. The reactions were then displayed on an SDS/PAGE gel, transferred to a membrane, and evaluated by Western blot analysis with antibiotin antibody that visualizes the peptide. All lanes shown are from a single gel. Lane 1, TnsB peptide B* incubated without TnsC with exposure to UV; lane 2, peptide B* incubated with TnsC without exposure to UV; lane 3, TnsC incubated with exposure to UV; lane 4, peptide B*2 incubated with TnsC with exposure to UV; lane 5, peptide B*1 incubated with TnsC with exposure to UV; lane 6, peptide B* incubated with TnsC with exposure to UV.
When activated by exposure to UV light, BPA can form a covalent linkage with protein residues within a ∼3-Å radius. This short reactive radius and the ability to switch BPA between relaxed and excited states by UV makes BPA crosslinking an excellent tool with which to identify direct protein–protein interactions. Here, three modified TnsB677–702 peptides were synthesized in which K699 was modified with biotin for detection of the peptide by Western blot analysis and BPA was substituted at Y683, Y688, and F693, generating peptides TnsB*1, TnsB*2, and TnsB*, respectively (Fig. 1A).
To look for TnsB peptide–TnsC interactions, we incubated TnsC (63 kDa) with the modified TnsB peptides in the presence of UV light and BSA, then displayed the samples by SDS/PAGE and analyzed the reaction products by Western blot analysis with a biotin antibody that detects the modified peptides. We observed no new species in the absence of TnsC (Fig. 1B, lane 1), the absence of UV (lane 2), or the absence of peptide (lane 3). Notably, however, we observed a new, much more slowly migrating species quite distinct from the peptide alone with each modified TnsB peptide after UV treatment (Fig. 1B, lanes 4–6), in which we believe that the TnsB peptide is crosslinked to TnsC. We suspect that the different amounts of the crosslinked TnsB peptide-TnsC species with the different TnsB peptides likely reflect different efficiencies of crosslinking when BPA is present at different positions in the peptide.
The C Terminus of TnsC Interacts with TnsB.
To localize the region of TnsC where interaction with the TnsB peptide occurs, we constructed several maltose-binding protein (MBP) fusions containing parts of TnsC, MBP-TnsC1–293, MBP-TnsC294–555, and MBP-TnsC361–555 and used them as substrates in TnsB* peptide crosslinking reactions (Fig. 2A). We found that MBP-TnsC294–555 (lane 9) and MBP-TnsC361–555 (lane 10) formed crosslinked complexes with peptide TnsB*, but MBP-TnsC1–293 did not (lane 8), localizing the TnsB–TnsC interaction to the C-terminal region of TnsC, TnsC361–555.
Fig. 2.
Crosslinking of a TnsB BPA-containing peptide to various MBP-TnsC fusions. BPA-containing TnsB peptide B* was incubated with MBP-TnsC fusions containing various segments of TnsC and exposed to UV light as indicated. The reactions were then displayed on an SDS/PAGE gel, transferred to a membrane, and analyzed by Western blot analysis with antibiotin antibody that visualizes the peptide. (A) TnsB peptide B* crosslinks MBP-TnsC361–555. Lane 1, peptide B* incubated without TnsC with exposure to UV light; lanes 2, 3, and 4, peptide B* incubated with MBP-TnsC derivatives TnsC1–293, TnsC294–555, and TnsC361–555, as indicated in the absence of UV; lanes 5, 6, and 7, MBP-TnsC derivatives TnsC1–293, TnsC294–555, and TnsC361–555, incubated as indicated in the presence of UV; lanes 8, 9, and 10, peptide B* incubated with MBP-TnsC derivatives TnsC1–293, TnsC294–555, and TnsC361–555 as indicated in the presence of UV. (B) TnsB peptide B* crosslinks MBP-TnsC451–555. Lanes 1, 2, and 3, peptide B* incubated with MBP-TnsC361–555, TnsC401–555, and TnsC451–555, as indicated in the absence of UV; lanes 4, 5, and 6, peptide B* incubated with MBP-TnsC361–555, TnsC401–555, and TnsC451–555 as indicated in the presence of UV. (C) The TnsC mutant MBP-TnsC361–555L475A/L476A crosslinks poorly with the TnsB peptide B*. Lanes 1, 2, and 3, peptide B* incubated with MBP-TnsC361–555, TnsC361–555P468A/M470A, and TnsC361–555L475A/L476A, as indicated in the absence of UV; lanes 4, 5, and 6, peptide B* incubated with MBP-TnsC361–555, TnsC361–555P468A/M470A, and TnsC361–555L475A/L476A, as indicated in the presence of UV.
Crosslinking analysis of MBP-TnsC fusions containing shorter C-terminal segments of TnsC revealed that peptide TnsB* also crosslinked to MBP-TnsC401–555 (Fig. 2B, lane 5) and MBP-TnsC451–555 (lane 6), localizing the region of TnsB interaction with TnsC to the C terminus of TnsC, TnsC451–555. We believe that the multiple crosslinked species observed in some experiments reflect degradation of some of the fusion proteins (Fig. 2B, lanes 5 and 6).
Proteolytic digestion of TnsC revealed that a protease-resistant fragment with TnsC411 as its amino terminus, as determined by N-terminal sequencing (Fig. S1A), which likely extends to near the C terminus at TnsC555. We found that TnsB and MBP-TnsC411–555 copurify when an intein fusion of TnsB is isolated by affinity chromatography on a chitin bead column (Fig. S1B). These findings support our conclusion that TnsB interacts with the C-terminal region of TnsC.
Mutation of Conserved Amino Acids in TnsC Blocks TnsB Peptide Crosslinking.
What amino acids within TnsC451–555 are critical for interaction with TnsB? Ronning et al. (22) described a cocrystal between the transposase subunit TnsA and the C-terminal fragment TnsC504–555, revealing that TnsC504–555 interacts with TnsA. Consideration of this structure led to the suggestion that a slightly longer TnsC fragment, TnsC495–555, would interact with TnsA and also with a short region of DNA adjacent to the TnsA cleavage site at the 5′ end of Tn7. This suggestion was supported by DNA-binding assays in which mutagenesis of the basic resides within TnsC494–504 abolished DNA binding by TnsA-TnsC494–555 (22).
Consequently, we hypothesized that the positions of peptide TnsB* crosslinking would lie within TnsC451–494. Alignment of the amino acid sequences of this region of various TnsCs revealed several highly conserved amino acid positions in the TnsC451–494 region (Fig. S2), including TnsC L475 and L476, which are conserved in all 24 sequences aligned. Whereas considerable crosslinking of peptide TnsB* to MBP-TnsC361–555 was observed (Fig. 2C, lane 4), little crosslinking was observed with TnsC361–555L475A/L476A (lane 6). In contrast, crosslinking of the double-mutant MBP-TnsC361–555P468A/L470A was similar to that of MBP-TnsC361–555 (lane 5). These findings suggest that the TnsC amino acids L475 and L476 play important roles in the interaction of the peptide TnsB* with TnsC.
Transposition in Vitro Is Greatly Reduced by Mutation of the TnsC TnsB-Binding Region.
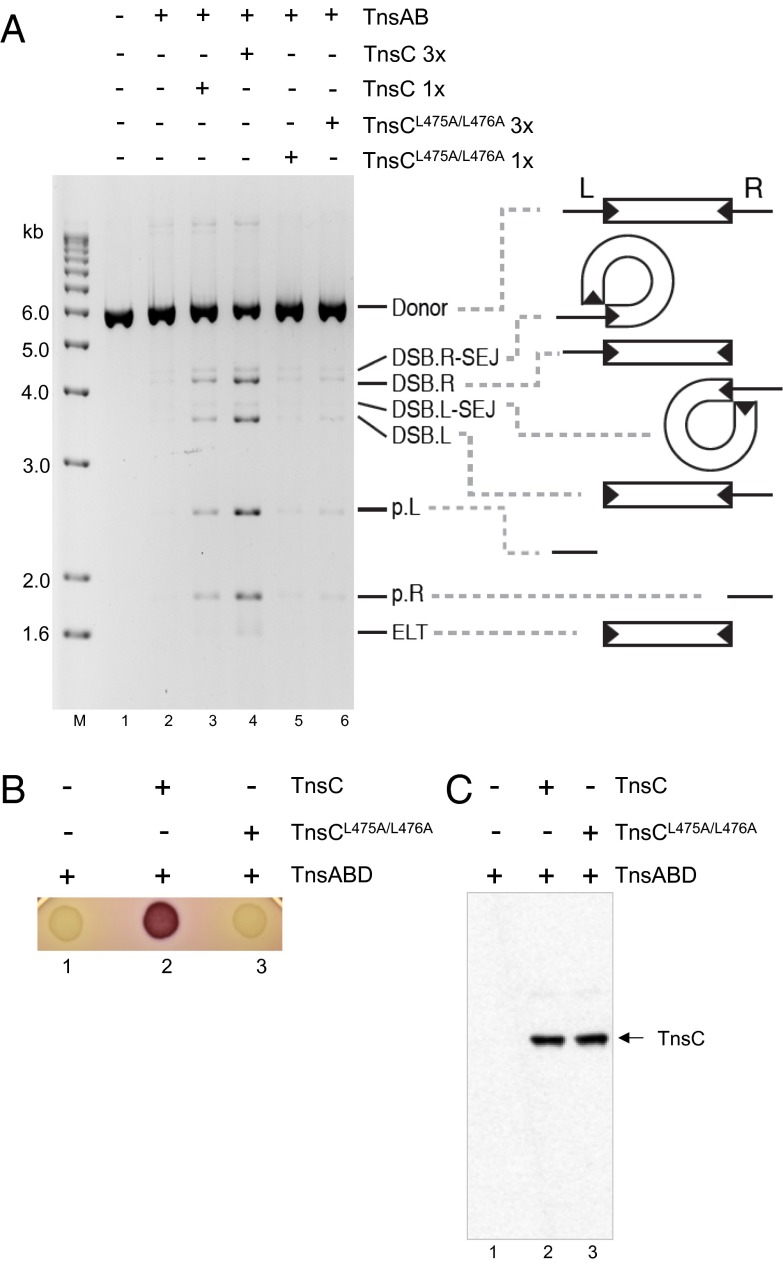
To examine the effect of the TnsCL475A/L476A mutation on transposition in vitro, we tested its activity in a TnsABC transposition system in which breakage and intramolecular joining by the TnsAB transposase is dependent on the presence of TnsC (Fig. 3A) (22). This TnsABC reaction was carried out in the presence of 20% (vol/vol) glycerol instead of the low glycerol conditions in our standard intermolecular TnsABC+D reactions (36). After incubation, the reactions were digested with a restriction enzyme that linearized the substrate DNA, then displayed on an agarose gel and stained with ethidium bromide. Recombination results in double-strand breaks (DSBs) at each end of the transposon, generating the transposon-containing species DSB.right (DSB.R), DSB.left (DSB.L), and excised linear transposon (ELT), along with the plasmid-only fragments p.L and p.R. Intramolecular joining of the exposed 3′OH end of a DSB product to its 5′ end results in circularization of the transposon to form a DSB.R-single end join (DSB.R-SEJ) and a DSB.L-single end join (DSB.L-SEJ).
Fig. 3.
The TnsC mutant TnsCL475A/L476A is not active in transposition in vitro or in vivo. (A) TnsCL475A/L476A does not stimulate TnsAB-dependent intramolecular breakage and joining in vitro. In intramolecular breakage and joining reactions, DSBs occur at either or both Tn7 ends to form DSB.L, DSB.R, and ELT species along with plasmid backbone fragments p.L and p.R. Intramolecular joining of the newly exposed Tn7 end 3′OH to its 5′ end results in an DSB.L.SEJ and a DSB.R-SEJ DSBs. A Tn7-containing plasmid was incubated with TnsAB and TnsC derivatives as indicated in the presence of high glycerol concentrations (20% vol/vol), digested with a restriction enzyme that cleaves once in the donor backbone, and displayed on an agarose gel. Lane 1, DNA without protein additions; lane 2, Tn7 plasmid incubated with TnsAB in the absence of TnsC; lanes 3 and 4, Tn7 plasmid incubated with TnsAB and 1× and 3× WT TnsC as indicated; lanes 5 and 6, Tn7 plasmid incubated with TnsAB and 1× and 3× mutant TnsCL475A/L476A as indicated. (B) The TnsC mutant TnsCL475A/L476A is not active in transposition in vivo. Tn7 transposition in vivo was assayed by identification of Lac+ (red) cells using MacConkey agar in a strain containing a miniTn7::lac element that lacks an internal promoter and is located on an F plasmid such that lac is not expressed from an external promoter. Transposition proteins TnsA, TnsB, and TnsD were supplied from a pACYC plasmid, and WT and TnsC TnsCL475A/L476A were supplied from a pUC-based plasmid. TnsABC+D transposition results in translocation of the miniTn7::lac to attTn7, where rare opposite-orientation insertions position the lac genes downstream of the glmS to yield Lac+ (red) cells. Lane 1, TnsABD + vector; lane 2, TnsABD + TnsC; lane 3, TnsABD + TnsCL475A/L476A. (C) TnsC levels in the indicated cultures were measured by Western blot analysis with TnsC antibody.
As expected, no recombination was detectable solely with TnsAB (Fig. 3A, lane 2). In the presence of TnsAB + WT TnsC (lanes 3 and 4), however, DSB.R, DSB.L, and a small amount of ELT products were observed, along with low levels of DSB.R-SEJ and DSB.L-SEJ intramolecular products. Notably, only very low levels of breakage occurred in the presence of TnsAB + mutant TnsCL475A/L476A (lanes 5 and 6), which is defective for TnsB–TnsC interaction. Thus, TnsB–TnsC interaction is critical for transposition in vitro.
Mutation of the TnsC TnsB-Binding Region Blocks Transposition in Vivo.
We also found that TnsCL475A/L476A does not support TnsABC+D transposition in vivo. We assayed transposition in vivo using a promoter-capture assay (8, 37). The Escherichia coli host strain contains pCW305, a plasmid that supplies TnsA, TnsB, and TnsD (3), and a tnsC plasmid that supplies TnsC and a miniTn7::lac donor plasmid, which lacks an internal lac promoter such that the strain is white (lac−) on MacConkey lactose plates when the miniTn7lac is not located downstream of an external promoter. When transposition occurs such that the miniTn7lac element is downstream of an external promoter, red color results. In this case, miniTn7lac insertion into attTn7 and the red color result from opposite-orientation miniTn7::lac insertion into attTn7 (37) (Fig. 3B, lane 2). Although TnsC can promote transposition in vivo in the presence of WT TnsA, TnsB, and TnsD, TnsCL475A/L476A cannot (lane 3). Western blot analysis revealed the presence of equivalent amounts of WT TnsC and TnsCL475A/L476A (Fig. 3C, compare lanes 2 and 3). Thus, TnsB–TnsC interaction mediated by the TnsCL475A/L476A region is also essential for transposition in vivo.
Mutation of the TnsC TnsB-Binding Region Does Not Block Formation of a TnsCD-attTn7 Target Complex.
We previously showed that TnsC and TnsD together form a complex with attTn7. TnsD binds asymmetrically to attTn7, protecting an ∼30-bp region extending from about attTn7+25 to attTn7+55 (the middle of the Tn7 insertion site being 0). Footprint analysis of the TnsCD-attTn7 complex showed that TnsC binds adjacent to TnsD, occupying the region extending from the edge of the TnsD region at about attTn7+25 to about attTn7−10. As shown in Fig. 4, lanes 2 and 6, both TnsD + WT TnsC and the TnsB-binding–defective mutant TnsCL475A/L476A can form a TnsCD-attTn7 complex. Thus, TnsCL475A/L476A is not defective in formation of a target complex.
Fig. 4.
Mutagenesis of TnsC inhibits TnsB-provoked dissociation of the TnsCD-attTn7 complex. TnsCD-attTn7 complexes containing a 32P end-labeled attTn7 fragment, TnsD and WT TnsC, or mutant TnsCL475A/L476A were formed, after which TnsB or the TnsB peptide B* was added to provoke dissociation of the TnsCD-attTn7 complexes. After crosslinking with glutaraldehyde, the reaction mixtures were displayed on an agarose gel. Lane 1, attTn7 DNA; lane 2, TnsCD-attTn7 complexes containing WT TnsC; lanes 3 and 4, TnsCD-attTn7 complexes containing WT TnsC, to which TnsB peptide B* was added at 1× and 5× as indicated; lane 5, TnsCD-attTn7 complexes containing WT TnsC, to which TnsB was added; lane 6, TnsCL475A/L476AD-attTn7 complexes containing TnsCL475A/L476A; lanes 7 and 8, TnsCL475A/L476AD-attTn7 complexes containing TnsCL475A/L476A, to which TnsB peptide B* was added at 1× and 5× as indicated; lane 9, TnsCL475A/L476AD-attTn7 complexes containing TnsCL475A/L476A, to which TnsB was added.
Mutation of the TnsC TnsB-Binding Region Blocks the TnsCD-attTn7 Complex Disassembly Provoked by TnsB.
One feature of Tn7 transposition is that it displays target immunity; that is, Tn7 inserts at reduced frequency into target DNAs that already contain the ends of Tn7, particularly the regions of the Tn7 ends containing the TnsB-binding sites (31, 32). We have suggested that Tn7 target immunity results from the TnsB-induced clearing of potential target DNAs of TnsC (30, 31), likely by direct interaction between TnsB and TnsC. As an example of this target DNA clearing process, we previously showed that incubation of a TnsCD-attTn7 complex with TnsB or the peptide TnsB677–694 results in ATP-dependent dissociation of the complex (30). ATP is a cofactor of TnsC, which is an ATP-dependent target DNA-binding protein and an ATPase (9, 12).
In the present work, we examined the effects of the TnsC mutations L475A and L476A that reduce TnsB-crosslinking to TnsC on the stability of a TnsCL475A/L476AD-attTn7 complex when challenged with TnsB. As shown in Fig. 4, disassembly of the TnsCL475A/L476AD-attTn7 complex did not occur after the addition of TnsB (compare lanes 5 and 9). This result supports the view that the TnsC amino acids L475 and L476 play a critical role in immunity and thus in the TnsB–TnsC interaction.
We also found that the TnsB peptide B* provokes dissociation of the TnsCD-attTn7 complex (compare lanes 2 and 4), but does not provoke disassembly of the TnsCL475A/L476AD-attTn7 complex (lanes 7 and 8). These results support the view that the biotin and BPA modifications of the peptide do not change its interaction with TnsC, as well as the view that TnsCL475A/L476A cannot interact with TnsB.
Mutation of the TnsC TnsB-Binding Region Blocks TnsC Stimulation of TnsB-Mediated Tn7 End Pairing.
We previously showed that TnsB alone can mediate the pairing of the Tn7 ends to form a paired end complex (PEC), which is critical for Tn7 recombination (18, 29). We show here that PEC formation is stimulated by the addition of TnsC to the pairing reactions (Fig. 5, compare lanes 2 and 4). Notably, however, PEC formation is not stimulated by the addition of the TnsC mutant TnsCL475A/L476A, which is defective in TnsC–TnsB interaction (lanes 5 and 6).
Fig. 5.
Mutation of the TnsB-binding region blocks TnsC stimulation of TnsB-mediated formation of the PEC. Pairing of the Tn7 ends to form a PEC was evaluated by incubation of a Tn7 plasmid in the presence of the end-binding protein TnsB and TnsC as indicated, crosslinking with glutaraldehyde, digestion with a restriction enzymes that cuts in the transposon backbone and the plasmid backbone to make separate Tn7L- and Tn7R-containing fragments. The PEC contains both the Tn7L and Tn7R fragments. Lane 1, DNA only; lane 2, PEC formed in the presence of TnsB; lanes 3 and 4, PEC formed in the presence of TnsB and 1× or 3× WT TnsC, as indicated; lanes 5 and 6, PEC formed in the presence of TnsB and mutant TnsCL475A/L476A and 3× mutant TnsCL475A/L476A, as indicated.
TnsC Interacts Directly with the Target Selector TnsD.
In a previous study, using yeast two-hybrid analysis, we identified an interaction between full-length TnsC1–555 and full-length TnsD1–508 (19). Deletion mapping revealed that TnsC1–293 and TnsD1–309 could interact as well. In the present work, we used affinity chromatography to ask whether TnsC1–85 and TnsD1–309 can interact directly, using this shorter segment of TnsC because it is distinct from the region of TnsC that contains AAA+ ATPase motifs (15). We looked for coelution of MBP-TnsC1–85 with TnsD1–309-His6 on purification and elution of TnsD1–309-His6 from a Ni2+ column. We used His6 antibody to identify TnsD1–309-His6 and MBP antibody to identify MBP-TnsC1–85. We found that the MBP-TnsC1–85 fragment did indeed coelute with TnsD1–309-His6 (Fig. 6). This finding provides evidence of a direct interaction between TnsC and TnsD.
Fig. 6.
TnsC1–85 interacts directly with TnsD. Cultures containing TnsD1–309-His6 and MBP-TnsC1–85 were mixed, and extracts were run over a Ni2+ resin column and then eluted by treatment with imidazole buffer. After SDS/PAGE of equivalent samples on a single gel, the elution profiles of TnsD1–309-His6 and MBP-TnsC1–85 were subjected to Western blot analysis with His6 or MBP antibody as indicated. M, size markers, protein marker. Lanes 1 and 8, crude extract; lanes 2 and 9, flow-through fractions; lanes 3, 4, 10, and 11, wash fractions; lanes 5, 6, 7, 12, 13, and 14, fractions eluted with elution buffer.
Discussion
Previous work led to the hypothesis that the transposase subunit TnsB, which binds specifically to the Tn7 ends (24, 25) and mediates breakage and joining at the transposon 3′ ends (26), and the target-binding transposition regulator TnsC interact in several processes critical to Tn7 transposition (14). One finding supporting the view that TnsB and TnsC interact in the transpososome that executes Tn7 transposition is detection of a TnsB-Tn7 end–TnsC target DNA complex in the presence of a glutaraldehyde crosslinker (18). TnsB also induces the ATP-dependent dissociation of TnsC from a target DNA containing the ends of Tn7, resulting in reduced Tn7 insertion into that target, i.e., target immunity (30, 31). We have previously identified the region of the TnsB C terminus essential to these interactions (18, 30), but the region of TnsC that interacts with TnsB had not been directly identified or defined until now.
In this work, we focused on directly identifying and defining the interaction between TnsC and TnsB. We have identified a region in the C terminus of TnsC that is essential for multiple TnsB–TnsC activities. To define the region of TnsC involved in the interaction with TnsB, we first developed a crosslinking assay in which a photoactivatable BPA-substituted TnsB peptide was crosslinked specifically to TnsC. By examining the crosslinking of the TnsB peptide to truncated TnsCs, we localized the TnsB-interacting region to the ∼100 C-terminal amino acids of TnsC, TnsC451–555. We also found that a TnsC C-terminal domain defined by protease sensitivity, TnsC411–555, copurifies with TnsB during affinity purification of TnsB.
Using mutagenesis guided by amino acid conservation in TnsC, we further found that the TnsCL475A/L476A mutant is defective in TnsB-C crosslinking and other TnsB–TnsC interactions. Notably, TnsCL475A L476A does not promote TnsABC intramolecular transposition in vitro under relaxed reaction conditions that include high glycerol concentrations, and does not promote TnsABC+D transposition in vivo. One reason for this transposition defect is that TnsCL475A L476A does not stimulate the TnsB-dependent pairing of the Tn7 ends. Furthermore, TnsB does not provoke TnsCL475A L476A dissociation from a TnsCD-attTn7 target complex, a reaction that underlies transposition target immunity. TnsCL475A L476A is not defective in formation of a TnsCD-attTn7 complex, however. Thus, multiple aspects of Tn7 transposition that depend on TnsB–TnsC interactions are defective with the TnsCL475A L476A mutant, suggesting that this region of TnsC is key for interaction with TnsB.
TnsC activates the Tn7 transposase by also interacting with the other subunit of the heteromeric transposase TnsA (13), which mediates breakage at the 5′ ends of the transposon (27). The interactions between TnsA and TnsC have been defined by structural analysis of a TnsA/TnsC504–555 cocrystal, and we have shown that a TnsC peptide containing this region can stimulate TnsAB intramolecular breakage and joining under permissive reaction conditions of high glycerol (22). Thus, the C terminus of TnsC contains the two regions that interact with the TnsAB transposase. Indeed, both of these regions are contained within the proteolytically defined domain TnsC411–555. An interesting possibility is that fusion of this TnsC transposase activation domain to a new specific DNA-binding domain may allow retargeting of Tn7 insertion to a new class of target sites.
It is important to note that activation of TnsAB by this domain does not occur in the TnsABC+D system unless TnsC is “activated” by interaction with TnsD-attTn7. We are intrigued by the possibility that the interaction of TnsC with TnsD-attTn7 results in a conformational change in TnsC that leads to exposure of the TnsC domain that activates the TnsAB transposase. It may be that TnsC-dependent TnsAB recombination can occur in the presence of high glycerol concentrations (20% vol/vol), because high glycerol facilitates a conformational change in TnsC that exposes the TnsAB activation domain.
We have shown that, along with its direct interaction with TnsA and TnsB, TnsC also interacts directly with TnsD, the target site selector protein that binds specifically to attTn7 (11, 19). Thus, both protein–protein interactions and binding of TnsC to attTn7 DNA, which likely is mediated by TnsC recognition of distortions in attTn7 DNA induced by the binding of TnsD to attTn7 (20, 21), are critical to Tn7 site-specific insertion into attTn7. It is intriguing that selection of attTn7 as a specific insertion site for Tn7 involves both protein–protein and protein–DNA interactions, and it will be interesting to further dissect the roles of these interactions in the highly site-specific insertion of Tn7 into attTn7.
Another unique aspect of Tn7 transposition is that insertion of Tn7 is not only site-specific, but also orientation-specific (38); the right end of Tn7 inserts adjacent to the host glmS gene in attTn7. Notably, the ends of Tn7 are structurally asymmetrical (24, 25); i.e., the left end of Tn7 contains three separated TnsB-binding sites, whereas the four TnsB sites in the right end of Tn7 are directly adjacent to one another. The ends of Tn7 are functionally asymmetrical as well (32); i.e., miniTn7 elements containing two right ends transpose, whereas elements containing two left ends do not. How is such asymmetry communicated at the molecular level? Our footprinting and stoichiometry analysis of the TnsABC+D posttransposition complex (23) is consistent with the view that the TnsACD-attTn7 target complex likely contains one protomer of TnsD and multiple copies of the TnsA2C2 heterotetramer. We hypothesize that directly at the point of insertion, there is a TnsA2C2 heterotetramer bounded on the glmS side by TnsD. Because TnsC interacts with TnsD-attTn7 both by direct TnsC–TnsD interaction (as we have shown here) and by TnsC recognition of TnsD-induced changes in attTn7 DNA (20, 21), we suggest that these TnsD-based interactions introduce an asymmetry into this TnsA2C2 heterotetramer that is critical to activating the Tn7 ends for asymmetric insertion, and that this asymmetry is communicated to the ends by interactions between TnsC and TnsB and/or TnsA, as well as the altered attTn7 structure.
Although we have now identified the molecular basis of the protein–protein interactions between the Tns proteins, much remains to be learned about the structure and function of various Tns nucleoprotein complexes.
Materials and Methods
Purification of Tns Proteins.
tnsC gene variants were PCR-amplified from pCW4, which contains tnsABCDE (3), using oligonucleotide pairs (Table S1). DNA segments were PCR-amplified with PicoMaxx High-Fidelity Master Mix (Stratagene) and an MJ Research PTC-200 Thermo Cycler (Bio-Rad). The PCR fragments were gel-purified with the Qiaquick Gel Extraction Kit (Qiagen), cloned into the pCYB1 intein vector (New England Biolabs) between the NdeI and EcoRI sites, and transformed into ER2566 (E. coli fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10–TetS)2 [dcm] R(zgb-210::Tn10–TetS) endA1 Δ(mcrC-mrr)114::IS10).
For TnsC purification, cells were grown to OD600nm 0.5–0.7 at 37 °C in LB broth and induced by addition of isopropyl-B-d-thiogalactopyranoside (IPTG) to a final concentration of 0.4 mM. The cultures were then grown for 16 h at 16 °C. All subsequent steps were performed at 4 °C. Cells were harvested by centrifugation and resuspended in 10 mL of column buffer (20 mM Tris pH 8.0, 0.5 M NaCl, and 1 mM EDTA). Cells were lysed by sonication, and cell debris was removed by centrifugation. The soluble material was filtered through a 0.45-μm filter (Millipore), loaded onto a column of chitin beads (New England Biolabs) that had been equilibrated with column buffer, and then shaken gently for 20 min. The column was incubated for 2 d at 4 °C in column buffer containing 50 mM DTT, after which the bound protein was eluted by washing column. Proteins were dialyzed into TnsC storage buffer [25 mM Hepes pH 8.0, 1.0 M NaCl, 2.5 mM DTT, 0.1 mM EDTA, 1 mM ATP, 10 mM CHAPS, and 10% (vol/vol) glycerol].
For isolation of the MBP-TnsC proteins, the tnsC variants were PCR-amplified using the primer sets listed in Table S1 and then cloned between the KpnI and EcoRI sites of the MBP fusion vector pMAL (New England Biolabs). Then 100 mL of TOP10 [E. coli mcrA Δ(mrr-hsdRMS-mcrBC)φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 GalE15 GalK16 rpsL(StrR) endA1] cultures containing MBP-TnsC fusion protein plasmids were grown at 37 °C to OD600nm 0.5–0.7 and induced with 0.3 mM IPTG for 16 h at 16 °C. All subsequent steps were performed at 4 °C. Cells were harvested by centrifugation and resuspended in 10 mL column buffer (20 mM Tris pH 8.0, 0.5 M NaCl, 1 mM EDTA, and 1 mM DTT) and lysed by sonication. The soluble material was filtered through a 0.45-μm filter (Millipore) before being loaded onto an amylose resin (New England Biolabs) column equilibrated with column buffer, followed by gentle shaking for 30 min. The column was then washed extensively with column buffer by gravity flow, and bound protein was eluted with column buffer containing 20 mM maltose. MBP-fused TnsC fragment proteins were dialyzed into TnsC storage buffer.
TnsA and TnsB proteins were purified as described previously (29), as was TnsD protein (23).
UV Crosslinking Assay.
The TnsB* peptide was synthesized by Anaspec, and TnsB*1 and TnsB*2 peptides were synthesized by NEO-peptide. The TnsB-TnsC crosslinking reaction was performed by preincubating 203 μM TnsB* peptide and 32 nM TnsC in a 100-μL final volume reaction containing 21.5 mM Na2HPO4, 1 mM Hepes pH 8.0, 36.8 mM NaCl, 4.6 mM ATP, 15 mM MgAc, 0.1 mM EDTA, 1.2 mM DTT, 1.9% (vol/vol) glycerol, 0.4 mM CHAPS, and 1 mg/mL BSA at 4 °C for 30 min, and then activating the crosslinker with a 360-nm UV light for 20 min after the warmup. Reactions with the MBP-TnsC protein fusions containined 203 μM TnsB* peptide and 32 nM MBP-TnsC in 21.5 mM Na2HPO4, 1.5 mM Hepes pH 8.0, 81.5 mM NaCl, 4.6 mM ATP, 15 mM MgAc, 0.1 mM EDTA, 1 mM DTT, 2% (vol/vol) glycerol, 0.6 mM CHAPS, and 1 mg/mL BSA. Proteins displayed on 4–12% NuPAGE SDS/PAGE gel (Invitrogen) were electrotransferred onto a PVDF membrane (Millipore) by a TurboBlotter (Bio-Rad) at 18V for 50 min. Western blot analysis probing for biotin on the peptide used a monoclonal mouse anti-biotin antibody (Santa Cruz Biotechnology) and ECL anti-mouse IgG HRP-linked whole antibody (GE Healthcare) for blotting. The membrane was soaked with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) for 1–2 min and then exposed to Kodak film.
In Vitro Intramolecular Transposition.
In vitro transposition reactions were performed in which end cleavage and intramolecular joining (22) were assayed with 0.25 nM pEMΔ (11), a 5.9-kb plasmid containing a 1.6-kb miniTn7KmR element (a kanamycin gene segment between the 166-bp Tn7L and 199-bp Tn7R ends), Tns proteins, and buffer including 20% (vol/vol) glycerol. The Tns proteins were 75 ng of TnsA, with 25 ng of TnsB or 30 ng of either WT TnsC or TnsCL475A/L476A added as indicated. The buffers contained 2.5 mM Tris pH 7.6, 25 mM Hepes pH 7.6, 2 mM DTT, 50 μg/mL BSA, 100 μg/mL tRNA, 25 μM ATP, 20% (vol/vol) glycerol, and 15 mM MgAc. The reactions were performed at 30 °C for 30 min in a final volume of 50 μL. DNA was extracted with phenol:chloroform:isoamylalcohol (25:24:1) (Invitrogen), precipitated with ethanol, digested with NdeI and RNase, and then displayed on a 1.0% (wt/vol) agarose Tris-borate-EDTA (TBE) gel.
Tn7 Transposition in Vivo.
Tn7 transposition was measured by a promoter capture assay (8, 37). pOXminiTn7::lac contains lac genes that have no internal promoter, and the element is not flanked by an external promoter in this plasmid. Thus, cells containing this plasmid are Lac−, i.e., white on MacConkey Lac plates, when the host strain was also lac−. In the presence of Tns proteins, the miniTn7::lac transposes such that the miniTn7lac is downstream of an external promoter, resulting in a Lac+ phenotype, i.e., red on MacConkey Lac plates. Transposition was carried out in CW51 [E. coli F− ara− arg− lac-proXIII recA56 NalR RifR(3)] containing pCW305, a pCW4 tnsABCDE derivative that contains a miniMu insertion in tnsC (3) and thus supplies TnsA, TnsB, TnsD, and WT TnsC or TnsCL475A/L476A from a pCYB1 plasmid. In these cells, the miniTn7::lac element inserts into attTn7, and the red color results from infrequent opposite orientation insertion such that the lac genes are downstream of E. coli glmS (37).
TnsCD-attTn7 Complex Dissociation Assay.
The TnsCD-attTn7 complex dissociation assay was performed as described previously (30). The attTn7 target fragment, 158 bp, containing attTn7 (−72 to +85), was PCR-amplified from pPK13 (20) and purified after being displayed on a 1.5% agarose gel in 1× Tris-acetate-EDTA (TAE) buffer (pH 8.0). The probe was labeled at its 5′ ends with 32P-γ-ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs), and purified with a Quick-Spin Column G-50 (Roche). For the reactions, 0.5 μg of poly(dI-dC) and 0.05 pmol labeled attTn7 DNA were incubated with 30 nM TnsD and 35 nM TnsC at 30 °C in a 20-μL reaction volume for target assembly. The reaction buffer contained 29 mM Hepes, 65 mM NaCl, 71 mM KCl, 0.5 mM CHAPS, 0.26 mM EDTA, 4.7 mM DTT, 2.3 mM ATP, and 5% (vol/ vol) glycerol. After TnsCD-attTn7 complex assembly, 1 μL of TnsB or B* peptide in TnsB buffer [25 mM Hepes pH 8.0, 0.5 M NaCl, 2 mM DTT, and 25% (vol/vol) glycerol] was added to a final concentration of 30 nM TnsB and 8 μM or 40 μM TnsB* peptide. Then 4 μL of 62.5 mM MgAc was added, increasing the reaction volume to 25 μL. Glutaraldehyde (Sigma-Aldrich) was then added to a final concentration of 0.01% (vol/vol), and the reaction was incubated for another 10 min at 30 °C. The reactions were electrophoresed through a 0.9% agarose gel in 1× TBE buffer (pH 8.0) at 50V for 1 h at 25 °C. Gels were vacuum-dried and viewed with a Molecular Dynamics PhophorImager and Typhoon (GE Healthcare).
PEC Assays.
The 20-μL PEC formation reactions (29) were carried out as in vitro transposition assays containing only the Tn7 containing plasmid, TnsB and TnsC proteins, and Ca2+ ions in place of Mg2+. The donor plasmid was pEMΔ (11), a 5.9-kb plasmid that contains a 1.6-kb miniTn7KmR element (a kanamycin gene segment between the 166-bp Tn7L and 99-bp Tn7R ends). As indicated, Tns proteins were incubated with 15 mM CaAc at 30 °C for 10 min, followed by incubation with glutaraldehyde (Sigma-Aldrich) to a final concentration of 0.01% (vol/vol). The crosslinker was quenched by the addition of 25 mM Tris pH 8.0 and 5 mM lysine for 10 min at room temperature. Then 8 units of PflMI (New England Biolabs) and 3 μL of New England Biolabs #3 buffer were added, followed by 1 h of digestion at 30 °C. The reactions were analyzed by 0.9% agarose gel electrophoresis in 0.5× TAE buffer at 50V for 100 min at 25 °C.
MBP-TnsC–TnsDHis6 Copurification Assays.
For these assays, 50-mL cultures of E. coli ER2566 pMAL-MBP-TnsC1–85 and 50 mL of BL21Star(DE3) pET101/D-TOPO-TnsD1–309His were grown at 37 °C to OD600nm 0.5–0.7 and induced with 0.3 mM IPTG for 16 h at 16 °C. The TnsD1–309His and MBP-TnsC1–85 cells were mixed, harvested by centrifugation, resuspended in 10 mL of binding buffer (20 mM Tris pH 7.9, 0.5 M NaCl, and 5 mM imidazole), and then lysed by sonication. Cell debris was removed by centrifugation. All subsequent purification steps were performed at 4 °C. The soluble material was filtered through a 0.45-μm filter (Millipore) before being loaded onto a column of Ni2+ Chelating Sepharose Fast Flow Resin (GE Healthcare) that had been equilibrated with binding buffer, followed by gentle shaking for 20 min. The column was washed in binding buffer and wash buffer (20 mM Tris pH 7.9, 0.5 M NaCl, and 50 mM imidazole), after which the bound protein was eluted by elution buffer (20 mM Tris pH 7.9, 0.5 M NaCl, and 250 mM imidazole). After the proteins were displayed on an SDS/PAGE gel and transferred as described above, the TnsD1–309His and MBP-TnsC1–85 proteins were identified by Western blot analysis. For TnsD1–309His, blotting was performed using His-probe (H-15) rabbit polyclonal antibodies (Santa Cruz Biotechnology) and ECL anti-rabbit IgG, HRP-linked whole antibody (GE Healthcare). For MBP-TnsC1–85, blotting was done using MBP mouse monoclonal antibodies (New England Biolabs) and ECL anti-mouse IgG, HRP-linked antibody (GE Healthcare). The exposure and detection procedures were the same as described above.
Supplementary Material
Acknowledgments
We thank the members of the N.L.C. laboratory for helpful discussions, Helen McComas for her assistance with the figures, Patti Kodeck for her assistance with the manuscript, and Joe Peters for his comments on the manuscript. This work was supported by National Institutes of Health Grants GM076425 (to N.L.C.) and GM007445 (to J.M.S.). N.L.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409869111/-/DCSupplemental.
References
- 1.Gueguen E, Rousseau P, Duval-Valentin G, Chandler M. The transpososome: Control of transposition at the level of catalysis. Trends Microbiol. 2005;11:543–549. doi: 10.1016/j.tim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Dyda F, Chandler M, Hickman AB. The emerging diversity of transpososome architectures. Q Rev Biophys. 2012;45(4):493–521. doi: 10.1017/S0033583512000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waddell CS, Craig NL. Tn7 transposition: Two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988;2:137–149. doi: 10.1101/gad.2.2.137. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Craig NL, Peters JE. Transposon Tn7. In: Roberts A, Mullany P, editors. Bacterial Integrative Mobile Genetic Elements. London: Landes Bioscience; 2011. pp. 1–25. [Google Scholar]
- 5.Craig NL. Tn7. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 423–456. [Google Scholar]
- 6.Wolkow CA, DeBoy RT, Craig NL. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 1996;10:2145–2157. doi: 10.1101/gad.10.17.2145. [DOI] [PubMed] [Google Scholar]
- 7.Parks AR, et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009;138(4):685–695. doi: 10.1016/j.cell.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stellwagen A, Craig NL. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein which activates the bacterial transposon Tn7. Genetics. 1997;145:573–585. doi: 10.1093/genetics/145.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stellwagen AE, Craig NL. Analysis of gain-of-function mutants of an ATP-dependent regulator of Tn7 transposition. J Mol Biol. 2001;305:633–642. doi: 10.1006/jmbi.2000.4317. [DOI] [PubMed] [Google Scholar]
- 10.Craig NL, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 11.Bainton RJ, Kubo KM, Feng J-N, Craig NL. Tn7 transposition: Target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell. 1993;72:931–943. doi: 10.1016/0092-8674(93)90581-a. [DOI] [PubMed] [Google Scholar]
- 12.Gamas P, Craig NL. Purification and characterization of TnsC, a Tn7 transposition protein that binds ATP and DNA. Nucleic Acids Res. 1992;20:2525–2532. doi: 10.1093/nar/20.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biery M, Lopata M, Craig NL. A minimal system for Tn7 transposition: The transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J Mol Biol. 2000;297:25–37. doi: 10.1006/jmbi.2000.3558. [DOI] [PubMed] [Google Scholar]
- 14.Stellwagen A, Craig NL. Mobile DNA elements: Controlling transposition with ATP-dependent molecular switches. Trends Biochem Sci. 1998;23:486–490. doi: 10.1016/s0968-0004(98)01325-5. [DOI] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaconas G, Harshey RM. Transposition of phage Mu DNA. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 384–402. [Google Scholar]
- 17.Harshey R, Jayaram M. The Mu transpososome through a topological lens. Crit Rev Biochem Mol Biol. 2006;41:387–405. doi: 10.1080/10409230600946015. [DOI] [PubMed] [Google Scholar]
- 18.Skelding Z, Sarnovsky R, Craig N. Formation of a nucleoprotein complex containing Tn7 and its target DNA regulates transposition initiation. EMBO J. 2002;21:3494–3504. doi: 10.1093/emboj/cdf347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra R, McKenzie GJ, Yi L, Lee CA, Craig NL. Characterization of the TnsD-attTn7 complex that promotes site-specific insertion of Tn7. Mob DNA. 2010;1(1):18. doi: 10.1186/1759-8753-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuduvalli P, Rao JE, Craig NL. Target DNA structure plays a critical role in Tn7 transposition. EMBO J. 2001;20:924–932. doi: 10.1093/emboj/20.4.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao JE, Miller PS, Craig NL. Recognition of triple-helical DNA structures by transposon Tn7. Proc Natl Acad Sci USA. 2000;97:3936–3941. doi: 10.1073/pnas.080061497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronning DR, et al. The carboxy-terminal portion of TnsC activates the Tn7 transposase through a specific interaction with TnsA. EMBO J. 2004;23:2972–2981. doi: 10.1038/sj.emboj.7600311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holder JW, Craig NL. Architecture of the Tn7 post-transposition complex: An elaborate nucleoprotein structure. J Mol Biol. 2010;401:167–181. doi: 10.1016/j.jmb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arciszewska LK, Craig NL. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 1991;19:5021–5029. doi: 10.1093/nar/19.18.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arciszewska LK, McKown RL, Craig NL. Purification of TnsB, a transposition protein that binds to the ends of Tn7. J Biol Chem. 1991;266:21736–21744. [PubMed] [Google Scholar]
- 26.Sarnovsky R, May EW, Craig NL. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 27.May EW, Craig NL. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996;272:401–404. doi: 10.1126/science.272.5260.401. [DOI] [PubMed] [Google Scholar]
- 28.Hickman AB, et al. Unexpected structural diversity in DNA recombination: The restriction endonuclease connection. Mol Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 29.Choi K, Li Y, Sarnovsky RJ, Craig NL. Direct interaction between the TnsA and TnsB subunits controls the heteromeric Tn7 transposase. Proc Natl Acad Sci USA. 2013;110(22):E2038–E2045. doi: 10.1073/pnas.1305716110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skelding Z, Queen-Baker J, Craig N. Alternative interactions between the Tn7 transposase and the Tn7 target DNA binding protein regulate target immunity and transposition. EMBO J. 2003;22:5904–5917. doi: 10.1093/emboj/cdg551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stellwagen AE, Craig NL. Avoiding self: Two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 1997;16(22):6823–6834. doi: 10.1093/emboj/16.22.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arciszewska LK, Drake D, Craig NL. Transposon Tn7: cis-acting sequences in transposition and transposition immunity. J Mol Biol. 1989;207:35–52. doi: 10.1016/0022-2836(89)90439-7. [DOI] [PubMed] [Google Scholar]
- 33.Adzuma K, Mizuuchi K. Target immunity of Mu transposition reflects a differential distribution of MuB protein. Cell. 1988;53:257–266. doi: 10.1016/0092-8674(88)90387-x. [DOI] [PubMed] [Google Scholar]
- 34.Shoelson S, Lee J, Lynch C, Backer J, Pilch P. BpaB25 insulins: Photactivatable analogues that quantitatively cross-link, radiolabel and activate the insulin receptor. J Biol Chem. 1993;268:4085–4091. [PubMed] [Google Scholar]
- 35.Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33(19):5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 36.Bainton R, Gamas P, Craig NL. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991;65:805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- 37.Hughes O. 1993. Host components of Tn7 transposition. PhD thesis (Univ California San Francisco)
- 38.Lichtenstein C, Brenner S. Unique insertion site of Tn7 in E. coli chromosome. Nature. 1982;297:601–603. doi: 10.1038/297601a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.