Abstract
Estimating viral diversity in infected patients can provide insight into pathogen evolution and emergence of drug resistance. With the widespread adoption of deep sequencing, it is important to develop tools to accurately calculate population diversity from very large datasets. Current methods for estimating diversity that are based on multiple alignments are not practical to apply to such data.
In this study, the authors report a novel method (Pairwise Alignment Positional Nucleotide Counting, PAPNC) for estimating population diversity from 454 sequence data. The diversity measurements determined using this method were comparable to those calculated by average pairwise difference (APD) of multiply aligned sequences using MEGA5. Diversities were estimated for 9 patient plasma HIV samples sequenced with Titanium 454 technology and by single-genome sequencing (SGS). Diversities calculated from deep sequencing using PAPNC ranged from 0.002 to 0.021 while APD measurements calculated from SGS data ranged proximately from 0.001 to 0.018, with the difference being attributable to PCR error (contributing background diversity of 0.0016 in a control sample). Comparison of APDs estimated from 100 sets of sequences drawn at random from 454 generated data and from corresponding SGS data showed very close correlation between the two methods with R2 of 0.96, and differing on average by about 1% (after correction for PCR error).
The authors have developed a novel method that is good for calculating genetic diversities for large scale datasets from next generation sequencing. It can be implemented easily as a function in available variation calling programs like SAM tools or haplotype reconstruction software for nucleotide genetic diversity calculation.
A Perl script implementing this method is available upon request.
Keywords: HIV-1, Viral population diversity calculation, Next generation sequencing
1. Introduction
Nucleotide diversity is often used to measure the degree of genetic variation in a population. In its original form proposed by Nei and Li, nucleotide diversity is defined by the average nucleotide differences per site between any two sequences (Nei and Li, 1979), i.e., Hamming distance (Hamming, 1950) in a genetics sense. Since then, several models have been developed, including Juke–Cantor, K80, and TN93, to allow for different nucleotide substitution mechanisms (Kumar, 2000; Tamura et al., 2011).
Knowledge of pathogen genetic diversity is important for understanding evolution and persistence of infections in vivo (Kearney et al., 2011). Numerous studies have investigated the diversity of intrapatient HIV-1 populations (Kearney et al., 2011; Nowak et al., 1996; Shankarappa et al., 1999; Troyer et al., 2005; Wolinsky et al., 1996). Shankarappa et al. showed that during the asymptomatic interval of HIV-1 infection, three phases of population genetics were observed. In phase 1, HIV-1 intrapatient population diversity and divergence increased linearly; in phase 2, while divergence continued to increase, diversity either declined or became stable; in phase 3, divergence and diversity were either stable or declined (Shankarappa et al., 1999). Kearney et al. reported that antiretroviral treatment of pigtail macaques infected with RT-SHIVmne did not reduce the viral diversity (Kearney et al., 2011). Troyer et al. suggested that HIV-1 replication efficiency may be related to genome diversity and that diversity may be a determining factor in AIDS disease progression (Troyer et al., 2005).
Genetic diversity of viral populations can be calculated easily with software like Molecular Evolutionary Genetics Analysis (MEGA5, version MEGA5.2.2) (Tamura et al., 2011). MEGA5 is a widely used software for molecular evolution analyses and proximately 390,000 copies have been downloaded worldwide (http://www.megasoftware.net) likely due to its wide array of molecular evolution functions, ease of use, and also its authors, who are well known molecular evolution researchers. However, currently it cannot handle large amounts of sequencing data produced by next generation sequencing. This issue motivated the authors to develop a simple method to calculate genetic diversity from large data sets.
The first step in calculating the nucleotide diversity of a population from a set of sequences is generation of a multiple sequence alignment. Many multiple alignment methods have been developed since the introduction of CLUSTALW in 1994 (Edgar and Batzoglou, 2006; Thompson et al., 1994). But multiple sequence alignment is still computationally intensive and can be very slow. One such program, MUSCLE is recommended for the task of aligning >500 sequences (Edgar and Batzoglou, 2006). A newer version of MUSCLE has improved accuracy but is only applicable to about 200 sequences (Katoh et al., 2005). Multiple sequence alignments generated by these methods require manual review and editing (Nuin et al., 2006) which is not possible with the large numbers of sequences obtained by deep sequencing.
With the widespread use of next generation sequencing technologies, including 454 pyrosequencing, Illumina, SOLiD, and others, large datasets of sequence information are being obtained, making current methods for generating multiple sequence alignments and then calculating genetic diversities impracticable. Many short sequence alignment methods for next generation sequencing have been developed (Li and Homer, 2010). Also, methods for reconstructing viral quasi-species or haplotypes and their frequencies in a population have been reported, for example, ShoRAH (Zagordi et al., 2011), QuRe (Prosperi and Salemi, 2012), QUASR (Watson et al., 2013), and ViSpA (Astrovskaya et al., 2011). Jabara et al. (2011) recently published a study in which they used primer IDs – sequences of 8 random nucleotides to label each input HIV cDNA molecule – and built consensus sequences from the 454 reads that shared an identical primer ID. Those consensus sequences thus represented each member of the HIV-1 quasi-species. It will be interesting to compare the result from this experimental study with the results from computational reconstructions described above.
While certainly viral quasi-species or haplotype distribution is a good measurement of viral population diversity, Hamming distance based viral genetic distance initially defined by Nei and Li (1979) is also useful in measuring population diversities. Indeed, the authors reported that average pairwise diversities of intra-patient HIV-1 populations were correlated with days of post seroconversion in the patients the authors studied (Kearney et al., 2009). Analysis to determine if this genetic diversity can be associated with the outcome of antiretroviral therapy in a clinical trial is underway (data not shown).
To calculate sequence diversity in terms of APD with data obtained from next generation sequencing technology, new methods will be useful. To date, the authors are not aware of any reports of such methods. In this study the authors report a new method that the authors call Pairwise Alignment Positional Nucleotide Counting (PAPNC), which allows calculation of nucleotide sequence diversity from a nucleotide count matrix, which can be generated easily and rapidly from large scale sequencing data.
2. Materials and methods
2.1. Samples and sequencing
Plasma samples were collected from 9 treatment-naïve, HIV-1 subtype B-infected patients participating in a research study at the National Institutes of Health (protocol # 00-I-0110). All participants provided written informed consent for viral genotyping.
HIV-1 RNA was extracted from patient plasma and cDNA was synthesized by reverse transcription (RT) using 0.2 µM of a common gene specific primer (Supplement Table 1), 2.5 mM MgCl2 and 200 U of Superscript III (Invitrogen Cat# 18080–044). Each RT reaction was allowed to proceed for 50 min at 50 °C. The resulting cDNA was quantified by qPCR using SYBR green fluorescence (Core Kit, Applied Biosystems Cat# 4306736) to detect and quantify the amplified products. A minimum of 20,000 copies of cDNA was used as template to amplify a 531 base pair fragment for 25 cycles under the condition of low recombination PCR (Shao et al., 2013) using unique 454 Titanium forward primers (Supplement Table 1). Each forward primer contained a sequencing primer designated “A”, a “key” sequence defined by Roche, a unique multiplex identifier (MID) and an HIV target specific sequence. A common reverse 454 Titanium primer that contained a sequencing primer designated “B”, the same “key” sequence, and an HIV sequence specific target sequence was also used. After amplification, all PCR products were gel purified and quantified by qPCR using the KAPA Library Quantification Kit (product # KK4802). Equal copy numbers of each sample were pooled, and sequenced with 454 Titanium according to the manufacturer’s protocol. HIV plasma diversities obtained by 454 were compared to those calculated by SGS of the same samples (Kearney et al., 2008; Palmer et al., 2005). For control, HIV-1 BH10 RNA transcripts were reverse transcribed and amplified with either standard PCR conditions (MID1) (Shao et al., 2013) or under low recombination PCR condition (MID2) (Shao et al., 2013) as described above for patient samples.
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jviromet.2014.03.008.
Initial 454 sequencing data process and error handling were described previously (Shao et al., 2013). Briefly any sequences less than 300 bases in length were discarded. Sequences from patients were pairwise aligned with blastn in the BLAST program of the 2.2.25 package to the HIV subtype B consensus sequences with the default blastn setting except that DUST filter was turned off, and both the gap opening and extension penalties were set at 2 (-F F -G 2 -E 2; http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html). A Perl script pipeline was constructed to parse the alignments to produce a nucleotide count matrix (Supplement Table 2) (Shao et al., 2013) that was used in diversity calculations. Indels and ambiguous base calls (quality score < Q20) were not used in the analysis.
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.jviromet.2014.03.008.
2.2. Method description
PAPNC is a method based on parsing the nucleotide count matrix (Supplement Table 2), which was produced by parsing pairwise alignment of sequencing reads with a reference sequence for determining the genetic diversity from the number of different nucleotides at each position. This nucleotide count matrix can be produced from an external program and used in PAPNC as well.
Specifically, the nucleotide diversity at site j (Dj) is:
where Aj, Cj, Gj, Tj are the numbers of each base at position j.
In terms of gap (deletion) handling, the authors took the approach of pairwise deletion instead of complete deletion (Tamura et al., 2011). That is, the authors only ignore the gaps involved in the comparison (pairwise deletion), instead of ignoring all the nucleotides at a site where there are gaps (complete deletion).
Diversity at a site is defined as Average Pairwise Distance, as described in the following section.
Perl scripts were written to implement this method to calculate diversity based on the following distance models.
2.2.1. Diversity based on p-distance
Diversity based on the p-distance model is expressed as Eq. (1):
| (1) |
where L is the length of the shorter sequence in a pair of sequences being aligned; J is nucleotide position in an alignment and site_pairs is the number of compared pairs of nucleotide sites in position j. Dj is the pairwise nucleotide distance at position j
where N is the number of total nucleotides at position j (ignoring gaps).
As Eq. (1) indicates, this method is implemented in the following way. Genetic distance (Dj)/site_pairs for each site is calculated and APD is calculated as the sum of Dj/site_pairs of all sites divided by the reference length. The distance models (Eqs. (2)–(4)) implemented as shown below were described by Yang (2006).
2.2.2. Diversity based on Juke–Cantor distance
| (2) |
where p = APD defined in Eq. (1)
2.2.3. Diversity based on K80 distance
| (3) |
where S = transition rate of all nucleotide positions
V = transversion rate of all nucleotide positions
Implementation: S and V are calculated for all aligned sites and used in Eq. (3) for diversity based on K80 distance model.
2.2.4. Diversity based on TN93 distance
| (4) |
where
S1 = transition rate of purine to purine of all nucleotide positions:
S2 = transition rate of pyrimidine to pyrimidine of all nucleotide positions:
V = transition rate of all nucleotide positions
πA, πC, πG, πT = rate of A, C, G, T of all nucleotide positions
Implementation: parameters S1, S2, V, πA, πC, πG, πT, πR, and πY are the sum of all the positions and used in Eq. (4) for diversity calculation based on TN93 model.
2.3. Sequencing error correction
It is known that 454 pyrosequencing of PCR products produce errors including both point mutations and indels at high rates (Gilles et al., 2011; Shao et al., 2013). Indel errors do not affect diversity calculations, but point mutation errors do. To minimize the impact of sequencing error on diversity calculation, two measures were taken: (1) in each 454 sequencing run, a plasmid with the same fragment of a known HIV-1 clone (HIV-1 BH10) identical to the samples was constructed and sequenced together with the samples. This set of sequences MID2 served as a background control; (2) the authors noticed that 454 pyrosequencing produced particular neighboring double mutations that were the result of consecutive insertion and deletion errors. For example,
 |
Which can be interpreted to result from a consecutive indel error:
 |
Double mutations that could be interpreted as the result of consecutive indels were not counted as true differences in diversity calculations.
2.4. Other software used
MEGA5 (version 5.2.2) (Tamura et al., 2011) was used in calculating overall mean distances. The model/method used is described in the text; pairwise deletion was selected for Gap/missing data treatment; all other parameters were default. Welch T test implemented in the Graphpad online application (http://www.graphpad.com/quickcalcs) was used for statistical analysis.
3. Results
3.1. Sequence data from 454 pyrosequencing and SGS
The authors performed both 454 pyrosequencing and single-genome sequencing (SGS) on the HIV-1 RT region starting approximately at codon 52 from HIV-1 genomes isolated from plasma of 9 patients. The number of SGS sequences obtained ranged from 12 (patient 1) to 46 (patient 3) (Table 1). The number of 454 sequences used for the analysis (longer than 300 bases) ranged from 1735 (patient 2, MID23) to 8097 (patient 1, MD22) (Table 1). The numbers of 454 reads from those patients were low, most likely due to low viral loads in those patients and due to the PCR conditions (25 cycles) used to reduce the artifactual recombination rate mediated by PCR (Shao et al., 2013) (noting that the diversity calculations are unaffected by recombination). In the same experimental run, 197,931 sequences from HIV-1 BH10 RNA transcripts (MID1) were obtained, which were prepared using standard PCR conditions (Shao et al., 2013). To control for overall background error, the same transcripts were amplified with low recommendation PCR conditions (identified as MID2) (Shao et al., 2013) (Table 1). Sequences obtained from 454 pyrosequencing and by SGS of the same samples were used to develop and validate PAPNC for calculating HIV diversity in patient plasma samples. For diversity calculations in this study, all the sequences from SGS and 454 were trimmed to 300 bases (from the last nucleotide of codon 51 through the first 2 nucleotides of codon 151 of RT). The authors focus explicitly on calculating genetic diversity with PAPNC which requires an input of nucleotide count matrix (an example of the matrix is shown in Supplement Table 2) which can be obtained by parsing pairwise alignment. But it can be also obtained by further transforming the variation detection output produced by software like SAM tools (Li et al., 2009), or VarScan (Koboldt et al., 2009).
Table 1.
Numbers of SGS and 454 sequences obtained and analyzed.
| PID | Number of SGS sequencesb |
454 ID | Number of 454 sequencesc |
|---|---|---|---|
| Controla | MID2 | 976 | |
| 1 | 12 | MID22 | 8097 |
| 2 | 22 | MID23 | 1735 |
| 3 | 46 | MID24 | 4895 |
| 4 | 37 | MID25 | 7168 |
| 5 | 23 | MID26 | 2997 |
| 6 | 23 | MID28 | 1811 |
| 7 | 23 | MID29 | 5176 |
| 8 | 22 | MID30 | 3997 |
| 9 | 24 | MID31 | 1975 |
Control: 100% wild type transcripts from cloned HIV-1, BH10 strain.
Number of SGS sequences obtained from each patient plasma sample.
Number of 454 Sequences obtained from each sample.
3.2. Validation of PAPNC with SGS sequences
PAPNC was implemented as a part of a Perl script and used to calculate nucleotide sequence diversities of the above described patient samples. Among various distance models used for diversity calculations, average pairwise p-distance (APD) is the most commonly used. To validate PAPNC, the diversities of HIV-1 sequences obtained by SGS from the 9 patients were calculated firstly. Diversities were calculated both by using PAPNC method and using MEGA5 based on a p-distance model (Table 2). Diversities obtained from PAPNC and MEGA5 were well correlated. The differences between these two methods ranged from 0 (patients 1, 5, 6 and 9) to 0.002 (patient 7). Welch T test showed that there was no significant difference between the numbers obtained with PAPNC and MEGA5 (p = 0.81). The authors then used SGS results to compare the diversities based on Juke–Cantor, K80, and TN93 models when calculated by PAPNC and MEGA5 (Table 3). Comparable diversities for all patients regardless of the model used were obtained.
Table 2.
Comparison of p-distance based diversities of single-genome sequences (SGS) measured by p-distance with Mega5 and with PAPNC.a,b
| PID | MEGA5 APD | PAPNC | Difference |
|---|---|---|---|
| 1 | 0.0043 | 0.0043 | 0.0000 |
| 2 | 0.0113 | 0.0107 | 0.0006 |
| 3 | 0.0065 | 0.0057 | 0.0008 |
| 4 | 0.0202 | 0.0183 | 0.0019 |
| 5 | 0.0111 | 0.0111 | 0.0000 |
| 6 | 0.0015 | 0.0014 | 0.0000 |
| 7 | 0.0115 | 0.0094 | 0.0021 |
| 8 | 0.0041 | 0.0038 | 0.0003 |
| 9 | 0.0120 | 0.0120 | 0.0000 |
Same SGS sequence data calculated with both Mega5 and PAPNC.
Welch T test, two-tailed P = 0.81.
Table 3.
Comparison of diversity by Jukes–Cantor, K80, and TN93 models of single-genome sequences (SGS) calculated using MEGA5 and PAPNC.a
| SGS samples | MEGA5 | PAPNC | ||||
|---|---|---|---|---|---|---|
| JC | K80 | TN93 | JC | K80 | TN93 | |
| 1 | 0.0044 | 0.0044 | 0.0044 | 0.0044 | 0.0044 | 0.0044 |
| 2 | 0.0114 | 0.0114 | 0.0011 | 0.0108 | 0.0108 | 0.0108 |
| 3 | 0.0066 | 0.0066 | 0.0066 | 0.0057 | 0.0057 | 0.0057 |
| 4 | 0.0206 | 0.0207 | 0.0208 | 0.0185 | 0.0185 | 0.0187 |
| 5 | 0.0113 | 0.0113 | 0.0114 | 0.0112 | 0.0112 | 0.0112 |
| 6 | 0.0015 | 0.0015 | 0.0015 | 0.0015 | 0.0015 | 0.0015 |
| 7 | 0.0115 | 0.0115 | 0.0116 | 0.0095 | 0.0095 | 0.0095 |
| 8 | 0.0041 | 0.0041 | 0.0041 | 0.0038 | 0.0038 | 0.0044 |
| 9 | 0.0121 | 0.0121 | 0.0122 | 0.0121 | 0.0121 | 0.0121 |
Same SGS sequence data calculated with both MEGA5 and PAPNC.
3.3. Impact of sequencing errors on diversity calculation
The results shown above demonstrated that the diversities calculated with PAPNC are comparable to the diversities calculated with MEGA5. It is known that SGS produces high quality sequencing reads with low PCR error (Palmer et al., 2005). Sequencing errors affect diversity calculations. It is also known that 454 pyrosequencing results in a high frequency of sequencing errors (Gilles et al., 2011; Shao et al., 2013). To measure the effect of 454 sequencing errors on diversity calculations, the authors studied the types and distributions of 454 sequencing errors.
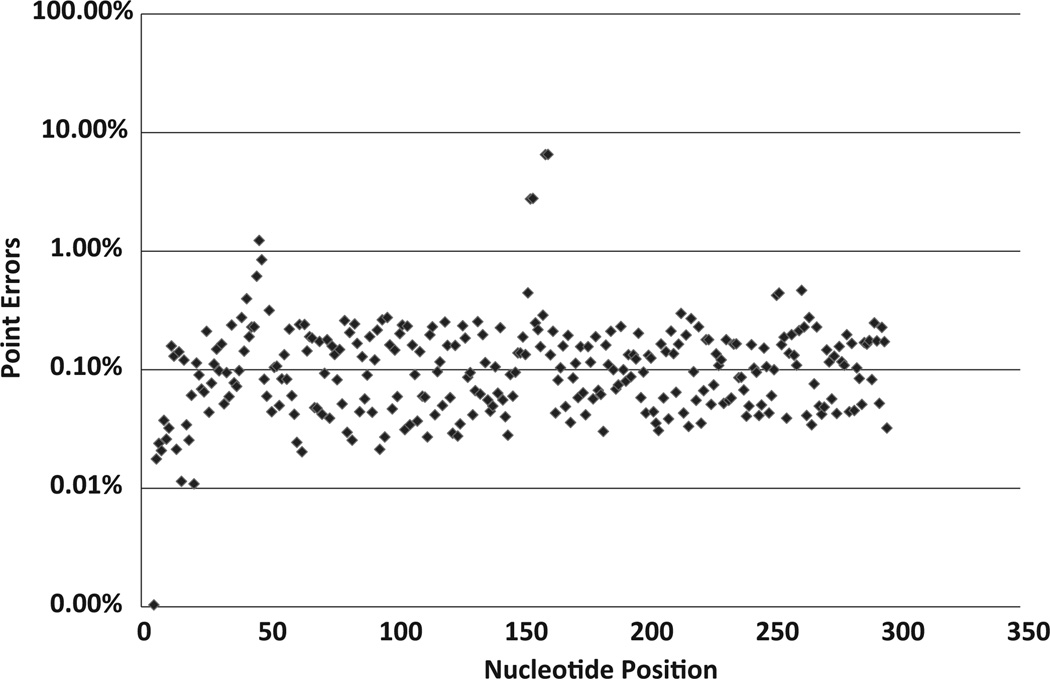
Generally, the authors observed that the majority of the point mutation errors were below 1% and the average point error was 0.18% per nucleotide position (Fig. 1). However, there were some positions with point error rates higher than 1%. The highest error rate was 6.5% at codon 103, which is in an A-rich region (AAAAAGAAAAAA) (Shao et al., 2013).
Fig. 1.
Point mutation error distribution in 454 sequences derived from cloned BH10 transcripts (MID1). The X-axis gives the nucleotide position of the sequences, with position 1 corresponding to the last nucleotide of codon 51 of RT. The Y-axis shows the percentage of point mutation errors at each position on a log scale.
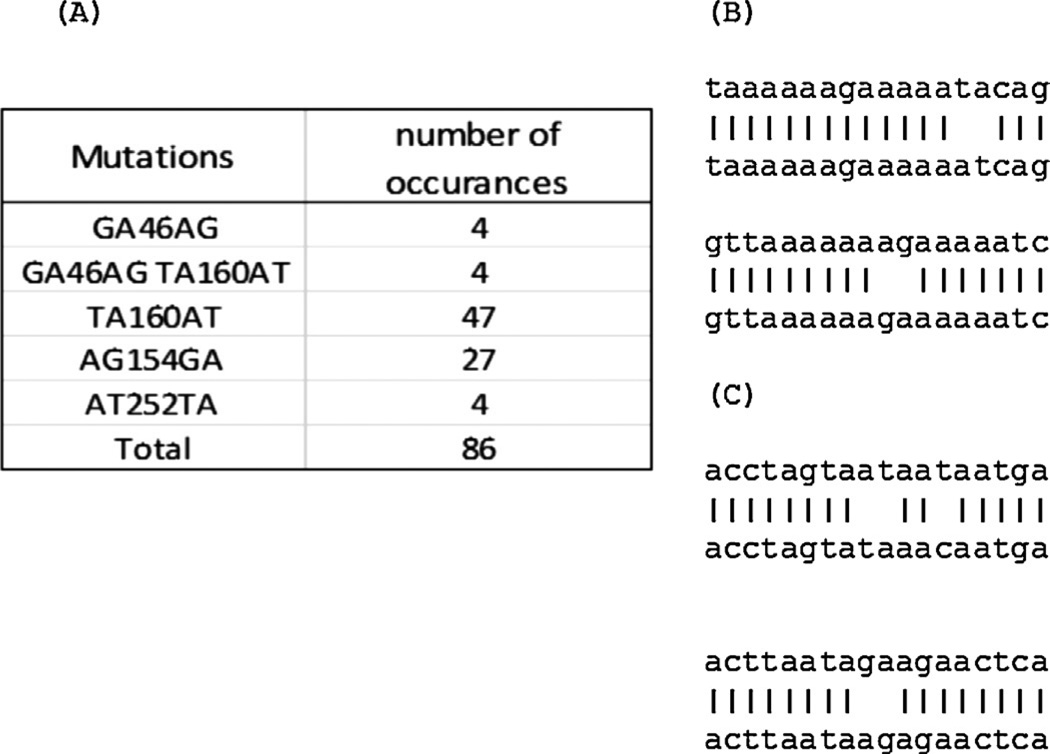
In addition to point mutations, which were controlled by using a control run (Shao et al., 2013). The authors also observed frequent “double mutations”, illustrated by the example in Fig. 2A. An “A” in the reference sequence at position 46 became a “G” in the sample sequence, and a “G” at position 47 in the reference sequence became an “A” in the sample sequence. Careful examination revealed that this double mutation was very likely caused by a deletion and an insertion in homopolymer runs flanking the affected bases, in this case, a deletion after position 45 and an insertion between positions 46 and 47 (Fig. 2B). Very few double mutations were observed in SGS sequences but they were frequently observed in 454 sequences. The diversities of SGS sequences calculated without discounting double mutations and the diversities of SGS sequences calculated after discounting double mutations were the same (compare Table 2 and Table 4). This analysis implies that the double mutations in 454 sequences were artifacts produced in the 454 sequencing process and did not occur in vivo. The authors found that such double mutation errors were generated at high frequencies by 454 sequencing. For example, in MID2, approximately 10% of sequences had such errors. Interestingly, these double mutations had symmetric patterns, for example, AT became TA, GA became AG (Fig. 3A). The most frequent double mutations the authors studied were TA → AT or GA → AG in an A-rich region of RT (Fig. 3B). It should be noted that while our method does not count the difference between a sample sequence and a reference sequence for diversity calculation, the selection of the reference may slightly affect nucleotide mapping in regions with indels and also this double mutation error detection. In this study, the authors used the consensus sequence of HIV-1 subtype B as the reference sequence. The diversity calculated using the consensus sequences of each patient produced by SGS was not identical, but the difference is very small, for example, the diversity of patient 1 (MID22) is 0.0100 (Table 4) with HIV-1 subtype B consensus and 0.0088 (data not shown) with the consensus of SGS sequences from this patient.
Fig. 2.
Double mutation errors caused by nearby indels. (A) A double mutation; (B) interpretation of the double mutation. The apparent double mutation is in italics and underlined.
Table 4.
| 454 samples | PID | 454 APD without correction | 454 APD corrected from double mutations | APD from MID2 (control) | 454 APD corrected from control | SGS APD |
|---|---|---|---|---|---|---|
| MID22 | 1 | 0.0100 | 0.0082 | 0.0016 | 0.0066 | 0.0043 |
| MID23 | 2 | 0.0184 | 0.0167 | 0.0016 | 0.0151 | 0.0107 |
| MID24 | 3 | 0.0098 | 0.0097 | 0.0016 | 0.0081 | 0.0057 |
| MID25 | 4 | 0.0248 | 0.0225 | 0.0016 | 0.0209 | 0.0183 |
| MID26 | 5 | 0.0127 | 0.0125 | 0.0016 | 0.0109 | 0.0111 |
| MID28 | 6 | 0.0061 | 0.0038 | 0.0016 | 0.0022 | 0.0014 |
| MID29 | 7 | 0.0160 | 0.0150 | 0.0016 | 0.0134 | 0.0094 |
| MID30 | 8 | 0.0085 | 0.0068 | 0.0016 | 0.0052 | 0.0038 |
| MID31 | 9 | 0.0179 | 0.0165 | 0.0016 | 0.0149 | 0.0120 |
Patient 454 sequences and MID2 control sequences were aligned with HIV-1 subtype B consensus sequence.
Welch’s T test, two-tailed between 454 APD without correction and SGS APD, p = 0.064; between 454 APD corrected for background and SGS APD, p = 0.400.
Fig. 3.
Double mutation distribution in cloned BH10 (MID2). (A) Double mutation patterns in MID2. The numbers are the nucleotide position of the inner mutation. The nucleotides before the position numbers are two nucleotides in MID sequences. The nucleotides after the position numbers are two corresponding nucleotides in the reference sequence; (B) examples of double mutations in a homopoly A region; and (C) examples of double mutations in a non-homopoly A region.
Table 4 shows the effect of sequencing errors on the diversity calculation. For each patient sample, SGS and 454 sequencing was performed and the viral diversity calculated with PAPNC without correcting for the 454 sequencing errors. The results show that there were differences between diversities calculated using 454 sequences and single-genome sequences for many of the patients even when both were calculated with PAPNC. For example, for patient 1, the diversity obtained from 454 sequences was 0.0100 and the diversity from SGS was approximately half of that at 0.0043. However, in some cases the differences were smaller. For example, for patient 9, the diversity from 454 was 0.0179 and from SGS was 0.0120. A Welch T test shows that the difference between the diversities obtained with 454 and with SGS when sequencing errors was not accounted for was not significant (p = 0.064). Nevertheless, in all cases the APD estimated from the 454 sequences were higher than that for SGS.
3.4. Diversity calculated from 454 sequences after sequencing error correction
As a part of the PAPNC method, the two types of errors that were identified in the 454 sequences were corrected. First, the authors eliminated the double mutations errors from the difference counting in the diversity calculation by replacing them with gaps. This correction reduced the diversities calculated from the 454 sequences (Table 4). For example, without any error correction, the diversity of 454 sequences from patient1 was 0.0100 (Table 4), dropping to 0.0082 after the double mutation correction (Table 4). Similarly, the uncorrected diversity from 454 sequences in patient 5 was 0.0127 (Table 4) and it became 0.0125 after the double mutation correction (Table 4). The authors next corrected the diversity estimated from 454 sequencing for point mutation errors arising during PCR and sequencing. The authors calculated the APD derived from transcripts prepared from the control HIV-1 BH10 clone (MID2). Any diversity in this sample must be due entirely to reverse transcription, PCR and/or sequencing errors and this value was used as our “background diversity”. After correcting for the background diversity (0.0016, Table 4), the in vivo diversities calculated from 454 and SGS generated data became closer, dropping from 20% to 7.8% averaged over all samples. A Welch T test showed that there were no significant differences between the 454 diversities and SGS diversities when sequencing errors were so corrected (two tailed p = 0.400).
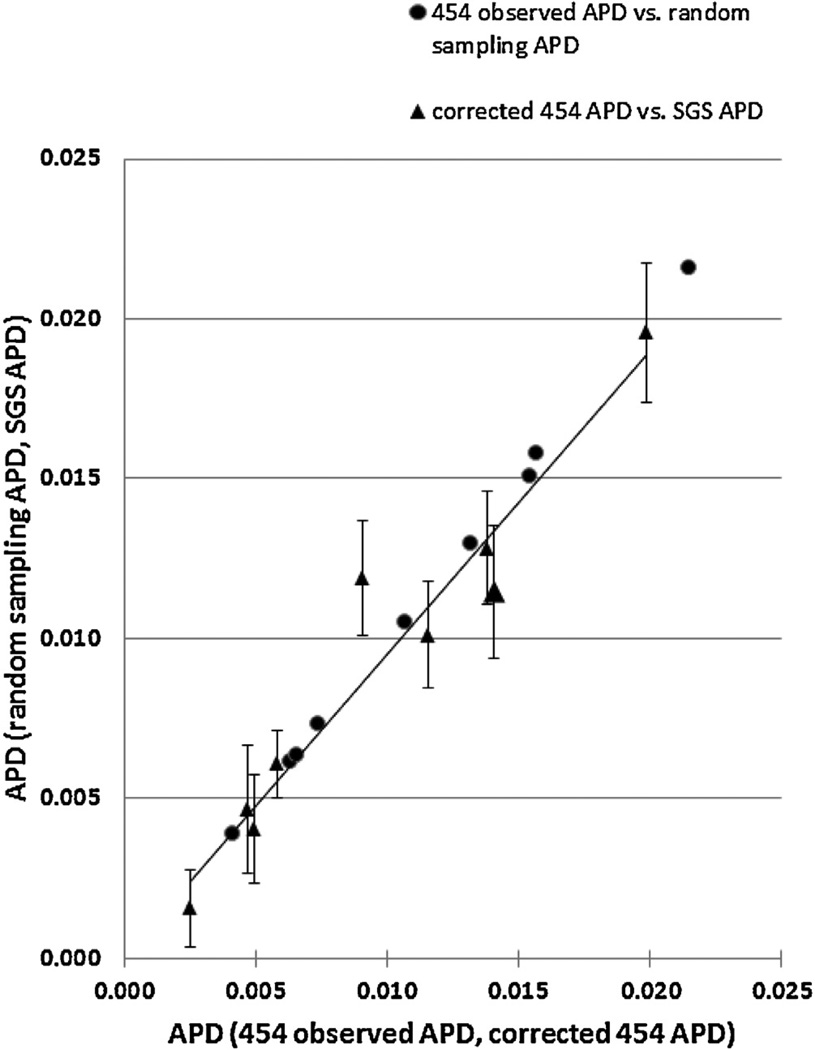
To study the diversity correlation between SGS and 454 further, the average diversity from 100 rounds of randomly picked 454 sequences for each sample were determined. The number of sequences chosen for each round was exactly the same as the number of SGS sequences obtained from that patient. For instance, there were 12 SGS sequences from patient 1; the authors calculated the average of diversities from 100 sets of 12 randomly selected 454 sequences. The results showed that there was good correlation between the average diversities from randomly picked 454 sequences and those from SGS (R2 = 0.9514, Fig. 4).
Fig. 4.
Correlation between SGS and corrected 454 diversities. Both X-axis and Y-axis are APD. The series represented by triangles is corrected 454 diversity vs. SGS diversity; the series represented by circles is 454 diversity corrected from double mutations vs. corrected diversity estimated from the average from 100 random sampling from 454. 454 observed APD and corrected 454 APD are on X-axis, and random sampling 454 APD and SGS APD are on Y-axis. The error bars are the standard deviations of random sampling 454 APD.
3.5. Diversity calculation from large numbers of sequences
Finally, the sequencing reads from MID1, a large scale dataset were used for diversity calculation (Supplement Table 1). MID1 was 100% wild type HIV-1 BH10 transcript RNA that was prepared with standard PCR conditions. From this sample, 197,931 sequencing reads with the length equal or longer than 300 bases were obtained. In this sample, 19,684 reads had at least one double mutation, accounting for approximately 10% of all the sequencing reads of this sample, similar to that in MID2. Table 5 shows the diversities calculated based on the 4 different distance models. After correcting for double mutations, diversities of 0.0027 based on p-distance and Juke–Cantor models, and 0.0025 based on K80 and TN93 models were obtained. Because this sample was a 100% wild type HIV-1 BH10 clone, this diversity was entirely contributed by background PCR and sequencing errors. Additionally, the authors also calculated diversity without correcting for double mutations. The authors obtained a value of 0.0040 based on p-distance and Juke–Cantor models and 0.0037 from K80 and TN93 models (Table 5). Therefore, double mutation errors alone contributed about 30–40% of the background diversity (0.0015 for p-distance and Juke–Cantor models, and 0.0012 for K80 and TN93 models), far in excess of what would be expected from the single point mutation frequency.
Table 5.
Diversities of MID1 using different distance models and the effect of double mutations on diversity calculation.
| Diversity by different distance models |
Without double mutations |
With double mutations |
|---|---|---|
| p-Distance | 0.0027 | 0.0040 |
| Juke–Cantor | 0.0027 | 0.0040 |
| K80 | 0.0025 | 0.0037 |
| TN93 | 0.0025 | 0.0037 |
4. Discussion
Nucleotide diversity defined initially by Nei and Li (1979) has been used to measure the genetic variations in viral populations (Kearney et al., 2011). Nucleotide diversities of sequences up to hundreds or thousands can be calculated easily with software like MEGA5 (Tamura et al., 2011). However, with wide usage of next generation sequencing including 454, Illumina, and SOLiD, the currently available software are not able to handle the huge number of the sequences those new technologies produce. To this end, the authors developed this new method PAPNC. The authors conclude that that the results from PAPNC are comparable to those calculated using MEGA5. The small differences between the results of PAPNC and MEGA5 are likely due to the calculation procedures. Normally, average pairwise diversity (APD) is calculated by firstly comparing every pair of sequences and counting the nucleotide differences based on multiple alignments. The sum of all the differences is then divided by the number of comparison pairs and then by the sequence length. With PAPNC, the authors calculated APD firstly by calculating diversity at each nucleotide site j (Dj) based on pairwise alignments to a reference sequence. Dj is then divided by the total number of pairs of nucleotides at position j. The sum of Dj is then divided by the sequence length (see Section 2.2.1 for details). Additionally, gap handling may be different. Notice that the nucleotide site-pairs at position j exclude gaps from this position. Therefore, the value of nucleotide site-pairs may vary at different positions. This probably is not true with MEGA5. Additionally, there might be sampling errors using SGS because fewer genomes were sequenced. Larger sets of data are much more accurate.
In terms of computation time, for PAPNC itself, the input is a nucleotide count matrix derived from sequencing reads (Supplement Table 2). The time needed to compute and process this matrix and calculate genetic diversities is less than one second for the matrix from a dataset of 197,931 sequences. The authors implemented it as a function in a 454 script pipeline. However, nucleotide count matrix needed for PAPNC can be easily obtained from transforming the output of mpileup function of SAM tools (Li et al., 2009) and SNP calling software like VarScan (Koboldt et al., 2009), or it can be added as an additional function for viral quasi-species reconstruction software to calculate not only the frequencies of individual members of a viral population quasi-species, but also the nucleotide genetic diversity of the population. However, precautions need to be taken if using SNP calling programs that are not specifically designed for retrovirus sequences because many of the SNP calling programs including SNPMix (Goya et al., 2010) treat a third allele at a given position as an error and discard it. The third allele in HIV can be biologically important. For example in codon 103 of HIV-1, the wild type is usually AAA (K), and the two common drug resistance mutations AAC and AAT (both encoding N), are often found together (Boltz et al., 2010). Additionally, the conventional definition of SNP being at least 1% is not suitable for HIV-1 studies because rare drug resistance mutation can be very important (Boltz et al., 2010).
In conclusions, the authors have developed a novel computational method, PAPNC, for determining population diversity from large scale next generation sequencing data. The authors have validated this method by comparing diversities calculated from single-genome sequences and those from 454 sequences using PAPNC. Accurate calculation requires that a background control using a cloned version of a related sequence be included for each 454 pyrosequencing run to correct for the contribution of diversity introduced by RT, PCR and 454 sequencing errors, and that sources of system-specific error, such as double mutations introduced by nearby 454-specific indel pairs, be determined and corrected. Only with such appropriate corrections can next generation sequencing data be used to provide accurate estimates of diversity of populations of sequences.
Supplementary Material
Acknowledgments
The authors thank Ann Wiegand and Natalia Volfovsky for useful discussions; Claudia Stewart and Alex Levitsky for 454 sequencing and data processing; Elizabeth Anderson for helping with the construction of figures, and Connie Kinna, Susan Jones, and Sue Toms for administrative support. Funding for this research was provided by the National Cancer Institute’s intramural Center for Cancer Research and in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. JMC was a research professor of the American Cancer Society, with support from the George Kirby Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Astrovskaya I, Tork B, Mangul S, Westbrooks K, Mandoiu I, Balfe P, Zelikovsky A. Inferring viral quasispecies spectra from 454 pyrosequencing reads. BMC Bioinform. 2011;12(Suppl. 6):S1. doi: 10.1186/1471-2105-12-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boltz VF, Maldarelli F, Martinson N, Morris L, McIntyre JA, Gray G, Hopley MJ, Kimura T, Mayers DL, Robinson P, Mellors JW, Coffin JM, Palmer SE. Optimization of allele-specific PCR using patient-specific HIV consensus sequences for primer design. J. Virol. Methods. 2010;164:122–126. doi: 10.1016/j.jviromet.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC, Batzoglou S. Multiple sequence alignment. Curr. Opin. Struct. Biol. 2006;16:368–373. doi: 10.1016/j.sbi.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Gilles A, Meglecz E, Pech N, Ferreira S, Malausa T, Martin JF. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goya R, Sun MG, Morin RD, Leung G, Ha G, Wiegand KC, Senz J, Crisan A, Marra MA, Hirst M, Huntsman D, Murphy KP, Aparicio S, Shah SP. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–736. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming RW. Error detecting and error correcting codes. Bell Syst. Tech. J. 1950;29:147–160. [Google Scholar]
- Jabara CB, Jones CD, Roach J, Anderson JA, Swanstrom R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20166–20171. doi: 10.1073/pnas.1110064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 2005;16:22–33. [PubMed] [Google Scholar]
- Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 2009;83:2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Palmer S, Maldarelli F, Shao W, Polis MA, Mican J, Rock-Kress D, Margolick JB, Coffin JM, Mellors JW. Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS. 2008;22:497–501. doi: 10.1097/QAD.0b013e3282f29478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Spindler J, Shao W, Maldarelli F, Palmer S, Hu SL, Lifson JD, KewalRamani VN, Mellors JW, Coffin JM, Ambrose Z. Genetic diversity of simian immunodeficiency virus encoding HIV-1 reverse transcriptase persists in macaques despite antiretroviral therapy. J. Virol. 2011;85:1067–1076. doi: 10.1128/JVI.01701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MNS. Molecular Evolution and Phylogenetics. 1st ed. Oxford, USA: Oxford University Press; 2000. [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Anderson RM, Boerlijst MC, Bonhoeffer S, May RM, McMichael AJ. HIV-1 evolution and disease progression. Science. 1996;274:1008–1011. doi: 10.1126/science.274.5289.1008. [DOI] [PubMed] [Google Scholar]
- Nuin PA, Wang Z, Tillier ER. The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinform. 2006;7:471. doi: 10.1186/1471-2105-7-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosperi MC, Salemi M. QuRe: software for viral quasispecies reconstruction from next-generation sequencing data. Bioinformatics. 2012;28:132–133. doi: 10.1093/bioinformatics/btr627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang XL, Mullins JI. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Boltz VF, Spindler JE, Kearney MF, Maldarelli F, Mellors JW, Stewart C, Volfovsky N, Levitsky A, Stephens RM, Coffin JM. Analysis of 454 sequencing error rate, error sources, and artifact recombination for detection of low-frequency drug resistance mutations in HIV-1 DNA. Retrovirology. 2013;10:18. doi: 10.1186/1742-4690-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J. Virol. 2005;79:9006–9018. doi: 10.1128/JVI.79.14.9006-9018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SJ, Welkers MR, Depledge DP, Coulter E, Breuer JM, de Jong MD, Kellam P. Viral population analysis and minority-variant detection using short read next-generation sequencing. Philos. Trans. R. Soc. Lond. B: Biol. Sci. 2013;368:20120205. doi: 10.1098/rstb.2012.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinsky SM, Korber BT, Neumann AU, Daniels M, Kunstman KJ, Whetsell AJ, Furtado MR, Cao Y, Ho DD, Safrit JT. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- Yang Z. Computational Molecular Evolution. Oxford, USA: Oxford Series in Ecology and Evolution, Oxford University Press; 2006. [Google Scholar]
- Zagordi O, Bhattacharya A, Eriksson N, Beerenwinkel N. ShoRAH: estimating the genetic diversity of a mixed sample from next-generation sequencing data. BMC Bioinform. 2011;12:119. doi: 10.1186/1471-2105-12-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.