Abstract
The APOE genotype is a known susceptibility factor for Alzheimer’s disease (AD). It is apparent that the presence of the APOE ε4 allele increases the risk for developing AD, lowers the age of onset in AD, and may influence the pathological burden seen in AD. In this study, we asked whether BACE1 levels differ by APOE genotype in the AD and non-demented (ND) brain. We isolated mid-frontal cortex (MFC) and mid-temporal cortex (MTC) from postmortem ND and AD subjects that were APOE ε3/3, ε3/4, ε4/4 carriers. All AD subjects met NINDS-ADRDA and NIA-Reagan criteria for a diagnosis of AD. The MFC and MTC were homogenized and the lysates underwent ELISA and Western blotting for BACE1. The ELISA revealed that total BACE1 levels were lower in the MFC of AD compared to ND subjects. Furthermore, in APOE ε4 carriers BACE1 levels were lower than ε3/3 carriers in the ND frontal cortex. No difference in BACE1 levels was observed in AD MFC and in ND and AD MTC tissues. The ELISA results were confirmed by Western blotting. Our data suggest that brain BACEl levels may be influenced by the apolipoprotein E genotype before the onset of AD, providing an alternative explanation for the lower amyloid beta 42 levels in CSF in ND and AD subjects.
Keywords: Alzheimer’s disease, APOE, BACE1, Brain, ELISA, Frontal cortex
INTRODUCTION
Alzheimer’s disease (AD) is the most prevalent form of dementia worldwide. One of the histological hallmarks of AD is the accumulation of amyloid beta (Aβ) peptides into extracellular dense-core senile plaques [1]. Amyloidogenesis results from the successive proteolysis of the membrane-tethered amyloid precursor protein (APP). First, APP is cleaved by a β-secretase, releasing a large soluble fraction of APP into the milieu. Second, the remaining APP C-terminal segment is cleaved in its trans-membrane domain by the γ-secretase complex to release extracellular Aβ peptides of different lengths [2], and an APP intracellular domain (AICD) which may act as a transcription regulator [3]. Recent evidence from transgenic animal and in vitro studies suggests that beta-site APP-cleaving enzyme 1 (BACE1) is the major β-secretase in the brain [4, 5] and the rate-limiting enzyme in amyloidogenesis [6, 7]. Several reports also suggested that BACE1 levels and activity are higher in the AD brain versus normal controls (Table 1), though these studies never included apolipoprotein E (APOE) alleles as confounding factors.
Table 1.
Summary of the main literature on brain BACE1 protein levels in Alzheimer’s disease post-mortem specimens.
| Reference | Technique (antibody used, BACE1 epitope)* | Brain Region | Observations |
|---|---|---|---|
| Holsinger et al., 2002 [8] | WB (anti-serum 00/6, C-term) | Frontal Cortex | AD (n=10) > ND (16) |
| Fukumoto et al., 2002 [9] | WB and ELISA (MAB5308, C-term; PA1-756, N-term) | Frontal Cortex | AD (52) > ND (22) |
| Temporal Cortex | AD (61) > ND (18), but not significant | ||
| Cerebellum | AD (57) = ND (26) | ||
| Li et al., 2004; Yang et al., 2003 [10, 11] | WB and ELISA (SECB1, ectodomain; SECB2, ectodomain) | Frontal Cortex | AD (39) > ND (40) |
| Hippocampus | AD (8) > ND (8) | ||
| Zhao et al., 2007 [12] | WB (BACE1-Cat, ectodomain) | Temporal Cortex | AD (3) > ND (3) |
| Ahmed et al., 2010 [13] | WB and ELISA (MAB5308, C-term; MBA931, ectodomain) | Frontal Cortex | AD (12) > ND (11) |
See Gonzales et al., 2011 [14], for antibody details and possible technical limitations; WB: Western blotting.
To date, the best susceptibility factor associated with sporadic AD is the APOE allele 4 (ε4). The APOE ε4 allele is inherited as one of three APOE alleles, termed ε2, ε3, and ε4, which have mean frequencies in the general population of 8%, 78%, and 14%, respectively [15]. Surprisingly, approximately 50% of Alzheimer’s disease patients are APOE ε4 allele carriers, and the degree of risk of dementia conferred by the APOE ε4 allele rises in a gene dose dependent manner [16], increasing with the number of APOE ε4 alleles inherited. APOE ε4 carriers have higher risks of developing sporadic AD [17] than non-carriers who tend to get the disease at a later age of onset [18]. Furthermore, the parenchymal and vascular amyloid neuropathology is greater in non-demented APOE ε4 carriers than in non-carriers [19].
Two mechanisms have been proposed to explain the high incidence of sporadic AD in individuals carrying the ε4 allele [20, 21]: 1- decreased clearance of Aβ out of the brain via the blood-brain barrier due to a lower Aβ-binding capability of ApoE4 versus ApoE2 and ApoE3 proteins; and 2- increased production of Aβ. The latter hypothesis is supported, in part, by the finding that lipid-poor ApoE4 protein increases the synthesis of Aβ in an APP-transfected neuronal cell line [22]. However, no link between APOE ε4 allele and amyloidogenesis enzymes has been reported in the brain. On the other hand, APOE ε4 has been included as a confounding factor in the context of CSF BACE1 levels and activity, which revealed no difference between ND and AD patients [23]. In the present study we asked whether brain BACE1 levels differ in ND and AD subjects carrying ε3/3, ε3/4, and ε4/4 APOE alleles. To answer this question, we used the immunoassay for total BACE1 levels that we have recently developed [14].
MATERIALS AND METHODS
Subjects
Study subjects were from the Banner Sun Health Research Institute (BSHRI) Brain Donation Program (BDP) which has been described in detail elsewhere [24]. In order to participate in the BDP, subjects sign Institutional Review Board-approved informed consents and undergo medical, neurologic, and neuropsychologic assessments. Outside medical records from primary care physicians, neurologists and other specialists are also reviewed extensively. For the present clinico-neuropathologic study, the BDP database was queried to identify all participants with Alzheimer’s disease by NINCDS-ADRDA ciriteria [25]. Subjects bearing any mutation in proteins of the amyloidogenesis cascade (such as APP, BACE1, presenilin1, etc) were excluded from the analysis. ND subjects were not suffering any other neurodegenerative disorder. The ND sample population was enriched in APOE4 carriers compared to the general population to explore an eventual APOE effect on brain BACE1 levels.
Neuropathological Assessment
All autopsies were performed by a certified neuropathologist at the BSHRI. Brain tissue is processed for neuropathological examination in a standardized protocol as previously described [24]. Briefly, paraffin blocks containing brain tissue were cut at 5 μm intervals and stained with hematoxylin-eosin for histological analysis. Additional paraffin sections containing tissue from the olfactory bulb, anterior medulla, anterior and mid-pons, amygdala with adjacent entorhinal and transentorhinal areas, middle frontal gyrus, middle temporal gyrus and inferior parietal lobule were immunostained for α-synuclein to identify Lewy bodies and Lewy-related neurites, using a method previously described [26, 27]. The diagnosis of AD was made when there was a clinical history of dementia and the histopathological assessment of the brain were consistent with the categories of “intermediate” or “high” as established by criteria outlined in a joint publication by the National Institute on Aging and the Reagan Institute (NIA-Reagan) [28].
Apolipoprotein E genotyping
DNA for APOE genotyping from autopsy cases was extracted from pieces of fixed cerebellum tissue. Tissue (100 mg) was digested with proteinase K (1 mg/ml) at 55°C and extracted multiple times with phenol/chloroform. DNA was recovered by isopropanol precipitation. From living patients, DNA was extracted from white blood cells (buffy coat) purified from heparinized blood by centrifugation through Histopaque (Sigma-Aldrich, St. Louis, MO) and using a genomic DNA purification kit (Bio-Rad Laboratories, Richmond, CA). For PCR reactions, 500 ng of DNA from each sample was used. PCR primers, amplification conditions employed, and identification of APOE genotypes by Hha I digestion of amplified material, were carried out according to published protocols [29]. Digested fragments were separated by electrophoresis through 9% acrylamide gels and identified by ethidium bromide staining.
ELISA and Western Blot
ELISA and BACE1 Western blot procedures have been reported recently [14]. Briefly, frozen post-mortem brain tissues from the mid-frontal and mid-temporal cortices (MFC and MTC, respectively) were homogenized in a lysis buffer (135 mM NaCl; 2.7 mM KCl; 10 mM Na2HPO4; 1.8 mM KH2PO4; pH 7.4; 1% Triton X110; 0.1% SDS; 0.05% NP-40; protease cocktail inhibitor [Complete mini tablets; Roche, Indianapolis, IN]). After centrifugation for 30 min at 10,000 x g at 4°C, the supernatants were collected and the protein concentration assessed by the DC protein assay (Bio-Rad Laboratories, Richmond, CA). For BACE1 ELISA, 50–100 μg of total protein were loaded per well and analyzed as described recently [14]. For Western blotting, 30–50 μg of total proteins were separated by SDS-PAGE and blotted onto PVDF membranes. Following blocking for 1h in 5% non-fat dry milk, the membranes were incubated overnight with mouse anti-BACE1 C-terminal antibody (#MAB5308; Millipore, Billerica, MA) diluted 1:2,500. Secondary antibody was HRP-conjugated goat anti-mouse (#SC-2055; Santa Cruz Biotechnologies, Santa Cruz, CA), and signal detection was carried out with ECL (Millipore, Billerica, MA) and exposure of autoradiographic films (Research Products International, Mt. Prospect, IL). The membranes were then stripped and reprobed with mouse anti-β actin (#A1978; Sigma-Aldrich, St. Louis, MO). Densitometry analysis was performed, and results were expressed as ratio of BACE1/actin signals. All Western blot experiments have been carried out in duplicate for each subject and tissue tested. For ELISA, all samples were analyzed in parallel in five independent experiments with duplicate wells each time. Figures were assembled in Acrobat Photoshop.
Statistical analysis
Two-sample t-tests were used to compare the AD and ND groups on BACE1 levels for both the frontal and temporal brain regions and also for brain weight. Additional two-sample t-tests were also carried out to discern differences between APOE ε4 carriers and non-carriers for both the ND and AD groups. One-way analysis of variance (ANOVA) was used to compare the individual APOE groups (ε3/3, ε3/4, ε4/4) on BACE1 levels for the frontal and temporal regions and also for brain weight.
RESULTS
The brain samples used in this study were from a total of 29 ND and 54 AD subjects, with the APOE allele distribution shown in Table 2. Analysis of post-mortem data demonstrated that brains weights were 14.8% lower in AD versus ND subjects (p<0.001). There was no difference in brain weights between the APOE genotypes in the ND and AD groups (p>0.05). In addition, ANOVA analyses indicated there was no difference in the subject age and post-mortem delay among all groups studied (p>0.05).
Table 2.
Demographics of the population used for brain BACE1 level measurements.
| APOE Alleles | Non-Demented
|
Alzheimer’s
|
||||
|---|---|---|---|---|---|---|
| ε3/3 (n=15) | ε3/4 (n=12) | ε4/4 (n=2) | ε3/3 (n=18) | ε3/4 (n=19) | ε4/4 (n=17) | |
| Expired Age (years) | 80.5 ± 17.4 | 83.8 ± 8.4 | 75.5 ± 7.8 | 84.6 ± 9.5 | 81.1 ± 10.1 | 82.0 ± 8.0 |
| Gender (M/F) | 10/5 | 11/1 | 0/2 | 6/12 | 9/10 | 9/8 |
| Post Mortem Delay (hours) | 2.8 ± 0.5 | 2.5 ± 0.5 | 2.5 ± 0.7 | 3.2 ± 2.7 | 3.2 ± 1.3 | 2.6 ± 0.7 |
| Braak Scores | 0–III | I–IV | I–II | IV–VI | IV–VI | IV–VI |
| Brain Weight (grams) | 1217 ± 128 | 1303 ± 92 | 1187 ± 123 | 1093 ± 117 | 1052 ± 80 | 1079 ± 162 |
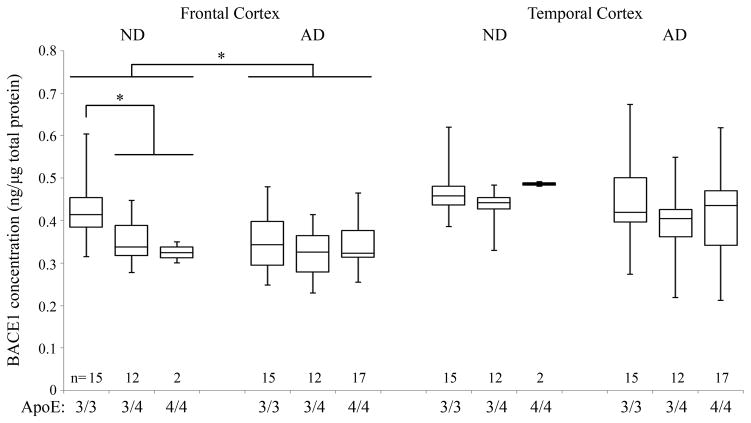
BACE1 levels were measured in the frontal and temporal cortices of all AD and ND subjects from Table 2 using a newly developed immunoassay which, compared to the assays reported previously (Table 1), detects total BACE1 (i.e. full-length and membrane-truncated isoforms) after capture with an antibody recognizing a conformational epitope in the vicinity of the BACE1 catalytic domain [14]. First, we analyzed BACE1 levels in all ND and AD brain samples independently of APOE status. Data indicated that total BACE1 levels were 12% lower in the AD frontal cortex compared to ND (p=0.003; Figure 1). In the temporal cortex a 6.5% decrease in BACE1 levels was obtained, though not statistically significant (p=0.14). These ELISA experiments have been carried out five times in total with duplicate wells each time to confirm the data.
Fig. 1.
BACE1 ELISA. Brain tissue from mid-frontal and mid-temporal cortices of ND and AD subjects carrying the APOE ε3/3, ε3/4, ε4/4 alleles were analyzed for total BACE1 levels by ELISA. BACE1 levels were significantly lower in AD compared to ND in the MFC. In addition, APOE ε4 carriers showed lower protein levels than ε3/3 subjects in MFC. No difference was found in ND MTC, and in AD MFC and MTC across the APOE groups. Boxes show the upper and lower quartiles, the central line is the median, and whiskers extend to the outliers. * p<0.005; n = number of subjects per group.
In order to determine whether the difference in BACE1 levels in the frontal cortex relates to APOE status, we then broke down the groups into APOE ε3/3, ε3/4, and ε4/4 allele carriers. Interestingly, this sub-analysis showed that BACE1 levels were 16.9% higher in APOE ε3/3 than in APOE ε4 carriers in the frontal cortex (Fig. 1). No difference by APOE status was observed in AD subjects. However, we observed that ND APOE ε4 carriers had similar BACE1 levels as AD subjects (Fig. 1). In the temporal cortex there was a 4.5% decrease in BACE1 levels in APOE ε4 carriers which was not statistically significant (p=0.27). No differences were observed across the APOE genotypes in the AD subjects in both frontal and temporal tissues.
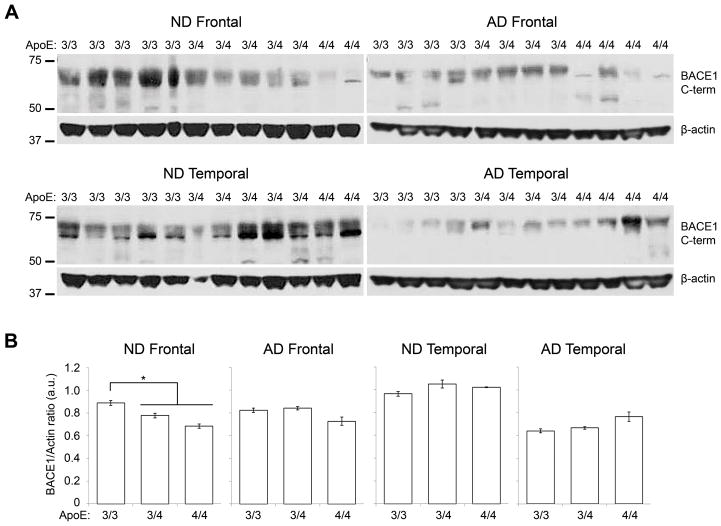
Next, we conducted BACE1 Western blots to validate the ELISA data. In this aim, we used a BACE1 C-terminus antibody that recognizes full-length BACE1 and is different from the antibodies used for the ELISA method. In the ND frontal cortex we observed bands of lower intensity in APOE ε3/4 and ε4/4 subjects compared to ε3/3 (Fig. 2A). Densitometry analysis with normalization to actin indicated that BACE1 levels in ND APOE ε3/3 subjects were 15.4% higher than APOE ε4 carriers (p=0.002), confirming the ELISA data. However, no significant differences were observed in ND temporal tissues, and AD frontal and temporal tissues across APOE genotypes (p>0.05), as obtained by ELISA.
Fig. 2.
BACE1 Western blot. (A) Representative Western blot of BACE1 (top images) and beta-actin (bottom images) in MFC (left) and MTC (right). APOE ε3/3 carriers showed higher BACE1 signals than APOE ε4 heterozygotes and homozygotes in ND samples. (B) Densitometry analysis of BACE1 signals in MFC (left) and MTC (right). BACE1 levels normalized to actin were significantly lower in APOE ε4 carriers (ε3/4 and ε4/4) than in non-carriers in ND frontal cortex. No significant difference was noted in the temporal cortex of ND subject and in AD groups. * p<0.05.
DISCUSSION
In the present study we investigated post-mortem BACE1 levels in relationship to APOE genotype in two areas of the neocortex where dense plaques accumulate in large numbers in AD subjects, i.e. the mid-frontal and mid-temporal cortices. Using two techniques detecting different pools of BACE1 (i.e. total BACE1 by ELISA, and full-length BACE1 by Western blot), we found that BACE1 levels in MFC tissues are lower in AD compared to ND. Further, we found that APOE genotype influences BACE1 levels in ND MFC. No difference was detected in AD MFC across APOE genotypes. Similarly, APOE genotypes had no significant effect on BACE1 levels in ND and AD MTC.
The observation that post-mortem BACE1 levels are significantly lower in the frontal lobe of ND APOE ε4-positive subjects is, to our knowledge, a novel finding. It may provide an alternative explanation to the low CSF Aβ42 levels found in ND APOE ε4 carriers. Several reports have indicated that ND and AD APOE ε4-positive subjects display lower CSF Aβ42 levels than APOE ε4-negative subjects [30–32]. The main explanation was that the ApoE4 variant promotes more Aβ deposition than ApoE2 and 3 [33]. However, a recent study combining brain imaging and CSF biomarkers found a significant correlation between cortical thickness, ventricular enlargement, CSF Aβ42 levels, and APOE status in ND and AD subjects [34]. Since BACE1 is principally expressed by neurons in the brain [35], if APOE ε4 carriers undergo faster neuronal loss that non-carriers this would result in lower brain BACE1 levels, as we observed here, and lower the production of soluble Aβ42 peptides released into the CSF. Furthermore, a decrease in BACE1 levels might also explain the fact that neuritic plaques correlate less with the progression of AD than neurofibrillary tangles. At early stages of the disease, high BACE1 levels would produce large amounts of Aβ that would rapidly deposit into neuritic plaques. But, when the neuronal loss is pronounced less Aβ is produced and the number and size of plaques increases less rapidly. Overall, combined with previous CSF Aβ data, our results indicate a possible APOE allelic effect on brain BACE1 levels and other AD biomarkers. Therefore, it would be interesting to conduct multicenter studies to analyze brain BACE1 levels and β-secretase activity using standardized methods (detecting total or transmembrane BACE1 isoforms, or both in parallel), sharing some of the samples for better comparisons, and including APOE genotype as confounder. Such multicenter studies would increase the power to detect APOE effects on AD biomarkers compared to individual studies, as well as increase the number of ND subjects homozygous for ε4 which are rare in individual repositories due the scarcity of such elderly individuals in the general population.
Our finding that ND subjects had higher BACE1 levels than AD in the frontal lobe is in contrast to most previous reports which showed an increase in AD in the same brain region (Table 1). Consequently, we repeated the ELISA experiments five times to confirm the data, and conducted Western blots in duplicate for further validation. Several technical and physiological points could be advanced to explain the difference. First, our ELISA method uses two antibodies directed at the extracellular domain of BACE1 [14] while the majority of previous studies used techniques detecting transmembrane, full-length BACE1 [9, 13]. Based on the fact that BACE1 can be shed from the plasma membrane [36] and on data collected from platelets [37], we believe that our method detects more soluble BACE1 isoforms than previous methods, though to date it is not known how much soluble BACE1 is present in the brain, whether its levels change during normal and pathological aging, and whether soluble BACE1 is able to cleave transmembrane APP in vivo. To investigate full-length BACE1 we carried out Western blots using the same antibody as used by others (i.e. Millipore #MAB5308, [9, 13]). Interestingly, our Western blot data confirmed the ELISA data, suggesting that most BACE1 isoforms are present at lower levels in the frontal lobe of the AD subjects in our sample population. Some studies have shown that the increase in brain BACE1 protein levels in AD was not corroborated by changes in mRNA levels in the frontal cortex [38], nor by BACE1 protein levels and activity in the CSF [23], both of which showed no difference between AD and ND. Therefore, it is a challenge to reach a consensus as whether BACE1 protein levels increase or decrease in the AD brain given the utilization of different methodologies (summarized in [14]), the existence of several isoforms of the protein (soluble vs. transmembrane), the brain regions studied, and cohorts with various post-mortem delays. Standardization of the methods would likely help resolving such issue. Second, it is possible that our results may be unique to the sample population used for this study. For example, while most studies showed an increase in the frontal cortex, results regarding BACE1 levels in the temporal cortex were much less clear (Table 1). Moreover, several studies have tested large cohorts to obtain significant results in the frontal cortex because interindividual variations were challenging [9, 11]. To test whether the data presented here are specific to our sample population we are planning to increase the sample size and compare our population to samples from other repositories. Nonetheless, our present data suggest that APOE genotype should be added as confounder in brain BACE1 levels and β-secretase activity analyses. Third, it is well documented that neuronal loss occurs during AD. For example, a decreased in NSE expression (marker of healthy neurons) in the frontal cortex was associated with disease severity [38]. Since BACE1 is principally expressed by neurons [35] one would anticipate BACE1 and Aβ levels to dramatically decrease when the neuronal loss is pronounced, though some compensatory mechanisms may develop such as BACE1 expression by glial cells under stress [39], hence maintaining Aβ levels, to a certain level during the progression of the disease. To note, we do not exclude that brain BACE1 levels are higher in AD than in ND subjects at early stages of the disease, though this hypothesis remains to explore.
Added to previous reports [30–32], our present data provide good evidence that the levels of several proteins involved in AD neuropathology are changed in APOE ε4-positive subjects. We anticipate that correlations with additional proteins will be identified in the future. However, the underlying mechanisms of these associations are still unclear. In addition, APOE effects may be more complicated than initially thought, as evidenced by the recent demonstration that APOE expression could be regulated by transcription modulators acting on upstream genes located on the same chromosomal region as APOE (TOMM40), and that such regulation might be brain region specific [40, 41]. Given the complex organization of the BACE1 promoter [42], such complex transcription regulation mechanisms may also happen for BACE1, though these remain to investigate. Nonetheless, the clinical and biological data acquired until now [31, 32, 34] suggest that APOE ε4 carriers develop AD neuropathological features earlier and progress faster than non-carriers, suggesting that the APOE ε4 allele accelerates neurodegeneration. To verify this hypothesis one could conduct longitudinal studies combining brain imaging and several biomarkers with baseline data acquired many years before what is currently done. Such paradigm could also help resolving the question of why, and maybe how, some APOE ε4 carriers do not develop dementia.
In conclusion, our data suggest that APOE alleles may regionally and differentially regulate BACE1 levels in the human brain. Further studies will include samples from subjects who suffered other dementia for specificity controls. Moreover, future multicenter analyzes of brain BACE1 levels by APOE genotype will answer the question of whether the data we presented here are unique to the cohort studied or whether they can be generalized to worldwide populations. Finally, our results provide some new hypotheses to explain the decrease in CSF Aβ42 levels in APOE ε4 carriers in ND and AD subjects, and for the large interindividual differences reported earlier for BACE1 levels in AD versus ND brain samples.
Acknowledgments
This project was supported by National Institute on Aging grants R01AG034155, P30AG019610-09, and 5P30AG019610-12. The Banner Sun Health Research Institute Brain and Body Donation Program is supported by the National Institute on Aging (P30 AG19610 Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901 and 1001 to the Arizona Parkinson’s Disease Consortium) and the Michael J. Fox Foundation for Parkinson’s Research.
Footnotes
CONFLICT OF INTEREST
None declared.
References
- 1.Selkoe DJ. Alzheimer’s disease: genotypes, phenotypes, and treatments. Science. 1997;275(5300):630–1. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 2.Brouwers N, Sleegers K, Van Broeckhoven C. Molecular genetics of Alzheimer’s disease: an update. Ann Med. 2008;40(8):562–83. doi: 10.1080/07853890802186905. [DOI] [PubMed] [Google Scholar]
- 3.Belyaev ND, Kellett KA, Beckett C, Makova NZ, Revett TJ, Nalivaeva NN, et al. The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway. J Biol Chem. 2010;285(53):41443–54. doi: 10.1074/jbc.M110.141390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4(3):231–2. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 5.McConlogue L, Buttini M, Anderson JP, Brigham EF, Chen KS, Freedman SB, et al. Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem. 2007;282(36):26326–34. doi: 10.1074/jbc.M611687200. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Zhou W, Tong Y, He G, Song W. Control of APP processing and Abeta generation level by BACE1 enzymatic activity and transcription. FASEB J. 2006;20(2):285–92. doi: 10.1096/fj.05-4986com. [DOI] [PubMed] [Google Scholar]
- 7.Cole SL, Vassar R. BACE1 structure and function in health and Alzheimer’s disease. Curr Alzheimer Res. 2008;5(2):100–20. doi: 10.2174/156720508783954758. [DOI] [PubMed] [Google Scholar]
- 8.Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer’s disease. Ann Neurol. 2002;51(6):783–6. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- 9.Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59(9):1381–9. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- 10.Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, et al. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer’s disease patients. Proc Natl Acad Sci U S A. 2004;101(10):3632–7. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9(1):3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O’Connor T, et al. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer’s disease pathogenesis. J Neurosci. 2007;27(14):3639–49. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed RR, Holler CJ, Webb RL, Li F, Beckett TL, Murphy MP. BACE1 and BACE2 enzymatic activities in Alzheimer’s disease. J Neurochem. 2010;112(4):1045–53. doi: 10.1111/j.1471-4159.2009.06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzales A, Decourt B, Walker A, Condjella R, Nural H, Sabbagh MN. Development of a specific ELISA to measure BACE1 levels in human tissues. J Neurosci Methods. 2011;202(1):70–6. doi: 10.1016/j.jneumeth.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utermann G, Langenbeck U, Beisiegel U, Weber W. Genetics of the apolipoprotein E system in man. Am J Hum Genet. 1980;32(3):339–47. [PMC free article] [PubMed] [Google Scholar]
- 16.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 17.Greenwood PM, Lambert C, Sunderland T, Parasuraman R. Effects of apolipoprotein E genotype on spatial attention, working memory, and their interaction in healthy, middle-aged adults: results From the National Institute of Mental Health’s BIOCARD study. Neuropsychology. 2005;19(2):199–211. doi: 10.1037/0894-4105.19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirier J. Apolipoprotein E in the brain and its role in Alzheimer’s disease. J Psychiatry Neurosci. 1996;21(2):128–34. [PMC free article] [PubMed] [Google Scholar]
- 19.Caselli RJ, Walker D, Sue L, Sabbagh M, Beach T. Amyloid load in nondemented brains correlates with APOE e4. Neurosci Lett. 2010;473(3):168–71. doi: 10.1016/j.neulet.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–44. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63(3):287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye S, Huang Y, Mullendorff K, Dong L, Giedt G, Meng EC, et al. Apolipoprotein (apo) E4 enhances amyloid beta peptide production in cultured neuronal cells: apoE structure as a potential therapeutic target. Proc Natl Acad Sci U S A. 2005;102(51):18700–5. doi: 10.1073/pnas.0508693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewers M, Zhong Z, Burger K, Wallin A, Blennow K, Teipel SJ, et al. Increased CSF-BACE 1 activity is associated with ApoE-epsilon 4 genotype in subjects with mild cognitive impairment and Alzheimer’s disease. Brain. 2008;131(Pt 5):1252–8. doi: 10.1093/brain/awn034. [DOI] [PubMed] [Google Scholar]
- 24.Beach TG, Sue LI, Walker DG, Roher AE, Lue LF, Vedders L, et al. The Sun Health Research Institute Brain Donation Program: Description and experience, 1987–2007. Cell and Tissue Banking. 2008;9(3):229–45. doi: 10.1007/s10561-008-9067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 26.Beach TG, Adler CH, Lue LF, Sue LI, Bachalakuri J, Henry-Watson J, et al. Unified staging system for Lewy body disorders: correlation with nigrostriatal degeneration, cognitive impairment and motor dysfunction. Acta Neuropathologica. 2009;117(6):613–34. doi: 10.1007/s00401-009-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beach TG, White CL, Hamilton RL, Duda JE, Iwatsubo T, Dickson DW, et al. Evaluation of alpha-synuclein immunohistochemical methods used by invited experts. Acta Neuropathologica. 2008;116(3):277–88. doi: 10.1007/s00401-008-0409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ball M, Braak H, Coleman P, Dickson D, Duyckaerts C, Gambetti P, et al. Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. Neurobiology of Aging. 1997;18(4):S1–S2. [PubMed] [Google Scholar]
- 29.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–8. [PubMed] [Google Scholar]
- 30.Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–13. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Shaw LM, Trojanowski JQ, et al. Effect of apolipoprotein E on biomarkers of amyloid load and neuronal pathology in Alzheimer disease. Ann Neurol. 2010;67(3):308–16. doi: 10.1002/ana.21953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prince JA, Zetterberg H, Andreasen N, Marcusson J, Blennow K. APOE epsilon4 allele is associated with reduced cerebrospinal fluid levels of Abeta42. Neurology. 2004;62(11):2116–8. doi: 10.1212/01.wnl.0000128088.08695.05. [DOI] [PubMed] [Google Scholar]
- 33.Fagan AM, Watson M, Parsadanian M, Bales KR, Paul SM, Holtzman DM. Human and murine ApoE markedly alters A beta metabolism before and after plaque formation in a mouse model of Alzheimer’s disease. Neurobiol Dis. 2002;9(3):305–18. doi: 10.1006/nbdi.2002.0483. [DOI] [PubMed] [Google Scholar]
- 34.Tosun D, Schuff N, Truran-Sacrey D, Shaw LM, Trojanowski JQ, Aisen P, et al. Relations between brain tissue loss, CSF biomarkers, and the ApoE genetic profile: a longitudinal MRI study. Neurobiol Aging. 2010;31(8):1340–54. doi: 10.1016/j.neurobiolaging.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai H, Wang Y, McCarthy D, Wen H, Borchelt DR, Price DL, et al. BACE1 is the major beta-secretase for generation of Abeta peptides by neurons. Nat Neurosci. 2001;4(3):233–4. doi: 10.1038/85064. [DOI] [PubMed] [Google Scholar]
- 36.Murayama KS, Kametani F, Araki W. Extracellular release of BACE1 holoproteins from human neuronal cells. Biochem Biophys Res Commun. 2005;338(2):800–7. doi: 10.1016/j.bbrc.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Decourt B, Walker A, Gonzales A, Malek-Ahmadi M, Liesback C, Davis KJ, et al. Can platelet BACE1 levels be used as a biomarker for Alzheimer’s disease? Proof-of-concept study. Platelets. 2012 doi: 10.3109/09537104.2012.688899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coulson DT, Beyer N, Quinn JG, Brockbank S, Hellemans J, Irvine GB, et al. BACE1 mRNA expression in Alzheimer’s disease postmortem brain tissue. J Alzheimers Dis. 2010;22(4):1111–22. doi: 10.3233/JAD-2010-101254. [DOI] [PubMed] [Google Scholar]
- 39.Rossner S, Lange-Dohna C, Zeitschel U, Perez-Polo JR. Alzheimer’s disease beta-secretase BACE1 is not a neuron-specific enzyme. J Neurochem. 2005;92(2):226–34. doi: 10.1111/j.1471-4159.2004.02857.x. [DOI] [PubMed] [Google Scholar]
- 40.Bekris LM, Galloway NM, Montine TJ, Schellenberg GD, Yu CE. APOE mRNA and protein expression in postmortem brain are modulated by an extended haplotype structure. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):409–17. doi: 10.1002/ajmg.b.30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekris LM, Lutz F, Yu CE. Functional analysis of APOE locus genetic variation implicates regional enhancers in the regulation of both TOMM40 and APOE. J Hum Genet. 2012;57(1):18–25. doi: 10.1038/jhg.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambamurti K, Kinsey R, Maloney B, Ge YW, Lahiri DK. Gene structure and organization of the human beta-secretase (BACE) promoter. FASEB J. 2004;18(9):1034–6. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]