Significance
Mammalian central neurons typically possess solitary appendages called primary cilia. These rod-shaped protrusions are enriched for signaling proteins, suggesting they respond to extracellular neuromodulators. Yet, neuronal ciliary signaling has never been observed and the roles of cilia in neuronal function are unclear. This study identifies Kiss1r, which plays a key role in reproductive function, as a novel ciliary receptor on gonadotropin-releasing hormone (GnRH) neurons. GnRH neurons possess multiple Kiss1r-positive cilia and the proportion of multiple cilia increases in parallel with pubertal maturation. Ablation of Kiss1r-positive cilia on GnRH neurons does not affect neuron migration or sexual maturation. However, kisspeptin-mediated increases in GnRH neuron firing rate are reduced in the absence of cilia. Thus, cilia enhance Kiss1r signaling on GnRH neurons.
Keywords: GPR54, neuronal primary cilia, electrophysiology
Abstract
Most central neurons in the mammalian brain possess an appendage called a primary cilium that projects from the soma into the extracellular space. The importance of these organelles is highlighted by the fact that primary cilia dysfunction is associated with numerous neuropathologies, including hyperphagia-induced obesity, hypogonadism, and learning and memory deficits. Neuronal cilia are enriched for signaling molecules, including certain G protein-coupled receptors (GPCRs), suggesting that neuronal cilia sense and respond to neuromodulators in the extracellular space. However, the impact of cilia on signaling to central neurons has never been demonstrated. Here, we show that the kisspeptin receptor (Kiss1r), a GPCR that is activated by kisspeptin to regulate the onset of puberty and adult reproductive function, is enriched in cilia projecting from mouse gonadotropin-releasing hormone (GnRH) neurons. Interestingly, GnRH neurons in adult animals are multiciliated and the percentage of GnRH neurons possessing multiple Kiss1r-positive cilia increases during postnatal development in a progression that correlates with sexual maturation. Remarkably, disruption of cilia selectively on GnRH neurons leads to a significant reduction in kisspeptin-mediated GnRH neuronal activity. To our knowledge, this result is the first demonstration of cilia disruption affecting central neuronal activity and highlights the importance of cilia for proper GPCR signaling.
Primary cilia are typically solitary nonmotile appendages that project from nearly every cell type in the mammalian body (1). They are specialized sensory organelles that incorporate a myriad of extracellular stimuli into signal transduction pathways to modulate cell physiology (2–4). Consequently, ciliary dysfunction can result in numerous human diseases, termed ciliopathies, which impact many organ systems (5). Ciliopathies are associated with certain neuropathologies, including structural malformations, hyperphagia-induced obesity, intellectual disability, and hypogonadism, thereby highlighting the importance of cilia for proper CNS development and function (3).
Most adult neurons in the mammalian brain possess a primary cilium that projects from its cell body. Specific signaling proteins are selectively targeted to and retained within neuronal cilia, which are restricted compartments and regulate entry and exit of proteins through multiple mechanisms (6, 7). These signaling proteins include type 3 adenylyl cyclase (AC3) (8), which converts ATP to cAMP, and the GPCRs, somatostatin receptor 3 (Sstr3) (9), serotonin receptor 6 (10, 11), melanin-concentrating hormone receptor 1 (Mchr1) (12, 13), dopamine receptor 1 (14), and neuropeptide Y receptors 2 and 5 (15). The functions of primary cilia are determined by the proteins that are enriched within them, thus, it is likely that neuronal cilia sense neuromodulators in the extracellular milieu and initiate signaling cascades. However, the precise roles of neuronal cilia remain unknown.
Identification of signaling proteins that are selectively targeted to neuronal cilia is a critical step in elucidating the functions of these organelles. We previously identified ciliary localization sequences in the third intracellular loop and carboxy tail of ciliary GPCRs and used these sequences to predict novel ciliary GPCRs (12, 14). One of these, the kisspeptin receptor (Kiss1r, also known as GPR54), was considered a strong candidate ciliary GPCR given that loss of Kiss1r leads to hypogonadotropic hypogonadism in humans and mice (16, 17) and hypogonadotropic hypogonadism is a feature of the ciliopathy Bardet-Biedl syndrome (18). Kiss1r is expressed in a large proportion of gonadotropin-releasing hormone (GnRH) neurons (19), a population of hypothalamic neurons that are central effectors driving the neuroendocrine reproductive axis. Treatment of Kiss1r-expressing GnRH neurons with kisspeptin increases the firing rate of the GnRH neurons and augments GnRH secretion. GnRH stimulates luteinizing hormone and follicle-stimulating hormone secretion, which in turn initiates puberty and supports adult sexual function.
Here, we show that Kiss1r is enriched in primary cilia on GnRH neurons in the mouse. Notably, GnRH neurons can possess multiple Kiss1r-positive cilia and the proportion of multiciliated GnRH neurons increases in parallel with sexual maturation. We also show that cilia are required for proper Kiss1r-mediated signaling on GnRH neurons. These results provide insight into the mechanism of Kiss1r signaling and demonstrate that loss of cilia on central neurons impairs neuronal signaling.
Results
Kiss1r Localizes to Cilia.
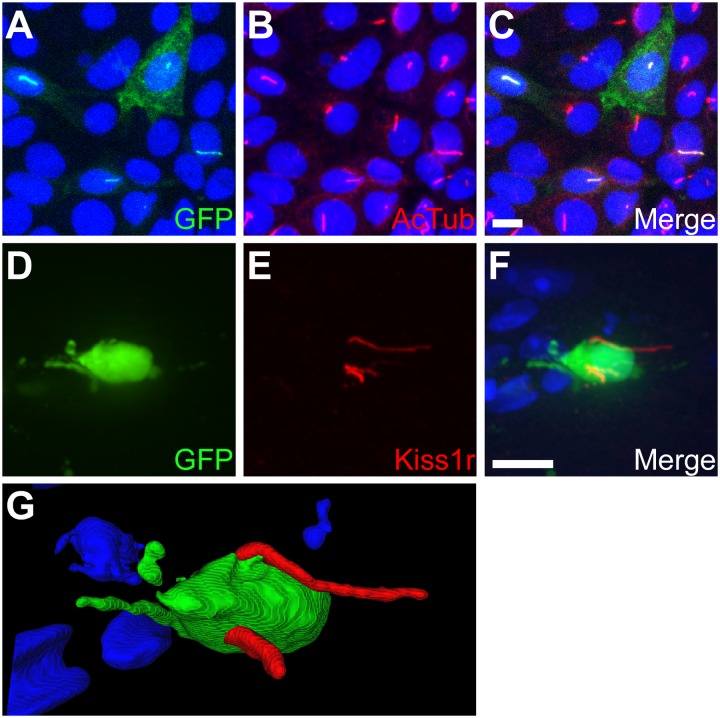
To test whether Kiss1r localizes to cilia, a construct encoding mouse Kiss1r fused at the carboxy-terminus to EGFP was expressed in inner medullary collecting duct (IMCD) cells. We have previously shown that ciliary GPCRs selectively localize to cilia when expressed in IMCD cells (12, 14). Notably, Kiss1r localized to cilia on IMCD cells (Fig. 1 A–C). We then tested whether Kiss1r localizes to neuronal cilia in tissue by labeling mouse brain slices with a custom antibody generated against Kiss1r. The specificity of the antibody was confirmed by immunofluorescence and immunoblotting (Fig. S1). In the mouse brain there are only ∼800 GnRH neurons, which are widely dispersed across the medial hypothalamus and basal forebrain (20). To facilitate identification of GnRH neurons we labeled brain slices from adult male and female transgenic mice expressing GFP under the control of the GnRH promoter (GnRH::GFP) (21). Interestingly, the majority of GnRH neurons in both male and female brains possessed Kiss1r-positive cilia, with some neurons possessing multiple Kiss1r-positive cilia (Fig. 1 D–G). Analysis of 3D renderings of GnRH neurons and colabeling for rootletin, which marks the ciliary base in neurons (22), confirmed that the Kiss1r-positive cilia were projecting from the same neuron (Fig. S2 A–D and Movies S1–S5).
Fig. 1.
Kiss1r localizes to primary cilia in vitro and in vivo. (A–C) Representative image of transiently transfected IMCD cells expressing Kiss1r fused at the C terminus to EGFP. (A) EGFP fluorescence (green) shows expression of Kiss1r. (B) Acetylated α-tubulin (AcTub; red) marks the cilia. (C) Merged image. (D–F) Representative image of the medial hypothalamus in adult GnRH::GFP mice. (D) GFP fluorescence (green) indicates a GnRH neuron. (E) Labeling for Kiss1r (red) shows the presence of multiple Kiss1r-positive cilia. (F) Merged image. (G) 3D rendering of the same neuron confirms the cilia project from the same cell. Nuclei are stained with DRAQ5 (blue). (Scale bars: 10 µm.)
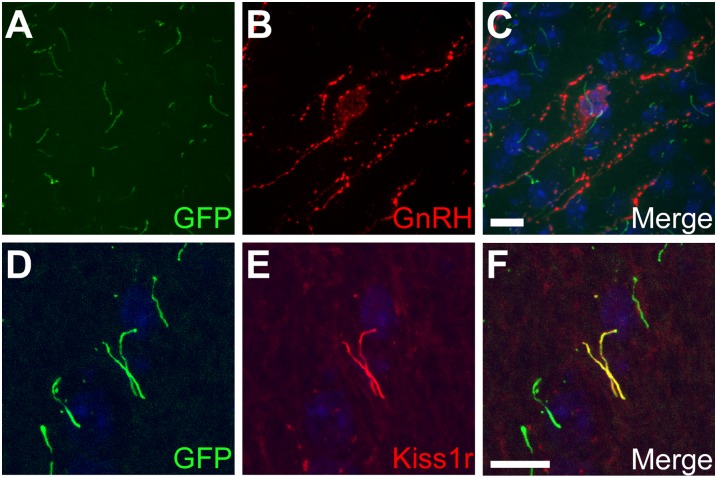
The presence of cilia on GnRH neurons was previously reported in several electron microscopy studies (23–25), with one study noting the presence of up to three cilia on some GnRH neurons (23). Nevertheless, we decided to confirm that Kiss1r was localizing to cilia on GnRH neurons. However, labeling with antibodies against proteins known to be enriched in neuronal cilia (i.e., AC3, Sstr3, Mchr1, and Arl13b (26)) failed to label cilia on GnRH neurons (Fig. S2E). To address this limitation, we generated brain slices from CiliaGFP mice, which are transgenic mice expressing Sstr3 fused to EGFP (27). Sstr3::EGFP is efficiently targeted to cilia across cell types and CiliaGFP mice provide a powerful tool for visualizing cilia in vivo or in vitro. Labeling sections from the medial hypothalamus of CiliaGFP brains with an antibody against GnRH revealed the presence of GnRH neurons projecting one or more GFP-positive cilia (Fig. 2 A–C). Moreover, labeling of corresponding sections with the Kiss1r antibody confirmed a subset of Sstr3::EGFP expressing cilia were positive for Kiss1r (Fig. 2 D–F). Multiple cilia were not detected on any surrounding non-GnRH neurons (Table S1), which is consistent with previous studies of neuronal cilia frequency (28–31) and suggests the presence of multiple cilia on central neurons is rare. Together, our results verify that GnRH neurons can be multiciliated and demonstrate that Kiss1r is selectively targeted to cilia on GnRH neurons.
Fig. 2.
GnRH neurons possess cilia that are positive for Kiss1r. (A–C) Representative image of the medial hypothalamus in adult CiliaGFP mice. (A) EGFP fluorescence (green) shows Sstr3 expression and ciliary localization. (B) Labeling for GnRH (red) indicates GnRH neurons. (C) Merged image confirms GnRH neurons are ciliated. (D–F) Representative image of the medial hypothalamus in adult CiliaGFP mice. (D) EGFP fluorescence (green) shows Sstr3 expression and ciliary localization. (E) Labeling for Kiss1r (red) confirms Kiss1r ciliary localization. (F) Merged image. Nuclei are stained with DRAQ5 (blue). (Scale bars: 10 µm.)
Kiss1r Multiciliation Increases Over Development.
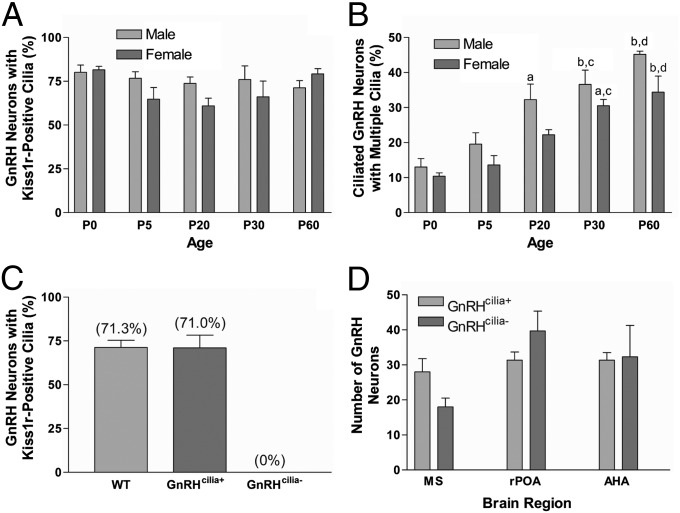
Given the important role of Kiss1r in sexual development, we tested whether Kiss1r ciliary localization is developmentally regulated. Brain slices of male and female GnRH::GFP mice from P0 to P60 were labeled for Kiss1r and the percentage of GnRH neurons possessing Kiss1r-positive cilia was quantified. The percentage of GnRH neurons displaying at least one Kiss1r-positive cilium was ∼75% in both sexes at birth and did not change significantly during postnatal development (Fig. 3A). However, quantifying the percentage of GnRH neurons possessing more than one Kiss1r-positive cilium revealed that the frequency of multiciliated GnRH neurons significantly increased during postnatal development (Fig. 3B). At birth, ∼10% of ciliated GnRH neurons in male and female mice possessed more than one Kiss1r-positive cilium but by P60 the percentages in male and female mice were 42% and 35%, respectively. This result indicates that the frequency of GnRH neurons possessing multiple Kiss1r-positive cilia increases during postnatal development in parallel with sexual maturation.
Fig. 3.
Quantification of GnRH cilia. (A) Percentage of GnRH neurons with one or more Kiss1r-positive cilia in the medial hypothalamus of P0-P60 male and female GnRH::GFP mice (n = 4 animals of each sex at all ages, with the exception of P0, n = 3). Note the percentages do not vary significantly between ages or sexes. (B) Percentage of ciliated GnRH neurons with more than one Kiss1r-positive cilium in the medial hypothalamus of P0-P60 male and female GnRH::GFP mice (n = 4 animals of each sex at all ages, with the exception of P0, n = 3). Note the proportion of multiciliated GnRH neurons increases significantly between P0 and P60. aSignificantly different from P0 (P < 0.01). bSignificantly different from P0 (P < 0.001). cSignificantly different from P5 (P < 0.01). dSignificantly different from P5 (P < 0.001). (C) Percentage of GnRH neurons with one or more Kiss1r-positive cilia in the medial hypothalamus of P60 male GnRH::GFP, GnRHcilia+, and GnRHcilia- mice (n = 3–4 animals for each genotype). Note there is no difference in the percentage of Kiss1r-positive cilia between GnRH::GFP and GnRHcilia+ mice, but Kiss1r-positive cilia are completely lacking in GnRHcilia- mice. (D) Number of GnRH neurons throughout the medial septum (MS), rostral preoptic area (rPOA), and anterior hypothalamic area (AHA) of P60 male GnRHcilia+ and GnRHcilia- mice (n = 3 animals for each genotype). Note there is no significant difference in the number of GnRH neurons in any region between GnRHcilia+ and GnRHcilia- mice. Values are expressed as mean ± SEM.
GnRH Neuronal Cilia are not Essential for Sexual Maturation.
As Kiss1r is enriched in cilia and the proportion of neurons with multiple Kiss1r-positive cilia increases during development, we next asked whether GnRH neuronal cilia are required for proper sexual maturation in mice. To selectively ablate cilia on GnRH neurons, mice carrying a conditional allele of Ift88 (32), which encodes an intraflagellar transport protein and is required for the formation and maintenance of neuronal cilia (32–34), were crossed with transgenic mice expressing Cre recombinase under the control of the GnRH promoter (35). These mice also carried the GnRH::GFP transgene to allow identification and analysis of GnRH neurons. GnRH expression begins during embryonic development and is detected as early as E11.5 (36). Control animals were heterozygous for a wild-type (wt) and floxed (flox) Ift88 allele (Ift88wt/flox) and positive for GnRH::Cre and GnRH::GFP. Experimental animals were heterozygous for a floxed and deleted (null) Ift88 allele (Ift88flox/null) and positive for GnRH::Cre and GnRH::GFP. Thus, in response to prenatal Cre expression, cilia on GnRH neurons would be unaffected in control animals (GnRHcilia+) due to the presence of the wild-type allele but lost in experimental animals (GnRHcilia-). Kiss1r labeling of brain slices from P60 mice confirmed that GnRHcilia+ animals possessed an equivalent number and distribution of Kiss1r-positive cilia on GnRH neurons to wild-type GnRH::GFP animals (Fig. 3C). Thus, loss of one Ift88 allele does not impact cilia on GnRH neurons. P0 GnRHcilia- mice rarely possessed Kiss1r-positive cilia (Fig. S3A), whereas P60 GnRHcilia- mice showed a complete lack of Kiss1r-positive cilia (Fig. 3C and Fig. S3 B and C), suggesting depletion of Ift88 in GnRH neurons effectively ablates cilia. Quantification of the number of GFP-positive neurons by region at P0 (Fig. S3D), when GnRH neurons are still migrating (37), and P60 (Fig. 3D), after migration is complete, was not significantly different between GnRHcilia+ and GnRHcilia- animals, indicating that GnRH neuron survival and/or migration is not affected by loss of cilia on these neurons.
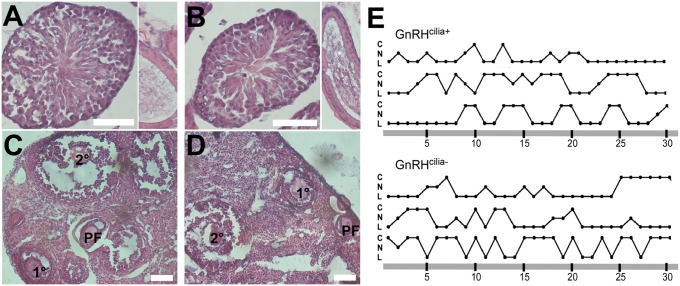
To test whether loss of Kiss1r-positive cilia on GnRH neurons causes defects in sexual maturation in mice, we quantified sex organ weights and day of vaginal opening, a measure of puberty onset in female mice. None of these measures was significantly different between GnRHcilia+ and GnRHcilia- animals (Table S2). Furthermore, histological analysis revealed mature sperm in the testes and seminiferous tubules of adult GnRHcilia- males (Fig. 4 A and B) and follicles in all stages of development in the ovaries of adult GnRHcilia- females (Fig. 4 C and D). Estrous cycle tracking in GnRHcilia+ and GnRHcilia- females revealed no obvious differences in cycling between the two groups (Fig. 4E). Finally, male and female GnRHcilia- mice were fertile and generated normal numbers and litter sizes. These results indicate that cilia on GnRH neurons are not required for reproductive function in mice.
Fig. 4.
GnRH cilia are dispensable for sexual maturation in male and female mice. (A and B) Representative testis (Left) and seminiferous tubule (Right) sections from P60 GnRHcilia+ (A) and GnRHcilia- (B) mice (n = 3 animals for each genotype) shows the presence of mature sperm in both genotypes. (C and D) Representative ovary section from P60 GnRHcilia+ (C) and GnRHcilia- (D) mice (n = 3 animals for each genotype) shows the presence of follicles at all stages of development. (Scale bars: 100 µm.) (E) Vaginal cytology of P60 GnRHcilia+ and GnRHcilia- mice show all stages of estrous cyclicity. C, cornified (estrous); L, leukocytic (metestrous and diestrous); N, nucleated (proestrous).
GnRH Cilia are Required for Proper Kiss1r-Mediated Signaling.
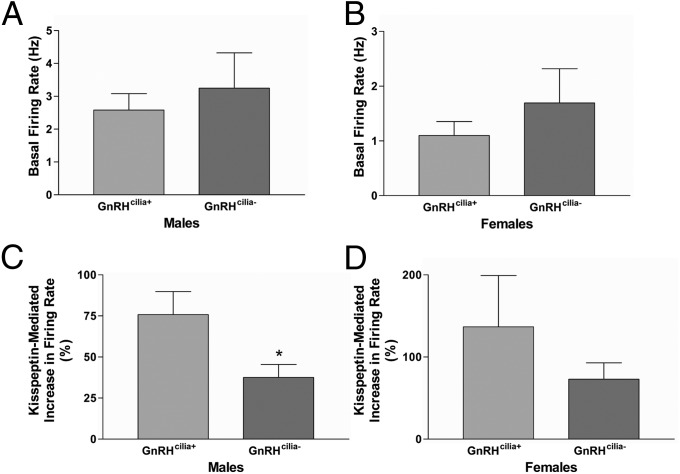
Because Kiss1r is normally targeted to and enriched within GnRH cilia, we then asked whether loss of cilia affects Kiss1r signaling. Application of the Kiss1r ligand, kisspeptin, increases the action potential firing rate of GnRH neurons (38) and this response is absent in GnRH neurons lacking Kiss1r (39). Thus, we performed extracellular loose-cell attached patch-clamp recording on acute sagittal brain slices from adult male and female GnRHcilia+ and GnRHcilia- animals. The female mice were ovariectomized to reduce potential variability from the estrous cycle. There was no difference in the baseline firing rate of GnRH neurons between male or female GnRHcilia+ and GnRHcilia- mice (Fig. 5 A and B) and application of 100 nM kisspeptin resulted in an increase in firing rate in the vast majority of GnRH neurons, consistent with previous results (38). Interestingly, GnRH neurons from male GnRHcilia- mice showed a significantly reduced kisspeptin-mediated increase in firing rate than GnRH neurons in GnRHcilia+ animals (Fig. 5C). The kisspeptin-mediated increase in firing rate was also reduced in GnRH neurons of female GnRHcilia- mice compared with GnRHcilia+ mice, but this difference was not statistically significant (Fig. 5D). To confirm that the decreased responsiveness to kisspeptin in male GnRHcilia- mice was not attributable to a decrease in Kiss1r expression, we performed real-time PCR analysis of hypothalamic RNA from adult male GnRHcilia+ and GnRHcilia- mice and found that Kiss1r was expressed at equivalent levels in both genotypes (Fig. S4). We also observed an equivalent kisspeptin-induced induction of Fos expression, a response that is mediated by Kiss1r (40), in GnRHcilia+ and GnRHcilia- male mice (Fig. S5). Together, our results suggest that in the absence of cilia Kiss1r is present and functional on the cell surface but cilia on GnRH neurons enhance kisspeptin-mediated Kiss1r signaling.
Fig. 5.
GnRH cilia are required for proper Kiss1r signaling. Basal firing rates of GnRH neurons from adult male (A) and ovariectomized female (B) GnRHcilia+ and GnRHcilia- mice. Note there is no significant difference in the basal firing rates between GnRHcilia+ and GnRHcilia- mice. Percentage increase in the firing rate of GnRH neurons after kisspeptin treatment in adult male (C) and ovariectomized female (D) GnRHcilia+ and GnRHcilia- mice. Note the increase in firing rate is significantly lower in male GnRHcilia- mice compared with GnRHcilia+ mice. Values are expressed as mean ± SEM. For males n = 10–13 neurons from 4 to 7 animals of each genotype. For females n = 9 neurons from 4 to 5 animals of each genotype. *Significantly different from GnRHcilia+ percentage (P = 0.02).
Discussion
Our results show that GnRH neurons possess cilia, which are enriched for Kiss1r. There are several theories regarding the possible functions of cilia on adult central neurons. Because cilia are enriched for signaling proteins, they may well function as sensory organelles. However, a direct role for cilia in neuronal signaling has not been observed. Here, we demonstrate that loss of Kiss1r-positive cilia on GnRH neurons causes a reduction in the response of GnRH neurons to kisspeptin in male mice, indicating that cilia enhance Kiss1r signaling. To our knowledge, this is the first demonstration that cilia play a role in GPCR signaling in central neurons. What is the role of cilia in Kiss1r signaling? One possibility is that Kiss1r normally signals on the ciliary membrane and ciliary localization concentrates the receptors to optimize ligand binding and signal propagation. There is evidence for both synaptic (41, 42) and volume (43) transmission of kisspeptin to GnRH neurons. Ciliary response to ligand should be involved in volume transmission as cilia project from the cell body and synaptic contacts have never been observed on neuronal cilia. Enriching and concentrating receptors in multiple long cilia protruding from the same neuron could thus increase the efficacy of kisspeptin binding in the extracellular space and enhance volume transmission. This model would be analogous to olfactory sensory neurons that project multiple primary cilia into the olfactory epithelium, which increases the sensory surface and the ability to detect and respond to odorants (44). Another possibility is that the signal generated by Kiss1r on the ciliary membrane is qualitatively different from the signal generated on the plasma membrane. The expression of Kiss1r on cilia may generate a unique signal based on coupling to different signaling transduction pathways and/or regulation of these signaling cascades. Loss of this signal may affect the overall response to kisspeptin. Alternatively, Kiss1r may not directly signal on the ciliary membrane, but rather, the cilium may act as a reservoir for the receptor and facilitate signaling. Regardless of the mechanisms, our results show that cilia are necessary for normal Kiss1r signaling on GnRH neurons.
We also demonstrate that GnRH neurons possess multiple Kiss1r-positive cilia and the proportion of multiciliated GnRH neurons increases over pubertal maturation. Because GnRH neurons become more responsive to the effects of kisspeptin over development (45), we suggest that the increase in Kiss1r-positive cilia per neuron may contribute to this process. Our results also show that ablation of cilia on GnRH neurons affects neither puberty nor adult reproductive function in mice; this is perhaps not surprising, because there is incredible redundancy in the Kiss1 signaling pathway in the brain, which supports reproductive function with only minute quantities of Kiss1 activity (46). In addition, there is evidence for Kiss1r signaling at the nerve terminals (47), which could overcome the loss of Kiss1r ciliary signaling. Given that GnRH release is a function of firing rate, we suggest that ablation of cilia at the cell body leads to a reduction in GnRH release at the nerve terminals but not below the threshold for sexual maturation.
We also demonstrate that cilia are not required for GnRH neuron migration. GnRH neurons originate in the olfactory placode/vomeronasal organ and migrate into the hypothalamus. This migration and targeting is a highly orchestrated process requiring a variety of factors and disruption of GnRH neuron migration is associated with deficits in reproductive function (48). Cilia have recently been shown to influence migration of interneurons of the developing cerebral cortex (49) and neurons migrating from the medial ganglionic eminence (50). We did not observe any alterations in either the number or location of GnRH neurons in newborn or adult GnRHcilia- mice, suggesting that cilia are not necessary for developmental migration of these cells.
Conditional disruption of Ift88 is a well established method for ablating cilia in a cell-type-specific manner (51) and has been effectively used to disrupt cilia on central neurons (32–34). Although we saw a complete lack of Kiss1r-positive cilia in adult GnRHcilia- mice, the fact that GnRH neurons are not positive for any canonical ciliary markers prevented us from validating the loss of cilia structure. Thus, it is feasible that disrupting Ift88 in GnRH neurons prevents Kiss1r ciliary localization without affecting cilia structure. Regardless, our results suggest that Kiss1r ciliary localization is necessary for proper kisspeptin-mediated signaling in male mice. It is also possible that loss of Ift88 has additional functional consequences that impact GnRH neuronal firing. However, it should be noted that the baseline firing rate was the same in GnRHcilia+ and GnRHcilia- neurons.
There are other possible links between cilia function and reproductive maturation. Hypogonadotropic hypogonadism is a typical clinical feature of Bardet-Biedl syndrome, a human ciliopathy. Another putative link is suggested by the finding that WDR11, which is mutated in patients with hypogonadotropic hypogonadism (52), is present within primary cilia (53). However, our results indicate that disruption of GnRH neuronal cilia structure is not sufficient to prevent normal sexual maturation in mice. It is conceivable that differences among species account for the discordant results; however, another possibility is that hypogonadotropic hypogonadism in humans with BBS reflects ciliary dysfunction on either multiple neuronal subtypes or other nonneuronal cells within the reproductive circuit. Alternatively, BBS and WDR11 proteins may have additional nonciliary functions that are required for reproductive maturation and function.
In summary, we have shown that Kiss1r is enriched in cilia on GnRH neurons, thereby implicating neuronal cilia in kisspeptin signaling and reproduction. We also demonstrate that disruption of cilia on GnRH neurons reduces Kiss1r signaling, indicating that cilia enhance Kiss1r signaling. Thus, cilia are likely to play a broad, hitherto unappreciated, role in the function of the brain.
Materials and Methods
Plasmid Construction.
The coding sequence for Kiss1r was amplified from cDNA generated from reverse-transcribed mouse whole brain RNA using the SuperScript First-Strand Synthesis RT-PCR kit (Invitrogen). The coding sequence was then cloned into the TA cloning vector pSTBlue-1 (Novagen), with primers at the C- and N-terminal regions designed for directional cloning. The Kiss1r construct was then subcloned into pEGFP-N (Clontech). All DNA constructs were sequence verified at the Nucleic Acid Shared Resource at Ohio State’s Comprehensive Cancer Center.
Cell Culture and Transient Transfections.
IMCD-3 cells (ATCC) were maintained in DMEM:F12 media supplemented with 10% (vol/vol) FBS, 1.2 g/L of sodium bicarbonate, and 0.5 mM sodium pyruvate (Invitrogen). Cells (n = 5 × 106) were electroporated with 10 µg DNA and plated at high density on glass coverslips. Cells were refed 16–18 h after transfection and harvested at 48 h after transfection by fixation in 4% (wt/vol) paraformaldehyde.
Mice and Tissue Preparation.
All animal procedures described are in accordance with institutional guidelines based on National Institutes of Health Standards, and were performed with Institutional Animal Care and Use Committee approval at the Ohio State University, University of California at San Diego, and the University of Alabama at Birmingham. All animals were maintained in a temperature and humidity controlled vivarium with 12 h light/dark cycle and given access to food and water ad libitum. Littermates were group housed by sex, no more than five to a cage, after weaning. CiliaGFP mice expressed Sstr3::EGFP systemically (27). GnRH::GFP mice were a gift of Suzanne M. Moenter (University of Michigan, Ann Arbor, MI). GnRH::Cre mice were a gift of Catherine Dulac (Harvard University, Cambridge, MA). P0 and P5 mice were anesthetized via isoflurane vapors and then decapitated. The brains were fixed 16–18h at 4 °C in 4% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer and cryoprotected in 30% (wt/vol) sucrose at 4 °C for at least 24 h. Brains from P20 and older mice were isolated and processed as described (13), with the exception that the mice were perfused and tissue was fixed with 4% (wt/vol) paraformaldehyde in 0.1 M phosphate buffer. All brains were sectioned on a freezing microtome at 60 microns. Ovaries and testes were collected from animals after perfusion, postfixed overnight at 4 °C in 4% (wt/vol) paraformaldehyde, and cryoprotected in 30% (wt/vol) sucrose. Testes and ovaries were weighed after cryoprotection. Testes were sectioned at 5 microns and ovaries were sectioned at 10 microns on a cryostat and mounted on Superfrost Plus slides (Fisher Scientific).
Immunofluorescence.
The Kiss1r antibody, an affinity purified rabbit polyclonal generated against amino acids 348–396 of mouse Kiss1r (Strategic Diagnostics), was used at 1:5,000. Mouse monoclonal anti-acetylated α-tubulin (T-6793; Sigma-Aldrich) was used at 1:1,000. Rabbit polyclonal anti-GnRH (Thermo Scientific) was used at 1:1,000. Secondary antibodies included; Alexa Fluor 546-conjugated goat anti-mouse IgG and Alexa Fluor 546-conjugated goat anti-rabbit IgG (Invitrogen). Nucleic acids were stained with DRAQ5 (Axxora). Immunofluorescence procedures have been described (54). Samples were imaged either on a Zeiss LSM 510 laser scanning confocal microscope at the Hunt–Curtis Imaging Facility in the Department of Neuroscience at OSU or an Andor Revolution WD spinning disk confocal imaging system at the Ohio State University Neuroscience Center Imaging Core. On the laser scanning confocal, z-stacks were acquired using 40×/1.3 NA and 63×/1.3 NA Oil DIC objectives and a step size of 0.43 µm. On the spinning disk confocal, z-stacks were acquired using a 100×/1.4 NA Plan Apo VC objective and an iXon Ultra 897 back-illuminated EMCCD camera (512 × 512 pixel array; 160 nm × 160 nm pixels) and a step size of 0.2 µm. 3D volume rendering of the image stacks was performed in MetaMorph software using the 4D viewer. The raw images were first processed using a 3 × 3 low pass “denoising” filter.
Neuron and Cilia Analysis.
Coronal brain sections from P20, P30, and adult animals were matched to the Franklin and Paxinos mouse atlas (55). GFP-positive (GFP+) neurons were examined at the medial septum (MS) on plate 22, rostral preoptic area (rPOA) on plate 27, and anterior hypothalamic area (AHA) on plate 32. Sections for P0 and P5 animals were matched to plates 9 and 10 of the coronal gestation day (GD) 18 atlas from the Schambra, Lauder and Silver prenatal mouse brain atlas (56). For each animal, two sections corresponding to the MS, rPOA, and AHA were analyzed. The number of GFP+ neurons in each region was averaged across animals to calculate the mean and SE. The number of GFP+ neurons in a particular region was determined by examination under the 20× objective. Once this number was recorded, the same region was examined under the 40× objective. Each GFP+ neuron was centered under the objective, and the filter set was switched to the 546 wavelength. If a Kiss1r-positive cilium emanated from the same area as the GFP+ cell body, it was considered positive for a Kiss1r cilium. If more than one cilium emanated from a single GFP+ cell body, the neuron was considered multiciliated. The number of ciliated GFP+ neurons was counted using a manual cell counter. The percentage of ciliated GnRH neurons was calculated by dividing the number of GFP+ neurons possessing one or more cilium by the total number of GFP+ neurons and multiplying by 100. The percentage of multiciliated GnRH neurons was calculated by dividing the number of GFP+ neurons possessing multiple cilia by the number of ciliated GFP+ neurons and multiplying by 100. Statistical analysis was performed using Graphpad Prism (Graphpad Software). Analysis of the percentages of GnRH neurons possessing Kiss1r-positive cilia across age and sex was performed using ANOVA with post hoc tukey test for multiple comparisons. Analysis of the percentages of GnRH neurons possessing Kiss1r-positive cilia between genotypes was performed using Student t test. A P value < 0.05 was considered significant.
Histology.
Hematoxylin/eosin staining of testes and ovaries sections was performed according to the manufacturer’s instructions (BBC Biochemical) kit directions. Stained sections were mounted with Permount (Fisher Scientific) and imaged on a Zeiss LSM 510 laser scanning confocal microscope using an Axiocam and Axiocam software.
Vaginal Opening and Estrous Cycle Determination.
Time of vaginal opening was determined by daily inspection of mice between 9 and 11:00 AM from P20 onward. Estrous cycle samples were collected daily between 9 and 11:00 AM from group housed animals via vaginal flush with 1× PBS. Samples were deposited on Superfrost Plus slides, allowed to dry and stained with a Kwik-Diff kit (Thermo Scientific) according to the manufacturer’s instructions. Slides were allowed to dry and examined on a Zeiss Axioskop 2 MOT light microscope by an individual blinded to the genotypes of the animals. Criteria for the different phases of the estrous cycle have been described previously (57).
Slice Preparation for Electrophysiology.
Procedures were adapted from a previous study (58). Briefly, adult mice (age 6–7 wk) were euthanized by decapitation, and whole brains were removed and immediately submerged in ice cold high-sucrose extracellular solution for 3 min [208 mM sucrose, 2 mM KCl, 26 mM NaHCO3, 10 mM glucose, 1.25 mM NaH2PO4, 2 mM MgSO4, 1 mM MgCl2, 10 mM Hepes, oxygenated (95% O2, 5% CO2), pH 7.4]. Cerebellar tissue was then removed, and 250 micron sagittal slices were cut from the tissue block containing the diagonal band of the preoptical area (DB-POA) using a manual advance vibroslicer (World Precision Instruments) in ice cold high-sucrose extracellular solution (oxygenated). Slices were immediately transferred upon isolation to an auxiliary chamber containing artificial CSF (aCSF) at 25 °C for 1.5–3.5 h before use in electrophysiology experiments [aCSF: 124 mM NaCl, 5 mM KCl, 2.6 mM NaH2PO4, 2 mM MgCl2, 2 mM CaCl2, 26 mM NaHCO3, 10 mM Hepes, 10 mM glucose, oxygenated (95% O2, 5% CO2), pH 7.4].
Electrophysiology Recording and Data Analysis.
Individual GnRH neurons in tissue slices were visually selected for recording by positive detection of GFP expression (Nikon eclipseE600FN epifluorescence microscope with 40× water-immersed objective). Loose cell-attached patch configuration in current-clamp mode (I = 0) was used to record extracellular action potential firing of the selected GnRH neurons. Recording pipettes with 2.5–4.0 MΩ (bath resistance) were filled with aCSF. At time of recording, seal resistance was between 18–100 MΩ. Recordings were obtained using an EPC10 amplifier (HEKA) with Patchmaster software (HEKA); the sampling frequency was 5 kHZ. Artificial CSF (25 °C) was oxygenated with 95% O2, 5% CO2 and perfused over slices at rate of 1.4 mL/min with a peristaltic pump (Peri-Star Pro World Precision Instruments). Basal firing rate was recorded for at least 3 min before treatment with 100 nM KiSS (112-121) Amide (Phoenix Pharmaceuticals) in aCSF for 4.5 min. Data were analyzed with Clampfit 9.2 software (Axon), and average firing rate was determined from the action potential events per time for each condition. “% Change in Firing Rate” was determined from the difference in average firing rates with peptide normalized to the average basal firing rate for each cell. Statistical significance was assessed with a two-tailed T test (unpaired data), a P value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Jackie Domire for contributions to this project, Dr. Tiansen Li [National Institutes of Health (NIH)/National Eye Institute] for the rootletin antibody, and the University of Alabama at Birmingham Hepatorenal Fibrocystic Kidney Disease Core Center (UAB RPKDCC, P30 DK074038) for providing the CiliaGFP mice. This work was supported by NIH/National Institute of General Medical Science research Project Grant R01 GM083120 (to K.M.), NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development research Project Grant R01 HD065856 (to A.S.K.), NIH/Office of Research Infrastructure Programs Shared Instrumentation Grant S10 OD010383 (to A.B.), and NIH/National Institute of Neurological Disorders and Stroke Center Core Grant P30 NS045758.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403286111/-/DCSupplemental.
References
- 1.Ishikawa H, Marshall WF. Ciliogenesis: Building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12(4):222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 2.Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19(13):R526–R535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green JA, Mykytyn K. Neuronal ciliary signaling in homeostasis and disease. Cell Mol Life Sci. 2010;67(19):3287–3297. doi: 10.1007/s00018-010-0425-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci. 2010;123(Pt 4):499–503. doi: 10.1242/jcs.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: Mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197(6):697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Najafi M, Calvert PD. Transport and localization of signaling proteins in ciliated cells. Vision Res. 2012;75:11–18. doi: 10.1016/j.visres.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop GA, Berbari NF, Lewis J, Mykytyn K. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J Comp Neurol. 2007;505(5):562–571. doi: 10.1002/cne.21510. [DOI] [PubMed] [Google Scholar]
- 9.Händel M, et al. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience. 1999;89(3):909–926. doi: 10.1016/s0306-4522(98)00354-6. [DOI] [PubMed] [Google Scholar]
- 10.Hamon M, et al. Antibodies and antisense oligonucleotide for probing the distribution and putative functions of central 5-HT6 receptors. Neuropsychopharmacology. 1999;21(2) Suppl:68S–76S. doi: 10.1016/S0893-133X(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 11.Brailov I, et al. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872(1-2):271–275. doi: 10.1016/s0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- 12.Berbari NF, Johnson AD, Lewis JS, Askwith CC, Mykytyn K. Identification of ciliary localization sequences within the third intracellular loop of G protein-coupled receptors. Mol Biol Cell. 2008;19(4):1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berbari NF, Lewis JS, Bishop GA, Askwith CC, Mykytyn K. Bardet-Biedl syndrome proteins are required for the localization of G protein-coupled receptors to primary cilia. Proc Natl Acad Sci USA. 2008;105(11):4242–4246. doi: 10.1073/pnas.0711027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domire JS, et al. Dopamine receptor 1 localizes to neuronal cilia in a dynamic process that requires the Bardet-Biedl syndrome proteins. Cell Mol Life Sci. 2011;68(17):2951–2960. doi: 10.1007/s00018-010-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loktev AV, Jackson PK. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Reports. 2013;5(5):1316–1329. doi: 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 16.de Roux N, et al. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100(19):10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seminara SB, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 18.Forsythe E, Beales PL. Bardet-Biedl syndrome. Eur J Hum Genet. 2013;21(1):8–13. doi: 10.1038/ejhg.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herbison AE, de Tassigny Xd, Doran J, Colledge WH. Distribution and postnatal development of Gpr54 gene expression in mouse brain and gonadotropin-releasing hormone neurons. Endocrinology. 2010;151(1):312–321. doi: 10.1210/en.2009-0552. [DOI] [PubMed] [Google Scholar]
- 20.Wu TJ, Gibson MJ, Rogers MC, Silverman AJ. New observations on the development of the gonadotropin-releasing hormone system in the mouse. J Neurobiol. 1997;33(7):983–998. doi: 10.1002/(sici)1097-4695(199712)33:7<983::aid-neu9>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Suter KJ, et al. Genetic targeting of green fluorescent protein to gonadotropin-releasing hormone neurons: Characterization of whole-cell electrophysiological properties and morphology. Endocrinology. 2000;141(1):412–419. doi: 10.1210/endo.141.1.7279. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, et al. Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia. 2012;1(1):21. doi: 10.1186/2046-2530-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jennes L, Stumpf WE, Sheedy ME. Ultrastructural characterization of gonadotropin-releasing hormone (GnRH)-producing neurons. J Comp Neurol. 1985;232(4):534–547. doi: 10.1002/cne.902320410. [DOI] [PubMed] [Google Scholar]
- 24.Kozlowski GP, Chu L, Hostetter G, Kerdelhué B. Cellular characteristics of immunolabeled luteinizing hormone releasing hormone (LHRH) neurons. Peptides. 1980;1(1):37–46. doi: 10.1016/0196-9781(80)90033-9. [DOI] [PubMed] [Google Scholar]
- 25.Witkin JW, Silverman AJ. Synaptology of luteinizing hormone-releasing hormone neurons in rat preoptic area. Peptides. 1985;6(2):263–271. doi: 10.1016/0196-9781(85)90050-6. [DOI] [PubMed] [Google Scholar]
- 26.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12(5):767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 27.O’Connor AK, et al. An inducible CiliaGFP mouse model for in vivo visualization and analysis of cilia in live tissue. Cilia. 2013;2(1):8. doi: 10.1186/2046-2530-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anastas SB, Mueller D, Semple-Rowland SL, Breunig JJ, Sarkisian MR. Failed cytokinesis of neural progenitors in citron kinase-deficient rats leads to multiciliated neurons. Cereb Cortex. 2011;21(2):338–344. doi: 10.1093/cercor/bhq099. [DOI] [PubMed] [Google Scholar]
- 29.Dahl HA. Fine structure of cilia in rat cerebral cortex. Z Zellforsch Mikrosk Anat. 1963;60:369–386. doi: 10.1007/BF00336612. [DOI] [PubMed] [Google Scholar]
- 30.Lafarga M, Hervás JP, Crespo D, Villegas J. Ciliated neurons in supraoptic nucleus of rat hypothalamus during neonatal period. Anat Embryol (Berl) 1980;160(1):29–38. doi: 10.1007/BF00315647. [DOI] [PubMed] [Google Scholar]
- 31.Mandl L, Megele R. Primary cilia in normal human neocortical neurons. Z Mikrosk Anat Forsch. 1989;103(3):425–430. [PubMed] [Google Scholar]
- 32.Davenport JR, et al. Disruption of intraflagellar transport in adult mice leads to obesity and slow-onset cystic kidney disease. Curr Biol. 2007;17(18):1586–1594. doi: 10.1016/j.cub.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berbari NF, et al. Leptin resistance is a secondary consequence of the obesity in ciliopathy mutant mice. Proc Natl Acad Sci USA. 2013;110(19):7796–7801. doi: 10.1073/pnas.1210192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chizhikov VV, et al. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27(36):9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon H, Enquist LW, Dulac C. Olfactory inputs to hypothalamic neurons controlling reproduction and fertility. Cell. 2005;123(4):669–682. doi: 10.1016/j.cell.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 36.Skynner MJ, Slater R, Sim JA, Allen ND, Herbison AE. Promoter transgenics reveal multiple gonadotropin-releasing hormone-I-expressing cell populations of different embryological origin in mouse brain. J Neurosci. 1999;19(14):5955–5966. doi: 10.1523/JNEUROSCI.19-14-05955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heger S, et al. Overexpression of glutamic acid decarboxylase-67 (GAD-67) in gonadotropin-releasing hormone neurons disrupts migratory fate and female reproductive function in mice. Endocrinology. 2003;144(6):2566–2579. doi: 10.1210/en.2002-221107. [DOI] [PubMed] [Google Scholar]
- 38.Pielecka-Fortuna J, Chu Z, Moenter SM. Kisspeptin acts directly and indirectly to increase gonadotropin-releasing hormone neuron activity and its effects are modulated by estradiol. Endocrinology. 2008;149(4):1979–1986. doi: 10.1210/en.2007-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirilov M, et al. Dependence of fertility on kisspeptin-Gpr54 signaling at the GnRH neuron. Nat Commun. 2013;4:2492. doi: 10.1038/ncomms3492. [DOI] [PubMed] [Google Scholar]
- 40.Kauffman AS, et al. The kisspeptin receptor GPR54 is required for sexual differentiation of the brain and behavior. J Neurosci. 2007;27(33):8826–8835. doi: 10.1523/JNEUROSCI.2099-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarkson J, Herbison AE. Postnatal development of kisspeptin neurons in mouse hypothalamus; sexual dimorphism and projections to gonadotropin-releasing hormone neurons. Endocrinology. 2006;147(12):5817–5825. doi: 10.1210/en.2006-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalló I, et al. Co-localisation of kisspeptin with galanin or neurokinin B in afferents to mouse GnRH neurones. J Neuroendocrinol. 2012;24(3):464–476. doi: 10.1111/j.1365-2826.2011.02262.x. [DOI] [PubMed] [Google Scholar]
- 43.Iijima N, Takumi K, Sawai N, Ozawa H. An immunohistochemical study on the expressional dynamics of kisspeptin neurons relevant to GnRH neurons using a newly developed anti-kisspeptin antibody. J Mol Neurosci. 2011;43(2):146–154. doi: 10.1007/s12031-010-9433-y. [DOI] [PubMed] [Google Scholar]
- 44.Jenkins PM, McEwen DP, Martens JR. Olfactory cilia: Linking sensory cilia function and human disease. Chem Senses. 2009;34(5):451–464. doi: 10.1093/chemse/bjp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han SK, et al. Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty. J Neurosci. 2005;25(49):11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popa SM, et al. Redundancy in Kiss1 expression safeguards reproduction in the mouse. Endocrinology. 2013;154(8):2784–2794. doi: 10.1210/en.2013-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patterson M, et al. Administration of kisspeptin-54 into discrete regions of the hypothalamus potently increases plasma luteinising hormone and testosterone in male adult rats. J Neuroendocrinol. 2006;18(5):349–354. doi: 10.1111/j.1365-2826.2006.01420.x. [DOI] [PubMed] [Google Scholar]
- 48.Wierman ME, Kiseljak-Vassiliades K, Tobet S. Gonadotropin-releasing hormone (GnRH) neuron migration: Initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front Neuroendocrinol. 2011;32(1):43–52. doi: 10.1016/j.yfrne.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higginbotham H, et al. Arl13b in primary cilia regulates the migration and placement of interneurons in the developing cerebral cortex. Dev Cell. 2012;23(5):925–938. doi: 10.1016/j.devcel.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baudoin JP, et al. Tangentially migrating neurons assemble a primary cilium that promotes their reorientation to the cortical plate. Neuron. 2012;76(6):1108–1122. doi: 10.1016/j.neuron.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 51.O’Connor AK, Kesterson RA, Yoder BK. Generating conditional mutants to analyze ciliary functions: The use of Cre-lox technology to disrupt cilia in specific organs. Methods Cell Biol. 2009;93:305–330. doi: 10.1016/S0091-679X(08)93015-6. [DOI] [PubMed] [Google Scholar]
- 52.Kim HG, et al. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2010;87(4):465–479. doi: 10.1016/j.ajhg.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishikawa H, Thompson J, Yates JR, 3rd, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22(5):414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green JA, Gu C, Mykytyn K. Heteromerization of ciliary G protein-coupled receptors in the mouse brain. PLoS ONE. 2012;7(9):e46304. doi: 10.1371/journal.pone.0046304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franklin KBJ, Paxinos G. 1997. The Mouse Brain in Stereotaxic Coordinates (Academic Press, San Diego) pp xxii, 186 pp.
- 56.Schambra UB, Lauder JM, Silver J. 1992. Atlas of the Prenatal Mouse Brain (Academic Press, San Diego), pp xxix, 327 pp.
- 57.Caligioni CS. Assessing reproductive status/stages in mice. Curr Protoc Neurosci. 2009;4(Appendix):4I. doi: 10.1002/0471142301.nsa04is48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C, Roepke TA, Kelly MJ, Rønnekleiv OK. Kisspeptin depolarizes gonadotropin-releasing hormone neurons through activation of TRPC-like cationic channels. J Neurosci. 2008;28(17):4423–4434. doi: 10.1523/JNEUROSCI.5352-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.